Abstract
It is currently well recognized that microRNA deregulation is an hallmark of human cancer, and how an aberrant expression of these tiny regulatory RNA molecules in several cell types is not just a random association, but it plays a causal role in different steps of the tumorigenic process, from the initiation and development to progression toward the acquisition of a metastatic phenotype.
Different regulatory mechanisms can control microRNA expression at a genetic or epigenetic level as well as involving the biogenesis machinery or the recruitment of specific transcription factors. The tumorigenic process implies a substantial alteration of these mechanisms, thus disrupting the equilibrium within the cell and leading to a global change in microRNA expression, with loss of oncosuppressor microRNAs and overexpression of oncomiRNAs.
Here we review the main mechanisms regulating microRNAs, and the consequences of their aberrant expression in cancer, with a glance at the possible implications at a clinical point of view.
Introduction
The interest and the knowledge on microRNA field has increased incredibly fast in the last few years, and it is currently well recognized how these small RNA molecules represent an essential part of the encoding genome, finely tuning gene expression and thus exerting a crucial role in all the most important processes and in different species, including vertebrates (1). microRNAs represent indeed an entire novel level of gene regulation that forced scientists to revise and somehow reorganize their view of the molecular biology.
Since the first discovery, remarkable advances in the understanding of microRNA biology have been made, including: the identification of hundreds of miRNA genes; the dissection of miRNA biogenesis pathways; the identification of numerous miRNA targets and the establishment of principles of target regulation; the study of their biological functions in physiological and pathological conditions.
microRNA biogenesis can be summarized in two main processing steps, taking place respectively in the nucleus and in the cytoplasm: mainly transcribed by RNA Polimerase II as long primary transcripts characterized by hairpin structures (pri-microRNAs), microRNAs are first processed into the nucleus by RNAse III Drosha into 70–100 nts long pre-microRNAs (2). However, there is alternative miRNA biogenesis pathway, the so-called miRtron pathway, which does not require the Drosha-mediated cleavage, since miRtrons (miRNAs localized within introns of protein-encoding or -non-encoding genes) are directly processed using the splicing machinery to generate pre-miRs (3,4,5).
The short stem plus a ~2-nt 3′ overhang of the originated precursor molecules are recognized by the Ran-GTP-dependent transporter Exportin 5, which mediates the translocation to the cytoplasm (6). Here the second cropping process (dicing) takes place, performed by the RNAse III enzyme Dicer (Dicer 1 in flies) associated to TRBP (TAR RNA-binding protein) or PACT (also known as PRKRA), and Argonaute (AGO1-4), which cleave the miRNA precursor hairpin generating a transitory miRNA/miRNA* duplex (approximately 22 nucleotides long), which includes the mature miRNA guide and the complementary passenger strand (miRNA*, star miRNA) (also named miR-3p/miR-5p). Whereas one of the two strands is selected as guide strand according to thermodynamic properties, the complementary one is usually subjected to degradation. The so called miRNA* was initially thought to be the strand subjected to degradation, instead more recent evidence suggests that it does not simply represent a non-functional bioproduct of miRNA biogenesis, but it can be selected as a functional strand and play significant biological roles (7).
This duplex is then loaded into the miRNA associated RNA-induced silencing complex (RISC), which preferentially includes the mature single-stranded miRNA molecule and AGO proteins, where they act as guiding molecules to deliver the complex to target mRNA. It seems that AGO is associated to Dicer in the dicing step as well as in the RISC assembly step. As a part of this complex, the mature miRNA is able to regulate gene expression at post-transcriptional level, binding for the most part through partial complementarity to target mRNAs, and mainly leading to mRNA degradation or translation inhibition, depending on the sequence complementarity between the small RNA and the target mRNA. The site-specific cleavage, commonly defined as RNA interference (RNAi), is actually restricted to miRNAs with a perfect or near-perfect match to the target RNA, and it is a very rare event in mammals. By contrast, in mammals the most frequent processes are enhanced mRNA degradation or translational inhibition, commonly associated respectively with higher or lower grade of mismatches in the miRNA/target sequences.
Considering the different rules regulating the interaction between a microRNA and its target mRNA, it is not surprising that each miRNA has the potential to target a large number of genes (8,9,10,11). On the other hand, approximately 60% of the mRNAs have one or more evolutionarily conserved sequences that are predicted to interact with miRNAs. Indeed, in silico analysis predicts that the 3′ UTR of a single gene is frequently targeted by several different miRNAs (8). Many of these predictions have been validated experimentally, suggesting that miRNAs might cooperate to regulate gene expression.
Overall, these data show the complexity and widespread regulation of gene expression by miRNAs, which should be taken into consideration when developing miRNA-based therapies.
microRNA expression regulation
Several mechanisms can control microRNA expression, and result to be altered in human diseases, including cancer (Figure 1).
