Abstract
Biofilms are composed of surface-attached microbial communities. A hallmark of biofilms is their profound tolerance of antimicrobial agents. While biofilm drug tolerance has been considered to be multifactorial, our findings indicate, instead, that bacteria within biofilms employ a classical regulatory mechanism to resist the action of antimicrobial agents. Here we report that the transcriptional regulator BrlR, a member of the MerR family of multidrug transport activators, plays a role in the high-level drug tolerance of biofilms formed by Pseudomonas aeruginosa. Expression of brlR was found to be biofilm specific, with brlR inactivation not affecting biofilm formation, motility, or pslA expression but increasing ndvB expression. Inactivation of brlR rendered biofilms but not planktonic cells grown to exponential or stationary phase significantly more susceptible to hydrogen peroxide and five different classes of antibiotics by affecting the MICs and the recalcitrance of biofilms to killing by microbicidal antimicrobial agents. In contrast, overexpression of brlR rendered both biofilms and planktonic cells more tolerant to the same compounds. brlR expression in three cystic fibrosis (CF) isolates was elevated regardless of the mode of growth, suggesting a selection for constitutive brlR expression upon in vivo biofilm formation associated with chronic infections. Despite increased brlR expression, however, isolate CF1-8 was as susceptible to tobramycin as was a ΔbrlR mutant because of a nonsense mutation in brlR. Our results indicate for the first time that biofilms employ a specific regulatory mechanism to resist the action of antimicrobial agents in a BrlR-dependent manner which affects MIC and recalcitrance to killing by microbicidal antimicrobial agents.
INTRODUCTION
Biofilms are composed of microorganisms attached to a solid surface and encased in an exopolysaccharide matrix of their own synthesis (17, 19, 44). One of the most important hallmarks of bacterial biofilms is their high tolerance to antimicrobial agents and components of the host immune system. This characteristic is considered the root of many persistent and chronic bacterial infections and renders biofilms extremely difficult to control in medical and industrial settings (18). Infections caused by bacterial biofilms are persistent and very difficult to eradicate and have been associated with a number of medical conditions, including periodontal disease, endocarditis, osteomyelitis, cystic fibrosis (CF), and indwelling-device-related biofilm infections (18). Bacteria living in biofilms can be up to 1,000 times more tolerant to antibacterial compounds than their planktonic counterparts. For instance, chlorine (as sodium hypochlorite), an oxidizing biocide that is considered to be one of the most effective antibacterial agents, has been shown to require a 600-fold higher concentration to kill biofilm cells of Staphylococcus aureus than that required to kill planktonic cells of the same species (41). Biofilm antimicrobial tolerance is distinct from commonly known mechanisms such as plasmid-borne resistance markers or resistance conferred by mutation (22, 27, 40, 43, 66), indicating that the mechanisms involved in biofilm resilience to antimicrobials may differ from the mechanisms responsible for antimicrobial resistance in planktonic bacteria.
Although several mechanisms have been postulated to explain reduced susceptibility to antimicrobials in bacterial biofilms, the current notion is that biofilm drug tolerance is multifactorial, as only a combination of different mechanisms could account for the level of resilience to antimicrobial agents observed in biofilm communities. The tenacious biofilm phenotype is believed to arise from a multiplicity of factors, including reduced metabolic and divisional rates (6, 7, 26, 64), starvation-induced growth arrest (52), the presence of persister cells that neither grow nor die in the presence of microbicidal antibiotics (12, 36, 37, 61, 63), and restricted penetration of a biofilm by antimicrobials (5, 14, 23, 40, 56, 65, 66, 70). However, recent reports suggest that bacteria within these microbial communities are physiologically distinct from planktonic bacteria, expressing specific protective factors such as multidrug efflux pumps and stress response regulons (7, 22, 27, 42, 43, 57, 58, 66, 67). Furthermore, quorum sensing (QS), required for the formation of the biofilm architecture (20), has been shown to play a role in drug tolerance. Biofilm bacteria in which QS was blocked either by mutation or by administration of QS inhibitory drugs were sensitive to treatment with tobramycin, in contrast to bacteria with functional QS systems (11). The findings indicated that biofilms themselves are not simply a diffusion barrier to these antibiotics but rather that bacteria within these microbial communities employ distinct mechanisms to resist the action of antimicrobial agents. This is further supported by findings that young Pseudomonas aeruginosa biofilm cells can be effectively eradicated with a combination of piperacillin and tobramycin, while old biofilm cells are less susceptible to these antibiotics (7), indicating that biofilm drug tolerance may coincide with the developmental stage or maturity of the biofilm (16). This is supported by recent findings suggesting that the formation of biofilms occurs in a regulated and stage-specific manner (2, 57, 58, 62).
We therefore hypothesized that biofilm tolerance of antimicrobial agents is part of a regulated developmental process and thus would require an identifiable set of genetic determinants. Here we found the transcriptional regulator BrlR (PA4878), a member of the MerR family of transcriptional regulators that activate the expression of multidrug transporters upon binding of the transporter substrate, to be expressed in a biofilm-specific manner and to be essential for the drug tolerance of P. aeruginosa biofilms. To our knowledge, this is the first description of a MerR-like regulator, expressed in a growth mode-dependent manner, playing a role in the antimicrobial tolerance of a Gram-negative bacterium. Our findings challenge the current dogma that biofilm drug tolerance is multifactorial in nature and distinct from mechanisms employed by planktonic bacteria. Instead, our findings suggest the existence of a classical, biofilm-specific mechanism of drug tolerance in P. aeruginosa.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
All of the bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa strain PAO1 was used as the parental strain. All planktonic cultures were grown in Luria-Bertani (LB) broth in shake flasks at 220 rpm. Biofilms were grown as described below at 22°C in 20-fold-diluted LB broth. Antibiotics were used at the following concentrations: 50 to 75 μg/ml gentamicin and 200 to 250 μg/ml carbenicillin for P. aeruginosa and 20 μg/ml gentamicin and 50 μg/ml ampicillin for E. coli. Where necessary, Escherichia coli cultures were grown in LB broth in the absence or presence of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 tonA | Invitrogen Corp. |
| BL21 | F− ompT hsdSB(rB− mB−) gal dcm rne131 (DE3) | Invitrogen Corp. |
| P. aeruginosa PAO1 | ||
| PAO1 | Wild type | B. H. Holloway |
| PAO1/pJN105 | PAO1 bearing empty pJN105 vector; Gmr | 55 |
| PAO1/pMJT1 | PAO1 bearing empty pMJT1 vector; Carbr | This study |
| PAO1/pJN-brlR | Arabinose-inducible expression of brlR; Gmr | This study |
| PAO1/pMJT-brlR-V5/6×His | Arabinose-inducible expression of V5/6×His-tagged brlR; Carbr | This study |
| ΔbrlR mutant | PAO1 ΔbrlR (PA4878) | This study |
| ΔbrlR/pJN-brlR mutant | Arabinose-inducible expression of brlR; Gmr | This study |
| ΔbrlR/pMJT1 mutant | ΔbrlR harboring empty pMJT1, vector control; Carbr | This study |
| ΔbrlR/pMJT-brlR-N mutant | Arabinose-inducible expression of N-terminal DNA binding domain of BrlR (BrlR-N) in ΔbrlR; Carbr | This study |
| CF1-2 | Isolate from CF patient | 53 |
| CF1-8 | Isolate from CF patient | 53 |
| CF1-8/pJN-brlR | Arabinose-inducible expression of brlR in CF1-8; Gmr | This study |
| CF1-8 ΔbrlR | CF1-8 harboring ΔbrlR (PA4878), Gmr | This study |
| CF1-13 | Isolate from CF patient | 53 |
| CF1-13 ΔbrlR | CF1-13 harboring ΔbrlR (PA4878); Gmr | This study |
| PAO1 attB::lacZ | pminiCTX-lacZ conjugated into PAO1, vector control; Tetr | This study |
| PAO1 attB::PbrlR-lacZ | pminiCTX-PbrlR-lacZ conjugated into PAO1; Tetr | This study |
| Plasmids | ||
| pCR2.1-TOPO | TA cloning vector; Kmr Apr | Invitrogen Corp. |
| pRK2013 | Helper plasmid for triparental mating; mob tra Kmr | 25 |
| pEX18Gm | Gene replacement vector; pUC18 MCS oriT sacB Gmr | 26 |
| pJN105 | Arabinose-inducible gene expression vector; pBRR-1 MCS araC-PBAD Gmr | 50 |
| pMJT1 | Arabinose-inducible gene expression vector; pUCP18 MCS araC-PBAD Amp/Carbr | 35 |
| pET101D | Vector for directional cloning and high level V5/6×His fusion protein expression; Ampr | Invitrogen Corp. |
| pminiCTX-lacZ | attB site-specific integration vector; Tetr | 10 |
| pET-brlR-V5/6×His | brlR cloned into pET101D | This study |
| pJN-brlR | brlR cloned into pJN105 | This study |
| pMJT-brlR-V5/6×His | brlR-V5/6×His cloned into pMJT1 | This study |
| pMJT-brlR-N | N-terminal DNA binding domain of BrlR cloned into pMJT1 | This study |
| pminiCTX-PbrlR-lacZ | brlR promoter reporter construct in mini-CTX-lacZ using PbrlR-lacZF/R primers; Tetr | This study |
Strain construction.
