Abstract
Long-term memory requires the activity-dependent reorganization of the synaptic proteome to modulate synaptic efficacy and consequently consolidate memory. Activity-regulated RNA translation can change the protein composition at the stimulated synapse. Cytoplasmic polyadenylation element-binding protein 3 (CPEB3) is a sequence-specific RNA-binding protein that represses translation of its target mRNAs in neurons, while activation of N-methyl-d-aspartic acid (NMDA) receptors alleviates this repression. Although recent research has revealed the mechanism of CPEB3-inhibited translation, how NMDA receptor signaling modulates the translational activity of CPEB3 remains unclear. This study shows that the repressor CPEB3 is degraded in NMDA-stimulated neurons and that the degradation of CPEB3 is accompanied by the elevated expression of CPEB3's target, epidermal growth factor receptor (EGFR), mostly at the translational level. Using pharmacological and knockdown approaches, we have identified that calpain 2, activated by the influx of calcium through NMDA receptors, proteolyzes the N-terminal repression motif but not the C-terminal RNA-binding domain of CPEB3. As a result, the calpain 2-cleaved CPEB3 fragment binds to RNA but fails to repress translation. Therefore, the cleavage of CPEB3 by NMDA-activated calpain 2 accounts for the activity-related translation of CPEB3-targeted RNAs.
INTRODUCTION
Synaptic plasticity, which is the ability of neuronal synapses to undergo morphological and functional changes in response to various stimuli, forms the underlying molecular basis of memory. Activity-induced plasticity-related protein (PRP) synthesis sustains long-lasting synapse changes that are crucial for establishing and consolidating long-term memory. Neurons employ three strategies to increase the synaptic levels of specific PRPs upon activation. PRPs are deposited at the stimulated synapses by capturing the trafficking molecules in the form of proteins or RNAs de novo synthesized from soma (20, 51). The newly delivered PRP RNAs are then translated locally at synapses (51). Alternatively, PRPs are produced through translational activation of preexisting dendritic dormant mRNAs (10, 13, 45). RNA-binding proteins play essential roles in the modulation of PRP production by regulating dendritic RNA transport, translation, and/or degradation (10, 13, 22, 45). Cytoplasmic polyadenylation element-binding protein 3 (CPEB3) is a sequence-specific RNA-binding protein in vertebrates that likely influences PRP synthesis and memory function for the following reasons. First, the Drosophila melanogaster homologue of CPEB3, Orb2, is required for the long-term conditioning of male courtship behavior (32). Clinical research has shown that a single-nucleotide polymorphism (SNP) (a T-to-C substitution) in intron 3 of CPEB3 gene affects human episodic memory. Homozygous carriers of the C allele of SNP have poorer performance in the delayed verbal memory recall tests (56). This C allele of SNP located in the CPEB3 ribozyme sequence, exhibits more than 2-fold rate of self-cleavage in vitro (47). Thus, a decrease in the CPEB3 protein level caused by increasing truncated CPEB3 mRNA with this C allele of SNP has been proposed (47) but not yet experimentally proven. Second, CPEB3 inhibits the translation of GluA1 and GluA2 RNAs (29, 41), which encode the subunits of a crucial glutamate receptor, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor, in neurons. The activation of N-methyl-d-aspartic acid receptor (NMDAR) abrogates the CPEB3-repressed translation of reporter RNAs, indicating that neuronal activity modulates CPEB3-controlled translation (29). Third, CPEB3 is a nucleocytoplasmic shuttling protein with predominant residence in the cytoplasm; upon stimulation with NMDA, CPEB3 accumulates in the nucleus where it inhibits Stat5b-dependent transcription (42). The transcription of epidermal growth factor receptor (EGFR) gene is regulated by Stat5b-CPEB3 interaction, and pharmacological intervention of EGFR's kinase activity in the mouse brain influences spatial memory performance in the Morris water maze tasks (42). Together, CPEB3 governs the syntheses of several PRPs, and its subcellular localization and translational ability are subject to control by NMDAR signaling. Although our recent study shows that CPEB3 impedes translation through interaction with the elongation factor eEF2 (12), it is unclear how NMDA stimulation activates CPEB3-controlled translation.
Since CPEB3 distributed throughout the cytoplasm accumulates in the nucleus within 30 min of NMDA treatment (42), neuronal activity could partition CPEB3's function between nucleocytoplasmic compartments to control gene expression. This follow-up study investigates whether nuclear accrued CPEB3 further downregulates EGFR gene transcription. Unexpectedly, EGFR expression was elevated because a significant degradation of CPEB3 occurred within 2 h after the stimulation of NMDARs. Consequently, the decreased repressor CPEB3 leads to up-synthesis of EGFR, first through translational activation followed by transcriptional activation (or derepression). Thus, the activity-regulated degradation of a translational repressor can induce selected PRP expression rather than simply functioning as an opposing process of protein synthesis. Growing evidence indicates that the activity-dependent protein degradation is as vital as protein synthesis to modulate synaptic plasticity. Other than the ubiquitin-proteasome degradation system, proteolytic cleavage enzymes such as tissue-type plasminogen activator, matrix metalloproteases, and calpains, are also important for neuronal plasticity (26, 55, 58). Using inhibitors and knockdown approaches, we have delineated that calpain 2 is the key protease responsible for NMDA-induced CPEB3 degradation in neurons. This activity-dependent proteolysis of CPEB3 leads to the synthesis of the plasticity-related protein, EGFR.
MATERIALS AND METHODS
Antibodies and reagents.
The antibodies used were EGFR (catalog no. sc-03), LRP130 (catalog no. sc-66845), lamin B (catalog no. sc-6217), and CAPN2 (catalog no. sc-7533) antibodies from Santa Cruz Biotechnology; CAPN1 (catalog no. MAB3104), αII-spectrin (catalog no. MAB1622), and synaptophysin (sy38; catalog no. MAB5258) antibodies from Millipore; and Flag epitope antibody (catalog no. F1804) from Sigma-Aldrich. The anti-CPEB3 RNA-binding domain serum was raised using the C-terminal 268 amino acids (aa) of human CPEB3 (hCPEB3) produced in Escherichia coli. Calpain 1 (catalog no. 208712) and calpain 2 (catalog no. 208718) were purchased from Calbiochem. With the exception of lactacystin (catalog no. 426100) and calpain inhibitor III, carbobenzoxy-valinyl-phenylalaninal (MDL 28170, catalog no. 208722), from Calbiochem, all other chemicals were from Sigma-Aldrich.
Cell culture, lentivirus infection, and pharmacological treatment.
All experimental protocols using mouse or rat brains for primary cultures were carried out following guidelines of the Institutional Animal Care and Utilization Committee. Cortical or hippocampal neurons isolated from embryonic day 19 mouse or rat brains were cultured in neurobasal medium with 1× B27 supplement (5, 30). HEK293T cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. Lentivirus particles were generated using Virapower packaging system (Invitrogen). For the knockdown experiments, hippocampal or cortical neurons cultured for 6 days in vitro (DIV 6) were infected with lentiviruses overnight and collected on DIV 11 for RNA or protein extraction. For the expression of myc-CPEB3-(Flag)3, the neurons cultured for DIV 9 were infected with the lentivirus overnight and used on DIV 13 for Western blotting. On the day of harvest, the reagents and concentrations of reagents used to treat neurons were as follows: 50 μM NMDA, 2 μg/ml actinomycin D (ActD), 50 μg/ml cycloheximide (CHX), 20 μM MG132, 20 μM calpain inhibitor III, 10 μM lactacystin, 50 μM MK-801, and 2 mM EGTA. The NMDA treatment was usually applied 15 min after the addition of transcription, translation, proteasome, or protease inhibitors. The neuronal cultures of DIV 11 to 13 were typically stimulated with a 3-min pulse of NMDA, rinsed once, and incubated in the culture medium with or without the indicated inhibitors for an additional 1 to 2 h. Except for the experiments in Fig. 1 and Fig. 5D, the hippocampal neurons of DIV 11 and the synaptosome preparations were incubated with NMDA-containing medium for the indicated time.
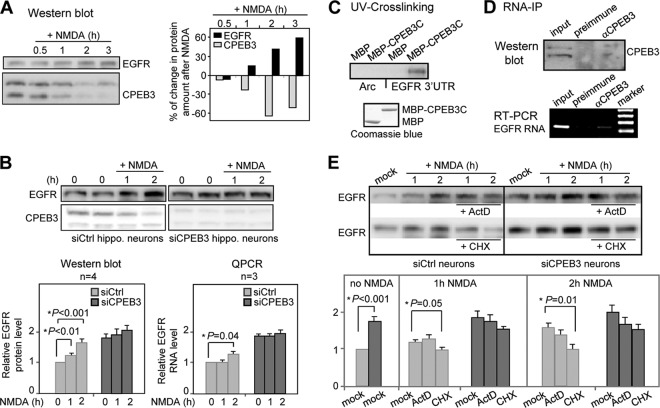
Fig 1.