FIGURE 1.
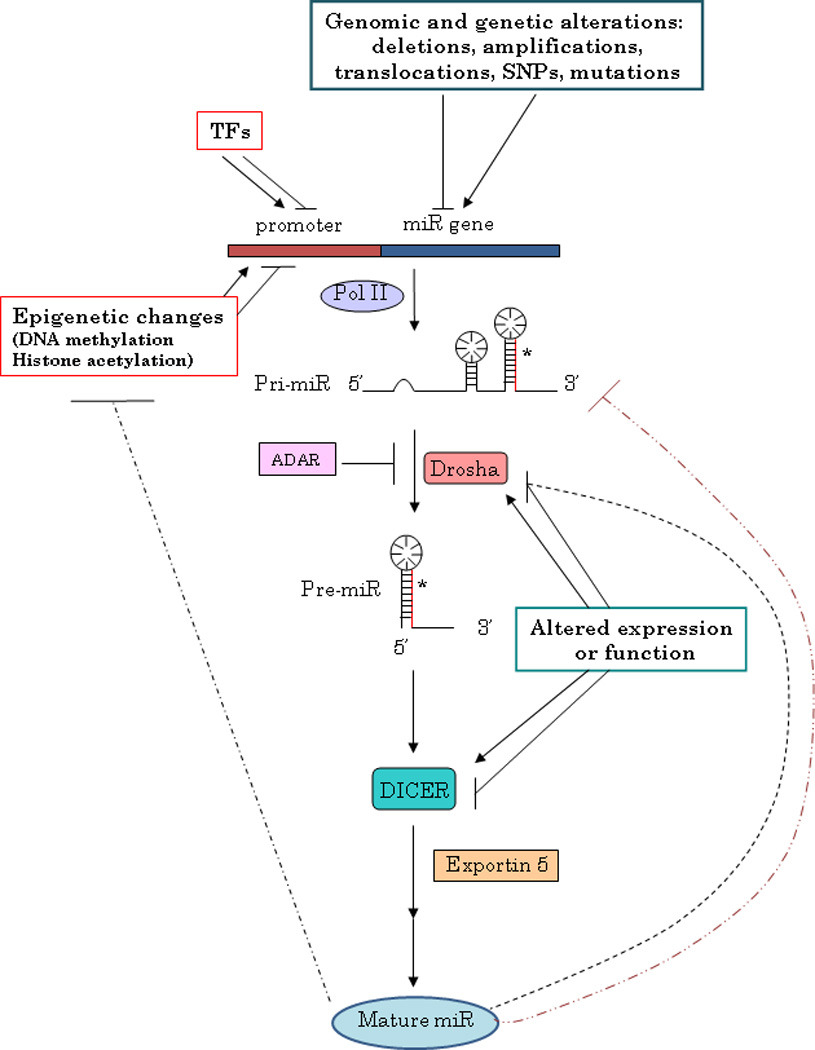
MicroRNA expression regulation.
Chromosomal abnormalities, as first suggested by the discovery of two microRNA genes, miR-15a and 16-1, in a region of the Chr. 13 frequently deleted in CLL (12), supported by the evidence that microRNAs are frequently located in regions of the genome involved in alterations in cancer (13), and then confirmed by several studies (12,14,15); mutations, as the inherited mutations in the primary transcripts of miR-15a and miR-16-1 responsible for reduced expression of the two microRNAs in vitro and in vivo in CLL (16); polymorphisms (SNPs), as described in lung cancer (17).
In addition to structural genetic alterations, microRNA expression can be also modulated as a consequence of defects in the microRNA biogenesis machinery. Indeed, it has been reported that deregulation of different cofactors can affect miRNA expression with important biological implications: DGR8 knock-out mice arrested early in development and have defects in ES cell proliferation and differentiation (18); Lin28, originally discovered as a heterochronic gene regulating developmental timing in worms, is able to block let-7 biogenesis and it is activated in many human tumors (15%), being particularly associated with less differentiated cancers (19). Moreover, adenosine deaminases acting on RNA (ADARs) can affect the expression of microRNAs recognizing adenosine residues, as reported for miR-142: the editing of pri-miR-142 results in the suppression of its processing by Drosha, degradation by a component of the RISC complex and reduced levels of the mature product (20).
microRNA processing can be also affected by other microRNAs, thus creating a complex level of reciprocal interaction and regulation, as very recently reported by Tang R and colleagues (21), who demonstrated that mouse miR-709 is predominantly located in the nucleus, where it directly binds to a miR-709 recognition element on pri-miR-15a/16-1 preventing its processing into pre-miR-15a/16-1 and thus leading to a suppression of miR-15a/16-1 maturation.
