Abstract
To better understand the biogeography and relationship between temperature and community structure within microbial mats, the bacterial diversity of mats at a slightly alkaline, sulfide-containing hot spring was explored. Microbial mats that developed at temperatures between 75–52°C were collected from an area of approximately 1 m2 in Nakabusa, Nagano, Japan. Bacterial 16S rRNA genes from these samples were examined by terminal restriction fragment length polymorphism (T-RFLP) and clone library analysis. T-RFLP profiles revealed 66 unique fragments (T-RFs). Based on total T-RFs observed in environmental profiles and clone libraries, a temperature effect on diversity was determined, with complexity in the community increasing as temperature decreased. The T-RF pattern indicated four distinct community assemblages related to temperature. Members of the Aquificales and particularly the sulfuroxidizing bacterium Sulfurihydrogenibium were present at all temperatures and were the dominant component of mats taken at 75–67°C. Sulfide oxidation, which persisted throughout the temperature gradient, was the presumed dominant pathway of primary production above 67°C. As temperature decreased, successive additions of anoxygenic and oxygenic phototrophs increased primary productivity, allowing for diversification of the community.
Keywords: biogeography, sulfur oxidizing bacteria, photosynthetic bacteria, thermophile, Aquificales
Understanding how the environment shapes microbial communities is a current challenge in microbial ecology (60, 65). Microbes play important roles in structuring ecosystems and in geochemical cycles (4, 8, 37); however, because microorganisms are extremely small, numerically abundant, markedly diverse and globally dispersed, an understanding of their biogeography has proven difficult (31, 49, 76), if it exists at all (15). Recent investigations have explored whether the rules that structure communities for larger organisms also hold true for microbes, e.g., Bryant et al. (6) and Green and Bohannan (20). In particular, identifying environmental parameters that shape the distribution of microbial taxa and communities has recently received much attention on continental (14, 51, 63), regional (6, 47, 74), local (13, 23, 61, 70, 82) and millimeter scales (44, 48, 83).
Microbial mats that develop in thermal springs have been proposed as suitable habitats to explore patterns of microbial abundance (52). The distribution of bacteria along hot spring outflows and the effects of temperature on these transitions on meter to kilometer scales has been reported for terrestrial hot springs and a subsurface geothermal water stream (13, 36, 52, 72). On such scales, several factors, not just temperature but also chemical components of the water, pH and distance from the source may also affect the succession of microbes. In contrast, clear temperature gradients of spring water in a limited area are observed at Nakabusa hot spring (73), which is a sulfidic, slightly alkaline (pH ranging from 8.3 to 8.9; sulfide concentration approximately 0.1 mM) geothermal spring found in Nagano Prefecture, Japan. The outflow emerges from seams in a sediment-control dam wall and several types of microbial mats develop on this wall. Differences in the volume of the individual outflow(s) create a spatially and temporally heterogeneous environment with areas within a 1 m radius ranging from 75°C to 50°C. These microbial mats are an ideal location to clarify the bacterial distribution along a temperature gradient.
Nakabusa hot spring is one of several well-studied thermal springs worldwide. Roseiflexus castenholzii, a filamentous photosynthetic bacterium lacking chlorosomes, was isolated from microbial mats on the dam wall at Nakabusa (27). However, only a few studies have explicitly explored the microbial diversity found here. Kubo et al. (43) determined the community members contributing to sulfur metabolism from 65°C at Nakabusa hot spring using clone library analyses. In a pioneering molecular ecological study in Nakabusa, Nakagawa and Fukui (57) surveyed microbial communities of mats that had developed at 6 temperatures (76 to 48°C) from 2 sites using denaturing gradient gel electrophoresis (DGGE) targeting 16S rRNA genes. They detected approximately 16 bacterial and archaeal taxa. The DGGE profiles indicated the existence of a major break in community composition between 60 and 66°C. Using DGGE, Kubo et al. (43) also determined clear differences between community members from 65 and 75°C. These previous studies, however, did not address quantitative questions of temperature-based changes in microbial communities.
The aim of the present study was to expand the current understanding of how temperature structures microbial communities at Nakabusa hot spring by sampling more temperatures in a smaller area than had been previously attempted (43, 57). To address this question, a broad-scale survey of molecular diversity of bacteria across a range of temperatures from 75°C to 52°C using terminal restriction fragment length polymorphism (T-RFLP) and sequence analysis of clone libraries of 16S rRNA genes amplified from environmental DNA was undertaken. Using statistical and phylogenetic analyses, this work quantitatively describes the patterns of individual taxa within the bacterial community structure and discusses the effects of temperature changes on the overall community structure and metabolism.
Materials and Methods
Sampling site and sampling
Nakabusa hot spring is located in Nagano Prefecture in Japan (36°23′15″ N, 137°45′00″ E, 1,500 m elevation). The hot spring is slightly alkaline, with a pH of 8.6±0.3. Mat samples were collected aseptically from various points on the dam wall on July 5th, 2008 at temperatures ranging from 75 to 52°C (Fig. S1). Mat samples were brought to the laboratory at room temperature in either 15 mL or 50 mL polypropylene tubes filled with hot spring water and without headspace. Once in the laboratory (about 4 h after sampling), subsamples were homogenized using a Polytron homogenizer (Kinematica, Littau-Lucerne, Switzerland) and aliquots of the mat samples ranging from 0.06 to 0.34 g were placed in 1.5 mL microcentrifuge tubes and frozen at −20°C until DNA extraction.
DNA extraction, PCR and cloning
Bulk DNA from mat samples was isolated using a modified chloroform phenol extraction protocol as described in (43). Briefly, samples were disrupted with freeze/thaw and bead-beating steps, and then further lysed using lysozyme and proteinase K. After adding NaCl and hexadecyl-trimethyl-ammonium bromide (CTAB) to 0.95 M and 1% w/v respectively, genomic DNA was extracted by successive chloroform-isoamyl alcohol and phenol-chloroformisoamyl alcohol steps and precipitated with isopropanol. Bacterial 16S rRNA genes were PCR-amplified for both clone libraries and T-RFLP analysis using the universal primers Ba27f and Ba907R (55, 78). PCR was performed in 25 μL volumes using ExTaq (Takara, Otsu, Japan). The manufacturer’s general reaction protocol was followed with 1 μL extracted genomic DNA and 0.5 μM of each primer was added. The PCR protocol was as follows: 94°C for 3 min followed by 25 cycles of 94°C for 30 s, 52°C for 45 s, 72°C for 90 s, and a final elongation step at 72°C for 5 min. PCR products were verified on ethidium bromide-stained 1.5% (w/v) agarose gels. PCR products were gel purified in 50 μL sterile water using the QIAquick gel extraction kit (Qiagen, Hilden, Germany).