Isogenic mutants were constructed by allelic replacement with sucrose counterselection as previously described (60) using the gene replacement vector pEX18Gm (33). Complementation and overexpression of brlR (PA4878) were accomplished by placing the respective genes under the control of an arabinose-inducible promoter in the pJN105 vector (50). The primers used for strain construction are listed in Table S1 in the supplemental material. Reverse transcriptase PCR (RT-PCR) was performed to confirm brlR expression in the ΔbrlR/pJN-brlR mutant and PAO1/pJN-brlR. When PAO1/pJN-brlR was grown in LB medium without arabinose, the brlR transcript was detectable at low levels (suggesting a leaky promoter or the presence of arabinose in LB medium) while brlR expression increased in the presence of 0.5% arabinose (see Fig. S1 in the supplemental material). Similar results were obtained with the ΔbrlR and ΔbrlR/pJN-brlR mutants (not shown). Under the condition tested, no transcript was detected in P. aeruginosa PAO1/pJN105 grown planktonically in the absence or presence of 0.5% arabinose (see Fig. S1).
Planktonic cell antibiotic susceptibility testing.
To determine the role of BrlR in antimicrobial susceptibility, P. aeruginosa strains grown planktonically in LB medium at 37°C to the exponential and stationary phases were treated with tobramycin (50 μg/ml) for 1 h or 0.3% hydrogen peroxide for 30 min and subsequently homogenized, serially diluted, and spread plated onto LB agar. Viability was determined via CFU counts. Susceptibility is expressed as log reduction. The MICs to tobramycin, norfloxacin, chloramphenicol, kanamycin, tetracycline, and trimethoprim were determined by 2-fold serial dilution in LB broth using 96-well microtiter plates. LB broth was used 10- and 20-fold diluted. The antibiotic concentrations used ranged from 0.02 to 200 μg/ml. The inoculum was ∼104 cells per well, and the results were read after overnight incubation at 37°C. The MIC was defined as the lowest antibiotic concentration that yielded no visible growth. To ensure overexpression of brlR, PAO1/pJN-brlR was grown in the presence of 0.5% arabinose. PAO1/pJN105 was used as a control.
Biofilm formation.
For biofilm antibiotic susceptibility testing, biofilms were grown in a continuous-flow tube reactor system (1 m of size 13 Masterflex silicone tubing; Cole Parmer, Inc.) with an inner surface area of 25 cm2 and in flow cells (BioSurface Technologies) which also allowed for the analysis of biofilm architecture as previously described (55, 58, 59). Biofilms were grown at 22°C in 20-fold-diluted LB medium. To ensure the overexpression of brlR, PAO1/pJN-brlR was grown in the presence of 0.1% arabinose. Quantitative analysis of confocal scanning laser microscopy images of flow cell-grown biofilms was performed with a Leica TCS SP5 confocal microscope and COMSTAT (32).
Biofilm antibiotic susceptibility assays.
Biofilms grown for 1 and 6 days under flowing conditions (0.1 ml/min) were treated for 1 h under flowing conditions with the following antimicrobial agents: tobramycin (50 to 150 μg/ml), norfloxacin (450 μg/ml), chloramphenicol (50 μg/ml), kanamycin (150 μg/ml), tetracycline (100 μg/ml), trimethoprim (150 μg/ml), and hydrogen peroxide (0.3%). Following exposure of biofilms to the respective antimicrobial agents, biofilms were harvested, homogenized, serially diluted, and spread plated onto LB agar. Hydrogen peroxide was neutralized by the addition of 100 mM sodium thiosulfate. Viability was determined via CFU counts. Susceptibility is expressed as log reduction. Under the conditions used, a total of 5 × 108 CFU were detected, on average, per biofilm tube reactor following 1 day of biofilm growth, which increased to 1 × 1010 and 7.2 ×1010 following 3 and 6 days of growth, respectively.
The MBC (minimal bactericidal concentration) for a biofilm is defined as the minimal concentration of antibiotic required to kill all of the bacteria in a preformed biofilm. To determine the MBC for a biofilm, a microtiter dish assay was used as described by Wozniak et al. (75), with a few modifications. Briefly, for the microtiter dish assay, overnight bacterial cultures were subcultured in fresh LB medium, the optical density was adjusted to 0.05, and the bacteria were added to the wells of microtiter dishes (25 μl per well with 150 μl medium). The plates were incubated for a total of 72 h with spent medium being removed and replaced with fresh medium every 12 h. Following 72 h of growth, the medium was replaced with medium containing the antibiotics and the plates were incubated for an additional 24 h. The antibiotic-containing medium was then removed and replaced with fresh antibiotic-free medium, the plates were incubated for an additional 24 h (during which time any viable cells in the biofilm grew and replenished the planktonic population), and the medium was finally assessed for viable cells. The antibiotic tobramycin was applied over a range of 0.3 to 400 μg/ml.
The biofilm MBC has been defined as the concentration at which no further increase in the log reduction is observed (47, 48, 73). To determine whether BrlR affects the biofilm MBC and resistance to killing, PAO1 and ΔbrlR mutant biofilms were grown for 3 days, after which time the medium was switched to the same medium containing increasing concentrations of tobramycin or norfloxacin ranging from 0.5 to 400 μg/ml. After 24 h of exposure to the antibiotic under continuous flow at 0.1 ml/min, biofilms were harvested and the surviving bacteria were enumerated. To determine recovery following 24 h of treatment, the medium was switched back to antibiotic-free medium. Following 24 h of incubation under flowing conditions, biofilms were harvested and the surviving bacteria were enumerated.
LacZ fusion analysis of brlR expression in P. aeruginosa grown attached in microtiter dishes.
The β-galactosidase activity of strains harboring the brlR promoter reporter was determined using the Miller assay (46) with the following modification. Instead of using total cells, β-galactosidase specific activity was determined using protein extracts obtained as previously described (62). An extinction coefficient for o-nitrophenyl-β-galactopyranoside cleavage at 420 nm of 4,500 nl/nmol/cm was used.
Quantitation of cell death in biofilm populations following treatment with tobramycin.
To quantify the differences in cell death in biofilm populations inactivated in or expressing brlR prior to and following tobramycin treatment, biofilms grown for 6 days were first stained with the LIVE/DEAD Bac Light stain and then separate images were acquired for SYTO 9- and propidium iodide (PI)-stained biofilm cells by using two distinct fluorescence channels. PI staining was used as an indicator of cell death. Biofilms were treated for 24 h with tobramycin. The resulting images were analyzed by COMSTAT.
Quantitative RT-PCR (qRT-PCR).
Isolation of mRNA and cDNA synthesis were carried out as previously described (3, 4, 55, 62). qRT-PCR was performed using the Eppendorf Mastercycler ep realplex (Eppendorf AG, Hamburg, Germany) and the KAPA SYBR FAST qPCR kit (Kapa Biosystems, Woburn, MA) with oligonucleotides listed in Table S1 in the supplemental material. mreB was used as a control. The stability of mreB levels were verified by determining 16S rRNA abundance using primers HDA1/HDA2 (45). Relative transcript quantitation was accomplished using the ep realplex software (Eppendorf AG) by first normalizing transcript abundance (based on the cycle threshold value) to mreB and then determining transcript abundance ratios. Melting curve analyses were employed to verify specific single product amplification.
Statistical analysis.
Student's t test was performed for pairwise comparisons of groups, and multivariate analyses were performed using a 1-way analysis of variance (ANOVA followed by a post-priori test using Sigma Stat software.
RESULTS
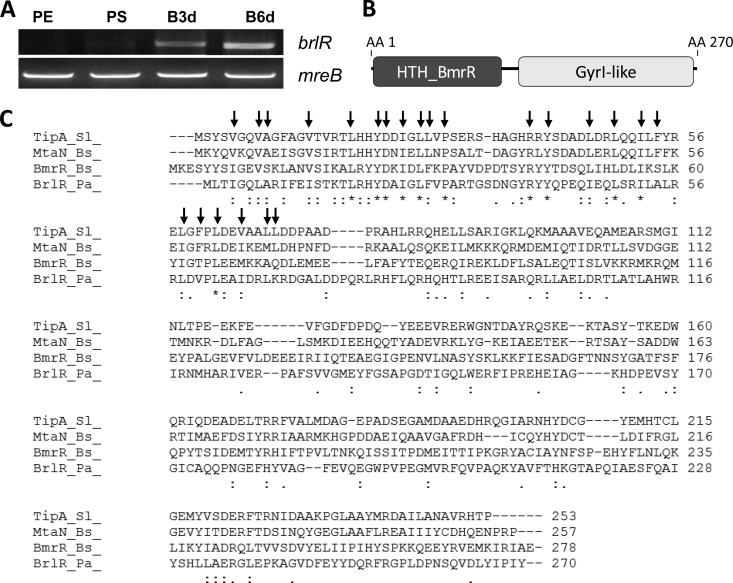
Previous analysis using a proteomic approach resulted in the identification of PA4878 encoding a probable transcriptional regulator (62). The protein was detectable by two-dimensional PAGE in cell extracts obtained from 1-, 3-, and 6-day-old P. aeruginosa PAO1 biofilms but absent from cell extracts obtained from planktonic cells. Subsequent analysis by RT-PCR confirmed that the gene is expressed in biofilms but absent from planktonic cells grown to the exponential and stationary phases (Fig. 1A). For reasons indicated below, we named the gene for PA4878 brlR (for biofilm resistance locus regulator).