NMDA-induced EGFR expression is mainly through CPEB3-controlled translation. (A) The Western blots of EGFR and CPEB3 using hippocampal neurons treated with 50 μM NMDA (+ NMDA) or not treated with NMDA for the indicated times (in hours). The changes in EGFR and CPEB3 protein levels after NMDA treatment are shown as a percentage of change compared to the signal obtained in unstimulated neurons. (B) The EGFR protein and RNA levels in control (siCtrl) and CPEB3 knockdown (siCPEB3) neurons treated with NMDA or not treated with NMDA for the indicated time were detected by Western blotting and RT-QPCR assay, respectively. The bar graphs at the bottom of the panel display the relative EGFR protein and RNA levels under different time treatments of NMDA with the signal from nontreated siCtrl neurons set at 1. The results were analyzed and expressed as means plus SEMs (error bars). An asterisk marks a statistical difference in the EGFR protein and RNA levels (two-tailed Student's t test) compared to those of nontreated cells. (C) The recombinant maltose-binding protein (MBP) and MBP fused to the RNA-binding domain of CPEB3 (MBP-CPEB3C) were UV cross-linked with 32P-labeled 3′-UTRs of Arc and EGFR RNAs, RNase A treated, and then analyzed by SDS-PAGE. The bottom panel shows Coomassie blue-stained proteins. (D) RNA immunoprecipitation (RNA-IP) using cortical neuronal lysate incubated with preimmune or CPEB3 IgG. EGFR RNA in the precipitated substances was detected by RT-PCR. αCPEB3, anti-CPEB3 antibody. (E) The Western blots of EGFR and CPEB3 using control (siCtrl) and CPEB3 knockdown (siCPEB3) neurons treated with NMDA or not treated with NMDA in the presence (+) or absence of 2 μg/ml actinomycin (ActD) or 50 μg/ml cycloheximide (CHX). The bar graph displays the relative EGFR protein level under different treatments with the signal from nontreated siCtrl neurons set at 1. The results from four independent experiments were analyzed and expressed as means plus SEMs. An asterisk denotes a statistical difference in the EGFR protein level (two-tailed Student's t test) compared to that of mock-treated cells.
Fig 5.
CPEB3 is degraded by NMDA-activated calpain 2 in the cytoplasmic and synaptic compartments. (A) Cultured hippocampal neurons of DIV 12 or 13 were treated with 50 μM NMDA for 3 min or not treated with NMDA and immunostained 30 min later with CAPN2 and CPEB3 antibodies. (B) The DIV 12 cortical neurons were treated with 50 μM NMDA for 3 min or not treated with NMDA and fractionated for cytosol and nuclear extracts at the indicated times (20 min or 1 or 2 h). The lysates were used for immunoblotting. (C) Low- and high-magnification views of CAPN2 and CPEB3 images are shown. The white arrowheads mark colocalized CAPN2 and CPEB3 in the dendrite. Bars, 20 μm (top) and 5 μm (bottom). (D) The postnuclear supernatant prepared from the rat brain was centrifuged at 12,000 × g (12k xg), and the supernatant (sup) and synaptosome-enriched pellet are shown below the blot. The synaptosomes were aliquoted and stimulated with 50 μM NMDA for the indicated times and harvested for Western blotting of CPEB3 and synaptophysin (sy38). The results from 5 independent synaptosome preparations were analyzed and expressed as means plus SEMs. An asterisk denotes a significant difference in the CPEB3 protein level (two-tailed Student's t test).
Plasmid construction.
The short hairpin RNA (shRNA) sequences, shRNA sequence 1 (GACTGTTGGCTCCTGGCTG) and shRNA sequence 2 (GAGTTCAACATCCTGTGGA), targeted against rat CAPN1 mRNA and shRNA sequence 1 (GAGAGAGCCATCAAGTACC) and shRNA sequence 2 (GAGAAGAAGGCTGACTACC) targeted against rat CAPN2 mRNA were cloned into the lentiviral vector pLL3.7-Syn (30). The shRNA clones, clone 1, TRCN0000087170 (GCTTTCAAATCTCTTGACAAA) and clone 2, TRCN0000087171 (CCACGTAGTCATTACTCTAAT) targeted against mouse Capns1 mRNA, were obtained from the RNAi Core Facility (RNAi stands for RNA interference) (Academia Sinica). The full-length and various truncated hCPEB3 DNA fragments were cloned into the pcDNA3.1-myc plasmid. The myc-CPEB3-(Flag)3 construct was first created in the pcDNA3.1-myc plasmid and then subcloned into pLL3.7-Syn in which the green fluorescent protein (GFP) coding region was replaced with the sequence of myc-CPEB3-(Flag)3. The open reading frames of CAPN1 and CAPN2 were PCR amplified from the rat neuronal cDNA and cloned into the pcDNA3.1-flag plasmid. The full-length coding sequence of hCPEB3 was cloned into the pMAL-c2 vector (New England BioLabs) to produce the maltose-binding protein (MBP)-CPEB3 recombinant protein. Rat EGFR 3′ untranslated region (3′-UTR) was PCR amplified from the hippocampal neuron cDNA with primers (5′-CGGAATTCCATTGAAGAGGCATTGTAC and 5′-CCGCTCGAGTGCTACTTTATTTTGTTATAAG) and cloned into the pcDNA3.1 plasmid.
The deletion mutants of EGFR 3′-UTR were generated by using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol.
Co-IP and RIP.
For coimmunoprecipitation (co-IP), the cultured neurons after DIV 14 treated with NMDA or not treated with NMDA or HEK293T cells expressing myc-tagged wild-type and mutant CPEB3 along with Flag-tagged CAPN1 or CAPN2 were lysed in the immunoprecipitation (IP) buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.2% NP-40, 1 mM EDTA, 1 mM dithiothreitol [DTT], 1× protease inhibitor cocktail, and 100 μg/ml RNase A). The neuronal lysates or 293T cell extracts were incubated with protein G beads (GE Healthcare) bound with anti-CPEB3 IgG or myc antibody, respectively, for 3 h at 4°C. The beads were then washed three times with 300 μl IP buffer, and the precipitated proteins were eluted with Laemmli sample buffer for Western blot analysis. For the RNA immunoprecipitation (RIP) assay, 1 × 107 cortical neurons were lysed in 800 μl of RIP buffer (diethyl pyrocarbonate [DEPC]-treated water containing 20 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 0.1% Triton X-100, 10% glycerol, 0.5 mM DTT, 1× protease inhibitor cocktail, and 40 U/ml RNase inhibitor) and centrifuged at 10,000 × g for 5 min. A portion (1/40th) of the resulting supernatant was saved for total RNA and protein isolation. The remaining solution was divided equally and incubated with protein G beads with preimmune IgG or anti-CPEB3 IgG for 2 h. The protein G beads were washed 6 times with 300 μl of RIP buffer. At the last wash, the beads were divided with one third of the beads eluted with Laemmli sample buffer for Western blotting, while the remaining beads were eluted with 4 M guanidine thiocyanate, followed by phenol-chloroform extraction and ethanol precipitation in the presence of 5 μg glycogen. The precipitated RNAs were used for reverse transcription-coupled PCR (RT-PCR). Primers designed to span exons 24 and 25 of EGFR mRNA are as follows: sense, 5′-CCGAGAGTTGATTCTCGAAT, and antisense, 5′-CGTCTTCCATGTCCTCCTC.
RNA extraction, cDNA synthesis, and Q-PCR.
Total RNA was extracted with TRIzol (Invitrogen). The cDNA was synthesized using oligo(dT) primer and ImProm-II reverse transcriptase (Promega). Quantitative PCR (Q-PCR) was conducted using the Universal Probe Library and Lightcycler 480 system (Roche). Data analysis was done using the comparative CT (threshold cycle value) method with the non-CPEB3 target, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA as the reference. The PCR primers are as follows: for Capns1, 5′-CGGTAGCCATGAACTTCCA and 5′-GCTGTAGAGATGCTCATTCAGGT; for CAPN1, 5′-GAGGCTGCAGGAACTACCC and 5′-ATAGTCGTCTGCGTCATCCA; for CAPN2, 5′-TCAGAAGGCTGTTTGCTCAG and 5′-GCGCTTGGCTAGAACTCTTC; and for GAPDH, 5′-GCCAAAAGGGTCATCATCTC and 5′-CACACCCATCACAAACATGG.
Immunostaining and confocal image acquisition.
The hippocampal neurons grown at a cell density of 15,000/cm2 on coverslips for 12 to 13 days in vitro were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) at room temperature for 15 min, permeabilized with 0.1% Triton X-100 for 5 min, blocked with 10% horse serum in PBS for 30 min, and then stained with CAPN2 and CPEB3 antibodies at 4°C overnight. After three washes with PBS, the coverslips were incubated with Alexa Fluor 488-labeled goat anti-rabbit and Alexa Fluor 594-labeled goat anti-mouse secondary antibodies for 1 h. The images were acquired with a Zeiss LSM 510 confocal microscope.