Clone libraries of selected samples (from 69, 63, 58, and 52°C) were constructed by ligating purified PCR products into the DynaExpress TA PCR cloning kit vector (BioDynamics, Tokyo, Japan). Escherichia coli JM109 competent cells were transformed chemically and grown overnight. PCR was performed as above but with 23 cycles on positive clones directly by touching the colony with a pipette tip and then dipping the tip into prepared PCR mix. Vector primer pairs were used according to the manufacturer’s instructions.
T-RFLP analysis
For T-RFLP analysis, the forward primer Ba27F was labeled with 6-carboxyfluorescein (FAM) at the 5′ end. For each DNA sample, PCR was performed as described above. Then, 2–4 μL of purified PCR product, or approximately 100 ng DNA from either environmental samples or clones were digested with 5 U MspI at 37°C for at least 16 h. For each digest, an aliquot of 0.3 μL (for environmental DNA) or 0.15 μL (for clones) was added to 15 μL 40:1 HiDi formamide: internal size standard (GeneScan 600 LIZ size standard; Applied Biosystems, Carlsbad, CA, USA). Labeled terminal-restriction fragments were denatured at 92°C for 1 min and immediately chilled on ice. Fragment profiles were determined using an ABI 3130xl capillary DNA sequencer in GeneScan mode. For environmental samples, fragments with a relative fluorescence signal >50 were retained for further analyses. To normalize T-RF profiles, the “standardization of DNA quantity between replicate profiles” approach described by Dunbar et al. (10) was used. Briefly, total florescence was calculated for each sample and a correction factor was applied so that all profiles had a total fluorescence equaling that of the sample with the lowest total fluorescence. After removing peaks that were corrected to <50 fluorescence units, the process was repeated three more times until no further peaks disappeared. T-RFLP data were converted to binary (presence/absence) matrices and within-sample proportional signal heights for summary and statistical analyses (7).
To identify individual T-RFs, clone libraries of samples from 75, 66, 60, and 56°C were constructed and analyzed in addition to the samples from 69, 63, 58, and 52°C as described above.
Sequencing, phylogenetic and cluster analyses
Partial 16S rRNA gene sequences of clones were obtained after PCR amplifications from E. coli colonies using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) with vector primers as sequencing primers. Sequences were run on an Applied Biosystems 3130xl capillary sequencer. Fragments were sequenced directly in both directions. The sequence data were visually edited and assembled using BioEdit Sequence Alignment Editor v.7.0.9.0 (24). Assembled sequences were screened for chimeras using Bellephron and Mallard (3, 33) and manually as needed by comparing partial sequence Basic Local Alignment Search Tool (BLAST) results (86) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Edited sequences were putatively identified using BLAST, with taxonomic affiliations crudely assigned by sequence identity to cultured isolates.
For phylogenetic analysis of Sulfurihydrogenibium-like sequences, sequence alignments were made with several other related taxa using ClustalX 2.0.5 with default gap penalties (30, 46). Alignments were visually inspected and gaps were excluded for phylogenetic analyses. Missing data were coded as missing. Maximum likelihood analyses were performed using the PhyML 3.0 on-line webserver (22). Likelihood analyses used the SPR and NNI tree searching option for 5 starting BIONJ trees (18) and were bootstrapped 100 times. Bayesian analysis was performed with MrBayes 3.1.2 (34, 69). Two independent runs of four chains (one cold) of Metropolis-coupled Markov chain Monte Carlo generations were run for 1×106 generations with trees sampled every 100 generations. Of these 10,000 sample trees, 2,500 were discarded as burn-in. Posterior probabilities were determined from a majority rule consensus tree calculated from the remaining trees. The nucleotide substitution model for both analyses was set to conform to the general time reversible model with gamma distribution and proportion of invariable sites (GTR+I+Γ) as indicated by AIC in jModeltest ver.0.0.1 (64). Parameter values were estimated by PHYML for the likelihood tree, and the default priors were set for Bayesian analysis.
Unweighted pair group method with arithmetic mean (UPGMA) cluster analysis was performed on the raw binary T-RF matrix using DendroUPGMA online (http://genomes.urv.cat/UPGMA/index.php) (17). Similarity coefficients and a distance matrix for the UPGMA dendrogram were calculated using the Dice similarity coefficient (9). For clone libraries, the Shannon diversity index (H′) and rarefaction curves were calculated using FastGroupII (71, 84).
Nucleotide sequence accession numbers
The nucleotide sequences reported in this study were deposited in the DDBJ/EMBL/GenBank database with the following accession numbers: JF826964–JF827017.
Results
T-RFLP, clone library and sequencing results
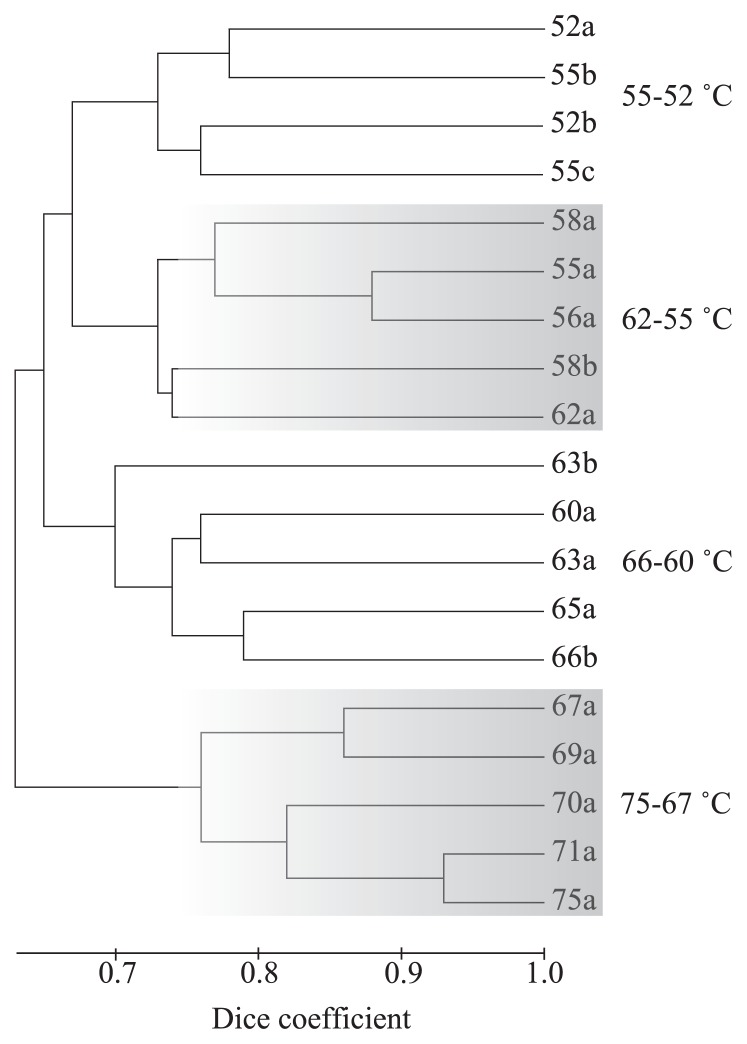
In all, 66 unique T-RFs were recovered from 19 environmental T-RFLP profiles sampled from 14 temperatures (75, 71, 70, 69, 67, 66, 65, 63, 62, 60, 58, 56, 55 and 52°C). The environmental T-RFs and their distributions are provided in Table S1. A significant inverse relationship between the sample temperature and the number of T-RFs was detected by linear regression, as shown in Fig. 1. As the sample temperature decreased the number of T-RFs increased. The UPGMA dendrogram using the raw binary T-RF matrix revealed four partitions related to temperature, with each group ranging from 75–67°C, 66–60°C, 62–55°C and 55–52°C (Fig. 2). Distance matrices for UPGMA calculated using the Jaccard similarity coefficient and Pearson’s r recovered near identical patterns (data not shown). These partitions could also be related to the shape and color of the mats, i.e., grey or white streamers at the highest temperatures, olive green mats of a few millimeters at around 65°C, and thick dark-green mats below 62°C. Visually, mats at 62–55°C were barely distinguishable from those at 55–52°C, but the dark-green mats, a part of which sometimes turned yellowish, tended to thicken as the temperature decreased.