Notably, changes in microRNA levels consequent an altered Drosha or Dicer activity have been reported in different tumor types (22,23,24). In particular, it seems that Dicer or Drosha silencing promotes cellular transformation and tumorigenesis in vivo: conditional loss of Dicer1 in the lung tissues of mice enhances the development of lung tumors in a K-ras mouse model (25). Finally, loss of Dicer and/or Drosha has also been inversely correlated with outcome in lung cancer (26), cancers of the ovarian epithelium (24), and more recently in other tumor types as nasopharyngeal carcinoma (27), neuroblastoma (28) and breast (29).
It may sound surprising that reprogramming of the whole ‘microRNome’, including both oncomiRNAs and tumor suppressor miRNAs, can lead to a specific anti-tumoral effect: how is the balance shifted in favor of a specific effect? This might be due to the possibility that most microRNAs seem to exert a role as oncosuppressors, and consequently are mostly dowregulated in human neoplasia (30).
A few reports, however, describe a positive correlation between Dicer expression and poor outcome in colorectal cancer (31) and in prostate cancer (32), or the overexpression of Drosha in cervical cancer (33), thus raising the important issue to validate this still debated question, and verify whether the effect of targeting the microRNA machinery might be tissue-related.
To complicate the scenario, a recent report by Piccolo’s group (34) describes a microRNA family, miR-103-107, able to empower the metastatic potential targeting Dicer and thus attenuating the global microRNA biosynthesis, with a particular effect mediated by the downregulation of miR-200 family, and the consequent switch to a more mesenchymal and aggressive phenotype.
The deregulated microRNA expression in cancer can also be due to epigenetic changes, as altered DNA methylation, as suggested by an extensive analysis of genomic sequences of miRNA genes, which have shown that approximately half of them are associated with CpG islands (35). One of the first reports proving that an altered methylation status can be responsible for the deregulated expression of microRNAs in cancer, as the silencing of putative tumor suppressor microRNAs, was the study by Saito and colleagues (36), who observed a strong upregulation of miR-127 upon treatment of a bladder cancer cell lines and human fibroblasts with DNMT inhibitor 5-Aza-2’-deoxycytidine. As expected, miR-127, able to target the proto-oncogene BCL-6, is characterized by a CpG island promoter, and is silenced in several cancer cells. With the same approach of unmask epigenetically silenced microRNAs inducing chromatin-remodeling by drug treatment, it has been demonstrated that miR-9-1 is hypermethylated and consequently down-modulated in breast cancer (37), as well as the clustered miR-34b and miR-34c in colon cancer (38).
Analyzing miRNA profiling of DNMT1- and DNMT3b-deficient colorectal cancer cells Lujambio and colleagues (39) identified another oncosuppressor microRNA which results to be hypermethylated, and thus silenced, in tumor: miR-124a, embedded in a large CpG island and able to target cyclin D kinase 6, which mediates the phosphorylation of RB tumor suppressor gene.
Conversely, the upmodulation of putative oncogenic microRNAs in cancer can be due to DNA hypomethylation, as shown in lung adenocarcinoma for let-7a-3 (40) or in epithelial ovarian cancer for miR-21 (41).
Methylation is not the only epigenetic mechanism that can affect microRNAs expression: Scott and colleagues (42) showed for the first time that histone deacetylase inhibition is followed by the extensive and rapid alteration of microRNA levels in a breast cancer cell line. After this first evidence, several reports have shown that HDACis alter microRNA expression in several human carcinomas including colon (43,44), and gastric (45).Very recently, Rhodes LV and colleagues (46) described how HDACi Trichostatin A (TSA) alter the miRNA signature of an apoptosis-resistant breast cancer cell line and reduces its clonogenic potential.
To complicate the scenario connecting microRNAs and epigenetics, microRNAs themselves can regulate the expression of components of the epigenetic machinery, creating a highly controlled feedback mechanism: miR-29 family directly targets the de novo DNA methyltransferases DNMT-3A and -3B, while indirectly, through regulation of the transactivator Sp1, the maintenance DNA methyl transferase DNMT1. Interestingly, introduction of miR-29s into lung cancers and AMLs results in reactivation of silenced tumor suppressors and inhibition of tumorigenesis (47,48). Loss of miR-290 cluster in Dicer-deficient mouse ES cells leads to the downregulation of DNMT3a, DNMT3b and DNMT1 through upmodulation of their repressor, RBL-2, proven target of miR-290 (49,50). miR-1, involved in myogenesis and related diseases, directly targets HDAC4 (51) and miRNA-9* down-modulates HDAC4 and HDAC5 (52).
Considering the reported evidence of how microRNA expression can be affected by changes in the epigenetic program, a possible therapeutic approach might be represented by the modulation of microRNA expression by targeting components of these regulatory networks. The existence of epigenetic drugs, such as DNA demethylating agents and histone deacetylase inhibitors, able to reverse an aberrant methylation or acetylation status, raises indeed the intriguing possibility to regulate microRNA levels, for example to restore the expression of tumor suppressor microRNAs, thus reverting a tumoral phenotype.
