Background: Stathmin-like domain (SLD) proteins from vertebrates bind two tubulin molecules.
Results: SLDs that bind tubulin with a programmed stoichiometry are characterized.
Conclusion: Rules are established to design (tubulin)x-SLD complexes, starting from a 1:1 stoichiometry.
Significance: This work provides new insights into stathmin family member function. The SLDs produced will be useful tools to study interactions of microtubule regulators with tubulin.
Keywords: Crystal Structure, Cytoskeleton, Microtubules, Protein Complexes, Protein Engineering, Structural Biology, Microtubule Dynamics, Stathmin-like Domain
Abstract
In cells, microtubule dynamics is regulated by stabilizing and destabilizing factors. Whereas proteins in both categories have been identified, their mechanism of action is rarely understood at the molecular level. This is due in part to the difficulties faced in structural approaches to obtain atomic models when tubulin is involved. Here, we design and characterize new stathmin-like domain (SLD) proteins that sequester tubulins in numbers different from two, the number of tubulins bound by stathmin or by the SLD of RB3, two stathmin family members that have been extensively studied. We established rules for the design of tight tubulin-SLD assemblies and applied them to complexes containing one to four tubulin heterodimers. Biochemical and structural experiments showed that the engineered SLDs behaved as expected. The new SLDs will be tools for structural studies of microtubule regulation. The larger complexes will be useful for cryo-electron microscopy, whereas crystallography or nuclear magnetic resonance will benefit from the 1:1 tubulin-SLD assembly. Finally, our results provide new insight into SLD function, suggesting that a major effect of these phosphorylatable proteins is the programmed release of sequestered tubulin for microtubule assembly at the specific cellular locations of members of the stathmin family.
Introduction
Microtubules are dynamic protein assemblies essential for cell morphogenesis, membrane trafficking, and cell division of eukaryotic cells. In vivo, typically 13 straight, parallel, protofilaments interact laterally to form a microtubule. Each protofilament is a longitudinal head-to-tail assembly of αβ tubulin heterodimers (hereafter referred to as tubulins). In vitro experiments with purified tubulin have demonstrated that microtubules switch stochastically between prolonged periods of assembly and disassembly, a phenomenon called dynamic instability (1). In vivo, microtubule dynamics is regulated by different classes of proteins. These include polymerases, depolymerases, microtubule stabilizing, and tubulin sequestering proteins (2–5).
The complex events underlying the regulation of microtubule assembly are difficult to observe structurally. This has been achieved on a few occasions, when sufficiently homogeneous samples were obtained. Two strategies have been used. The first exploited the properties of tubulin sequestering proteins to produce homogeneous complexes whose crystal structure has been determined (6, 7). Alternatively, tubulin assemblies have been analyzed using cutting edge transmission electron microscopy (TEM).3 These studies focused on microtubules (8), microtubules decorated with globular proteins (9–11), and a few non-microtubular tubulin species (e.g. Ref. 12). But, in most cases, due to the heterogeneity of the assemblies present in solutions of tubulin and of its complexes, obtaining crystals that diffract to atomic resolution remains challenging. Moreover, because of the limitations of the lifetime of the sample in the electron beam (13) and because extensive averaging of images of identical species is not possible, the study of such heterogeneous assemblies by cryo-TEM is also restricted to low resolutions that hardly go beyond the dimensions of globular domains. The availability of new stable and well defined tubulin complexes, including single sequestered heterodimers, would offer new options for crystallization or allow TEM images to be collected that could then be averaged. This would therefore greatly facilitate the study of tubulin assembly regulation structurally and also biochemically.