Synaptosome preparation and nucleocytosolic fractionation.
The freshly isolated rat brain was homogenized in buffer containing 0.32 M sucrose, 40 mM HEPES (pH 7.0), 4 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× protease inhibitor (Roche) and then centrifuged at 1,000 × g for 10 min. The resulting supernatant was collected and centrifuged at 12,000 × g for 30 min. The synaptosome-enriched pellet was gently resuspended in Krebs-Ringer phosphate buffer (123 mM NaCl, 3 mM KCl, 0.4 mM MgCl2, 0.5 mM NaH2PO4, 0.25 mM Na2HPO4, and 1 mg/ml glucose [pH 7.4]) (48). The synaptosome solution was aliquoted and either stimulated by the addition of 50 μM NMDA and 2.2 mM CaCl2 at 37°C for the designated time or not stimulated and then lysed in Laemmli sample buffer for Western blot analysis. Cortical neurons of DIV 12 in 100-mm dishes were treated with 50 μM NMDA for 3 min and then incubated for the indicated time before nucleocytosolic fractionation using a ProteoJET cytoplasmic and nuclear protein extraction kit (Fermentas) according to the manufacturer's protocol.
Recombinant protein purification and in vitro calpain-mediated cleavage assay.
The recombinant MBP-CPEB3 produced in E. coli was purified according to the established method for purification of MBP and MBP-CPEB3C (the RNA-binding domain [RBD] of CPEB3 fused to maltose-binding protein) (29). To prepare the Jurkat S100 lysate, Jurkat cells were homogenized in hypotonic buffer (20 mM HEPES [pH 7.5], 10 mM potassium acetate, and 1.5 mM magnesium acetate) and centrifuged at 100,000 × g for 1 h. The collected supernatant (S100) was used as a source of calpains for the in vitro cleavage assay (40). The [35S]methionine/cysteine-labeled substrates, p53 and CPEB3, were synthesized using TNT coupled reticulocyte lysate system (Promega) and then incubated with the S100 lysates in the presence or absence of 0.2 mM CaCl2, 2 mM EGTA, and/or 20 μM calpain inhibitor III at 37°C for 1 h. The resulting mixtures were separated on SDS-polyacrylamide gels, and the radioactive signals were monitored by the phosphorimager Typhoon FLA 4100 system (GE Healthcare). The MBP-CPEB3 protein was digested with the indicated units of calpain 1 or calpain 2 in buffer containing 40 mM HEPES (pH 7.0), 150 mM NaCl, and 5 mM DTT supplemented with 1 or 3 mM CaCl2, respectively. The reaction mixtures with or without 20 μM calpain inhibitor III or 5 mM EGTA were incubated at 37°C for 30 min prior to separation on SDS-polyacrylamide gels, followed by Coomassie blue staining.
Gel retardation and UV-cross-linking RNA-binding assay.
The 1904 RNA probe identified previously as the CPEB3-binding sequence and the 3′-UTRs of Arc and EGFR RNAs were labeled by in vitro transcription with [α-32P]UTP (29). For cross-linking with Arc or EGFR 3′-UTR probe, 20-μl reaction mixtures containing 105 cpm of labeled RNA, 50 μg heparin, 1 μg of recombinant protein (MBP or MBP-CPEB3C), and yeast tRNA in 10 mM HEPES (pH 7.4), 50 mM KCl, 1 mM MgCl2, 10% glycerol, and 0.5 mM DTT were kept on ice for 10 min and then irradiated with 1,200 J of UV (254-nm) light for 15 min. The UV-cross-linked samples were treated with 200 ng of RNase A at 37°C for 10 min and then resolved by SDS-PAGE. For the experiments using calpain-cleaved mixtures, the reaction mixtures containing one of the recombinant proteins, 0.5 μg of MBP or 2 μg of MBP-CPEB3, along with the presence or absence of 0.5 U calpain 2 and/or 5 mM CaCl2 were incubated in buffer containing 10 mM HEPES (pH 7.0), 50 mM KCl, 10% glycerol, and 0.5 mM DTT at 37°C for 30 min and then mixed with 1.2 × 105 cpm of [α-32P]UTP-labeled 1904 RNA probe. The RNA-protein mixtures were kept on ice for 20 min before being resolved by TBE-PAGE (PAGE with Tris-borate-EDTA [TBE]) for the gel retardation experiment or UV cross-linked, RNase A treated and separated on SDS-polyacrylamide gels.
DNA or RNA transfection and reporter assay.
HEK293T cells were transfected with plasmids encoding myc-tagged CPEB3 variants along with Flag-tagged CAPN1 or CAPN2 using Lipofectamine 2000 (Invitrogen). The RNAs used for transfection were synthesized with the mMessage mMachine T3 and T7 Ultra kits (Invitrogen) following the manufacturer's protocol. Hippocampal neurons cultured for 10 to 11 days were cotransfected with 2.2 pmol of β-galactosidase RNA, myc-tagged full-length CPEB3 (684 aa) or with the C terminus (aa 428 to 684) of CPEB3 RNA along with 0.05 μg of firefly luciferase RNA appended to the EGFR 3′-UTR and 0.05 μg of Renilla luciferase RNA using the TransMessenger Transfection reagent (Qiagen). For NMDA stimulation, the neurons were transfected with 0.2 μg of firefly luciferase RNA, 0.05 μg of Renilla luciferase RNA, and 1.5 μg of myc-CPEB3 RNA. The neurons after 3 h transfection were incubated for an additional 3 h with 50 μM NMDA or without NMDA before lysis for the dual-luciferase assay (Promega).
Mapping the calpain-cleaved site of CPEB3.
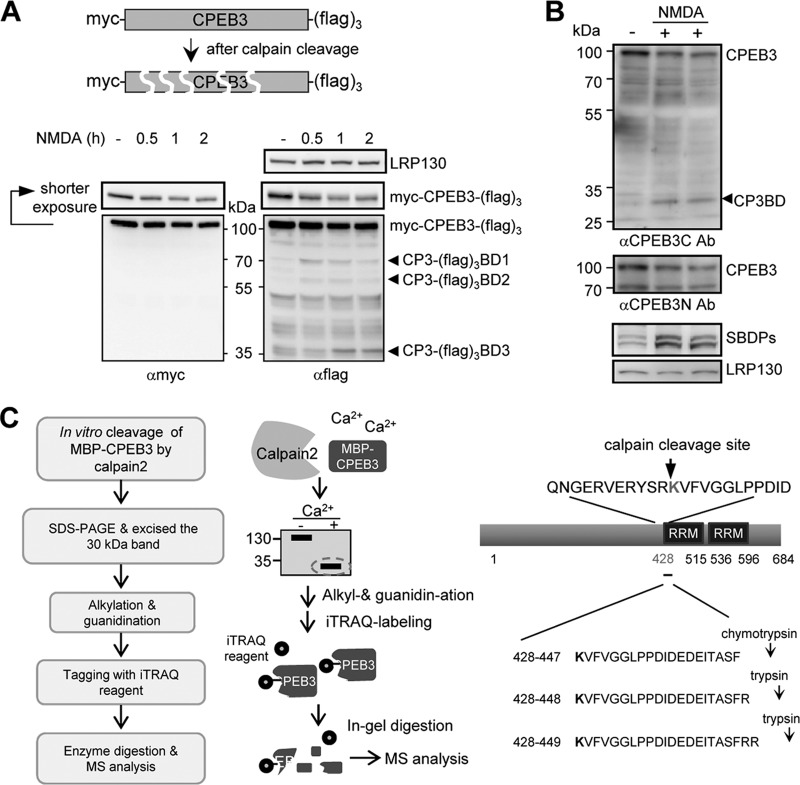
Twenty micrograms of MBP-CPEB3 was in vitro proteolyzed by calpain 2, and the cleavage products were divided in half, resolved on SDS-polyacrylamide gels, and stained with Coomassie blue. The two gel pieces containing the 30-kDa CPEB3 cleavage fragment were excised and washed with 25 mM triethylammonium bicarbonate (TEABC) and 50% acetonitrile (ACN) 3 to 5 times until they were destained. The gels were rinsed twice with 100% ACN and dried in a Speed-Vac. To prevent reformation of disulfide linkages between cysteine residues, the gel was reduced with 20 mM DTT in 50 mM TEABC buffer at 60°C for 1 h and then S-alkylated with 55 mM iodoacetamide in 50 mM TEABC buffer in the dark at room temperature for 45 min. The resulting gels were rinsed first with 50% ACN and then with 100% ACN and dried in a Speed-Vac. To block iTRAQ (isobaric tag for relative and absolute quantitation) labeling on the ε-amino group of lysine, the gels were guanidinated with O-methylisourea hemisulfate (15 mg dissolved in 90 μl of 7 M NH4OH) at 65°C for 10 min and then conjugated with iTRAQ reagent 114 at room temperature for 1 h with gentle vortices. The conjugated gels were digested with trypsin or chymotrypsin in 50 mM TEABC buffer at 37°C for 16 h. The digested peptides were dried and analyzed by reverse-phase liquid chromatography followed by tandem mass spectrometry (LC-MS/MS).