Demethylating agents as Decitabine and 5-azacytidine, for example, are currently approved for the treatment of myelodysplastic syndrome, although they have shown activity in many other malignancies, including AML (53). These drugs are known to inhibit DNA methyltransferases, resulting in tumor suppressor gene re-expression. As previously described, miRNAs have also been shown to be actively re-expressed after treatment with these drugs and to largely contribute to the therapeutic effects of these compounds. Even though it is tempting to suggest that many of the biological effects of these drugs may be mediated by the re-expression of non-coding RNAs, this still needs to be verified. In particular, as previously underlined concerning the possibility to interfere with the biogenesis machinery, it is mandatory to verify the consequences of a global reprogramming of miRNA expression.
Finally, a deregulation of miRNA expression can be a result of increased or decreased transcription due to an altered transcription factor activity.
MiR-34a family of miRNAs, for instance, has been shown to be directly induced by the tumor suppressor p53, and to be partially responsible of the phenotype induced by this oncosuppressor (54,55).
Vice versa, oncogenes can also affect microRNA expression, and a clear example is represented by the oncoprotein MYC, which is able to both induce oncogenic microRNAs, as the miR-17-92 cluster, and negatively regulate transcription of tumor suppressor miRNAs, such as let-7 (56) and miR-29 family members (57).
An interesting regulatory loop has been demonstrated between ZEB1 transcription factor and miR-200 family: EMT inducers ZEB1 and ZEB2 are direct target of miR-200 family members (58), and in turn, ZEB1 has been shown to directly repress miR-200c and miR-141 transcription (59).
Another relevant example is represented by miR-221 and -222, negatively regulated by ERα and able, in turn, to directly target the receptor (60). Therefore, silencing ERα, for example by methylation or by dysregulating miR-221 and miR-222 through the activation of pathways, such as MET pathway, involved in oncogenesis, results in the constitutive activation of miR-221 and miR-222 and inhibition of the tumour suppressors p27, p57, PTEN and TIMP3, thus contributing to the development of the invasive phenotype characteristic of frankly malignant cells (61). This regulatory feedback loop seems to be involved in the development of ERα–breast cancers (62).
Nevertheless, despite the advances in our understanding of the mechanisms causing miRNA deregulation, the daunting task still remains the elucidation of the biological role of miRNAs in the initiation and in the development of cancer.
microRNA function: what is new under the sun
It is currently well recognized how microRNAs are not only deregulated in human cancer, but how they play a causal role, functioning as either tumor suppressors or oncogenes by targeting different steps of the tumorigenesis process, either occurrence, development or progression to a metastatic phenotype. Indeed, cancers develop sophisticated networks of biological activities that contribute to their ability to develop and, in some cases, evade treatment. This complex program relies on the communication between multiple cell types, including both the primary tumor as well as the stromal cells, and can be summarized in 6 essential characteristics of cancer progression: self–sufficiency in growth signals, insensitivity to anti-growth signals, evading apoptosis, limitless replicative potential, sustained angiogenesis, and tissue invasion and metastasis (63). microRNAs can affect any of these processes, as demonstrated by gain of-function and loss-of-function experiments, in combination with target prediction analyses.
miRNAs acting as tumor suppressors target oncoproteins with crucial roles in various cancer pathways, as the ones reported in the first pioneer studies: miR-15a-16-1, targeting BCL-2 (64) and let-7, targeting RAS (65) and MYC (66), whereas miRNAs with oncogenic properties negatively regulate tumor suppressor proteins, as the well described miR-21, which targets the tumor suppressors phosphatase and tensin homologue (PTEN) and programmed cell death 4 (67,68).