Stathmin and stathmin-like domains (SLDs) prevent the formation of microtubules (5, 14). The SLDs from vertebrates have been best studied; they bind two tubulins arranged longitudinally, head-to-tail, in protofilament-like complexes (see Fig. 1A) (15, 16). These complexes are homogeneous and stable, but their size (∼200 kDa) is at the lower end of the range of species that may be studied at high resolution by cryo-TEM (17). SLDs from Drosophila can bind up to four tubulins, in a dynamic association (18). No SLD has been identified that sequesters efficiently a single tubulin, although several attempts at designing such proteins have been made (19, 20). Because vertebrate SLDs allow the binding of other regulatory proteins to their complexes with tubulin (21), they appear to be a useful starting point for the development of stable, well defined, assemblies of tubulin that could be used to study the regulation of microtubule assembly, both biochemically and structurally, including by electron microscopy. But to do so, stable complexes comprising three or four heterodimers should be engineered to be of a size large enough for this methodology to be conveniently applied. The smaller version of these complexes, comprising one tubulin, would extend the range of tubulin complexes that may be crystallized for higher resolution studies beyond T2R, the ternary complex of two tubulin heterodimers with the SLD of the RB3 protein (RB3SLD). Such platforms will provide stable entities to which regulatory proteins may bind. They may also be used to study the interaction with tubulin of small molecule compounds (6).
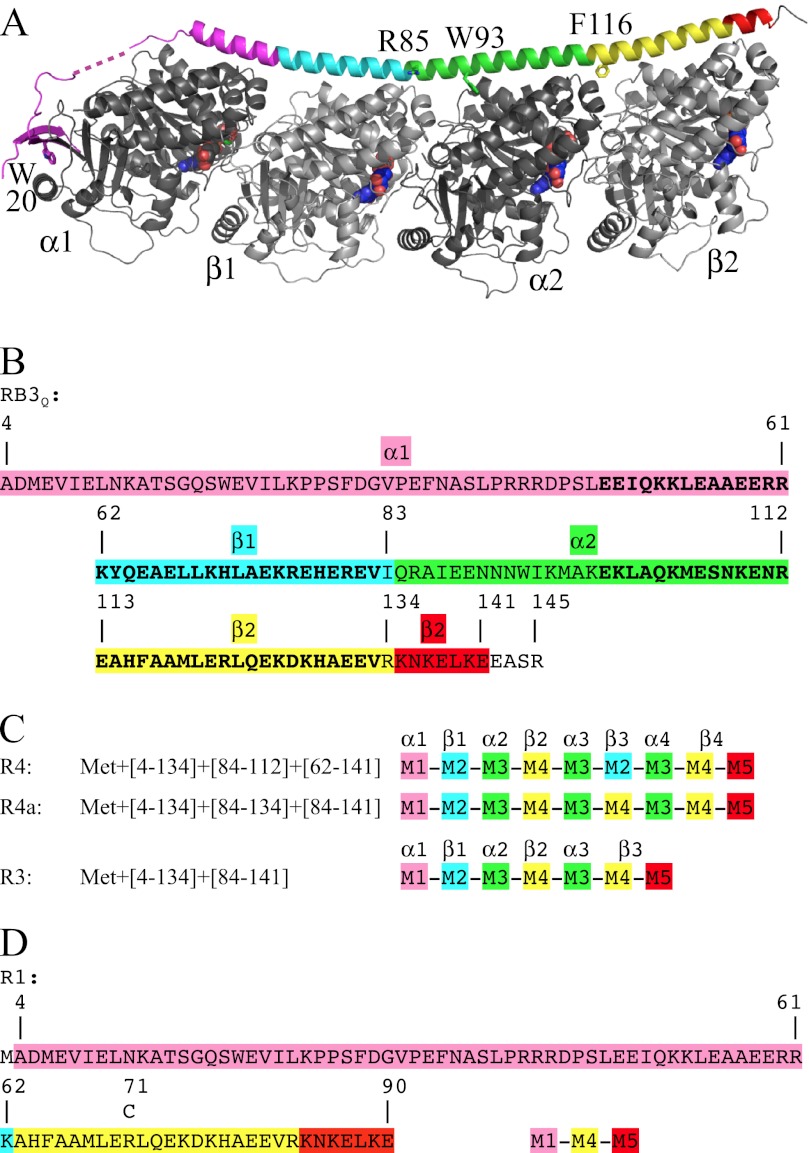
FIGURE 1.