RESULTS
NMDA-increased EGFR synthesis requires the degradation of repressor CPEB3.
Because NMDAR signaling modulates the nucleocytoplasmic distribution and translational ability of CPEB3 and because the pharmacological intervention of the CPEB3's target, EGFR, in the mouse brain influences spatial memory performance (29, 42), we investigated whether NMDA stimulation can regulate EGFR expression in a CPEB3-dependent manner. Hippocampal neurons cultured for 11 days in vitro (DIV 11) were treated with 50 μM NMDA or were not treated with NMDA and harvested for Western blot analysis. Interestingly, the EGFR protein level was elevated in response to NMDA treatment, and this increase was inversely correlated with the level of CPEB3 in neurons (Fig. 1A). If downregulating CPEB3 elevates EGFR, this activity-related EGFR synthesis would be absent in the CPEB3 knockdown (KD) neurons. The control (siCtrl) and CPEB3 KD (siCPEB3) hippocampal neurons were stimulated with NMDA or were not stimulated and collected for Western blot and reverse transcription-coupled quantitative PCR (RT-QPCR) assays. The EGFR protein and RNA levels from 3 or 4 independent experiments were averaged and plotted as bar graphs with the amounts in the nontreated siCtrl neurons arbitrarily set at 1 (Fig. 1B). Similar to previous findings (42), the EGFR protein and RNA levels were upregulated in CPEB3 KD neurons (Fig. 1B). As expected, NMDA-increased EGFR expression was not obvious in CPEB3 KD neurons (Fig. 1B). When CPEB3 is appended to the nuclear localization sequence of simian virus 40 (SV40) large T antigen to increase its nuclear residence, this nuclear form of CPEB3 impedes Stat5-dependent transcription more than CPEB3 does (42). This implies that the NMDA-induced nuclear accumulation of CPEB3 should further inhibit EGFR gene transcription. Nonetheless, because of the long half-life of EGFR mRNA (>4 h) in neurons (42) and apparent degradation of CPEB3 in 2 h, NMDA stimulation eventually upregulates the EGFR RNA level instead of downregulating it (Fig. 1B). Notably, the increased EGFR RNA level at 2 h was preceded by the elevated EGFR protein level at 1 h. Because the 3′-UTR of the EGFR transcript contains U-rich sequences, CPEB3 may bind to and inhibit the translation of EGFR mRNA. The increased EGFR protein in the CPEB3 KD neurons may have, in part, resulted from the translational activation (or derepression) of EGFR RNA. To test the binding, the radiolabeled 3′-UTRs of EGFR and Arc (a nontarget control) RNAs were subjected to UV cross-linking with the RNA-binding domain (RBD) of CPEB3 fused to the maltose-binding protein (MBP-CPEB3C). In addition, neuronal lysates were incubated with preimmune or CPEB3 IgG, and the presence of EGFR RNA in the immunoprecipitates was determined by reverse transcription-coupled PCR (RT-PCR). Both in vitro (Fig. 1C) and in vivo (Fig. 1D) binding assays confirmed the interaction of CPEB3 with the EGFR RNA. Thus, CPEB3 inhibits EGFR synthesis at the translational level.
Elevated EGFR, observed within 2 h of NMDA treatment, was primarily a result of increased translation of EGFR RNA, because cycloheximide (CHX) hinders induction more efficiently than actinomycin D (ActD) does (Fig. 1E). Despite the fact that NMDAR signaling enhances nuclear retention of CPEB3 (42), a substantial decrease in the CPEB3 level eventually upregulates EGFR transcription, probably by reducing the nuclear pool of CPEB3 (Fig. 1B). This result agrees with the ActD treatment trend toward inhibition of NMDA-induced EGFR synthesis at the 2-h time point (Fig. 1E). Thus, NMDA-induced EGFR expression occurs first through translational control and is likely caused by degrading the repressor CPEB3. This is because the presence of proteasome inhibitor MG132, but not CHX and ActD, prevents the decline of CPEB3 in NMDA-treated neurons (see Fig. S1A in the supplemental material). Moreover, CPEB3 degradation is signaled through NMDA receptors because the NMDAR antagonist, MK-801, also blocks degradation (see Fig. S1B). In the absence of NMDA, the 2-h treatment of ActD, CHX, or MG132 has no obvious effect on CPEB3 and EGFR protein levels (see Fig. S1C).
NMDAR signaling induces calpain 2-dependent cleavage of CPEB3.
To understand the CPEB3 degradation mechanism and to examine whether this degradation could be observed in neurons stimulated with NMDA without accompanied excitotoxicity, the neurons were treated with a 3-min pulse of 20 to 50 μM NMDA, a protocol used to induce AMPA receptor endocytosis and chemical long-term depression (c-LTD) (4, 6, 34). The 50 μM pulse of NMDA caused more profound CPEB3 degradation than 20 μM (see Fig. S2A in the supplemental material), so we adopted the 3-min pulse of 50 μM NMDA stimulation for the following experiments. The stimulated neurons were incubated for various times before harvesting lysates for immunoblotting (Fig. 2A) or RNA for RT-QPCR (Fig. 2B) to measure the CPEB3 protein or RNA levels, respectively. The treated neurons were also labeled with propidium iodide to assess their viability. CPEB3 protein, but not RNA, was reduced within 1 to 2 h after a pulse of NMDA stimulation (Fig. 2A and B). The reduction in CPEB3 protein level was not caused by neuronal death from stimulation. The 3-min treatment caused less than 5% neuronal death after 2 h of incubation. In contrast, the 2-h NMDA incubation induced around 30% neuronal death (see Fig. S2B in the supplemental material). Using the 3-min pulse of 50 μM NMDA, we observed decreased CPEB3 level and elevated EGFR expression 2 h after stimulation (Fig. 2C). Moreover, CPEB3 degradation is not proteasome dependent, because only MG132, but not lactacystin, prevented the disappearance of CPEB3 in NMDA-stimulated neurons (Fig. 2D). MG132 inactivates not only proteasome but also other cysteine proteases, such as cathepsins and calpains (33). Thus, we pretreated neurons with calpain inhibitor III, a specific blocker of calpain 1 and calpain 2, prior to NMDA stimulation. This inhibitor blocked NMDA-triggered CPEB3 degradation (Fig. 2E). The chelation of extracellular calcium with EGTA also impeded CPEB3 degradation, presumably hindering the calcium influx through NMDA receptors (Fig. 2F). Thus, these results indicate that calpain 1 and calpain 2 are the most likely candidates responsible for CPEB3 degradation.
Fig 2.
Blockade of calpain activity inhibits NMDA-induced CPEB3 degradation. (A and B) The cortical neurons at DIV 11 or 12 were treated with 50 μM NMDA for 3 min and then harvested at the indicated times for Western blotting with CPEB3 and leucine-rich PPR motif-containing protein (LRP130) antibodies (A) or for RT-QPCR to measure the relative CPEB3 RNA level (B). (C) Similar to panel A but the lysates were also used for Western blotting of CPEB3, EGFR, and LRP130. The results from 4 independent experiments were analyzed and expressed as means plus SEMs. An asterisk marks a statistical difference in CPEB3 and EGFR protein levels with or without 3 min of stimulation of NMDA (two-tailed Student's t test). (D) The cortical neurons were pretreated without (mock) or with 20 μM MG132 or 10 μM lactacystin for 15 min prior to the presence (+) or absence (−) of NMDA stimulation (3-min pulse). The neurons were harvested 2 h after for Western blot analysis. (E and F) Similar to panel D, except 20 μM calpain inhibitor III and 2 mM EGTA were used to treat neurons before the pulse stimulation of NMDA.