Fig. 1.
Regression between temperature and total number of T-RFs for each sample collected. T-RF signals were normalized by sample to the lowest sample value to correct for variations in loading between samples and for an unbiased estimate of richness. Temperature has a significant effect on the number of T-RFs observed (R2=0.6276, t=6.97 (17); P<0.0001). Raw values also showed a similar relationship, but with a lower R2 value (0.39).
Fig. 2.
UPGMA cluster analysis of T-RF patterns for all samples analyzed. Sample number indicates temperature (°C).
The composition of clone libraries from four temperatures (69, 63, 58 and 52°C), each representing one of the four main by-temperature clusters identified in UPGMA analysis from Fig. 2, is shown in Table 1. At the highest temperatures, the mats were dominated by 16S rRNA gene sequences similar to the sulfur-oxidizing bacterium Sulfurihydrogenibium azorense Az-Fu1 (97–98% identity). The corresponding T-RF (=95) was found at all temperatures examined (Table 1 and Table S1). In addition, another Aquificales similar to the sulfur- and hydrogen-oxidizing Hydrogenobacter sp. GV1-4 (97% identity, T-RF=125) was detected. The aerobic heterotroph Thermus kawarayensis (98% identity, T-RFs=117 and 116) and the anaerobic heterotroph Eubacterium sp. (96% identity, T-RF=158) were also observed and widely distributed to lower temperatures.
Table 1.
Clone number and Shannon diversity index at different temperatures
| T-RF (bp) | No. of clones | Accession no. | Closest cultured match and identity | Putative phylum or class | Select predicted metabolic characteristics (reference) | |||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| 52°C | 58°C | 63°C | 69°C | |||||
| 95a | 1 | 2 | 7 | 35 | JF826981 | Sulfurihydrogenibium azorense Az-Fu1 97% | Aquificales | Sulfur/H2 oxidation (2) |
| 125 | 1 | 1 | JF826993 | Hydrogenobacter sp. GV1-4 97% | Aquificales | Sulfur/H2 oxidation (38) | ||
| 117 | 2 | 3 | JF826970 | Thermus kawarayensis 98% | Deinococcus/Thermus | Aerobic heterotroph (45) | ||
| 116 | 1 | JF82690 | Thermus kawarayensis 98% | Deinococcus/Thermus | Aerobic heterotroph (45) | |||
| 158 | 1 | 1 | 3 | JF826973 | Eubacterium sp. (OS type L) 96% | Firmicutes | Fermentation (77) | |
| 557 | 1 | 1 | JF827016 | Caldisericum exile AZM16c01 81% | Caldiserica | Fermentation (54) | ||
| 262 | 1 | JF827015 | Fervidobacterium sp. YNP 98% | Thermotogae | Fermentation (32) | |||
| 453 | 4 | JF827017 | Fervidobacterium nodosum Rt17-B1 98% | Thermotogae | Fermentation (32) | |||
| 265 | 1 | JF826986 | Fervidobacterium riparium 1445t 99% | Thermotogae | Fermentation (32) | |||
| 118 | 1 | JF826983 | Thermotogales sp. SRI-15 95% | Thermotogae | Fermentation (66) | |||
| 300 | 7 | JF826982 | Dictyoglomus sp. 1512 98% | Dictyoglomi | Fermentation (42) | |||
| 69 | 7 | 7 | 16 | JF826969 | Chloroflexus aggregans DSM 9485 98% | Chloroflexi | Anoxygenic phototroph (25) | |
| 63 | 1 | 3 | JF826984 | Chloroflexus aggregans DSM 9485 98% | Chloroflexi | Anoxygenic phototroph (25) | ||
| 83 | 1 | 3 | 2 | JF826976 | Ignavibacterium album 86% | Chlorobi | Fermentation (35) | |
| 65 | 1 | JF826988 | Caldimicrobium rimae 95% | Thermodesulfobacteria | Sulfur reduction (53) | |||
| 154 | 1 | JF826985 | Caldimicrobium rimae 95% | Thermodesulfobacteria | Sulfur reduction (53) | |||
| 298 | 1 | 1 | JF826987 | Thermodesulfovibrio hydrogeniphilus Hbr5 94% | Nitrospirae | Sulfate reduction (28) | ||
| 490 | 9 | 4 | JF826965 | Thermosynechococcus elongatus BP-1 99% | Cyanobacteria | Oxygenic phototroph (29) | ||
| 446 | 5 | JF826977 | Meiothermus sp. L462 100% | Deinococcus/Thermus | Aerobic heterotroph (85) | |||
| 148 | 2 | 1 | JF826974 | Meiothermus sp. L462 95% | Deinococcus/Thermus | Aerobic heterotroph (85) | ||
| 145 | 1 | JF827014 | Acidobacteria bacterium KBS 96 93% | Acidobacteria | Aerobic heterotroph (11) | |||
| 141 | 1 | JF826972 | Persephonella sp. 124-5-R1-1 85% | Aquificales | H2 oxidation (19) | |||
| 191 | 1 | JF826979 | Bellilinea caldifistulae 89% | Chloroflexi | Fermentation (80) | |||
| 513 | 3 | JF826978 | Bellilinea caldifistulae 90% | Chloroflexi | Fermentation (80) | |||
| 111 | 3 | JF826967 | Roseiflexus castenholzii DSM 13941 99% | Chloroflexi | Anoxygenic phototroph (27) | |||
| 61 | 1 | JF827013 | Roseiflexus castenholzii DSM 13941 99% | Chloroflexi | Anoxygenic phototroph (27) | |||
| 109 | 5 | JF826964 | Chloroflexi bacterium GNS-1 85% | Chloroflexi | Fermentation (21) | |||
| 426 | 3 | JF827011 | Dehalococcoides sp. 11a5 85% | Chloroflexi | Anaerobic H2 oxidation (50) | |||
| 173 | 2 | JF827012 | Elioraea tepidiphila TU-7 96% | Alphaproteobacteria | Aerobic heterotroph (1) | |||
| 77 | 2 | JF826966 | Thiobacillus aquaesulis 92% | Betaproteobacteria | Sulfur oxidation (79) | |||
| 450 | 1 | JF826968 | Alcaligenaceae bacterium BL-169 95% | Betaproteobacteria | Aerobic heterotroph (5) | |||
| Total | 39 | 33 | 46 | 44 | ||||
| bH′ | 2.52 | 2.39 | 1.99 | 0.81 | ||||
Several sequences for this T-RF are also listed in Fig. 3,
Shannon diversity index for 52°C includes clones for 3 unidentified TRFs=107, 156 and 191.