The design of RB3SLD-based constructs for binding tubulin with a predefined stoichiometry. A, the T2R structure in which RB3SLD is colored according to the modules used in the design of the new SLDs. The RB3SLD according to which the modules are defined (RB3Q) contains four mutations that are modeled in the structure, F20W, K85R, F93W, and L116F (numbering is in reference to stathmin), plus the additional C14A mutation. The linker between the N-terminal β hairpin and the C-terminal helix, starting at residue Leu47, contains the least ordered region of RB3SLD; part of it is shown as a dashed line. Figs. 1A, 3, and 5 were generated using PyMOL (44). B, the RB3Q sequence colored according to the same modules as in A. The residues of the two stretches of the internal repeat (Glu48–Val82 and Glu99–Val133) are highlighted in boldface type. C, the design in terms of the modules defined in panel B of SLDs engineered to bind three (R3) or four tubulin heterodimers (R4 and R4a). In A–C, the tubulin subunits interacting with the modules we defined are indicated. D, sequence of R1 designed to bind one tubulin molecule. The position of the residue mutated to cysteine (R71C) and used to label R1 for affinity measurements is indicated.
Here, we describe the design of SLD-based proteins that make stable complexes with tubulin. The structural characterization of a complex comprising four tubulin heterodimers demonstrates that it consists of longitudinally assembled molecules that have the same overall structure as tubulin in T2R, strongly suggesting that this applies to all SLD-mediated tubulin assemblies. Moreover, the high resolution structure of a single sequestered tubulin shows that its interactions with the SLD are identical to those in T2R, thus validating the rules we established for the design of SLDs binding a predefined number of heterodimers assembled longitudinally.
EXPERIMENTAL PROCEDURES
Gene Synthesis, Cloning, Protein Overexpression, and Purification
R4 and R4a genes were purchased from Genscript (Piscataway, NJ). R3 was synthesized according to the method of Stemmer et al. (22). R1 was obtained from a plasmid coding for an RB3SLD variant by a modified overlap extension PCR method (23). Its sequence is displayed in Fig. 1. All these constructs have been cloned between the NcoI and XhoI sites in a pET28 plasmid carrying a kanamycin resistance gene and a promoter inducible by isopropyl β-d-1-thiogalactopyranoside. Proteins were overexpressed in Escherichia coli BL21 DE3 Star, in LB medium supplemented with kanamycin, using 0.5 mm isopropyl β-d-1-thiogalactopyranoside to induce an expression period of 3 h at 37 °C. Purification was as described (6) except that a first step of nucleic acid precipitation by spermine (24) was added and that the heating step was omitted for R4 and R4a. The concentration of purified SLD was determined by measuring the absorbance at 280 nm, taking advantage of the presence of tryptophan residues in these constructs, as opposed to wild type RB3SLD, whose absorbance at 280 nm is very weak. A mass spectrometry analysis of R4 showed it has the expected molecular mass, taking into account the removal of the N-terminal methionine and a subsequent Nα-acetylation as is the case for RB3SLD (25). Tubulin was purified from sheep brain by two cycles of assembly-disassembly in a high molarity Pipes buffer (26). Before use, an additional cycle of assembly-disassembly was performed to remove inactive protein. The designed ankyrin repeat protein (DARPin) used in this study and named D2 was selected in the same screen as the D1 DARPin and was produced and purified similarly (27).