Both calpain 1 and calpain 2 are heterodimeric calcium-activated cysteine proteases that consist of an identical small regulatory subunit, Capns1, and unique 80-kDa large catalytic subunits, CAPN1 and CAPN2, respectively (Fig. 3A) (23). These CAPN1 and CAPN2 subunits have high sequence homology with identical domain arrangement. The protease domain II of CAPN consists of two subdomains (IIa and IIb) with its substrate binding cleft between the two subdomains. The Ca2+-induced structural changes of calpain are prerequisites for forming the active catalytic triad (Cys115-His272-Asn296 in CAPN1 and Cys105-His262-Asn286 in CAPN2) to cleave the substrate (27). The regulatory subunit Capns1 binds to the domain IV of CAPN and is required for the full activity of both calpains (Fig. 3A), as calpain 1 and calpain 2 activities do not appear in Capns1 knockout cells (3). The mouse cortical neurons infected with the lentiviral particles (control [siCtrl]) or with the shRNAs against the common small subunit, Capns1 (siCapns1#1 and siCapns1#2), were harvested for RNA isolation, and the knockdown efficiency was evaluated by RT-QPCR to detect the level of Capns1 RNA (Fig. 3B). To confirm that calpains 1 and 2 are involved in NMDA-induced CPEB3 degradation, the control and Capns1 KD neurons were treated with NMDA or not treated with NMDA and then used for immunoblotting of CPEB3 along with the loading control, leucine-rich PPR motif-containing protein (LRP130), and the calpain-targeted control, αII-spectrin (Fig. 3C). The cytoskeletal protein αII-spectrin, which regulates plasma membrane dynamics and the docking of cell surface receptors (49), is cleaved into the 150-kDa and 145-kDa αII-spectrin breakdown products (SBDPs) by activated calpains 1 and 2 (43). The NMDA-induced CPEB3 decay and SBDP production in Capns1 KD neurons were hindered, presumably due to the reduced proteolytic activity of both calpains. We next examined which calpain (or both) participated in CPEB3 degradation by knocking down the catalytic subunit, CAPN1 or CAPN2, since no inhibitor is available to specifically block the individual calpain. Similar to the experiments in Fig. 3, the knockdown efficiencies of CAPN1 and CAPN2 were examined by RT-QPCR (Fig. 4A). The deficiency of CAPN2, but not CAPN1, impaired the proteolysis of CPEB3 and αII-spectrin (Fig. 4B). The Western blot results from four independent experiments were averaged and displayed as means plus standard errors of the means (SEMs) (Fig. 4B). Despite the presence of CAPN1 RNA in neurons (Fig. 4A), we could not detect CAPN1 protein using hippocampal and cortical neuronal lysates (data not shown). Previous research shows that the RNA level of CAPN1 is 50-fold lower than that of CAPN2 in neurons and that CAPN1 can be detected in the lysates prepared from fibroblasts but not neurons by immunoblotting (8). The RT-QPCR measurement in this study also shows that CAPN2 RNA is expressed approximately 100-fold more than CAPN1 RNA, assuming that the amplification efficiencies of primers used for the PCR of CAPN1 and CAPN2 cDNAs are comparable. Thus, calpain 2 is the main protease responsible for degrading CPEB3 in NMDA-treated neurons. Calpain 2 can be activated by the influx of calcium through NMDA receptors. Alternatively, the interaction of CPEB3 and calpain 2 might enhance upon NMDA stimulation to accelerate CPEB3 degradation. Using neurons treated with NMDA or not treated with NMDA in the presence of calpain inhibitor III to block CPEB3 degradation, the coimmunoprecipitation (co-IP) assay revealed that NMDAR signaling did not regulate the association of CPEB3 and calpain 2 (Fig. 4C). This indicates that the activation of calpain 2 by elevated calcium accounts for NMDA-induced CPEB3 degradation. The co-IP assays using the 293T cells expressing various myc-tagged truncated CPEB3 mutants along with Flag-tagged CAPN1 or CAPN2 show that amino acids 216 to 317 of CPEB3 are required to interact with CAPN1 (see Fig. S3A in the supplemental material) and CAPN2 (Fig. 4D) to a similar degree. This region of CPEB3 is also required for its translational repression activity (12), so we could not generate a nondegradable CPEB3 mutant with repressor function. Because a domain IV-deleted CAPN2 mutant was coprecipitated with myc-CPEB3 (see Fig. S3B), the binding of CPEB3 to CAPN1 and CAPN2 was not mediated through the regulatory subunit Capns1.
Fig 3.
NMDA-induced CPEB3 degradation is ameliorated in the Capns1 knockdown neurons. (A) Schematic diagram showing the domain structure of the catalytic subunits of calpain 1 and calpain 2, CAPN1 and CAPN2, respectively. The small regulatory subunit, Capns1, binds to the domain IV of CAPN. The catalytic triad, Cys-His-Asn, located in domain II, is essential for protease activity. a.a., amino acids. (B) The two shRNAs, siCapns1#1 and siCapns1#2, targeted against the mouse Capns1 were introduced into mouse cortical neurons using lentiviral infection. The Capns1 mRNA level in control (siCtrl) and knockdown (siCapns1) neurons was analyzed by RT-QPCR. (C) The siCtrl and siCapns1 neurons were treated with a 3-min pulse of 50 μM NMDA or not treated with NMDA and then collected 1 or 2 h later for Western blot analysis using CPEB3, LRP130, and αII-spectrin (SBDPs, αII-spectrin breakdown products) antibodies. The results were analyzed and expressed as means plus SEMs. An asterisk marks a statistical difference in the CPEB3 protein level (two-tailed Student's t test).
Fig 4.
Calpain 2 proteolyzes CPEB3 in the NMDA-treated neurons. (A) The two shRNAs, siCAPN1#1 (or siCAPN2#1) and siCAPN1#2 (or siCAPN2#2), targeted against the rat CAPN1 (or CAPN2) were introduced into rat neurons using lentiviral infection. The CAPN1 and CAPN2 RNA levels in the siCtrl, siCAPN1, and siCAPN2 neurons were detected by RT-QPCR from two independent measurements. The results were expressed as means plus SEMs. (B) The control and various knockdown neurons treated with a 3-min pulse of 50 μM NMDA or not treated with NMDA were analyzed 2 h later by Western blotting. The results were expressed as means plus SEMs. An asterisk denotes a significant difference in the CPEB3 protein level (two-tailed Student's t test). (C) The cultured cortical neurons were treated with 20 μM calpain inhibitor III along with NMDA stimulation or without NMDA stimulation. The cell lysates were pulled down with control or CPEB3 IgG and probed with CAPN2 and CPEB3 antibodies. IP, immunoprecipitation; IB, immunoblotting. (D) The top diagram denotes the various CPEB3 truncated mutants. The 293T cell lysates containing Flag-tagged CAPN2 along with myc-tagged full-length (wild-type [wt]) or truncated mutant CPEB3 were precipitated with the myc antibody and immunoprobed with myc and Flag antibodies. RRM, RNA recognition motif; Zif, zinc finger.
CPEB3 degradation by NMDA-activated calpain 2 occurs in the cytoplasmic and synaptic compartments.
Because NMDA also triggers nuclear accumulation of CPEB3 (42), we investigate whether calpain 2-mediated CPEB3 degradation takes places in the cytoplasm or nucleus. The distribution of CAPN2 and CPEB3 is mainly located in the cytoplasm; however, NMDA signaling causes accumulation of CPEB3 but not CAPN2 in the nucleus (Fig. 5A). This suggests that CPEB3 is proteolyzed by calpain 2 in the cytoplasm. Although the nuclear level of CPEB3 increased within 20 to 30 min after the pulse of NMDA treatment (Fig. 5A and B), not only the cytoplasmic level but also the nuclear level of CPEB3 was eventually downregulated at 1 to 2 h (Fig. 5B). CPEB3 and CAPN2 were distributed in dendrites and partially colocalized (Fig. 5C, arrowheads). Thus, we biochemically isolated synaptosomes and examined whether NMDA stimulation could induce CPEB3 degradation in the synaptic compartment (Fig. 5D). The Western blots show that the level of CPEB3 but not synaptophysin (sy38) declined in the NMDA-stimulated synaptosomes. The results from five independent experiments were displayed as means plus SEMs (Fig. 5D).
CPEB3 is cleaved by calpain 1 and calpain 2 in vitro.
Calpain does not recognize specific amino acid sequences but usually cleaves substrate proteins at linker regions that connect globular functional domains, which effectively alters their properties (52). To identify any functional motif of CPEB3 left after cleavage that could not be detected by the antibody, we used the Jurkat S100 cytoplasmic extract as a source of calpains to trace the degradation pattern of [35S]Met/Cys-labeled CPEB3 (11, 46). The radiolabeled CPEB3 and the calpain substrate p53 (40) were incubated with the Jurkat S100 lysate in the presence or absence of calcium. When coincubated with the calcium-added S100 lysate, p53 was proteolyzed to yield two major fragments of approximately 50 and 41 kDa in size (Fig. 6A). Similar to p53, CPEB3 was cleaved multiple times to produce prominent proteolytic fragments with a size of approximately 28 to 30 kDa by the calpain activity in the S100 extract because the cleavage of CPEB3 was inhibited in the presence of calcium chelator EGTA and calpain inhibitor III (Fig. 6A).
Fig 6.