In addition to classical tumor suppressor or oncogene functions, miRNAs have been implicated also in cell migration and metastasis, either as promoters, as miR-10b (69) and miR-21 (70) or inhibitors, as miR-126 and miR-335 (71), miR-34a (72), miR-200 family (58) and miR-205 (73). Moreover, one of the crucial steps of the metastatic process is represented by neo-angiogenesis, which allows cells to reach and disseminate through the systemic circulation. microRNAs can control tumor progression also at this level, either promoting or inhibiting the proliferation of endothelial cells. miR-221 and miR-222 repress proliferative and angiogenetic properties of c-Kit in endothelial cells, whereas hypoxic reduction of miR-16, miR-15b, miR-20a and miR-20b, directly targeting VEGF, supports the angiogenic process (74). Angiogenesis can be also promoted by miR-210, activated by hypoxia and directly repressing endothelial ligand Ephrin A3 (75), and by miR-17-92 cluster, which sustains MYC angiogenic properties through repression of connective tissue growth factor (CTGF) and the anti-angiogenic adhesive glycoprotein thrombospondin 1 (TSP1) (76), also targeted by miR-27b and let-7f (77). Notably, the same group who identified a set of human microRNAs robustly suppressing breast cancer metastasis (71) has more recently revealed that endogenous miR-126 non-cell-autonomously regulates endothelial cell recruitment to metastatic breast cancer cells, in vitro and in vivo. It suppresses metastatic endothelial recruitment, metastatic angiogenesis and metastatic colonization through coordinate targeting of IGFBP2, PITPNC1 and MERTK, pro-angiogenic genes and biomarkers of human metastasis (78). Thus, miRNAs can exert their function influencing interaction between different cell types. Another example is represented by the acquirement of a metastatic phenotype following miR-320 loss in cancer-associated fibroblasts: miR-320 is indeed a crucial component of a PTEN-controlled tumor-suppressive axis in stromal fibroblasts, and loss of PTEN and miR-320 induces an oncogenic secretome that reprograms the tumor microenvironment to promote invasion and angiogenesis (79). However, despite the increasing body of in vitro and in vivo evidence supporting the involvement of microRNAs in cancer development or progression, to date just few microRNA engineered animal models have been developed: through knock out or transgene introduction, these animal models can provide the genetic demonstration of the causative involvement of a specific microRNA in a biological phenomenon. miR-17-92 cluster and miR-155, both overexpressed in lymphoproliferative disorders, including lymphomas and leukemia (80,81), were the first examples of miRNAs with oncogenic activity validated in engineered animal models. Infection of murine hematopoietic stem cells with a retrovirus carrying the miR-17-92 cluster accelerated the development of lymphomas in Myc transgenic mice (81). Transgenic mice overexpressing miR-17-92 cluster in B cells were discovered to develop lymphoproliferative disease and autoimmunity (82). The higher rate of proliferation and the lower rate of activation-induced cell death of lymphocytes in these mice were partially attributed to the direct targeting of the anti-apoptotic genes Bim and Pten by miR-17-92 cluster. Moreover, Ventura and colleagues showed that mice deficient for miR-17-92 cluster die shortly after birth with lung hypoplasia and a ventricular septal defect (83). Finally, Mu and colleagues determined that deletion of the complete miR-17-92 cluster slows Myc-induced oncogenesis (84).
Notably, overexpression of miR-155 alone in the lymphoid compartment was sufficient to cause cancer and did not require any other cooperative mutation or oncogene expression. miR-155 transgenic mice developed polyclonal lymphoid proliferation followed by acute lymphocytic lymphoma or leukemia (85). This was the first report that the dysregulation of a single miRNA can lead to malignancy. More recently, Slack’s group (86) has shown that mice conditionally expressing miR-21 develop a pre-B malignant lymphoid-like phenotype, thus demonstrating that miR-21 is a genuine oncogene.
Considering the different rules regulating miRNA/target interaction, and the evidence that microRNAs can target multiple molecules, it is unlikely that miRNAs will be responsible for a specific phenotype by aiming at a single target. Instead, it is thought that miRNAs engage in complex interactions with the machinery that controls the transcriptase and concurrently target multiple mRNAs. This is probably the most intriguing rational supporting the idea of using microRNAs as anticancer drugs.
However, recent reports have shed more light into the complex mechanisms regulating microRNA function on target mRNAs. Indeed, microRNAs mainly recognize complementary sequences in the 3’ untraslated regions (UTRs) of their target mRNAs, however more recent studies have reported that they can also bind to the 5’UTR or the ORF (87,88,89,90) and, even more surprisingly, they can upregulate translation upon growth arrest conditions (91).
The discovery of other functional non-coding RNAs, interconnected with each other, has revealed a network of regulatory molecules definitely more complicated than expected.
One of the first studies reporting the existence of other non-coding RNAs involved in tumorigenesis and connected to microRNAs was reported by our group (92): more in detail, Calin and colleagues observed that a large fraction of genomic ultraconserved regions (UCRs) encode a particular set of ncRNAs whose expression is altered in human cancers, and which can be regulated by microRNAs.
A more recent report by Pandolfi’s group (93) has introduced the revolutionary concept that miRNA effect on mRNA containing common miRNA recognition elements (MREs) can be affected by ceRNAs (competing endogenous RNAs): RNA transcripts, both protein coding and non-coding, can compete for miRNA binding, thus co-regulating each other.
Beside the existence of other RNAs able to interfere with miRNA function, other mechanisms can affect their regulatory action on target molecules: one example is represented by the evidence that mRNAs can present or develop specific alterations to escape miRNA control. Different studies have indeed reported the existence of oncogenic mRNAs carrying mutations or SNPs in their 3’UTR allowing them to avoid miRNA binding and consequent negative control, as demonstrated for example for let-7 and RAS interaction in lung cancer, where a SNP in the let-7 binding site on RAS 3’UTR alters RAS expression and is associated with higher occurrence risk (94). Another very interesting report is the study published by Sandberg R and colleagues (95), who discovered how proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. It would be of extreme interest to evaluate the selection for oncogenes with shortened 3’UTRs in different tumor types.