FIG 1.
BrlR is a member of the MerR family of transcriptional regulator functioning as multidrug transporter activators. (A) brlR transcripts are not detected in planktonic cells grown to the exponential (PE) and stationary (PS) phases but are present in biofilms of various ages (B3d, 3-day biofilm; B6d, 6-day biofilm). mreB was used as a control. Transcript abundance was visualized after 30 cycles by separating the transcripts on a 1% agarose gel. (B) BrlR domain organization. HTH_BmrR, helix-turn-helix DNA binding domain of the MerR BmrR transcription regulator; GyrI-like, GyrI-like small-molecule binding domain; AA, amino acid position. (C) Alignment of BrlR (BrlR_Pa_) with other known MerR-like transcriptional regulators, including TipA from S. lividans (TipA_Sl_), MtaN from B. subtilis (MtaN_Bs_), and BmrR from B. subtilis (BmrR_Bs_). Colons denote residues with strong conservation (score of >0.5 in PAM250 matrix), and periods denote residues with weak conservation (score of ≤0.5). Asterisks denote identical amino acids. Arrows denote residues with side chains contributing to the core of the DNA binding protein (29).
BrlR is a member of the MerR family.
BLAST analysis revealed BrlR to be homologous to regulatory proteins belonging to the MerR family of transcriptional regulators. Members of this family include proteins involved in regulating resistance to oxidative stress, heavy metals, and antibiotics (13, 68, 69). Moreover, BrlR was found to be most similar to those MerR transcriptional regulators that function as multidrug transporter activators, including Mta, BltR, and BmrR of Bacillus subtilis and TipA of Streptomyces lividans, which activate the expression of multidrug transporters upon binding of the transporter substrate (9, 13, 54, 77). These multidrug gene regulators have in common homologous N-terminal DNA binding domains but differ in their variable C-terminal modulation or “coactivator” binding domains involved in modulating the transcriptional activation of MerR proteins and/or their target genes in response to an inducer(s) (Fig. 1B) (30, 31, 54). Similar to other MerR regulators, the homology of BrlR to BmrR, TipA, and Mta was limited to the DNA binding domain, with BrlR being 93% similar to TipA, 92% similar to BmrR, and 91% similar to MtaN. In addition, amino acid residues that contribute to the core of the DNA binding domain of MtaN are conserved in BrlR (Fig. 1C) (29, 49). Due to the similarity of BrlR to MerR multidrug transporter activators, we hypothesized that BrlR plays a role in resistance. However, while MerR transcriptional activators have been extensively studied with respect to their mechanisms of activation and regulation, they have not been linked to biofilms and biofilm drug tolerance.
Inactivation of brlR has no effect on the susceptibility of P. aeruginosa planktonic cells to antimicrobial agents.
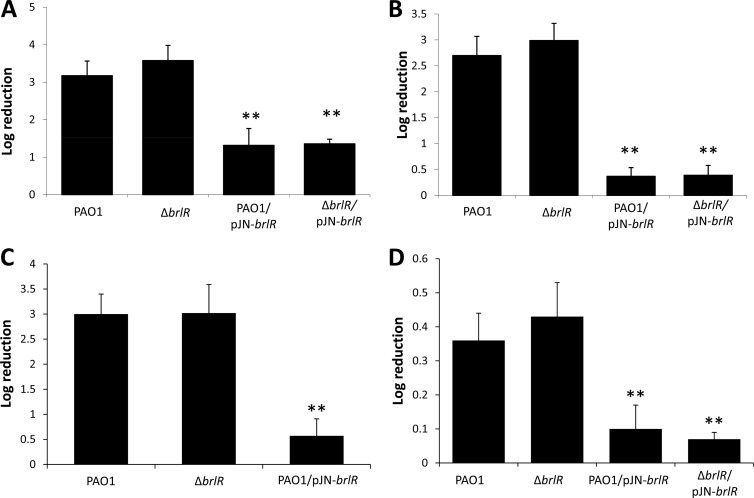
To determine whether BrlR plays a role in antimicrobial tolerance, we generated a ΔbrlR deletion mutant using an unmarked, nonpolar deletion strategy (33, 60). Inactivation of brlR did not affect the susceptibility of P. aeruginosa cells grown planktonically to exponential phase to tobramycin (50 μg/ml) and 0.3% hydrogen peroxide (Fig. 2A and C). Similarly, no difference in log reduction was observed for P. aeruginosa wild-type and ΔbrlR mutant cells grown to stationary phase when treated with tobramycin or hydrogen peroxide (Fig. 2B and D).
FIG 2.
Inactivation of brlR does not alter the susceptibility of planktonic cells to antimicrobial agents. Shown are the susceptibilities of planktonic cells grown to the exponential (A, C) or stationary (B, D) phase to 50 μg/ml tobramycin (A, B) and 0.3% hydrogen peroxide (C, D). Planktonic cells were treated for 30 min with hydrogen peroxide and for 1 h with tobramycin. **, significantly different (P < 0.001) from PAO1 as indicated by ANOVA and SigmaStat.
While inactivation of brlR had no effect on the susceptibility of planktonic cells to antimicrobial agents, overexpression of brlR rendered planktonic cells grown to exponential phase more tolerant to tobramycin. Cells overexpressing brlR remained significantly more tolerant to tobramycin, as indicated by the decrease in log reduction of PAO1/pJN-brlR compared to that of PAO1 (Fig. 2A). Similar results were obtained for the ΔbrlR and ΔbrlR/pJN-brlR mutants grown to stationary phase (Fig. 2B). Likewise, cells overexpressing brlR were also significantly less susceptible to 0.3% hydrogen peroxide (Fig. 2C and D). The difference in susceptibility was not due to differences in growth, as deletion and overexpression of brlR had no effect on P. aeruginosa grown in broth (see Fig. S2 in the supplemental material).
Antimicrobial tolerance is the ability of a microorganism to grow in the presence of an elevated level of an antimicrobial agent, as indicated by an increased MIC. To further determine the contribution of BrlR to antibiotic tolerance, MIC studies were carried out. In particular, five different classes of antibiotics, including norfloxacin, chloramphenicol, tetracycline, tobramycin, kanamycin, and trimethoprim, were used. MICs were determined by 2-fold serial broth dilution in LB medium using 96-well microtiter plates and an inoculum of ∼104 cells per well. For all of the antibiotics tested, no difference in the MIC, which is defined as the lowest antibiotic concentration yielding no visible growth, was observed when PAO1 and the ΔbrlR mutant were compared (Table 2). While inactivation of brlR had no effect on the MICs of various antibiotics, the MICs for strains expressing brlR (PAO1/pJN-brlR and the ΔbrlR/pJN-brlR mutant) were 6-fold higher for tobramycin and chloramphenicol, and 4-fold higher for tetracycline, trimethoprim, and norfloxacin (Table 2). The findings strongly support the biofilm-specific expression of brlR and further suggest that BrlR contributes to the antimicrobial tolerance of P. aeruginosa, probably by altering the MIC.
Table 2.
Overexpression of brlR in planktonic cells correlated with 4- to 6-fold MIC increasesa
| Strain or parameter | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| Tob | Tet | Cm | Trim | Nor | |
| PAO1 | 1.25 | 10 | 6.25 | 12.5 | 0.25 |
| ΔbrlR mutant | 1.25 | 10 | 6.25 | 12.5 | 0.25 |
| PAO1/pJN-brlR | 10 | 40 | 50 | 50 | 1 |
| ΔbrlR mutant/pJN-brlR | 10 | 40 | 50 | 50 | 1 |
| Fold change | 6 | 4 | 6 | 4 | 4 |
All strains were grown in 20-fold-diluted LB medium. Tob, tobramycin; Tet, tetracycline; Cm, chloramphenicol; Trim, trimethoprim; Nor, norfloxacin.
BrlR plays a role in the drug tolerance of P. aeruginosa biofilms.