Size Exclusion Chromatography (SEC) and Size Exclusion Chromatography Coupled to Multiangle Laser Light Scattering (SEC-MALLS)
For SEC experiments, samples of tubulin alone or mixed with RB3SLD, R1, R3, or R4 were analyzed on a Superdex S200 column (GE Healthcare) equilibrated with 15 mm Pipes-K, pH 6.8, 0.4 mm MgCl2, and 0.2 mm EGTA (low salt buffer) or with the same buffer containing in addition 60 mm KCl (higher salt buffer). For SEC-MALLS analyses, SEC was carried out on a Prominence HPLC system (Shimadzu) using a KW804 column (Shodex) run in the higher salt buffer. 30-μl samples at 20 or 40 μm tubulin concentrations and containing variable amounts of SLDs were run at a 0.5 ml/min flow rate. Detection was performed using a three-detector static light-scattering apparatus (MiniDAWN TREOS, Wyatt Technology, equipped with a quasi-elastic light-scattering module) and a refractometer (Optilab rEX, Wyatt Technology). Molecular weight calculations were performed with the ASTRA V software (Wyatt Technology) using a dn/dc value of 0.183 ml/g.
Electron Microscopy (EM)
A 5-μl (0.15 mg/ml) tubulin-R4 sample containing Protein A conjugated with 5-nm colloidal gold particles (Cell Microscopy Center, University Medical Center, Utrecht, The Netherlands) as fiducial markers was applied to a glow-discharged carbon-coated Cu 200-mesh grid, allowed to adsorb for 30 s, washed twice with water, and negatively stained for 20 s with 0.5% (w/v) uranyl formate. Specimens were examined in a Fei Tecnai 20 TEM operated at 200 kV. Double tilt tomograms were recorded with a magnification at the detector plane of ×69,000 with a Gatan CCD camera through Inspect3D with the specimen supported by a Fishione double tilt holder. The sample was tilted from −62 to +62 ° in 0.5 ° steps. Tomographic reconstructions were made with IMOD (28). The Slicer tool of IMOD was used to combine multiple slices in the beam direction.
Kinetic Analysis of the Tubulin-R1 Interaction
An R71C mutant of R1 was obtained by standard molecular biology techniques and produced and purified as the wild type protein. It was reduced with dithiothreitol and then reacted with acrylodan. Excess acrylodan and protein aggregates were removed by gel filtration on a Superdex S200 column. The resulting protein, named R1*, was used for fluorescence studies (λex, 290 nm; λem, 510 nm). The dissociation constant (Kd) was determined using a fixed concentration of R1* titrated against an increasing amount of tubulin at 20 °C in a Cary Eclipse spectrofluorimeter (Varian). The buffer used consisted of 25 mm Pipes-K, pH 6.8, 0.2 mm EGTA, 0.5 mm MgCl2, and 10 μm GDP. The data were fitted to a 1:1 binding isotherm with Equation 1,
 |
where ΔFluo is the variation of the fluorescence signal, Fluomax is the fluorescence at saturating concentration of tubulin, [T] and [R1*] are the concentrations of tubulin and labeled R1, respectively, and Kd is the dissociation constant.
The dissociation rate constant koff was determined using a Hi-Tech KinetAsyst stopped-flow system (TgK Scientific) at 20 °C in the same buffer. A 30 nm R1* and 50 nm tubulin solution was mixed with either 2.1 or 4.2 μm R1. R1* suffered from some photobleaching in these conditions (data not shown); this was taken into account by fitting the fluorescence decrease with the following mono-exponential decay function,
where Fluo is the fluorescence signal, Fluomin is fluorescence at infinite time, ΔFluo is the amplitude of the fluorescence variation, kobs corresponds to koff at saturating concentration of R1, and b is the photobleaching term.
The association rate constant kon was also determined using the same stopped-flow apparatus. We added at least a 6.7-fold excess of tubulin to a fixed concentration of R1* (30 nm), which made the binding reaction pseudo-first-order. The kobs value was fitted with the following exponential equation,
where Fluomin is the fluorescence at time 0. kon was extracted from the plot of apparent rate constant kobs as a function of the tubulin concentration.