From bench to clinics: how far are we with microRNAs?
The potential of miRNA signatures to differentiate tumors in comparison with their normal counterpart, to discriminate between different subgroups of tumors and to predict outcome or response to therapy have focused scientist attention on these small molecules as potential clinical biomarkers, either diagnostic, predictive or prognostic.
Interestingly, it has been observed that primary tumors and metastasis from the same tissue show a similar pattern of microRNAs expression (96). Being a more accurate classifier than mRNA expression studies, miRNA profiling has thus revealed the potential to solve one of the most demanding issues in cancer diagnostic: the origin of metastasis of unknown primary tumors.
Another major issue in clinics is clearly represented by the need of biomarkers for an early diagnosis, which is usually associated with the best prognosis. microRNAs have revealed a great potential as new potential early diagnosis biomarkers, as shown in ductal adenocarcinoma, where overexpression of miR-205 and miR-21 precede phenotypic changes in the ducts (97). Moreover, miRNAs can be reliably extracted and detected from different biological fluids, as blood (either total blood, plasma or serum) (98,99), from circulating exosomes (100), urine (101, saliva (102,103) and even sputum (104,105), and it has been reported that the profile of circulating miRNA of individuals affected by different neoplasias reflects the pattern observed in the tumor tissues (106). This evidence suggests the fascinating possibility of using circulating microRNAs as easily detectable tumor biomarkers, especially for early diagnosis (107,108,109,110).
Concerning the possibility to use miRNAs as prognostic markers to predict outcome, several groups have successfully addressed this issue: after the first evidence in CLL, where a unique microRNA signature was associated with prognostic factors and disease progression in CLL (111) and lung cancer, where miR-155 overexpression and let-7a downregulation were able to predict poor disease outcome (112), several other reports have supported the significance of microRNAs as prognostic biomarkers (113,114).
Even though outcome prediction is certainly relevant, the prediction of response to specific therapies is of even greater clinical value, since it might be useful for a more accurate selection of patients potentially responsive to a specific therapy. miR-21, for example, is sufficient to predict poor response to adjuvant chemotherapy in adenocarcinomas (115) and in pancreatic cancer patients treated with gemcitabine (116).
The correlation between microRNA expression and response to specific therapies has also suggested their promising potential as therapeutic adjuvant, even though this hypothesis mostly derives from in vitro studies of gain or loss of function, where candidate miRNAs are initially identified in tumor cell lines with different degrees of resistance to specific therapeutic drugs and then targeted in order to overcome drug resistance (117,118,119,120). Beside chemotherapy, microRNAs can also improve the responsiveness to targeted therapies, as anti-estrogenic therapies (121,122) to Tyrosin Kinase Inhibitors (123).
However, although significant advances have been made for the future role of miRNAs in diagnostics, there have been far fewer reported successes in the development of miRNAs for use in therapy. Indeed, even though a number of reports have described the possibility to reintroduce (124, 125) or inhibit (126,127,128,129,130,131,132) microRNAs (reviewed by Iorio and Croce) (133), there are still many issues that need to be addressed for an effective translation in clinics, as the development of efficient methods of a specific drug delivery, and the accurate prevision of putative unwanted off target effects.
Conclusions and future perspectives
The past decade has witnessed an explosion of research focused on small non-coding RNAs: conserved among the species and involved in every biological process examined, these tiny RNA molecules have been demonstrated to be crucial regulators of gene expression.
Cancer is defined by abnormal and uncontrolled cell division, a phenotype that arises from the alteration of different mechanisms, leading not only to the misregulation of several protein coding genes, but also to a global change in miRNA profile. Being microRNAs major regulators of gene expression, with roles in nearly every area of cell behavior, development and survival, and able to regulate multiple targets acting as oncogenes or tumor suppressor genes, it is not surprising that their altered expression contributes to a substantial cell re-organization and is causally involved in so many different human tumors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest and Source of Funding: CM Croce is supported by grants of the National Cancer Institute; Iorio MV is supported by Start Up AIRC Grant.