CPEB3 is cleaved by calpain 1 and calpain 2 in vitro. (A) The [35S]Met/Cys-radiolabeled p53 and CPEB3 were synthesized using the reticulocyte lysate system and then mixed with the Jurkat cell cytoplasmic extract in the presence or absence of 0.2 mM CaCl2 along with other indicated chemicals (EGTA, dimethyl sulfoxide [DMSO], and calpain inhibitor III [Cal inh III]) at 37°C for 1 h. The radioactive mixtures were separated on SDS-polyacrylamide gels and analyzed by a phosphorimager. The arrowheads indicate the positions of major breakdown fragments after cleavage. (B) The recombinant maltose-binding protein (MBP)–CPEB3 was incubated with or without purified calpain 1 or calpain 2 in the Ca2+-containing buffer along with other indicated reagents at 37°C for 1 h. The mixtures were separated by SDS-PAGE and stained with Coomassie blue. The asterisks denote various breakdown products, CAPN1BD, CAPN2 BD, and CP3BD, derived from CAPN1, CAPN2, and CPEB3, respectively.
Calpain 1 and calpain 2 are also called μ- and m-calpain, respectively, because of the differential requirement in the calcium concentration needed to activate the protease activity in vitro. However, these calpains generally show no discrimination in terms of substrate specificity (23). To test whether the knockdown of CAPN2 but not CAPN1 weakened NMDA-induced CPEB3 degradation was simply because of the abundance of CAPN2 in neurons, we used purified calpain 1, calpain 2, and recombinant CPEB3 protein for the in vitro cleavage assay. To increase the solubility of CPEB3, the protein was appended with MBP at the N terminus (Fig. 6B). Because MBP is not the substrate of calpains (see Fig. S4A in the supplemental material), the cleaved product at the size of 43 kDa is MBP. In addition, the catalytic subunits of activated calpain 1 and calpain 2 were autolyzed to produce the 37- and 40-kDa breakdown fragments, CAPN1BD and CAPN2BD, respectively (see Fig. S4B). Both calpains proteolyzed MBP-CPEB3 to yield a major breakdown product of 30 kDa, CP3BD (Fig. 6B). To compare the proteolytic efficiency of CPEB3 by calpain 1 and calpain 2, MBP-CPEB3 was incubated with increasing doses of enzymes, and the remaining MBP-CPEB3 was monitored by Western blotting using CPEB3 antibody. Calpain 1 cleaved CPEB3 with a slightly better efficiency than calpain 2 did (see Fig. S4C), confirming that calpain 2 is the major calpain responsible for cleaving CPEB3 simply because of its abundance in neurons.
The calpain-cleaved CPEB3 fragment binds to RNA but fails to repress translation.
In most cases, calpains proteolyze their substrates in a limited manner and produce large segments with intact domains. Since the CPEB3 antibody recognizes the N terminus of CPEB3 and cannot detect the 30-kDa breakdown product, CP3BD, in the Western blots, this cleaved fragment might correspond to the C-terminal RNA-binding domain with a calculated molecular mass of approximately 29 kDa. Thus, we used neurons infected with the lentivirus expressing myc-CPEB3 appended with triplicate Flag tags at the C terminus, myc-CPEB3-(Flag)3, so the N-terminal and C-terminal breakdown fragments in NMDA-treated neurons could be monitored by myc and Flag antibodies, respectively. Unlike the myc antibody, which recognized only gradually reduced full-length myc-CPEB3-(Flag)3, the Flag antibody detected several NMDA-induced proteolyzed products, CP3-(Flag)3BD1, -BD2, and -BD3 (Fig. 7A). As time increased, the 35-kDa CP3-(Flag)3BD3 accumulated as the major end product in vivo (Fig. 7A), with a size similar to the 30-kDa in vitro-proteolyzed CP3BD fragment (Fig. 6) with the addition of 5 kDa of triplicate Flag tags. Thus, we used the RBD of CPEB3 as the antigen to generate the C-terminus-specific antibody, anti-CPEB3C antibody. Unlike the original antibody (anti-CPEB3N antibody), the anti-CPEB3C antibody detected not only the declined level of full-length CPEB3, but also the accompanying elevated amount of 30-kDa breakdown fragment after the activation of NMDARs (Fig. 7B), confirming that this cleaved product was indeed generated endogenously.
Fig 7.
Identification of the calpain cleavage site at the extreme C terminus of CPEB3. (A) The neurons infected with the lentivirus expressing myc-CPEB3-(Flag)3 were treated with NMDA and harvested at the indicated times. The neuronal lysates were immunoblotted with LRP130, myc, and Flag antibodies. The arrowheads show the positions of three breakdown fragments, CP3-(Flag)3BD1, -BD2, and -BD3. (B) The NMDA-treated neuronal lysates were analyzed by Western blotting using antibodies recognizing the N or C terminus of CPEB3 (anti-CPEB3N antibody [αCPEB3N Ab] or anti-CPEB3C), SBDPs, and LRP130. (C) Work flow used to determine the cleavage site on CPEB3. The calpain 2 cleaved C-terminal fragment of MBP-CPEB3 separated by SDS-PAGE was isolated and alkylated, guanidinated, and then labeled with the iTRAQ reagent on the gel. The labeled gel was digested with trypsin or chymotrypsin, which cleaved bonds at the carboxyl sides of lysine/arginine residues or tyrosine/tryptophan/phenylalanine residues, respectively. The collected peptides were analyzed by tandem mass spectrometry. The iTRAQ-conjugated peptides are listed on the right.
To determine the cleavage site (i.e., the most N-terminal amino acid of the 30-kDa proteolytic fragment), the CP3BD produced in vitro (Fig. 6B) was isolated on gels, and the most N-terminal amino acid was labeled with the iTRAQ (isobaric tag for relative and absolute quantitation) reagent. Figure 7C shows a schematic flow chart of this experiment. Because the iTRAQ reagent is conjugated to primary amines, it is possible to modify not only the α-amino group in the most N-terminal amino acid but also the ε-amino group in the lysine side chain. To prevent the labeling on the ε-amino group of lysine, the gels were first guanidinated with O-methylisourea hemisulfate, followed by iTRAQ labeling, trypsin or chymotrypsin digestion, and tandem mass spectrometry analysis. The major iTRAQ-conjugated tryptic or chymotryptic peptides listed in Fig. 7C indicate that the calpain-cutting site is located between Arg427 and Lys428. Thus, the 30-kDa CP3BD includes the entire RBD and should bind to RNA just like MBP-CPEB3C does (Fig. 1C), which contains amino acids 426 to 684 of CPEB3. To test this notion, MBP-CPEB3 with or without calpain 2 cleavage was employed for gel retardation and UV-cross-linking RNA-binding assays (Fig. 8A). Both assays denote that the product (CP3BD) cleaved by calpain 2 (Fig. 8A) or calpain 1 (see Fig. S5 in the supplemental material) bind to the RNA. Unlike myc-CPEB3, myc-CP3BD (aa 428 to 684) did not repress translation in the RNA reporter assay (Fig. 8B). Using the RNAs containing the whole length of EGFR 3′-UTR or various deleted mutants of EGFR 3′-UTR for the luciferase reporter assay (see Fig. S6A in the supplemental material) and the UV-cross-linking RNA-binding analysis (see Fig. S6B), CPEB3 appeared to bind and function at the multiple regions in the EGFR 3′-UTR. Nonetheless, NMDA stimulation decreased 20% of ectopically expressed myc-CPEB3 and concomitantly increased 15% translation of the luciferase-EGFR 3′-UTR reporter RNA (Fig. 8C). Since the RBD alone cannot inhibit translation due to lack of the eEF2-interacting motif (12, 29), NMDA-induced cleavage of CPEB3 accounts for the translational activation (or derepression) of some, if not all, of its target RNAs (Fig. 8D).
Fig 8.
The cleavage of CPEB3 by calpain 2 abolishes its translational repression activity. (A) The recombinant MBP or MBP-CPEB3 was incubated with calpain 2 in the absence or presence of Ca2+, the mixtures were incubated with the 32P-labeled 1904 RNA probe. One half of the RNA-protein mixture was used for the gel retardation assay (top gel). The other half was UV cross-linked and RNase A digested, followed by SDS-PAGE separation. The gel was stained with Coomassie blue and exposed to a phosphorimager. The RNA-protein complexes are labeled with one or two asterisks. (B) RNA reporter assay. The hippocampal or cortical neurons of DIV 10 or 11 were transfected with mRNAs encoding β-galactosidase (β-gal), myc-CPEB3 or myc-CP3BD, along with the firefly luciferase appended to the EGFR 3′-UTR and Renilla luciferase. (C) Similar to panel B, the transfected neurons were stimulated with 50 μM NMDA or not stimulated with NMDA. The neurons were harvested 3 h later for Western blotting and luciferase assays. The normalized luciferase activity (firefly/Renilla) was calculated. The results were analyzed and expressed as means plus SEMs. An asterisk marks a statistical difference (two-tailed Student's t test). The intensities of immunodetected signals analyzed by the ImageJ software are displayed as relative ratios at the bottoms of blots. (D) Schematic model of CPEB3-governed translation. The binding of CPEB3 to the 3′-UTR of plasticity-related protein (PRP) RNA, such as EGFR, reduces the translation through its interaction with eEF2. NMDA-induced EGFR RNA translation is in part caused by the calpain 2-dependent cleavage of CPEB3, which presumably allows eEF2 to resume its maximal GTPase activity and enhances the translation of EGFR RNA.