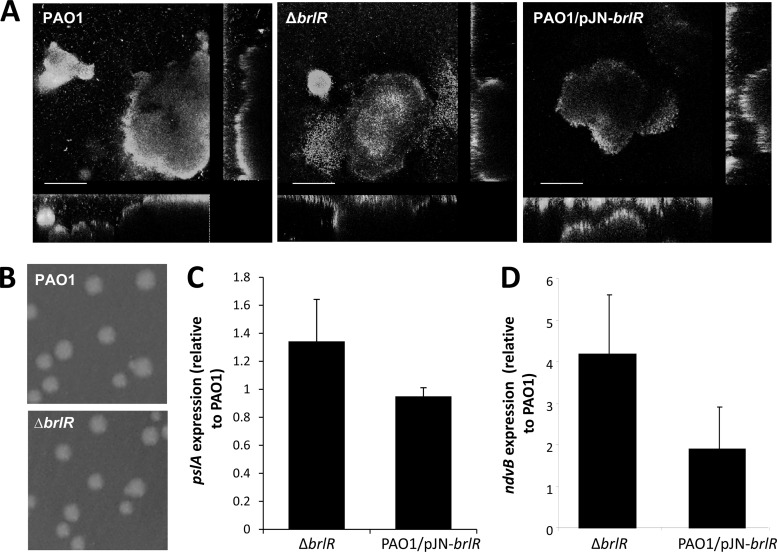
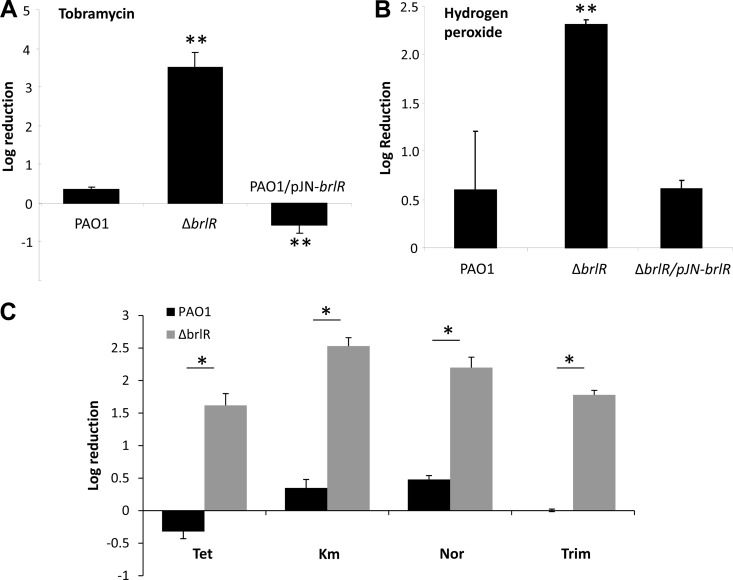
While MerR transcriptional activators have been extensively studied with respect to their mechanisms of activation and regulation, they have not been linked to biofilms and biofilm drug tolerance. We therefore asked whether BrlR plays a role in biofilms. Inactivation of brlR did not affect the attachment, formation, or architecture of P. aeruginosa biofilms as determined by confocal microscopy, COMSTAT analysis, and CFU determination (Fig. 3A; Table 3). However, while still capable of forming biofilms with the characteristic P. aeruginosa architecture, ΔbrlR mutant biofilms did not develop high-level biofilm-specific drug tolerance, as evidenced by the finding that treatment of mature, 6-day-old P. aeruginosa wild-type and ΔbrlR mutant biofilms with tobramycin (20 μg/ml) for 1 h resulted in a 0.2-log reduction of wild-type biofilms but a 1.8-log reduction of ΔbrlR mutant biofilms, respectively (not shown). Similar results were obtained when 1-day-old wild-type and ΔbrlR mutant biofilms were treated with tobramycin (100 μg/ml) for 1 h, resulting in a 0.4-log reduction of wild-type biofilms but a 3.5- to 4-log reduction of ΔbrlR mutant biofilms, respectively (Fig. 4A; see Fig. S3 in the supplemental material).
FIG 3.
Inactivation of brlR does not alter P. aeruginosa architecture or colony morphology or pslA but does affect ndvB expression. (A) Biofilm architecture of P. aeruginosa PAO1 strains with brlR inactivation or overexpression. Biofilms were stained with the LIVE/DEAD Bac Light viability stain (Invitrogen Corp.). Bars = 100 μm. Inactivation of brlR has no effect on colony morphology (B) or pslA expression (C). (D) Increased ndvB expression in strains with brlR inactivated.
Table 3.
COMSTAT analysisa of biofilm structure of P. aeruginosa PAO1 and strains with brlR inactivation or overexpression
| Strain | Total biomass (μm3/μm2) | Surface coverage (%) | Avg thickness (μm) | Max thickness (μm) | Roughness coefficient |
|---|---|---|---|---|---|
| PAO1 | 14.0 ± 3.5 | 81.0 ± 16.1 | 18.2 ± 3.7 | 87.8 ± 5.7 | 1.2 ± 0.1 |
| PAO1/pMJT1 | 12.7 ± 0.8 | 78.7 ± 4.8 | 18.3 ± 2.6 | 82.7 ± 18.5 | 1.2 ± 0.3 |
| ΔbrlR mutant | 13.7 ± 0.9 | 81.4 ± 22.9 | 16.5 ± 27.9 | 90.3 ± 22.9 | 0.9 ± 0.2 |
| PAO1/pMJT-brlR-V5/6×His | 15.9 ± 2.8 | 63.7 ± 9.0b | 19.8 ± 5.3 | 88.7 ± 19.4 | 0.6 ± 0.3b |
COMSTAT analysis was done for biofilms grown in triplicate by using at least six images per replicate. Values are means and standard deviations.
Significantly different from 144-h-old PA14 (P ≤ 0.05) as determined by ANOVA.
FIG 4.
BrlR contributes to drug tolerance of biofilm cells. (A) Inactivation of brlR resulted in 1-day biofilms with significantly increased susceptibility to tobramycin (100 μg/ml). Overexpression of brlR restored their susceptibility to tobramycin to the wild-type level. (B) Inactivation of brlR resulted in 1-day biofilms with significantly increased their susceptibility to 0.3% hydrogen peroxide. Expression of brlR in the ΔbrlR mutant restored its susceptibility to hydrogen peroxide to the wild-type level. (C) Increased susceptibility of the ΔbrlR mutant biofilm formed after 1 day was observed with tetracycline (Tet; 100 μg/ml), kanamycin (Km; 150 μg/ml), norfloxacin (Nor; 450 μg/ml), and trimethoprim (Trim; 150 μg/ml). Biofilms were treated for 1 h. **, significantly different (P < 0.001) from PAO1 as indicated by ANOVA and SigmaStat. *, significantly different (P < 0.001) from PAO1 as indicated by t test.
The increased susceptibility of ΔbrlR mutant biofilms was not limited to tobramycin. Instead, inactivation of brlR coincided with a significant increase in log killing upon treatment with norfloxacin, trimethoprim, tetracycline, kanamycin (Fig. 4C), and chloramphenicol (not shown) compared to that of wild-type biofilms. Moreover, ΔbrlR mutant biofilms were more susceptible to treatment with hydrogen peroxide (Fig. 4B).
Since ΔbrlR mutant biofilms were more susceptible to antimicrobial agents than PAO1 biofilms were and overexpression of brlR increased MICs but reduced the susceptibility of planktonic cells to various antimicrobial agents (Fig. 2; Table 2), we hypothesized that increased expression of brlR will also render biofilms more tolerant. In most cases, no reduction in CFU counts was observed for PAO1 overexpressing brlR (PAO1/pJN-brlR). Instead, an increase in CFU counts was detected (Fig. 4A). Expression of brlR in ΔbrlR mutant biofilms restored drug tolerance to wild-type levels (Fig. 4B; data not shown).
Together, the findings indicate that BrlR contributes to drug tolerance only under biofilm growth conditions, suggesting that BrlR is a regulator responsible for the recalcitrant phenotype of P. aeruginosa biofilms.
BrlR contributes to drug tolerance in a manner independent of growth, colony morphology, Psl production, and periplasmic glucans.
Differences in drug susceptibility have been attributed to differences in growth, colony morphology, and the production of Psl polysaccharide (15, 24, 38). Curves of growth in liquid media suggested that the difference in susceptibility between the wild-type and ΔbrlR mutant strains was not a result of a general increase in the growth rate (see Fig. S2 in the supplemental material). Moreover, no difference in colony morphology or Psl polysaccharide production following inactivation or overexpression of brlR was observed (Fig. 3B and C). Moreover, no difference in motility (swimming swarming, twitching) was observed (not shown). Similar results were obtained by Wagner et al. (74). In addition, the authors did not detect differences in elastase, protease, or rhamnolipid production between a ΔbrlR mutant and the wild type (74). Periplasmic glucans have been reported to sequester tobramycin and other aminoglycosides in the periplasm (42). ndvB, which is required for the synthesis of periplasmic glucans, was therefore tested as an indicator of differential production of periplasmic glucans using qRT-PCR. ndvB expression was detected in strain PAO1, the ΔbrlR mutant, and cells overexpressing brlR, with the ΔbrlR mutant expressing the highest level of ndvB (Fig. 3D).
BrlR plays a role in P. aeruginosa biofilm resistance to killing.