References
- 1.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. Epub 2007 Jun 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. Epub 2007 Jun 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhayani MK, Calin GA, Lai SY. Functional relevance of miRNA* sequences in human disease. Mutat Res. 2011 Nov 6; doi: 10.1016/j.mrfmmm.2011.10.014. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. Epub 2005 Apr 3. [DOI] [PubMed] [Google Scholar]
- 10.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–D153. doi: 10.1093/nar/gkm995. Epub 2007 Dec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. Epub 2008 Oct 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19:2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- 16.Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, Fredrickson T, Landgraf P, Ramachandra S, Huppi K, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, Chen Y, Shen H. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. Epub 2007 Jan 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. Epub 2008 Feb 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. Epub 2005 Dec 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang R, Li L, Zhu D, Hou D, Cao T, Gu H, Zhang J, Chen J, Zhang CY, Zen K. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: evidence for a microRNA hierarchy system. Cell Res. 2012;22:504–515. doi: 10.1038/cr.2011.137. Epub 2011 Aug 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura T, Canaani E, Croce CM. Oncogenic All1 fusion proteins target Drosha-mediated microRNA processing. Proc Natl Acad Sci USA. 2007;104:10980–10985. doi: 10.1073/pnas.0704559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. Epub 2009 Nov 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, Takahashi T. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo X, Liao Q, Chen P, Li X, Xiong W, Ma J, Li X, Luo Z, Tang H, Deng M, et al. The microRNA-processing enzymes: Drosha and Dicer can predict prognosis of nasopharyngeal carcinoma. J Cancer Res Clin Oncol. 2011 Sep 28; doi: 10.1007/s00432-011-1058-1. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY, Diccianni MB, London WB, Chang CH, Yu AL. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010;70:7841–7850. doi: 10.1158/0008-5472.CAN-10-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grelier G, Voirin N, Ay AS, Cox DG, Chabaud S, Treilleux I, Léon-Goddard S, Rimokh R, Mikaelian I, Venoux C, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101:673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J, Getz G, Miska EA, varez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 31.Faber C, Horst D, Hlubek F, Kirchner T. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur J Cancer. 2011;47:1414–1419. doi: 10.1016/j.ejca.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muralidhar B, Winder D, Murray M, Palmer R, Barbosa-Morais N, Saini H, Roberts I, Pett M, Coleman N. Functional evidence that Drosha overexpression in cervical squamous cell carcinoma affects cell phenotype and microRNA profiles. J Pathol. 2011;224:496–507. doi: 10.1002/path.2898. Epub 2011 May 18. [DOI] [PubMed] [Google Scholar]
- 34.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 35.Weber B, Stresemann C, Brueckner B, Lyko F. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6:1001–1005. doi: 10.4161/cc.6.9.4209. [DOI] [PubMed] [Google Scholar]
- 36.Saito Y, Suzuki H, Tsugawa H, Nakagawa I, Matsuzaki J, Kanai Y, Hibi T. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene. 2009;28:2738–2744. doi: 10.1038/onc.2009.140. Epub 2009 Jun 8. [DOI] [PubMed] [Google Scholar]
- 37.Lehmann U, Hasemeier B, Christgen M, Muller M, Romermann D, Langer F, Kreipe H. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 38.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 39.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 40.Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, Sultmann H, Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- 41.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Ménard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 42.Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 43.Shin S, Lee EM, Cha HJ, Bae S, Jung JH, Lee SM, Yoon Y, Lee H, Kim S, Kim H, Lee SJ, Park IC, Jin YW, An S. MicroRNAs that respond to histone deacetylase inhibitor SAHA and p53 in HCT116 human colon carcinoma cells. Int J Oncol. 2009;35:1343–1352. doi: 10.3892/ijo_00000452. [DOI] [PubMed] [Google Scholar]
- 44.Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 45.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 46.Rhodes LV, Nitschke AM, Segar HC, Martin EC, Driver JL, Elliott S, Nam SY, Li M, Nephew KP, Burow ME, Collins-Burow BM. The histone deacetylase inhibitor trichostatin A alters microRNA expression profiles in apoptosis-resistant breast cancer cells. Oncol Rep. 2012;27:10–16. doi: 10.3892/or.2011.1488. Epub 2011 Oct 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. Epub 2009 Oct 22 Cancer Res, 70, 4528-38. Epub 2010 May 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, Li E, Serrano M, Millar S, Hannon G, Blasco MA. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, rtus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 51.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roccaro AM, Sacco A, Jia X, Azab AK, Maiso P, Ngo HT, Azab F, Runnels J, Quang P, Ghobrial IM. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood. 2010;116:1506–1514. doi: 10.1182/blood-2010-01-265686. Epub 2010 Jun 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galm O, Herman JG, Baylin SB. The fundamental role of epigenetics in hematopoietic malignancies. Blood Rev. 2006;20:1–13. doi: 10.1016/j.blre.2005.01.006. Epub 2005 Feb 23. [DOI] [PubMed] [Google Scholar]