Crystallization and Structure Determination
Crystallizations were performed at 293 K by vapor diffusion with the hanging drop method, using 1 ml of well solution and drops formed by 0.8 μl of proteins and 0.8 μl of the crystallization buffer. The tubulin-R4 complex, at a 30 mg/ml concentration in 15 mm Pipes-K, pH 6.8, 0.1 mm GDP, 0.3 mm MgCl2, and 0.2 mm EGTA, was crystallized at pH 6.8 using a mix of polyethylene glycol 20,000 (PEG 20,000) and NaCl as precipitants. Most crystals diffracted poorly and anisotropically, leaving ambiguities in the space group and cell parameters. Only one crystal diffracted to almost 4 Å resolution; diffraction data were measured at 100 K on ID29 (European Synchroton Radiation Facility, Grenoble, France). This crystal was obtained with a crystallization buffer consisting of 1.7 m NaCl and 7% (w/v) PEG 20,000. Before being flash-cooled in liquid nitrogen, it was transferred to the same solution buffered with 15 mm Pipes-K, pH 6.8, and supplemented with 0.1 mm GDP, 1 mm MgCl2, 0.5 mm EGTA, and 20% (v/v) glycerol. The data were processed in the C2 space group with XDS (29) (Table 1). The structure was determined by molecular replacement using Amore (30), with αβ tubulin as a search model (Protein Data Bank code 3RYC (7)). The correlation coefficient between calculated and observed data increased with the number of tubulin heterodimers found (41.9, 49.5, 53.9, and 58.5%). Rigid body refinement in which α and β subunits were refined separately resulted in a correlation coefficient of 61%. The R4 starting model was generated from RB3SLD and T2R. Owing to the moderate resolution of the data, only a few refinement steps with Buster (31) were performed.
TABLE 1.
Data collection and refinement statistics
| T4R4 | TR1-D2 | |
|---|---|---|
| Data collectiona | ||
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 639.7, 66.1, 128.1 | 95.1, 75.6, 155.7 |
| α, β, γ | 90.0°, 92.0°, 90.0° | 90.0°, 96.3°, 90.0° |
| Resolution (Å) | 50.0-4.17 (4.28-4.17) | 48.5-2.64 (2.78-2.64) |
| Rsym | 0.112 (0.876) | 0.089 (0.627) |
| I/σI | 8.2 (1.6) | 10.5 (2.1) |
| Completeness (%) | 98.6 (90.8) | 98.7 (97.5) |
| Multiplicity | 3.7 (3.4) | 3.7 (3.7) |
| Refinement | ||
| Resolution (Å) | 4.17 | 2.64 |
| No. reflections | 40,332 | 31,979 |
| Rwork/Rfree | 0.241/0.261 | 0.177/0.201 |
| No. atoms | ||
| Protein | 28,901 | 8446 |
| Ligands/ions | 244 | 180 |
| Waters | 0 | 102 |
| B-factors | ||
| Protein | 183 | 60.2 |
| Ligands/ions | 141 | 65.0 |
| Waters | 49.2 | |
| Coordinate error (Å)b | 1.417 | 0.317 |
| r.m.s.d.c from ideal values | ||
| Bond lengths (Å) | 0.010 | 0.010 |
| Bond angles | 1.20° | 1.20° |
a In both cases, data were collected on a single crystal. There is one complex per asymmetric unit. Values in parentheses are for highest resolution shell.
b Values shown were estimated from Luzzati plots.
c r.m.s.d. indicates root mean square deviation.
In the case of the tubulin-R1 complex (TR1), the C-terminal tail of tubulin was cleaved by subtilisin (7, 32) before tubulin-R1 (TR1) complex formation. TR1 was further complexed with the D2 DARPin. The ternary TR1-D2 complex at 15 to 20 mg/ml, in the same buffer as the one of tubulin-R4 was crystallized in a buffer consisting of 15% (v/v) PEG 550 monomethyl ether and 0.1 m Mes-K, pH 6.5. For data collection, the PEG concentration was raised to 30% before the crystals were flash-cooled in liquid nitrogen. A 2.65 Å data set was collected at 100 K at the Proxima1 beam line (Soleil Synchrotron, Saint Aubin, France) and processed with XDS (29). The intensities were scaled and merged using Scala (33). The structure was solved by molecular replacement with Amore (30) using the structure of a tubulin-D2 complex4 as a search model. The structure was refined using the Buster program (31) with iterative model building in Coot (34). Data processing and refinement statistics are summarized in Table 1. Accessible surface areas were calculated with the Areaimol program (33).