As the temperature decreased, sequences of photosynthetic anoxygenic bacteria appeared as a dominant component, i.e., Chloroflexus aggregans DSM 9485 (97–99% identity, T-RFs= 63 and 69). T-RF=69 was a major component at 63, 58 and 52°C. Two T-RFs (T-RFs=65 and 154), similar to the sulfur-reducing bacterium Caldimicrobium sp. were observed, as was the sulfate-reducing Thermodesulfovibrio sp. (T-RF=298). Clone libraries indicated that these sulfur reducers were found in small numbers (i.e., Caldimicrobiumlike clones were ~4% of the library at 63°C and Thermodesulfovibrio-like clones were ~2% of the libraries at both 63 and 52°C; Table 1). Sequences with low identity to Chlorobi (T-RF=83) also appeared; they were distributed to 52°C. The appearance of several members of the Thermotogae and Dictyoglomi, probable fermenters, also characterized the temperature region around 63°C. One of these, T-RF=265, related to Fervidobacterium sp. (62), was detected at 63, 60 and 55°C (Table S1). T-RFLP analysis also detected members of Deinococcus/Thermus and Acidobacteria (T-RFs=123, 148 and 379) below 66°C. In addition, a variety of unidentified T-RFs specific to 62 or 63°C were detected (Table S1).
At 65°C T-RF=490, similar to the oxygenic phototroph “Thermosynechococcus (Synechococcus) elongatus” BP-1 of the Synechococcus C1 lineage (63), appeared as an additional primary producer (99% identity); although based on the proportion of the total T-RF signal and clone library analysis, it was not widely distributed above 58°C. This T RF was distributed from 65 to 52°C (Table 1 and S1). A decrease in the population of Sulfurihydrogenibium sp. (T-RFs= 95 and 270) and an increase in those of C. aggregans (T-RFs=63 and 69) and “Thermosynechococcus” (T-RF=490) along the temperature gradient were also shown by changes in the proportional peak intensity of these T-RFs along the gradient (Fig. S2). In addition to the appearance of “Thermosynechococcus”, the aerobic heterotroph Meiothermus sp. was observed (T-RF=446) at 58°C. From 58°C, several low-identity fragments were identified belonging to the Aquificales and Chloroflexi (i.e., Persephonella-like and Bellilinia-like sequences).
The clone library of the sample taken at 52°C indicated the existence of the anoxygenic phototroph Roseiflexus castenholzii DSM 13941 (99% identity, T-RFs=61 and 111). T-RF=111 was widely observed at 62, 58, 55 and 52°C by T-RFLP analyses (Table S1). At cooler temperatures (i.e., below 58°C), several additional groups of Chloroflexi were detected as a major component of the clone libraries.
Phylogenetic analysis of Sulfurihydrogenibium sp
T-RF=95, related to Sulfurihydrogenibium sp., was distributed throughout the temperatures tested. A phylogenetic analysis was performed on clones from Nakabusa with this T-RF. Maximum likelihood and Bayesian analyses were performed on an alignment of 35 taxa that included 18 Nakabusa clones recovered from 8 temperatures in the present study that all shared T-RF=95. The maximum likelihood tree is shown in Fig. 3. All Nakabusa clones from this study clustered together in the phylogeny with other environmental sequences from this spring (NHS-01, NAB10 and NKB1-2) and two sequences (NAK9 and NAK14) from Nakanoyu hot spring, which is approximately 25 km from Nakabusa (40, 43, 57, 81). A second feature of this phylogeny was that all taxa/clones within the tree were grouped together by geographic location. The Nakabusa and Nakanoyu clones plus an additional Japanese isolate (Sulfurihydrogenibium subterraneum HGM-K1; from the subsurface hot-aquifer water at Hishikari gold mine) were monophyletic, as were the clones and isolates from North America (YO3AOP1, pCOF_65.7_G9 and SS-5) and East Russia (UZ1-1C1 and UZ1-1C2; 12, 23, 56, 75). The single European isolate Sulfurihydrogenibium azorense Az-Fu1 did not affiliate closely with any other group.
Fig. 3.
Maximum likelihood phylogenetic tree of Sulfurihydrogenibium spp. based on partial 16S rRNA gene sequences. Clones from this study are in bold. Genbank accession numbers follow each sequence name. Bootstrap support >50 for ML and Bayesian posterior probabilities >0.7 are given at the nodes supported by both measures. Hydrogenothermus marinus and Persephonella sp. EPR486 were used as outgroup taxa.
Discussion
Nakabusa hot spring is a promising site to clarify the factors determining microbial community composition and how microbial communities develop, since a spatially heterogeneous environment is observed within areas approximately 1 m in radius. Although numerous studies have characterized the transitions of microbial taxa across temperature gradients at thermal springs (13, 52, 57, 72), only a few have quantitatively analyzed these changes (e.g., 52). Based on their own clone library analyses and the results from several studies for thermal springs worldwide, Skirnisdottir et al. (72) observed that, in general community complexity increased inversely with temperature. Using barcoded pyrotag sequencing, Miller et al. (52) observed an increase in community complexity with respect to temperature on several hundred meter to kilometer scales. They also proposed that temperature plays a more important role than geography for determining species composition at thermal springs. The present study offers strong support of this idea; here, it was detected that temperature strongly shapes both community assembly and increasing diversity on the centimeter to meter scale. For Nakabusa hot spring, transitions of community composition across different temperatures have been previously observed (43, 57, 73), but the increase of diversity and the structuring of the community along the temperature gradient have not been reported.