DISCUSSION
This study identifies calpain 2-dependent cleavage of the repressor CPEB3 as a means to achieve NMDA-induced translational activation (derepression) of its target RNAs. This finding agrees with a previous study showing that NMDA-stimulated translational activation of CPEB3-targeted reporter RNA does not require an AAUAAA polyadenylation element, implying that it does not promote cytoplasmic polyadenylation like CPEB1 (29). Recently, we have identified that CPEB2 and CPEB3 employ a distinct mechanism to repress translation at elongation (12). Thus, CPEB1-mediated polyadenylation-induced translation initiation (44) [i.e., the elongated poly(A) tails of RNAs are bound by more poly(A)-binding proteins, which subsequently recruit eIF4G to compete with maskin or neuroguidin for the binding to eIF4E], is unlikely to alleviate CPEB3-repressed translation. Nonetheless, a recent study has implicated that monoubiquitination of CPEB3 by the E3 ligase, neuralized 1, switches CPEB3 from a translational repressor to activator to increase the poly(A) length of GluA1 and GluA2 RNAs (41). However, it is unclear whether neuronal activity regulates this modification, which lysine residue of CPEB3 is conjugated with ubiquitin, how the monoubiquitinated CPEB3 promotes polyadenylation of GluA1 and GluA2 RNA, and whether this modification interferes with the interaction of CPEB3 and eEF2. Because GluA1 is also subject to calpain-mediated cleavage in response to NMDAR activation (9, 21, 60), we measured the protein level of EGFR instead of GluA1 or GluA2 in this study to avoid the AMPA receptor degradation issue.
Calpains regulate diverse physiological functions by cleaving various substrates in a limited manner. In many cases, the cleaved products execute alternative functions (14). In the central nervous system, NMDA receptor signaling-mediated calcium influx induces the activation of calpains (2, 16), which in turn processes downstream substrates. Postsynaptic substrates of calpains include αII-spectrin, postsynaptic density 95, Kv4.2 potassium channel, β-catenin, and the AMPA receptors (1, 35–37, 60), which are important molecules in regulating synaptic transmission and memory formation. Calpains under controlled activation (i.e., the influx of calcium sufficient to activate a few molecules of calpains) are vital for synaptic plasticity (24, 59), whereas sustained calcium overload hyperactivates all calpain molecules, resulting in neuronal death-associated pathological events such as brain injury and ischemia (7, 19, 31). In several neurodegenerative mouse models, the inhibition of calpain activity ameliorates neuronal death and improves behaviors (15, 25, 39). This study shows that a short pulse of NMDA, a paradigm used to induce c-LTD, and sustained NMDA incubation accompanied with excitotoxicity, all induce calpain activity to proteolyze CPEB3. Moreover, NMDA-induced CPEB3 degradation can take place in the synaptic compartments. Therefore, the activity-dependent CPEB3-mediated translational control may be important under physiological and pathological conditions.
Calpain 1 and calpain 2 are present in almost all mammalian cells and are considered the major calpain forms expressed in neurons. The absence of fully specific calpain inhibitors has prevented unequivocal proof of a particular physiological role for individual calpain family members. Early studies using inhibitors such as leupeptin, calpain inhibitor I (ALLN [N-acetyl-Leu-Leu-Asn]), and calpain inhibitor II (ALLM [N-acetyl-Leu-Leu-methioninal]) have demonstrated that calpain activity is involved in certain forms of long-term potentiation (LTP) and LTD induction (16, 17, 28, 50). However, these inhibitors used in the earlier works also inhibit proteasome and/or cathepsins. Thus, the genetic knockout approach can demonstrate the role of individual calpains in vivo. Both CAPN2 (18) and Capns1 (3) knockout mouse are embryonic lethal. In contrast, the CAPN1 null mice are viable and have normal theta burst-evoked LTP and fear conditioning learning (24). The lack of neuronal phenotypes in CAPN1 knockout animals is probably because of the low abundance of CAPN1, since the knockdown of CAPN2 has more profound effect on proteolysis of calpain substrates, such as CPEB3 and αII-spectrin, in neurons. Mice with the loxp-flanked conditional allele of Capns1 (54) and CAPN2 (53) are now available, so the brain-specific CAPN2 knockout mice will be useful tools for assessing calpain 2's role in learning and memory.
Because many calpain substrates are important molecules for neuronal function and plasticity, the depletion of calpain activity in knockout mice should have a profound effect on plasticity and memory. Here, we demonstrate that calpain can also influence translation-dependent plasticity by cleaving the translational repressor CPEB3 to activate the target RNA translation. Although CPEB3 also functions as a transcription repressor of the EGFR gene, the NMDA stimulation activates EGFR expression first at the translational level for two reasons. The distribution of calpain 2 is mainly located in the cytoplasm; however, NMDA signaling accumulates CPEB3 but not calpain 2 in the nucleus. The increased cytoplasmic degradation and nuclear accumulation of CPEB3 result in a faster decline of CPEB3 level in the cytoplasm than in the nucleus. Other translation regulators, such as dicer and eIF4G1, have also been identified as calpain substrates (38, 57). This study identifies the underlying mechanism of NMDA-induced CPEB3 degradation as a prerequisite to activating CPEB3-targeted RNA translation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sheau-Yann Shieh for the p53 construct and Woan-Yuh Tarn for the cDNA3.1-flag plasmid. We appreciate Fu-Ann Lee in the institutional core for conducting LC-MS/MS analysis and the RNAi Core in Academia Sinica for the Capns1 shRNA clones.
This work was supported by grants from the National Science Council (NSC 99-2311-B-001-020-MY3), National Health Research Institute (NHRI-EX101-9814NI), and Academia Sinica (AS-100-TP-B09) in Taiwan. The RNAi Core, which is supported by the National Research Program for Genomic Medicine grants, is also gratefully acknowledged.