While the hallmark characteristic of biofilm populations is their increased antimicrobial tolerance, recent reports suggest that biofilm drug tolerance describes an increased resistance of biofilm cells to killing (39, 63). To address the question of whether BrlR affects resistance to killing, we examined the effect of brlR expression on attached cells by using MBC assays. Biofilms were formed in microtiter dishes and treated for 24 h with tobramycin, and then the number of viable cells remaining following treatment was determined. However, no difference between the wild-type and ΔbrlR mutant strain responses to tobramycin treatment was noted. The finding suggested that BrlR either does not contribute to the resistance of biofilm cells to killing or, alternatively, that brlR is not expressed when bacteria are grown attached in microtiter plates, which is contrary to brlR expression in biofilms formed under flowing growth conditions (Fig. 4). We therefore employed chromosomal transcriptional fusion constructs and determined the β-galactosidase activity of attached cells grown in microtiter plates. Compared to a vector control, no increase in β-galactosidase activity was found using Miller assays (see Fig. S4 in the supplemental material), further supporting the finding that brlR is not expressed in microtiter plate-grown attached cells.
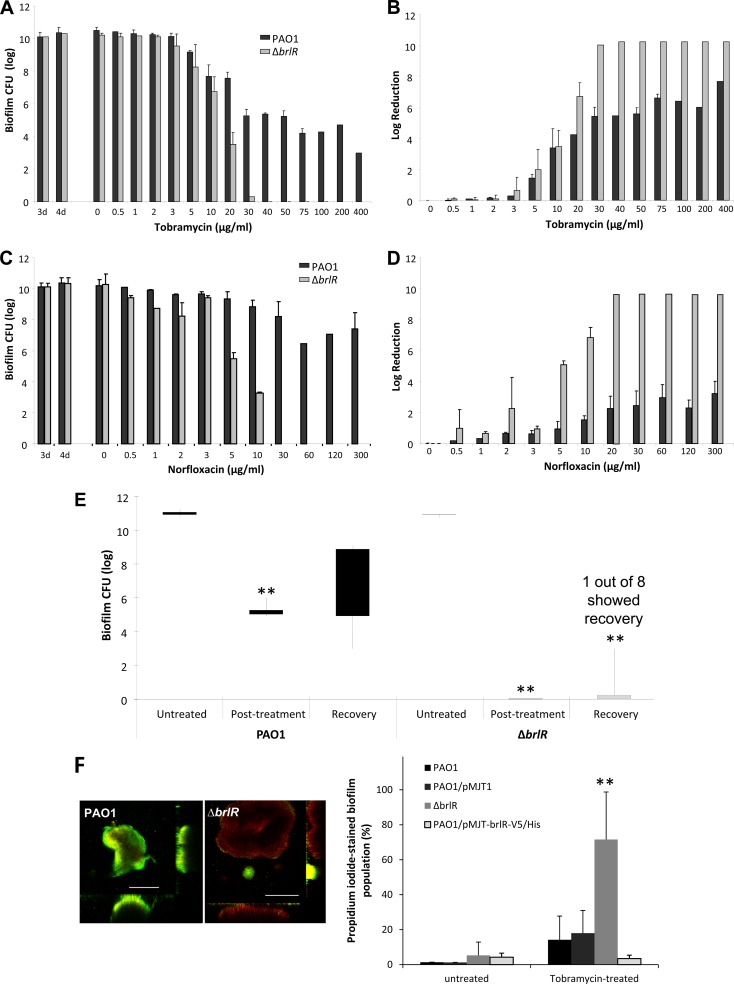
To further address the question of whether BrlR affects biofilm MBC or, alternatively, resistance to killing, we made use of tube reactor-grown biofilms, shown to express brlR, and the two antibiotics tobramycin and norfloxacin. The P. aeruginosa wild-type and ΔbrlR mutant strains were grown for 3 days, after which time the medium was switched to the same medium containing increasing concentrations of tobramycin ranging from 0.5 to 400 μg/ml. After 24 h of exposure to the antibiotic under continuous flow, biofilms were harvested and the surviving bacteria were enumerated. The biofilm MBC has been defined as the concentration at which no further increase in log reduction is observed (47, 48, 73). With P. aeruginosa wild-type biofilms, no further increase in log reduction was observed at concentrations higher than 75 μg/ml tobramycin following 24 h of treatment. Higher concentrations resulted in neither an increased log reduction nor complete killing of P. aeruginosa wild-type biofilms (Fig. 5A). In contrast, complete killing of ΔbrlR mutant biofilms was accomplished following treatment with tobramycin concentrations exceeding 30 μg/ml (Fig. 5A). Similar results were obtained when biofilms were treated with increasing concentrations of norfloxacin (Fig. 5C and D). Total killing of ΔbrlR biofilm cells was achieved at 20 μg/ml of norfloxacin, while PAO1 biofilms maintained a steady level of survivors through the highest concentrations tested (Fig. 5C).
FIG 5.
BrlR regulates resistance to killing in P. aeruginosa biofilms. P. aeruginosa PAO1 and ΔbrlR mutant biofilms were grown for 3 days and subsequently treated for 24 h under continuous flowing conditions before the recovery and enumeration of surviving cells. (A) Biofilm susceptibility to tobramycin as determined by viable CFU counts. CFU counts were obtained from biofilm tube reactors having an inner surface area of 25 cm2. The number of viable ΔbrlR mutant cells was below the detection limit at the highest concentrations of tobramycin tested. (B) Biofilm susceptibility to tobramycin as determined by log reduction. Total killing of ΔbrlR mutant biofilm cells was achieved at 40 μg/ml of tobramycin. In contrast, P. aeruginosa PAO1 biofilms maintained a steady level of persisting survivors at concentrations higher than 40 μg/ml of tobramycin. (C, D) Biofilm susceptibility to norfloxacin was determined similarly. Error bars denote standard deviations. (E) Inactivation of brlR eliminates resistance to killing of P. aeruginosa biofilms. Biofilms (3 days old) formed by wild-type PAO1 and the ΔbrlR mutant were treated with 50 μg/ml of tobramycin under continuous flowing conditions for 24 h and then allowed to recover under flowing conditions in the absence of tobramycin for 24 h. Viable cells were collected and enumerated after the recovery period. CFU counts were obtained from biofilm tube reactors having an inner surface area of 25 cm2. Lines indicate the difference between the highest and lowest numbers of viable cells recovered, while bars represent the distribution between the mean and median of the replicates. **, significantly different (P < 0.001) from untreated biofilms. (F) Biofilms formed after 5 days by wild-type PAO1, PAO1/pMJT1, the ΔbrlR mutant, and the overexpresser PAO1/pMJT-brlR-V5/6×His were treated with tobramycin under continuous flow for 24 h. The effect of tobramycin was examined using LIVE/DEAD Bac Light viability stain. Images for PAO1 and the ΔbrlR mutant were acquired with the same microscope settings. The percentages of the biofilm population stained with PI prior to and following treatment with tobramycin were quantitated using COMSTAT. **, significantly different (P < 0.001) from PAO1 as indicated by ANOVA and SigmaStat. White bars, 100 μm.
The findings indicated that the tobramycin and norfloxacin MBCs for biofilms of P. aeruginosa defective in brlR expression were ∼4-fold lower than for wild-type P. aeruginosa biofilms. In addition, P. aeruginosa wild-type biofilms were overall more resistant to killing by tobramycin and norfloxacin than ΔbrlR mutant biofilms, from which no viable cells were recovered at concentrations exceeding the biofilm MBCs.
Considering that biofilms have been described as up to 1,000 times more resistant to antimicrobial agents than their planktonic counterparts, the biofilm MBCs obtained here appear to be low. However, we believe this to be related to the manner in which the antibiotics are delivered to the biofilms under the conditions tested, as biofilms are continuously exposed to a certain concentration of antibiotics under flowing conditions. This is supported by the finding that P. aeruginosa strains harboring a gentamicin resistance cassette (e.g., PAO1/pJN105) are capable of growing in the presence of ≥75 μg/ml gentamicin in batch culture but are unable to grow and form biofilms at concentrations exceeding 3 μg/ml under the flowing conditions used for biofilm MBC determinations. It is important to note that at this concentration (3 μg/ml), 100% of the bacteria recovered from the biofilms are gentamicin resistant, indicating that differences in susceptibility under the conditions tested are not due to loss of the plasmid pJN105. Taking into account the increased susceptibility (>25-fold) of otherwise recalcitrant P. aeruginosa strains under flowing conditions, the ability to resist a concentration of 75 μg/ml suggests a >600-fold greater drug tolerance of P. aeruginosa when it is grown as a biofilm than when it is in the planktonic mode of growth.
brlR expression contributes to survival following antibiotic challenge.