Here, the first quantitative analysis of bacterial diversity across a broad temperature gradient (75–52°C) at Nakabusa hot spring revealed a clear relationship between temperature and community composition. Temperature had a clear effect on the number of T-RFs (Fig. 1). Fig. 1 shows strong evidence for an apparently linear temperature effect on diversity; richness increased by roughly 2–3 T-RFs per 5°C decrease. The Shannon diversity index for the individual clone libraries increased inversely to the source temperature of the library (Table 1). Likewise, rarefaction analysis of these libraries indicated that for libraries of comparable sample size, richness estimates were higher in clone libraries from lower temperatures (Fig. S3). In contrast, no clear trends were observed for oxidation-reduction potential (ORP), dissolved oxygen (DO) and sulfide across this temperature gradient (not shown). As would be expected, sulfide concentrations appeared to decline as distance from the source of hot spring waters increased, but the thickness of the mats frequently meant that concentrations of sulfide were maintained within the mats (approximately 0.1 mM).
The cluster analysis shown in Fig. 2 indicates a major break in community composition at 60–62°C, consistent with the previous report of a similar break between 60–66°C in Nakabusa (57). Additional breaks were also observed; in total, four distinct community assemblages were identified related to temperature. The break between the highest temperature zone and the 66 to 60°C zone can be characterized by the appearance of an anoxygenic phototroph, i.e., Chloroflexus aggregans. C. aggregans provides a second pathway for carbon fixation within the mat from 52 to 66°C in addition to the chemolithotrophic sulfur oxidation performed by members of the Aquificales. Furthermore, the filamentous cell morphology and motility of C. aggregans may contribute to the structure of the mats as suggested previously (26, 43). We propose that the appearance of C. aggregans causes an increase in the productivity and thickness of the mats (~3 mm), and provides a variety of niches for diverse species of anaerobic bacteria (Table 1).
Succession to the next coolest by-temperature community assemblage found from 62 to 55°C may be in part due to the presence of the oxygenic phototroph, related to “Thermosynechococcus elongatus” BP-1. This group becomes a major component of the mats at temperatures below 58–60°C (Table 1), but was observed in T-RF profiles up to 65°C (Table S1). This upper range for cyanobacteria in Nakabusa is consistent with previously reported in situ pigment analysis by absorbance spectrophotometry, but is higher than previous molecular analyses had reported (57, 73). The ability of cyanobacteria to utilize water as an electron source seems to further increase productivity. Additionally, cyanobacteria produce oxygen and extracellular organic compounds which may allow for the association of a variety of heterotrophs, including several species of aerobic bacteria, to exist within the thick mats (Table 1). Temperatures below 55°C were characterized by the further appearance of a variety of heterotrophs. In this way, transitions to cooler communities were largely characterized by the successive addition of chemolithotrophs, anoxygenic phototrophs, and oxygenic phototrophs (Table 1, Table S1 and Fig. S2). Based on previously reported genomic information, these transitions also indicated the successive addition of carbon fixation pathways, i.e., the reductive TCA cycle by Sulfurihydrogenibium (68), 3-hydroxypropionate pathway by Chloroflexus (41), and reductive pentose phosphate cycle by “Thermosynechococcus elongatus” (58). This addition of different autotrophic pathways provides a diversity of metabolic intermediates and expands the overall ability of the community to fix carbon under different environmental conditions. These factors may combine to expand available niches in the mats and to increase the matter and energy flux in the system, thus facilitating the observed increase in diversity and biomass. In natural ecosystems, it has been reported that productivity has a positive linear relationship with biodiversity on a local scale (16). These results strongly support this idea. Possible mechanisms of diversification concomitant with productivity are as follows: high productivity thickens the mat, the thick mat structure provides additional niches, and multiple primary producers lead to the diversification of available organic compounds. Furthermore, such biomass may allow for the presence in the mat community of bacteria (T-RFs=446 and 148) similar to proteolytic bacteria that produce cell-lytic enzymes (59, 85).
Ex situ anaerobic incubation of mats collected in Nakabusa from 75 to 65°C have shown biological sulfide production (43, 57). In addition to members of the Thermodesulfobacteria, (T-RFs=65, 154), which were previously identified as potential sulfide producers in Nakabusa (57), the clone library at 63°C also identified T-RF=298 as a candidate sulfate-reducing bacteria. These T-RFs were also detected at lower temperatures (Table S1). An anaerobic niche which is required for sulfate-reducing bacteria was likely provided by aerobic respiration of Sulfrihydrogenibium within the mats at 63°C. The sulfide produced by sulfate reducers may be simultaneously oxidized to sulfate and elemental sulfur (S0) by aerobic chemolithotrophy of Sulfrihydrogenibium and anaerobic photoautotrophy of C. aggregans, respectively, as suggested by Kubo et al. (43).
There was evidence of endemism in Japan for Sulfurihydrogenibium (Fig. 3), with all clones from Nakabusa, including clones NHS-01, NAB10 and NKB1-2 from previous studies clustering together (40, 43, 57). Such geographic diversification is of great interest to elucidate the ecological roles and diversification of this group. In contrast to an earlier finding that Aquificales were only present from 66°C and above (57), these results also indicate that Sulfurihydrogenibium sp. (T-RF=95) was present throughout the temperature gradient (Table 1 and Table S1). Kimura et al. (39) reported the high productivity of microbial mats dominated by Sulfurihydrogenibium sp. at Nakabusa hot spring. Continuous supply of electron donors from hot spring water may support the growth of Sulfurihydrogenibium sp. in a wide range of temperatures. Previous reports have revealed different temperature optima for isolated strains and divergence in gene sequences in this group across thermal gradients (23, 67). Takacs-Vesbach et al. has shown that in Yellowstone National Park this genus has a biogeography defined by different calderas (74). In Nakabusa hot spring, phylogenetic analysis of sequences belonging to T-RF=95 showed no evidence of structuring by temperature (Fig. 3). The conserved nature of the 16S rRNA locus may obscure the real relationships between temperature and Sulfurihydrogenibium diversity. Genetic and physiological identification of locally adapted ecotypes of Sulfurihydrogenibium in the narrow area at Nakabusa hot spring (Fig. 3) is worthy of further investigation.
Here it is shown that the microbial community at Nakabusa hot spring contains an amazing diversity; 13 bacterial phyla were identified across a temperature range of 23°C. In comparison, insufficient DNA fragments were recovered for Archaea, even when using targeted primers. This suggests that the archaeal population was much lower than that of Bacteria. From the deeply branching bacterial phylum Aquificales to more recently diverged photoheterotrophic and photoautotrophic bacteria, the successive appearance of a diverse species of bacteria was characterized by the addition of species rather than by the replacement of species (Table 1 and Table S1). The decrease in temperature allowed an increasing variety of bacterial species to grow; however, bacteria also appeared to be able to co-exist with the bacterial members proliferating from higher temperatures. Elucidation of the spatial and metabolic relationships between these groups inevitably gives us a deeper insight into the development and structuring of microbial ecosystems.