Footnotes
Published ahead of print on 18 June 2012.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Abe K, Takeichi M. 2007. NMDA-receptor activation induces calpain-mediated beta-catenin cleavages for triggering gene expression. Neuron 53:387–397 [DOI] [PubMed] [Google Scholar]
- 2. Adamec E, Beermann ML, Nixon RA. 1998. Calpain I activation in rat hippocampal neurons in culture is NMDA receptor selective and not essential for excitotoxic cell death. Brain Res. Mol. Brain Res. 54:35–48 [DOI] [PubMed] [Google Scholar]
- 3. Arthur JS, Elce JS, Hegadorn C, Williams K, Greer PA. 2000. Disruption of the murine calpain small subunit gene, Capn4: calpain is essential for embryonic development but not for cell growth and division. Mol. Cell. Biol. 20:4474–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ashby MC, et al. 2004. Removal of AMPA receptors (AMPARs) from synapses is preceded by transient endocytosis of extrasynaptic AMPARs. J. Neurosci. 24:5172–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banker G, Goslin K. 1988. Developments in neuronal cell culture. Nature 336:185–186 [DOI] [PubMed] [Google Scholar]
- 6. Beattie EC, et al. 2000. Regulation of AMPA receptor endocytosis by a signaling mechanism shared with LTD. Nat. Neurosci. 3:1291–1300 [DOI] [PubMed] [Google Scholar]
- 7. Bevers MB, et al. 2010. RNAi targeting micro-calpain increases neuron survival and preserves hippocampal function after global brain ischemia. Exp. Neurol. 224:170–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bevers MB, et al. 2009. Knockdown of m-calpain increases survival of primary hippocampal neurons following NMDA excitotoxicity. J. Neurochem. 108:1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bi X, et al. 1997. Characterization of calpain-mediated proteolysis of GluR1 subunits of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionate receptors in rat brain. J. Neurochem. 68:1484–1494 [DOI] [PubMed] [Google Scholar]
- 10. Bramham CR, Wells DG. 2007. Dendritic mRNA: transport, translation and function. Nat. Rev. Neurosci. 8:776–789 [DOI] [PubMed] [Google Scholar]
- 11. Carillo S, et al. 1994. Differential sensitivity of FOS and JUN family members to calpains. Oncogene 9:1679–1689 [PubMed] [Google Scholar]
- 12. Chen PJ, Huang YS. 2011. CPEB2-eEF2 interaction impedes HIF-1alpha RNA translation. EMBO J. 31:959–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. 2009. Translational control of long-lasting synaptic plasticity and memory. Neuron 61:10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Croall DE, Ersfeld K. 2007. The calpains: modular designs and functional diversity. Genome Biol. 8:218 doi:10.1186/gb-2007-8-6-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crocker SJ, et al. 2003. Inhibition of calpains prevents neuronal and behavioral deficits in an MPTP mouse model of Parkinson's disease. J. Neurosci. 23:4081–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. del Cerro S, et al. 1994. Stimulation of NMDA receptors activates calpain in cultured hippocampal slices. Neurosci. Lett. 167:149–152 [DOI] [PubMed] [Google Scholar]
- 17. Denny JB, Polan-Curtain J, Ghuman A, Wayner MJ, Armstrong DL. 1990. Calpain inhibitors block long-term potentiation. Brain Res. 534:317–320 [DOI] [PubMed] [Google Scholar]
- 18. Dutt P, et al. 2006. m-Calpain is required for preimplantation embryonic development in mice. BMC Dev. Biol. 6:3 doi:10.1186/1471-213X-6-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Faddis BT, Hasbani MJ, Goldberg MP. 1997. Calpain activation contributes to dendritic remodeling after brief excitotoxic injury in vitro. J. Neurosci. 17:951–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frey U, Morris RG. 1997. Synaptic tagging and long-term potentiation. Nature 385:533–536 [DOI] [PubMed] [Google Scholar]
- 21. Gellerman DM, Bi X, Baudry M. 1997. NMDA receptor-mediated regulation of AMPA receptor properties in organotypic hippocampal slice cultures. J. Neurochem. 69:131–136 [DOI] [PubMed] [Google Scholar]
- 22. Giorgi C, et al. 2007. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell 130:179–191 [DOI] [PubMed] [Google Scholar]
- 23. Goll DE, Thompson VF, Li H, Wei W, Cong J. 2003. The calpain system. Physiol. Rev. 83:731–801 [DOI] [PubMed] [Google Scholar]
- 24. Grammer M, Kuchay S, Chishti A, Baudry M. 2005. Lack of phenotype for LTP and fear conditioning learning in calpain 1 knock-out mice. Neurobiol. Learn. Mem. 84:222–227 [DOI] [PubMed] [Google Scholar]
- 25. Granic I, et al. 2010. Calpain inhibition prevents amyloid-beta-induced neurodegeneration and associated behavioral dysfunction in rats. Neuropharmacology 59:334–342 [DOI] [PubMed] [Google Scholar]
- 26. Hegde AN. 2010. The ubiquitin-proteasome pathway and synaptic plasticity. Learn. Mem. 17:314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hosfield CM, Elce JS, Davies PL, Jia Z. 1999. Crystal structure of calpain reveals the structural basis for Ca(2+)-dependent protease activity and a novel mode of enzyme activation. EMBO J. 18:6880–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hrabetova S, Sacktor TC. 1996. Bidirectional regulation of protein kinase M zeta in the maintenance of long-term potentiation and long-term depression. J. Neurosci. 16:5324–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang YS, Kan MC, Lin CL, Richter JD. 2006. CPEB3 and CPEB4 in neurons: analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 25:4865–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang YS, Richter JD. 2007. Analysis of mRNA translation in cultured hippocampal neurons. Methods Enzymol. 431:143–162 [DOI] [PubMed] [Google Scholar]
- 31. Jang YN, et al. 2009. Calpain-mediated N-cadherin proteolytic processing in brain injury. J. Neurosci. 29:5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keleman K, Kruttner S, Alenius M, Dickson BJ. 2007. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat. Neurosci. 10:1587–1593 [DOI] [PubMed] [Google Scholar]
- 33. Lee DH, Goldberg AL. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397–403 [DOI] [PubMed] [Google Scholar]
- 34. Lee HK, Kameyama K, Huganir RL, Bear MF. 1998. NMDA induces long-term synaptic depression and dephosphorylation of the GluR1 subunit of AMPA receptors in hippocampus. Neuron 21:1151–1162 [DOI] [PubMed] [Google Scholar]
- 35. Lei Z, Deng P, Li Y, Xu ZC. 2010. Downregulation of Kv4.2 channels mediated by NR2B-containing NMDA receptors in cultured hippocampal neurons. Neuroscience 165:350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu X, Rong Y, Baudry M. 2000. Calpain-mediated degradation of PSD-95 in developing and adult rat brain. Neurosci. Lett. 286:149–153 [DOI] [PubMed] [Google Scholar]
- 37. Lu X, Rong Y, Bi R, Baudry M. 2000. Calpain-mediated truncation of rat brain AMPA receptors increases their Triton X-100 solubility. Brain Res. 863:143–150 [DOI] [PubMed] [Google Scholar]
- 38. Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. 2005. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 94:896–905 [DOI] [PubMed] [Google Scholar]
- 39. Nimmrich V, et al. 2008. Inhibition of calpain prevents N-methyl-d-aspartate-induced degeneration of the nucleus basalis and associated behavioral dysfunction. J. Pharmacol. Exp. Ther. 327:343–352 [DOI] [PubMed] [Google Scholar]
- 40. Pariat M, et al. 1997. Proteolysis by calpains: a possible contribution to degradation of p53. Mol. Cell. Biol. 17:2806–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pavlopoulos E, et al. 2011. Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell 147:1369–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peng SC, Lai YT, Huang HY, Huang HD, Huang YS. 2010. A novel role of CPEB3 in regulating EGFR gene transcription via association with Stat5b in neurons. Nucleic Acids Res. 38:7446–7457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pike BR, et al. 2004. Accumulation of calpain and caspase-3 proteolytic fragments of brain-derived alphaII-spectrin in cerebral spinal fluid after middle cerebral artery occlusion in rats. J. Cereb. Blood Flow Metab. 24:98–106 [DOI] [PubMed] [Google Scholar]
- 44. Richter JD. 2007. CPEB: a life in translation. Trends Biochem. Sci. 32:279–285 [DOI] [PubMed] [Google Scholar]
- 45. Richter JD, Klann E. 2009. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 23:1–11 [DOI] [PubMed] [Google Scholar]
- 46. Ruiz-Vela A, Gonzalez de Buitrago G, Martinez AC. 1999. Implication of calpain in caspase activation during B cell clonal deletion. EMBO J. 18:4988–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. 2006. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 313:1788–1792 [DOI] [PubMed] [Google Scholar]
- 48. Sha D, et al. 2008. Role of mu-calpain in proteolytic cleavage of brain L-glutamic acid decarboxylase. Brain Res. 1207:9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Siman R, Baudry M, Lynch G. 1985. Regulation of glutamate receptor binding by the cytoskeletal protein fodrin. Nature 313:225–228 [DOI] [PubMed] [Google Scholar]
- 50. Staubli U, Larson J, Thibault O, Baudry M, Lynch G. 1988. Chronic administration of a thiol-proteinase inhibitor blocks long-term potentiation of synaptic responses. Brain Res. 444:153–158 [DOI] [PubMed] [Google Scholar]
- 51. Steward O, Wallace CS, Lyford GL, Worley PF. 1998. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21:741–751 [DOI] [PubMed] [Google Scholar]
- 52. Suzuki K, Hata S, Kawabata Y, Sorimachi H. 2004. Structure, activation, and biology of calpain. Diabetes 53(Suppl 1):S12–S18 [DOI] [PubMed] [Google Scholar]
- 53. Takano J, et al. 2011. Vital role of the calpain-calpastatin system for placental-integrity-dependent embryonic survival. Mol. Cell. Biol. 31:4097–4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tan Y, et al. 2006. Conditional disruption of ubiquitous calpains in the mouse. Genesis 44:297–303 [DOI] [PubMed] [Google Scholar]
- 55. Tomimatsu Y, Idemoto S, Moriguchi S, Watanabe S, Nakanishi H. 2002. Proteases involved in long-term potentiation. Life Sci. 72:355–361 [DOI] [PubMed] [Google Scholar]
- 56. Vogler C, et al. 2009. CPEB3 is associated with human episodic memory. Front. Behav. Neurosci. 3:4 doi:10.3389/neuro.08.004.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vosler PS, et al. 2011. Ischemia-induced calpain activation causes eukaryotic (translation) initiation factor 4G1 (eIF4GI) degradation, protein synthesis inhibition, and neuronal death. Proc. Natl. Acad. Sci. U. S. A. 108:18102–18107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wright JW, Harding JW. 2009. Contributions of matrix metalloproteinases to neural plasticity, habituation, associative learning and drug addiction. Neural Plast. 2009:579382 doi:10.1155/2009/579382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu HY, et al. 2005. Regulation of N-methyl-d-aspartate receptors by calpain in cortical neurons. J. Biol. Chem. 280:21588–21593 [DOI] [PubMed] [Google Scholar]
- 60. Yuen EY, Gu Z, Yan Z. 2007. Calpain regulation of AMPA receptor channels in cortical pyramidal neurons. J. Physiol. 580:241–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.