- 54.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. Epub 2009 Feb 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mott JL, Kurita S, Cazanave SC, Bronk SF, Werneburg NW, Fernandez-Zapico ME. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J Cell Biochem. 2010;110:1155–1164. doi: 10.1002/jcb.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 59.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO. 2008;9:582–589. doi: 10.1038/embor.2008.74. Epub 2008 May 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, Iorio MV, Li M, Volinia S, Alder H, Nakamura T, Nuovo G, Liu Y, Nephew KP, Croce CM. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102:706–721. doi: 10.1093/jnci/djq102. Epub 2010 Apr 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Derynck R, Akhurst RJ, Balmain A. TGF-β signaling in tumor suppression and cancer progression. Nature Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 63.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 64.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 66.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 67.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. Epub 2007 May 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 69.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 70.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 71.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 73.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 75.Pulkkinen K, Malm T, Turunen M, Koistinaho J, Yla-Herttuala S. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 2008;582:2397–2401. doi: 10.1016/j.febslet.2008.05.048. [DOI] [PubMed] [Google Scholar]
- 76.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 78.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2011;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 79.Khew-Goodall Y, Goodall GJ. Stromal miR-320 keeps an oncogenic secretome in check. Nat Cell Biol. 2012;14:124–125. doi: 10.1038/ncb2431. [DOI] [PubMed] [Google Scholar]
- 80.Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K, Alder H, Nakamura T, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. Epub 2010 Aug 8. [DOI] [PubMed] [Google Scholar]
- 87.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 88.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. Epub 2007 May 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5' untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. Epub 2010 Oct 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qin W, Shi Y, Zhao B, Yao C, Jin L, Ma J, Jin Y. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One. 2010;5:e9429. doi: 10.1371/journal.pone.0009429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. Epub 2007 Nov 29. [DOI] [PubMed] [Google Scholar]
- 92.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G, Alder H, Rassenti L, Volinia S, Schmittgen TD, Kipps TJ, Negrini M, Croce CM. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 93.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. Epub 2011 Jul 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel R, Kulkarni T, Homer R, Zelterman D, Kidd KK, Zhu Y, Christiani DC, Belinsky SA, Slack FJ, Weidhaas JB. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. Epub 2008 Mar 23. [DOI] [PubMed] [Google Scholar]
- 97.du Rieu MC, Torrisani J, Selves J, Al Saati T, Souque A, Dufresne M, Tsongalis GJ, Suriawinata AA, Carrère N, Buscail L, Cordelier P. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010;56:603–612. doi: 10.1373/clinchem.2009.137364. Epub 2010 Jan 21. [DOI] [PubMed] [Google Scholar]
- 98.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. Epub 2008 Jul 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. Epub 2011 May 12. [DOI] [PubMed] [Google Scholar]
- 100.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 101.Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, Warnecke JM, Sczakiel G. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2009;28:655–661. doi: 10.1016/j.urolonc.2009.01.027. Epub 2009 Apr 17. [DOI] [PubMed] [Google Scholar]
- 102.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, Wong DT. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. Epub 2009 Aug 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. Epub 2009 Jul 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xie Y, Todd NW, Liu Z, Zhan M, Fang H, Peng H, Alattar M, Deepak J, Stass SA, Jiang F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67:170–176. doi: 10.1016/j.lungcan.2009.04.004. Epub 2009 May 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu Z, Fang H, Zhang J, Katz RL, Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. Epub 2008 Mar 3. [DOI] [PubMed] [Google Scholar]
- 107.Heneghan HM, Miller N, Kelly R, Newell J, Kerin MJ. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010;15:673–682. doi: 10.1634/theoncologist.2010-0103. Epub 2010 Jun 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 109.Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. 2010;23:1157–1164. doi: 10.1038/modpathol.2010.111. Epub 2010 Jun 4. [DOI] [PubMed] [Google Scholar]
- 110.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabro E, Croce CM, Pastorino U, Sozzi G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 112.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 113.Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–585. doi: 10.1136/gut.2008.175497. Epub 2009 Nov 30. [DOI] [PubMed] [Google Scholar]
- 114.Caramuta S, Egyházi S, Rodolfo M, Witten D, Hansson J, Larsson C, Lui WO. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–2070. doi: 10.1038/jid.2010.63. Epub 2010 Apr 1. [DOI] [PubMed] [Google Scholar]
- 115.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A, Falcone A, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. Epub 2010 May 11. [DOI] [PubMed] [Google Scholar]
- 117.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 118.Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152–2159. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 119.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 120.Chen F, Zhu HH, Zhou LF, Wu SS, Wang J, Chen Z. Inhibition of c-FLIP expression by miR-512-3p contributes to taxol-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2010;23:1457–1462. doi: 10.3892/or_00000784. [DOI] [PubMed] [Google Scholar]
- 121.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 122.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. Epub 2008 Aug 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, Ménard S, Croce CM, Tagliabue E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- 124.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 125.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 127.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 128.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 129.Ørom UA, Kaupinnen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 130.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008a;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 131.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. Epub 2007 Aug 30. [DOI] [PubMed] [Google Scholar]
- 133.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. Epub 2012 Feb 20. [DOI] [PMC free article] [PubMed] [Google Scholar]