Supplementary Material
Acknowledgements
This work was supported in part by a research grant (2009–2011) of the Institute for Fermentation, Osaka, Japan.
References
- 1.Albuquerque L, Rainey FA, Nobre MF, da Costa MS. Elioraea tepidiphila gen. nov., sp. nov., a slightly thermophilic member of the Alphaproteobacteria. Int J Syst Evol Microbiol. 2008;58:773–778. doi: 10.1099/ijs.0.65294-0. [DOI] [PubMed] [Google Scholar]
- 2.Aguiar P, Beveridge TJ, Reysenbach A-L. Sulfurihydrogenibium azorense, sp. nov., a thermophilic hydrogen-oxidizing microaerophile from terrestrial hot springs in the Azores. Int J Syst Evol Microbiol. 2004;54:33–39. doi: 10.1099/ijs.0.02790-0. [DOI] [PubMed] [Google Scholar]
- 3.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azam F. Microbial control of oceanic carbon flux: the plot thickens. Science. 1998;280:694–696. [Google Scholar]
- 5.Bowman KS, Moe WM, Rash BA, Bae H-S, Rainey FA. Bacterial diversity of an acidic Louisiana groundwater contaminated by dense nonaqueous-phase liquid containing chloroethanes and other solvents. FEMS Microbiol Ecol. 2006;58:120–133. doi: 10.1111/j.1574-6941.2006.00146.x. [DOI] [PubMed] [Google Scholar]
- 6.Bryant JA, Lamanna C, Morlon H, Kerkhoff AJ, Enquist BJ, Green JL. Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc Natl Acad Sci USA. 2008;105:11505–11511. doi: 10.1073/pnas.0801920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Culman SW, Gauch HG, Blackwood CB, Thies JE. Analysis of T-RFLP data using analysis of variance and ordination methods: A comparative study. J. Microbiol Methods. 2008;75:55–63. doi: 10.1016/j.mimet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 8.van der Heijden MGA, Bardgett RD, van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. 2008;11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- 9.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 10.Dunbar J, Ticknor LO, Kuske CR. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl Environ Microbiol. 2001;67:190–197. doi: 10.1128/AEM.67.1.190-197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichorst SA, Kuske CR, Schmidt TM. Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl Environ Microbiol. 2011;77:586–596. doi: 10.1128/AEM.01080-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrera I, Longhorn S, Banta AB, Liu Y, Preston D, Reysenbach A-L. Diversity of 16S rRNA gene, ITS region and aclB gene of the Aquificales. Extremophiles. 2007;11:57–64. doi: 10.1007/s00792-006-0009-2. [DOI] [PubMed] [Google Scholar]
- 13.Ferris MJ, Ward DM. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 16.Fukami T, Morin PJ. Productivity-biodiversity relationships depend on the history of community assembly. Nature. 2003;424:423–426. doi: 10.1038/nature01785. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Vallvé S, Palau J, Romeu A. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol Biol Evol. 1999;16:1125–1134. doi: 10.1093/oxfordjournals.molbev.a026203. [DOI] [PubMed] [Google Scholar]
- 18.Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- 19.Götz D, Banta A, Beveridge TJ, Rushdi AI, Simoneit BRT, Reysenbach A-L. Persephonella marina gen. nov., sp. nov. and Persephonella guaymasensis sp. nov., two novel, thermophilic, hydrogen-oxidizing microaerophiles from deep-sea hydrothermal vents. Int J Syst Evol Microbiol. 2002;52:1349–1359. doi: 10.1099/00207713-52-4-1349. [DOI] [PubMed] [Google Scholar]
- 20.Green J, Bohannan BJM. Spatial scaling of microbial biodiversity. Trends Ecol Evol. 2006;21:501–507. doi: 10.1016/j.tree.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 21.Grégoire P, Fardeau M-L, Joseph M, et al. Isolation and characterization of Thermanaerothrix daxensis gen. nov., sp. nov., a thermophilic anaerobic bacterium pertaining to the phylum “Chloroflexi”, isolated from a deep hot aquifer in the Aquitaine Basin. Syst Appl Microbiol. 2011;34:494–497. doi: 10.1016/j.syapm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 23.Hall JR, Mitchell KR, Jackson-Weaver O, Kooser AS, Cron BR, Crossey LJ, Takacs-Vesbach CD. Molecular characterization of the diversity and distribution of a thermal spring microbial community by using rRNA and metabolic genes. Appl Environ Microbiol. 2008;74:4910–4922. doi: 10.1128/AEM.00233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 25.Hanada S, Hirashi A, Shimada K, Matsuura K. Chloroflexus aggregans sp. nov., a filamentous phototrophic bacterium which forms dense cell aggregates by active gliding movement. Int J Syst Evol Microbiol. 1995;45:676–681. doi: 10.1099/00207713-45-4-676. [DOI] [PubMed] [Google Scholar]
- 26.Hanada S, Shimada K, Matsuura K. Active and energy-dependent rapid formation of cell aggregates in the thermophilic photosynthetic bacterium Chloroflexus aggregans. FEMS Microbiol Lett. 2002;208:275–279. doi: 10.1111/j.1574-6968.2002.tb11094.x. [DOI] [PubMed] [Google Scholar]
- 27.Hanada S, Takaichi S, Matsuura K, Nakamura K. Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int J Syst Evol Microbiol. 2002;52:187–193. doi: 10.1099/00207713-52-1-187. [DOI] [PubMed] [Google Scholar]
- 28.Haouari O, Fardeau M-L, Cayol J-L, Fauque G, Casiot C, Elbaz-Poulichet F, Hamdi M, Ollivier B. Thermodesulfovibrio hydrogeniphilus sp. nov., a new thermophilic sulphate-reducing bacterium isolated from a Tunisian hot spring. Syst Appl Microbiol. 2008;31:38–42. doi: 10.1016/j.syapm.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Herdman H, Castenholz RW, Waterbury JB, Rippka R. Form-genus XIII. Synechococcus. In: Boone DR, Castenholz RW, Garity GM, editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Vol. 1. Springer; New York: 2001. pp. 508–512. [Google Scholar]
- 30.Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 31.Horner-Devine MC, Carney KM, Bohannan BJM. An ecological perspective on bacterial biodiversity. Proc. R. Soc. London, Ser B. 2004;271:113–122. doi: 10.1098/rspb.2003.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber R, Stetter KO. Genus II, Fervidobacterium Patel, Morgan and Daniel 1985b, 535VP. In: Boone DR, Castenholz RW, Garity GM, editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Vol. 1. Springer; New York: 2001. pp. 375–377. [Google Scholar]