While wild-type biofilm cells survived exposure to tobramycin and norfloxacin at concentrations exceeding the biofilm MBCs, no viable cells were recovered from ΔbrlR mutant biofilms. The findings suggested a role for BrlR in resistance to killing by antimicrobial agents. To determine whether tobramycin treatment indeed resulted in the killing of ΔbrlR mutant biofilms, wild-type and ΔbrlR mutant biofilms treated with 50 μg/ml tobramycin for 24 h were allowed to recover for an additional 24 h under flowing conditions in the absence of tobramycin. Following recovery, the P. aeruginosa PAO1 biofilm population increased from ∼105 cells/ml to up to ∼108 cells/ml. ΔbrlR mutant biofilms, however, demonstrated very little recovery. No viable cells were detected in seven out of eight trials, with one ΔbrlR mutant biofilm showing 6 × 102 CFU/ml following recovery (Fig. 5E). Taken together, our findings strongly support a role for BrlR in biofilm drug tolerance by affecting the biofilm MBC and resistance to killing.
Effect of brlR expression on killing of biofilm bacteria.
To visualize the effect of brlR inactivation on viability following antimicrobial treatment, biofilms were grown in flow cells and subsequently stained with the LIVE/DEAD Bac Light viability stain to distinguish cells that are alive from those considered to be dead. Confocal images of biofilms thus stained were acquired prior to and following treatment with tobramycin for 24 h. Prior to tobramycin treatment, the overall percentages of the biofilm population stained with PI were similar in wild-type, ΔbrlR mutant, and brlR-overexpressing biofilms (Fig. 5F). Tobramycin treatment, however, resulted in a significantly increased population of dead cells located in the interior of microcolonies of ΔbrlR mutant biofilms (Fig. 5F). The effect of tobramycin treatment on PAO1 biofilms was less pronounced, with very few cells staining with PI. In contrast, no change in PI staining was observed in biofilms overexpressing brlR (Fig. 5F).
Increased brlR expression contributes to drug tolerance in CF clinical isolates.
To determine if BrlR-dependent drug-tolerant phenotypes are selected during chronic infections, we next asked whether differential brlR expression and BrlR-dependent tolerance are detectable in clinical isolates. We therefore selected three clinical P. aeruginosa isolates that were previously recovered from a single patient suffering from CF, namely, CF1-2, CF1-8, and CF1-13 (53). Analysis of these three CF isolates by Southern blot analysis using a 741-bp PstI-NruI fragment derived from the upstream region of exotoxin A demonstrated that they originated from the same parental strain (53). Similar results were obtained by serotyping using Difco antisera (53). On the basis of their colony morphology on agar, the three clinical isolates have been described as classical, rough, and mucoid. While no significant difference in growth in liquid, motility, pyocyanin production, or other biochemical reactions was found by biotyping, the isolates differed with respect to their antibiograms obtained by using Kirby-Bauer disk susceptibility assays. Isolate CF1-8 was found to be susceptible to chloramphenicol and trimethoprim-sulfamethoxazole, while CF1-2 was susceptible to only chloramphenicol (53). The antibiogram of CF1-13 was not determined.
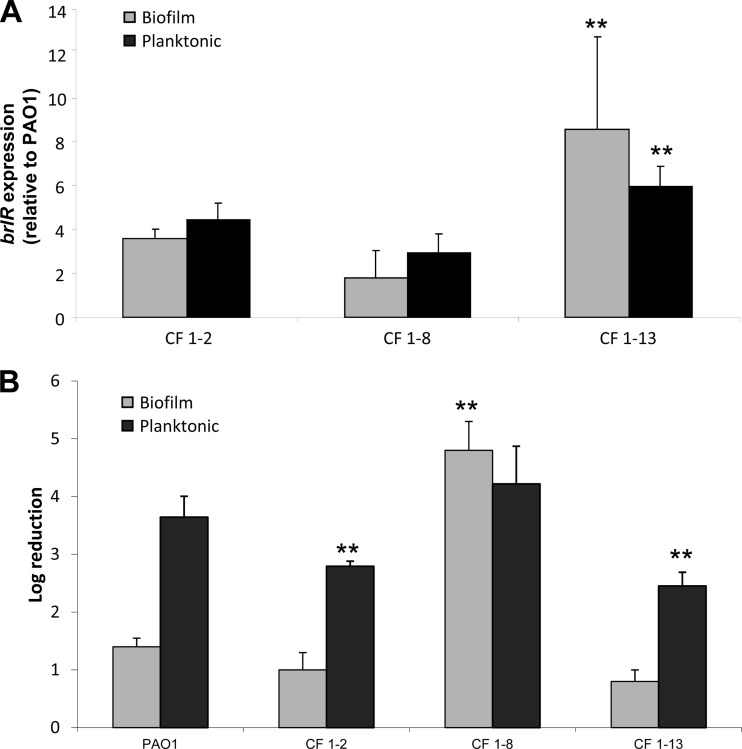
We first determined brlR expression by qRT-PCR. In contrast to brlR expression in P. aeruginosa PAO1, its expression in these three strains was detectable under both planktonic and biofilm growth conditions and was up to 8-fold higher than that in P. aeruginosa PAO1 biofilms, with the highest expression levels detected in mucoid isolate CF1-13 (Fig. 6A).
FIG 6.
While brlR is overexpressed in clinical CF isolates under both planktonic and biofilm growth conditions, isolate CF1-8 is more susceptible to tobramycin under biofilm growth conditions. (A) brlR transcript abundance was determined by qRT-PCR in clinical isolates grown as biofilms for 3 days and in planktonic culture. Transcript levels were normalized to PAO1. (B) Susceptibility determination of P. aeruginosa PAO1 and clinical CF isolates grown planktonically and as biofilms (1 day). Planktonic cells were treated with 50 μg/ml tobramycin for 1 h, while biofilms were treated with 100 μg/ml tobramycin for 1 h under flowing conditions. **, significantly different (P < 0.001) from PAO1 as indicated by ANOVA and SigmaStat.
Treatment of clinical isolates CF1-2 and CF1-13 grown to exponential phase with tobramycin indicated a susceptibility lower than that of PAO1 (Fig. 6B). Similar results were obtained following tobramycin treatment of biofilms for 1 h (Fig. 6B). Despite increased brlR expression levels, however, no difference in the susceptibility of clinical isolate CF1-8 under planktonic growth conditions and that of PAO1 was noted. Instead, biofilms of isolate CF1-8 were found to be significantly more susceptible to tobramycin than P. aeruginosa PAO1 biofilms and to be as susceptible as ΔbrlR mutant biofilms (Fig. 4 and 6B).
To further determine the contribution of BrlR to the antimicrobial tolerance phenotypes of the three clinical isolates, MIC studies were done with norfloxacin, chloramphenicol, tobramycin, kanamycin, and tetracycline. Regardless of the antibiotics tested, no difference between the MICs for PAO1 and isolate CF1-8 were observed (Table 4; see Fig. S5 in the supplemental material), even though brlR expression levels were higher in this isolate than in PAO1 under planktonic conditions. In contrast, the MICs for clinical isolates CF1-2 and CF1-13 were, on average, 2- to 10-fold higher than those for PAO1, depending on the antimicrobial agents tested (Table 4; see Fig. S5 in the supplemental material).
Table 4.
MICs for clinical isolates grown in 10-fold-diluted LB mediuma
| Strain | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| Cm | Tob | Tet | Km | Nor | |
| PAO1 | 150 | 6.25 | 25 | 100 | 0.39–0.78 |
| CF1-2 | >150 (R)b | 12.5 | 25 | >100 (R) | 3.13 |
| CF1-8 | 150 | 6.25 | 6.25 | 100 | 0.39–0.78 |
| CF1-8/pJN-brlR | >150 (R) | 25 | NDc | ND | 3.75 |
| CF1-13 | 150 | 50 | 25 | >100 (R) | 1.56 |
Cm, chloramphenicol; Tob, tobramycin; Tet, tetracycline; Km, kanamycin; Nor, norfloxacin.
R, resistant.
ND, not determined.
BrlR in CF clinical isolate CF1-8 is nonfunctional.
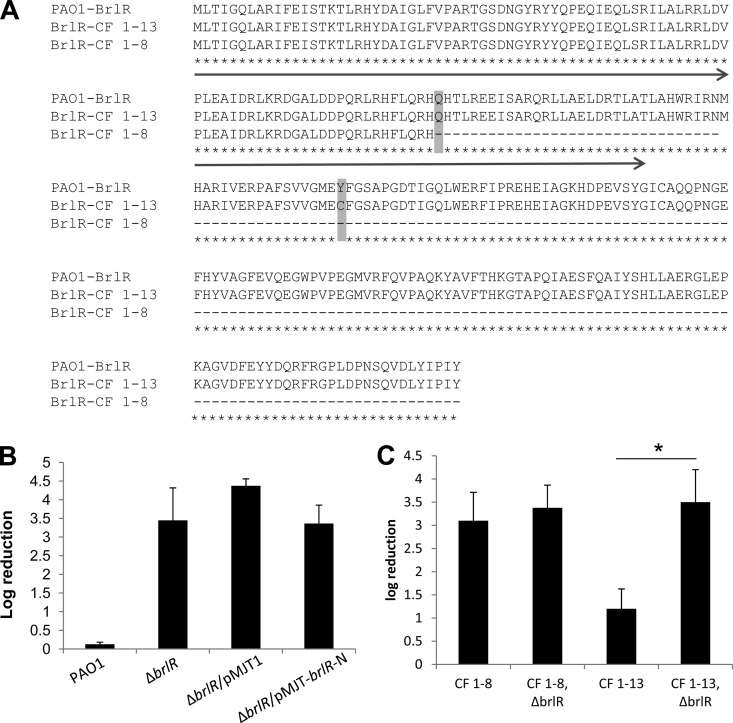
The finding that the clinical isolate CF1-8 was comparable in susceptibility to the ΔbrlR mutant despite elevated brlR expression raised the question of whether the brlR gene in this strain encodes a functional BrlR. We therefore cloned and sequenced the brlR gene from clinical isolates CF1-8 and CF1-13 and compared the sequence to brlR obtained from PAO1. The analysis revealed a nonsense mutation (CAG → UAG) in the CF1-8 sequence at amino acid position 88 (262 bp) that results in translation termination at the end of the N-terminally located DNA binding domain (Fig. 1B and 7A).