RESULTS
Five Modules in Its Sequence Account for the RB3SLD Interaction with the Two Tubulins To which It Binds
To design modular SLD-based constructs that bind any given number of tubulin heterodimers, we used an RB3SLD sequence as a starting point because this protein makes the most stable complex with tubulin among SLDs of other stathmin family members (14). For increased stability, in particular at higher ionic strength, RB3SLD was mutated at four positions compared with the protein used in our previous structural analyses (see supplemental data). The resulting mutant is termed RB3Q. The boundaries in the RB3Q sequence of the regions that interact with each tubulin heterodimer were established based on an internal repeat conserved in all SLD sequences (18, 35). Inspection of T2R structures has indeed shown that the two repeats correspond to two tubulin heterodimer binding regions (16, 36). In RB3SLD, the limits of these repeats, which are displayed in boldface type in Fig. 1B, are Glu48–Val82 and Glu99–Val133 (16) (numbering is in reference to stathmin (14)). The residue immediately downstream the second repeat is an arginine (Arg134), which is close to tubulin acidic residues (36); we therefore included it in the second RB3Q tubulin-binding region. For consistency, the corresponding residue downstream the first repeat (Ile83) was included in the first region (Fig. 1B). To gain more flexibility in the design, we divided each of these two regions into two tubulin subunit binding modules, using a high resolution (2.1 Å) T2R structure (7). This defines four modules (depicted in different colors in Fig. 1). The N-terminal module (module 1 or M1) comprises residues 4 to 61; it interacts with α1, the “first” α subunit in T2R. The following three modules, M2 to M4 (residues 62–83, 84–112, and 113–134, respectively), interact with β1, α2, and β2. When completing this attribution, we noticed that the region