- 33.Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 34.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- 35.Iino T, Mori K, Uchino Y, Nakagawa T, Harayama S, Suzuki K. Ignavibacterium album gen. nov., sp. nov., a moderately thermophilic anaerobic bacterium isolated from microbial mats at a terrestrial hot spring and proposal of Ignavibacteria classis nov., for a novel lineage at the periphery of green sulfur bacteria. Int J Syst Evol Microbiol. 2010;60:1376–1382. doi: 10.1099/ijs.0.012484-0. [DOI] [PubMed] [Google Scholar]
- 36.Inagaki F, Takai K, Hirayama H, Yamato Y, Nealson KH, Horikoshi K. Distribution and phylogenetic diversity of the subsurface microbial community in a Japanese epithermal gold mine. Extremophiles. 2003;7:307–317. doi: 10.1007/s00792-003-0324-9. [DOI] [PubMed] [Google Scholar]
- 37.Ishii S, Ikeda S, Minamisawa K, Senoo K. Nitrogen cycling in rice paddy environments: past achievements and future challenges. Microbes Environ. 2011;26:282–292. doi: 10.1264/jsme2.me11293. [DOI] [PubMed] [Google Scholar]
- 38.Kawasumi T, Igarashi Y, Kodama T, Minoda Y. Hydrogenobacter thermophilus gen. nov., sp. nov., an extremely thermophilic, aerobic, hydrogen-oxidizing bacterium. Int J Syst Bacteriol. 1984;34:5–10. [Google Scholar]
- 39.Kimura H, Mori K, Nashimoto H, Hanada S, Kato K. In situ biomass production of a hot spring sulfur-turf microbial mat. Microbes Environ. 2010;25:140–143. doi: 10.1264/jsme2.me09181. [DOI] [PubMed] [Google Scholar]
- 40.Kimura H, Sugihara M, Kato K, Hanada S. Selective phylogenetic analysis targeted at 16S rRNA genes of thermophiles and hyperthermophiles in deep-subsurface geothermal environments. Appl Environ Microbiol. 2006;72:21–27. doi: 10.1128/AEM.72.1.21-27.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klatt CG, Bryant DA, Ward DM. Comparative genomics provides evidence for the 3-hydroxypropionate autotrophic pathway in filamentous anoxygenic phototrophic bacteria and in hot spring microbial mats. Environ Microbiol. 2007;9:2067–2078. doi: 10.1111/j.1462-2920.2007.01323.x. [DOI] [PubMed] [Google Scholar]
- 42.Kochetkova TV, Rusanov II, Pimenov NV, Kolganova TV, Lebedinsky AV, Bonch-Osmolovskaya EA, Sokolova TG. Anaerobic transformation of carbon monoxide by microbial communities of Kamchatka hot springs. Extremophiles. 2011;15:319–325. doi: 10.1007/s00792-011-0362-7. [DOI] [PubMed] [Google Scholar]
- 43.Kubo K, Knittel K, Amann R, Fukui M, Matsuura K. Sulfur-metabolizing bacterial populations in microbial mats of Nakabusa hot spring, Japan. Syst Appl Microbiol. 2011;34:293–302. doi: 10.1016/j.syapm.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Kunin V, Raes J, Harris JK, et al. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol Syst Biol. 2008;4:198. doi: 10.1038/msb.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurosawa N, Itoh YH, Itoh T. Thermus kawarayensis sp. nov., a new member of the genus Thermus, isolated from Japanese hot springs. Extremophiles. 2004;9:81–84. doi: 10.1007/s00792-004-0419-y. [DOI] [PubMed] [Google Scholar]
- 46.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 47.Lau MCY, Aitchison JC, Pointing SB. Bacterial community composition in thermophilic microbial mats from five hot springs in central Tibet. Extremophiles. 2009;13:139–149. doi: 10.1007/s00792-008-0205-3. [DOI] [PubMed] [Google Scholar]
- 48.Ley RE, Harris JK, Wilcox J, et al. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl Environ Microbiol. 2006;72:3685–3695. doi: 10.1128/AEM.72.5.3685-3695.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martiny JBH, Bohannan BJM, Brown JH, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 50.Maymó-Gatell X, Chien Y, Gossett JM, Zinder SH. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- 51.Miller SR, Castenholz RW, Pedersen D. Phylogeography of the thermophilic cyanobacterium Mastigocladus laminosus. Appl Environ Microbiol. 2007;73:4751–4759. doi: 10.1128/AEM.02945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller SR, Strong AL, Jones KL, Ungerer MC. Bar-coded pyrosequencing reveals shared bacterial community properties along the temperature gradients of two alkaline hot springs in Yellowstone National Park. Appl Environ Microbiol. 2009;75:4565–4572. doi: 10.1128/AEM.02792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miroshnichenko ML, Lebedinsky AV, Chernyh NA, Tourova TP, Kolganova TV, Spring S, Bonch-Osmolovskaya EA. Caldimicrobium rimae gen. nov., sp. nov., an extremely thermophilic, facultatively lithoautotrophic, anaerobic bacterium from the Uzon Caldera, Kamchatka. Int J Syst Evol Microbiol. 2009;59:1040–1044. doi: 10.1099/ijs.0.006072-0. [DOI] [PubMed] [Google Scholar]
- 54.Mori K, Yamaguchi K, Sakiyama Y, Urabe T, Suzuki K. Caldisericum exile gen. nov., sp. nov., an anaerobic, thermophilic, filamentous bacterium of a novel bacterial phylum, Caldiserica phyl. nov., originally called the candidate phylum OP5, and description of Caldisericaceae fam. nov., Caldisericales ord. nov. and Caldisericia classis nov. Int J Syst Evol Microbiol. 2009;59:2894–2898. doi: 10.1099/ijs.0.010033-0. [DOI] [PubMed] [Google Scholar]
- 55.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakagawa S, Shtaih Z, Banta A, Beveridge TJ, Sako Y, Reysenbach A-L. Sulfurihydrogenibium yellowstonense sp. nov., an extremely thermophilic, facultatively heterotrophic, sulfuroxidizing bacterium from Yellowstone National Park, and emended descriptions of the genus Sulfurihydrogenibium, Sulfurihydrogenibium subterraneum and Sulfurihydrogenibium azorense. Int J Syst Evol Microbiol. 2005;55:2263–2268. doi: 10.1099/ijs.0.63708-0. [DOI] [PubMed] [Google Scholar]
- 57.Nakagawa T, Fukui M. Phylogenetic characterization of microbial mats and streamers from a Japanese alkaline hot spring with a thermal gradient. J Gen Appl Microbiol. 2002;48:211–222. doi: 10.2323/jgam.48.211. [DOI] [PubMed] [Google Scholar]
- 58.Nakamura Y, Kaneko T, Sato S, et al. Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 2002;9:123–130. doi: 10.1093/dnares/9.4.123. [DOI] [PubMed] [Google Scholar]