FIG 7.
Isolate CF1-8 harbors a nonsense mutation in BrlR. (A) Alignment of BrlR sequences of PAO1, CF1-8, and CF1-13. CF1-8 harbors a point mutation resulting in a premature stop codon at amino acid position 88. CF1-13 harbors a mutation resulting in an amino acid substitution (Y→C). The arrow indicates the location of the N-terminal DNA binding domain of BrlR. (B) Complementation of the ΔbrlR mutant with the N-terminal DNA binding site of BrlR alone (BrlR-N) does not restore the susceptibility to tobramycin of this strain to the wild-type level. (C) Inactivation of brlR in isolate CF1-13 increases the susceptibility of this strain to tobramycin, while inactivation of brlR in isolate CF1-8 has no effect. Strains were grown planktonically to the exponential phase and treated with 50 μg/ml tobramycin for 1 h. *, significantly different (P < 0.001) from CF1-13 as indicated by t test.
Considering that MtaN from B. subtilis, which is composed of only the N-terminal DNA-binding and dimerization domains of MerR family member Mta, is a constitutive, transcriptionally active, 109-residue truncation mutant protein (29, 49), we next asked whether the N-terminal DNA-binding domain of BrlR alone is capable of restoring the drug tolerance phenotype of ΔbrlR mutant biofilms by complementing the mutant with the N-terminal DNA binding domain of BrlR (ΔbrlR/pMJT-brlR-N). While truncated BrlR was found to be as soluble as intact BrlR (data not shown), the complemented ΔbrlR mutant (ΔbrlR/pMJT-brlR-N) was as susceptible as a ΔbrlR mutant in the absence or presence of the empty vector (Fig. 7B; see Fig. S3 in the supplemental material).
The findings indicate that the truncated BrlR protein is not functional. Moreover, the findings indicate that clinical isolate CF1-8 harbors a nonfunctional BrlR which, despite elevated levels of brlR expression, is not capable of contributing to the recalcitrant phenotype of this strain. The finding was further supported by the observation that overexpression of brlR in clinical isolate CF1-8 resulted in 2- to 4-fold increases in the MICs of norfloxacin, chloramphenicol, tobramycin, kanamycin, and tetracycline (Table 4).
Inactivation of brlR renders CF clinical isolate CF1-13 but not CF1-8 more susceptible to tobramycin.
To further confirm a role for BrlR in the drug tolerance phenotypes of these clinical isolates, isogenic ΔbrlR mutants of clinical isolates CF1-8 and CF1-13 were constructed and tested for susceptibility to tobramycin. Inactivation of brlR in clinical isolate CF1-8 resulted in no difference in susceptibility between the mutant and parental strains (Fig. 7C). In contrast, inactivation of brlR rendered clinical isolate CF1-13 significantly more susceptible than the parental strain (Fig. 7C).
DISCUSSION
Biofilm bacteria exhibit a profound tolerance of various antimicrobial agents, rendering biofilm cells 10- to 1,000-fold less susceptible than the same bacteria grown in a planktonic (free-floating) culture (34). The underlying cause of this resilience has been considered to be multifactorial and unlike mechanisms that confer resistance to cells grown planktonically. Here we demonstrate that the antimicrobial tolerance of P. aeruginosa biofilms is dependent on BrlR, a member of the MerR family of transcriptional regulators. BrlR was found to be closely related to regulators that play a role in tolerance to antibiotics in Gram-positive bacteria, including BmrR, BltR, and MtaN from B. subtilis and TipA from S. lividans. A common feature of these regulators is activation of the expression of multidrug transporters upon binding of the transporter substrate. BmrR is induced upon exposure to rhodamine and tetraphenylphosphonium, BltR is induced upon exposure to rhodamine, and TipA is induced upon exposure to thiostrepton (1, 9, 13, 30, 51, 72). Inducers of the global activator Mta are not known (29, 30). While further research is required to determine the exact mechanism by which BrlR confers protection from antimicrobial agents, including the identification of downstream targets and/or potential multidrug efflux pumps, it is apparent that BrlR is unique among the MerR regulators, as BrlR only plays a role in the antimicrobial tolerance of biofilms. To our knowledge, BrlR is the first MerR regulator identified in a Gram-negative bacterium that has been linked to antibiotic tolerance, and unlike previously described MerR proteins, it is expressed in a growth mode-specific manner.
While the recalcitrance of biofilms to antimicrobials has often been compared to the tolerance of planktonic cells grown to stationary phase, our findings indicate that different mechanisms are at work. This is apparent from the observed difference in susceptibility when the ΔbrlR mutant is grown as a biofilm and planktonically to stationary phase, with the recalcitrance of the biofilm being distinct from the antimicrobial tolerance seen during the stationary phase. Considering that the formation of persister cells has been attributed to the tolerance phenotypes of both stationary-phase and biofilm-grown bacterial cells, our findings suggest that the function of BrlR is independent of mechanisms contributing to persistence. Moreover, while our findings do not exclude previously described mechanisms such as slow growth or increased Psl or glucan production contributing to biofilm drug tolerance (the relationship of which to BrlR-mediated tolerance will be the subject of future studies), we demonstrate the presence of a regulatory component to biofilm drug tolerance.
Resistance is the ability of a microorganism to grow in the presence of an elevated level of an antimicrobial agent, as indicated by an increased MIC. Many reports have shown that by this convention, biofilm cells do not display growth in the presence of an antimicrobial agent at concentrations inhibitory to planktonic cell growth (40, 63). Instead, the reported “resistance” describes an increased tolerance of cells to killing (40, 63). Here we demonstrate that BrlR contributes to the tenacity exhibited by P. aeruginosa biofilms against antimicrobial compounds in two different ways. For one, BrlR lowered the susceptibility to hydrogen peroxide and five different classes of antibiotics by increasing the MICs up to 6-fold. In addition, BrlR contributes significantly to the recalcitrance of biofilms to eradication by microbicidal antimicrobials, as biofilms of bacteria with brlR inactivated were completely killed by microbicidal antimicrobials at concentrations inhibitory to planktonic cell growth. Thus, our findings strongly suggest that biofilm drug tolerance is due not only to increased recalcitrance to eradication by microbicidal antimicrobials but also to the ability of biofilms to grow in the presence of increased concentrations of microbicidal antimicrobials.
Trunk et al. (71) reported increased brlR expression under oxygen-limiting conditions. While brlR expression levels increased only 2.3-fold, the finding suggested that oxygen limitation present in biofilms is a possible reason for the biofilm-specific expression of brlR. Likewise, a 2.3-fold increase in brlR levels was noted upon the addition of pyocyanin to P. aeruginosa PA14 grown exponentially (21). Here, increased brlR expression was detectable in P. aeruginosa PAO1 biofilms and in three clinical isolates under both planktonic and biofilm growth conditions. While the mechanism that results in biofilm-specific brlR expression remains to be elucidated, this finding suggests a selection for increased expression of brlR in vivo. This raises the question of why BrlR has not been found to contribute to biofilm antimicrobial tolerance in previous studies (8, 12, 23, 24, 28, 40, 42, 76). On the basis of our findings, BrlR appears to be active only under biofilm-specific growth conditions and not under planktonic growth conditions. Moreover, most studies make use of MBC assays with microtiter plate-grown attached cells. Under these growth conditions, brlR expression was not detectable in P. aeruginosa, suggesting a requirement for the transition to specific biofilm maturation stages (perhaps irreversible attachment) for the induction of brlR expression.
In summary, we show that the tolerance of biofilm to antimicrobial agents is a regulated process and at least in part dependent on BrlR, which is expressed in a biofilm-specific manner. While our findings do not exclude the contribution of previously described mechanisms, including slow growth and diffusion limitation, to antibiotic tolerance of biofilms, our findings of a genetic determinant playing an important role in biofilm drug tolerance suggest the existence of a classical, biofilm-specific mechanism of drug tolerance in P. aeruginosa and may open up avenues for therapeutics of biofilm-related complications in medical, industrial, and environmental settings.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Vasil at University of Colorado School of Medicine for providing clinical isolates CF1-2, CF1-8, and CF1-13.
This work was supported by a grant from the National Institutes of Health (R01 AI080710).