binding to β2 (113–141, i.e. 29 residues) is significantly longer than the one we had attributed based on the internal repeats and therefore defined a fifth module, M5 (residues 135–141), which caps the β2 binding region. M5 contributes significantly to T2R stability as the removal of the last seven residues of RB3SLD (residues 139–145) is detrimental to the stability of the complex, whereas a stop codon at position 142 is not (data not shown). We have used these five modules to design SLDs for binding tubulin with a programmed stoichiometry.
RB3SLD-based Constructs Lead to Programmed Binding of Three and Four Tubulins
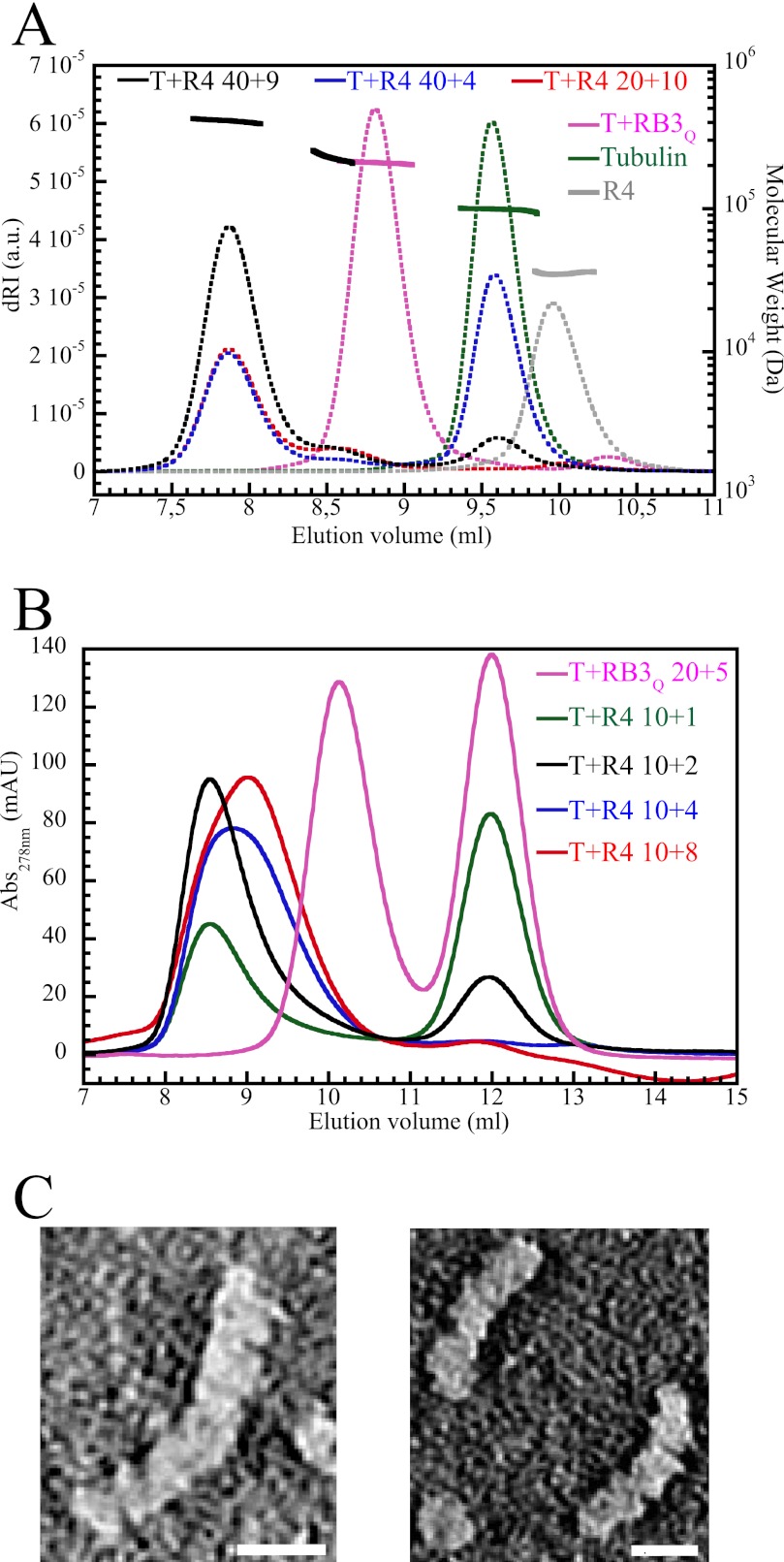
To produce a tubulin-SLD 4:1 complex, we designed a construct (R4) consisting of modules M1 to M4 (interacting with two heterodimers in T2R), followed by M3 (interacting with an α subunit) and ending with the M2–M5 sequence (interacting with a β subunit and a heterodimer) (Fig. 1C). R4 was produced, purified, and characterized both biochemically and structurally. We used SEC-MALLS to determine the mass of the complexes R4 forms with tubulin. In SEC-MALLS conditions, independent of the tubulin-R4 ratio in the sample, the major peak of the complex does not shift, indicating a constant tubulin-R4 stoichiometry (Fig. 2A). The mass determined for the species in this peak (410 kDa, Table 2) is in reasonable agreement with that of a 4:1 tubulin-R4 complex (T4R4; theoretical mass, 428 kDa). The mass of the controls (tubulin and T2R) is also fully consistent with the expected values. A second peak is observed (at about 8.6 ml), whose magnitude increases somewhat along with the concentration of R4 but remains small. Because of the small size of this peak and because it is not well separated from the T4R4 peak, the determination of the mass of the corresponding species is not accurate. Nevertheless, it fits best with a complex containing two tubulins.
FIGURE 2.