- 59.Nobre MF, da Costa MS. Genus II, Meiothermus Nobre, Trüper and da Costa 1996b, 605VP. In: Boone DR, Castenholz RW, Garity GM, editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Vol. 1. Springer; New York: 2001. pp. 414–420. [Google Scholar]
- 60.Okabe S, Oshiki M, Kamagata Y, et al. A Great Leap forward in Microbial Ecology. Microbes Environ. 25:230–240. doi: 10.1264/jsme2.me10178. [DOI] [PubMed] [Google Scholar]
- 61.Onodera Y, Nakagawa T, Takahashi R, Tokuyama T. Seasonal change in vertical distribution of ammonia-oxidizing archaea and bacteria and their nitrification in temperate forest soil. Microbes Environ. 2010;25:28–35. doi: 10.1264/jsme2.me09179. [DOI] [PubMed] [Google Scholar]
- 62.Otaki H, Everroad RC, Matsuura K, Haruta S. Production and consumption of hydrogen in hot spring microbial mats dominated by a filamentous anoxygenic photosynthetic bacterium. Microbes Environ. 2012;27:293–299. doi: 10.1264/jsme2.ME11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Papke RT, Ramsing NB, Bateson MM, Ward DM. Geographical isolation in hot spring cyanobacteria. Environ Microbiol. 2003;5:650–659. doi: 10.1046/j.1462-2920.2003.00460.x. [DOI] [PubMed] [Google Scholar]
- 64.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 65.Prosser JI, Bohannan BJM, Curtis TP, et al. The role of ecological theory in microbial ecology. Nat Rev Microbiol. 2007;5:384–392. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- 66.Reysenbach A-L. Family I. Thermotogaceae fam. nov. In: Boone DR, Castenholz RW, Garity GM, editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Vol. 1. Springer; New York: 2001. pp. 370–388. [Google Scholar]
- 67.Reysenbach A-L, Banta A, Civello S, et al. Aquificales in Yellowstone National Park. In: Inskeep W, McDermitt T, editors. Geothermal Biology and Geochemistry in Yellowstone. National Park Thermal Biology Institute; Bozeman: 2005. pp. 129–142. [Google Scholar]
- 68.Reysenbach A-L, Hamamura N, Podar M, et al. Complete and draft genome sequences of six members of the Aquificales. J Bacteriol. 2009;191:1992–1993. doi: 10.1128/JB.01645-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 70.Segawa T, Takeuchi N, Ushida K, Kanda H, Kohshima S. Altitudinal changes in a bacterial community on Gulkana Glacier in Alaska. Microbes Environ. 2010;25:171–182. doi: 10.1264/jsme2.me10119. [DOI] [PubMed] [Google Scholar]
- 71.Shannon CE. A mathematical theory of communication. Bell System Tech J. 1948;27:379–423. 623–656. [Google Scholar]
- 72.Skirnisdottir S, Hreggvidsson GO, Hjörleifsdottir S, Marteinsson VT, Petursdottir SK, Holst O, Kristjansson JK. Influence of sulfide and temperature on species composition and community ctructure of hot spring microbial mats. Appl Environ Microbiol. 2000;66:2835–2841. doi: 10.1128/aem.66.7.2835-2841.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sugiura M, Takano M, Kawakami S, Toda K, Hanada S. Application of a portable spectrophotometer to microbial mat studies: Temperature dependence of the distribution of cyanobacteria and photosynthetic bacteria in hot spring water. Microbes Environ. 2001;16:255–261. [Google Scholar]
- 74.Takacs-Vesbach C, Mitchell K, Jackson-Weaver O, Reysenbach A-L. Volcanic calderas delineate biogeographic provinces among Yellowstone thermophiles. Environ Microbiol. 2008;10:1681–1689. doi: 10.1111/j.1462-2920.2008.01584.x. [DOI] [PubMed] [Google Scholar]
- 75.Takai K, Hirayama H, Sakihama Y, Inagaki F, Yamato Y, Horikoshi K. Isolation and metabolic characteristics of previously uncultured members of the order Aquificales in a subsurface gold mine. Appl Environ Microbiol. 2002;68:3046–3054. doi: 10.1128/AEM.68.6.3046-3054.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Torsvik V, Øvreås L, Thingstad TF. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- 77.Wade WG. Genus I Eubacterium Prevot 1938, 294AL. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB, editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Vol. 3. Springer; New York: 2009. pp. 865–890. [Google Scholar]
- 78.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wood AP, Kelly DP. Isolation and physiological characterisation of Thiobacillus aquaesulis sp. nov., a novel facultatively autotrophic moderate thermophile. Arch Microbiol. 1988;149:339–343. [Google Scholar]
- 80.Yamada T, Imachi H, Ohashi A, Harada H, Hanada S, Kamagata Y, Sekiguchi Y. Bellilinea caldifistulae gen. nov., sp. nov. and Longilinea arvoryzae gen. nov., sp. nov., strictly anaerobic, filamentous bacteria of the phylum Chloroflexi isolated from methanogenic propionate-degrading consortia. Int J Syst Evol Microbiol. 2007;157:2299–2306. doi: 10.1099/ijs.0.65098-0. [DOI] [PubMed] [Google Scholar]
- 81.Yamamoto H, Hiraishi A, Kato K, Chiura HX, Maki Y, Shimizu A. Phylogenetic evidence for the existence of novel thermophilic bacteria in hot spring sulfur-turf microbial mats in Japan. Appl Environ Microbiol. 1998;64:1680–1687. doi: 10.1128/aem.64.5.1680-1687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoshinaga I, Amano T, Yamagishi T, Okada K, Ueda S, Sako Y, Suwa Y. Distribution and diversity of anaerobic ammonium oxidation (anammox) bacteria in the sediment of a eutrophic freshwater lake, Lake Kitaura, Japan. Microbes Environ. 2011;26:189–197. doi: 10.1264/jsme2.me10184. [DOI] [PubMed] [Google Scholar]
- 83.Youssef NH, Couger MB, Elshahed MS. Fine-scale bacterial beta diversity within a complex ecosystem (Zodletone Spring, OK, USA): The role of the rare biosphere. PLoS ONE. 2010;5:e12414. doi: 10.1371/journal.pone.0012414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu Y, Breitbart M, McNairnie P, Rohwer F. FastGroupII: A web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics. 2006;7:57. doi: 10.1186/1471-2105-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang X-Q, Zhang W-J, Wei B-P, Xu X-W, Zhu X-F, Wu M. Meiothermus cateniformans sp. nov., a slightly thermophilic species from north-eastern China. Int J Syst Evol Microbiol. 2010;60:840–844. doi: 10.1099/ijs.0.007914-0. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.