Footnotes
Published ahead of print 22 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Ahmed M, Borsch CM, Taylor SS, Vazquez-Laslop N, Neyfakh AA. 1994. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506–28513 [PubMed] [Google Scholar]
- 2. Allegrucci M, et al. 2006. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 188:2325–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allegrucci M, Sauer K. 2007. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J. Bacteriol. 189:2030–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allegrucci M, Sauer K. 2008. Formation of Streptococcus pneumoniae non-phase-variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J. Bacteriol. 190:6330–6339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderl JN, Franklin MJ, Stewart PS. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44:1818–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderl JN, Zahller J, Roe F, Stewart PS. 2003. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 47:1251–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anwar H, Strap JL, Chen K, Costerton JW. 1992. Dynamic interactions of biofilms of mucoid Pseudomonas aeruginosa with tobramycin and piperacillin. Antimicrob. Agents Chemother. 36:1208–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bagge N, et al. 2004. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and β-lactamase and alginate production. Antimicrob. Agents Chemother. 48:1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baranova NN, Danchin A, Neyfakh AA. 1999. Mta, a global MerR-type regulator of the Bacillus subtilis multidrug-efflux transporters. Mol. Microbiol. 31:1549–1559 [DOI] [PubMed] [Google Scholar]
- 10. Becher A, Schweizer HP. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29:948–952 [DOI] [PubMed] [Google Scholar]
- 11. Bjarnsholt T, et al. 2005. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373–383 [DOI] [PubMed] [Google Scholar]
- 12. Brooun A, Liu S, Lewis K. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44:640–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brown NL, Stoyanov JV, Kidd SP, Hobman JL. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145–163 [DOI] [PubMed] [Google Scholar]
- 14. Campanac C, Pineau L, Payard A, Baziard-Mouysset G, Roques C. 2002. Interactions between biocide cationic agents and bacterial biofilms. Antimicrob. Agents Chemother. 46:1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Colvin KM, et al. 2011. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264 doi: 10.1371/journal.ppat.1001264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Connell JL, et al. 2010. Probing prokaryotic social behaviors with bacterial “lobster traps.” mBio 1(4):e00202-10 doi:10.1128/mBio.00202-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 18. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 19. Davey ME, O'Toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davies DG, et al. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298 [DOI] [PubMed] [Google Scholar]
- 21. Dietrich LEP, Price-Whelan A, Petersen A, Whiteley M, Newman DK. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61:1308–1321 [DOI] [PubMed] [Google Scholar]
- 22. Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Drenkard E. 2003. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5:1213–1219 [DOI] [PubMed] [Google Scholar]
- 24. Drenkard E, Ausubel FM. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740–743 [DOI] [PubMed] [Google Scholar]
- 25. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fux CA, Wilson S, Stoodley P. 2004. Detachment characteristics and oxacillin resistance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 186:4486–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilbert P, Maira-Litran T, McBain AJ, Rickard AH, Whyte FW. 2002. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microb. Physiol. 46:202–256 [PubMed] [Google Scholar]
- 28. Gillis RJ, et al. 2005. Molecular basis of azithromycin-resistant Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 49:3858–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Godsey MH, Baranova NN, Neyfakh AA, Brennan RG. 2001. Crystal structure of MtaN, a global multidrug transporter gene activator. J. Biol. Chem. 276:47178–47184 [DOI] [PubMed] [Google Scholar]
- 30. Godsey MH, Zheleznova Heldwein EE, Brennan RG. 2002. Structural biology of bacterial multidrug resistance gene regulators. J. Biol. Chem. 277:40169–40172 [DOI] [PubMed] [Google Scholar]
- 31. Grkovic S, Brown MH, Skurray RA. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heydorn A, et al. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407 [DOI] [PubMed] [Google Scholar]
- 33. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 34. Hoyle BD, Costerton JW. 1991. Bacterial resistance to antibiotics: the role of biofilms. Prog. Drug Res. 37:91–105 [DOI] [PubMed] [Google Scholar]
- 35. Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Invest. 117:877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13–18 [DOI] [PubMed] [Google Scholar]
- 37. Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Khan W, et al. 2010. Aminoglycoside resistance of Pseudomonas aeruginosa biofilms modulated by extracellular polysaccharide. Int. Microbiol. 13:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322:107–131 [DOI] [PubMed] [Google Scholar]
- 40. Lewis K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luppens SB, Rombouts FM, Abee T. 2002. The effect of the growth phase of Staphylococcus aureus on resistance to disinfectants in a suspension test. J. Food Prot. 65:124–129 [DOI] [PubMed] [Google Scholar]
- 42. Mah T-F, et al. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310 [DOI] [PubMed] [Google Scholar]
- 43. Mah T-F, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39 [DOI] [PubMed] [Google Scholar]
- 44. Marshall KC. 1988. Adhesion and growth of bacteria at surfaces in oligotrophic habitats. Can. J. Microbiol. 34(4):503–506 [Google Scholar]
- 45. McBain AJ, et al. 2003. Microbial characterization of biofilms in domestic drains and the establishment of stable biofilm microcosms. Appl. Environ. Microbiol. 69:177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miller J. 1972. Experiments in molecular genetics, p 352–355 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 47. Monzón M, Oteiza C, Leiva J, Amorena B. 2001. Synergy of different antibiotic combinations in biofilms of Staphylococcus epidermidis. J. Antimicrob. Chemother. 48:793–801 [DOI] [PubMed] [Google Scholar]
- 48. Moriarty TF, Elborn JS, Tunney MM. 2007. Effect of pH on the antimicrobial susceptibility of planktonic and biofilm-grown clinical Pseudomonas aeruginosa isolates. Br. J. Biomed. Sci. 64:101–104 [DOI] [PubMed] [Google Scholar]
- 49. Newberry KJ, Brennan RG. 2004. The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J. Biol. Chem. 279:20356–20362 [DOI] [PubMed] [Google Scholar]
- 50. Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203 [DOI] [PubMed] [Google Scholar]
- 51. Neyfakh AA. 2001. The ostensible paradox of multidrug recognition. J. Mol. Microbiol. Biotechnol. 3(2):151–154 [PubMed] [Google Scholar]
- 52. Nguyen D, et al. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ogle JW, Janda JM, Woods DE, Vasil ML. 1987. Characterization and use of a DNA probe as an epidemiological marker for Pseudomonas aeruginosa. J. Infect. Dis. 155:119–126 [DOI] [PubMed] [Google Scholar]
- 54. Paulsen IT. 2003. Multidrug efflux pumps and resistance: regulation and evolution. Curr. Opin. Microbiol. 6:446–451 [DOI] [PubMed] [Google Scholar]
- 55. Petrova OE, Sauer K. 2009. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 5:e1000668 doi:10.1371/journal.ppat.1000668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Picioreanu C, van Loosdrecht MCM, Heijnen JJ. 2001. Two-dimensional model of biofilm detachment caused by internal stress from liquid flow. Biotechnol. Bioeng. 72:205–218 [PubMed] [Google Scholar]
- 57. Sauer K, Camper AK. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sauer K, et al. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186:7312–7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schweizer HP, Hoang TT. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15–22 [DOI] [PubMed] [Google Scholar]
- 61. Shah D, et al. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53 doi:10.1186/1471-2180-6-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Southey-Pillig CJ, Davies DG, Sauer K. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187:8114–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Spoering AL, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sternberg C, et al. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stewart PS. 1996. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob. Agents Chemother. 40:2517–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138 [DOI] [PubMed] [Google Scholar]
- 67. Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187–209 [DOI] [PubMed] [Google Scholar]
- 68. Storz G, Tartaglia L, Ames B. 1990. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248:189–194 [DOI] [PubMed] [Google Scholar]
- 69. Summers AO. 1992. Untwist and shout: a heavy metal-responsive transcriptional regulator. J. Bacteriol. 174:3097–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thormann KM, et al. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 188:2681–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Trunk K, et al. 2010. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ. Microbiol. 12:1719–1733 [DOI] [PubMed] [Google Scholar]
- 72. Vazquez-Laslop N, Zheleznova EE, Markham PN, Brennan RG, Neyfakh AA. 2000. Recognition of multiple drugs by a single protein: a trivial solution of an old paradox. Biochem. Soc. Trans. 28(4):517–520 [PubMed] [Google Scholar]
- 73. Villain-Guillot P, Gualtieri M, Bastide L, Leonetti J-P. 2007. In vitro activities of different inhibitors of bacterial transcription against Staphylococcus epidermidis biofilm. Antimicrob. Agents Chemother. 51:3117–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wagner VE, Li L-L, Isabella VM, Iglewski BH. 2007. Analysis of the hierarchy of quorum-sensing regulation in Pseudomonas aeruginosa. Anal. Bioanal. Chem. 387:469–479 [DOI] [PubMed] [Google Scholar]
- 75. Wozniak DJ, et al. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. U. S. A. 100:7907–7912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang L, Mah T-F. 2008. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J. Bacteriol. 190:4447–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zheleznova EE, Markham PN, Neyfakh AA, Brennan RG. 1999. Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell 96:353–362 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.