R4 forms mainly a T4R4 complex with tubulin. A, SEC-MALLS analysis. The differential refractive index (dRI, arbitrary unit (a.u.) on the left axis, dotted lines) and molecular mass (displayed as solid lines for the regions of interest, i.e. for the chromatographic peaks, with the scale on the right axis) are plotted as a function of the column elution volume. The samples analyzed were as follows: tubulin (40 μm, green), R4 (60 μm, gray), T2R (40 μm tubulin and 30 μm RB3Q, magenta), and tubulin-R4 (20:10 μm, red; 40:4 μm, blue; 40:9 μm, black). The molecular masses of tubulin-R4 complexes are only displayed in the case of the 40:9 μm sample. B, gel filtration profiles obtained with a low salt buffer. Samples (100 μl) containing 10 μm tubulin and increasing concentrations of R4 (1 μm, green curve; 2 μm, black; 4 μm, blue; and 8 μm, red) were injected on the column. As a control, a sample containing 20 μm tubulin and 5 μm RB3Q was also analyzed (magenta). mAU, milliabsorbance units. C, electron micrographs of negatively stained tubulin-R4 complexes. Species comprising four tubulin heterodimers (left) predominate, whereas complexes with three tubulins are also identified (right). Their dimensions (∼55 Å × 355 Å and 55 Å × 265 Å, respectively) are consistent with those of a smaller SLD complex comprising two tubulins (15). Scale bar, 100 Å.
TABLE 2.
Mass determination of tubulin-SLD complexes by SEC-MALLS
| Mass (kDa) | T:R4 |
T:RB3Q | Tubulin | R4 | ||
|---|---|---|---|---|---|---|
| 40:4 μm | 40:9 μm | 20:10 μm | ||||
| Theoreticala | 428 (T4R4), 228 (T2R4) | 409b, 237c | 217 (T2R) | 100 | 28.4 | |
| Determined by SEC-MALLS | 411b, 222c, 99d | 410b, 228c, 101d | 209 | 99 | 35 | |
| Rh (nm) | 7.7b | 7.9b | 7.7b | 5.5 | 3.8 | NDe |
a Assuming 100 kDa for tubulin.
b Main peak of the complex.
c Small peak of the complex.
d Uncomplexed tubulin.
e ND, not determined; Rh, hydrodynamic radius.
Tubulin was also analyzed by classical gel filtration in the presence of varying amounts of R4 (Fig. 2B). Consistent with the SEC-MALLS observations, a species of Stokes radius larger than that of T2R is formed, most probably indicative of the complex that contains four tubulins. As long as there are more than four tubulin heterodimers per R4 in the sample, the largest Stokes radius species elutes as a well defined and sharp peak. Increasing the concentration of R4 beyond this ratio has two consequences. First, the free tubulin peak disappears. Second, the shape of the fast migrating peak may change, depending on the buffer used for the analysis. In the higher salt buffer used for SEC-MALLS (data not shown) and in a similar one (supplemental Fig. S1), the results are identical to those obtained in SEC-MALLS. By contrast, in a low salt buffer, this peak broadens and its maximum shifts slightly toward lower Stokes radii (Fig. 2B).
We also visualized directly, by electron microscopy, the tubulin complexes formed in presence of R4. Micrographs confirm the presence of complexes with four tubulin molecules, whereas a smaller amount of oligomers with three tubulins can also be seen (Fig. 2C), together with a few presumably uncomplexed tubulin molecules (e.g. see bottom left of the right panel of Fig. 2C). Free heterodimers have been commonly observed in micrographs of tubulin-SLD mixtures (e.g. see Ref. 18); they might originate from dissociation of the complex during EM sample preparation. To summarize, complexes of 2:1 and 3:1 stoichiometries form between tubulin and R4. These are detected by SEC-MALLS (T2R4, Fig. 2A) or, in the case of T3R4, seen by EM (Fig. 2C) and suggested by the SEC experiments (Fig. 2B), implying that R4 molecules compete with each other for tubulin binding. But our results show that T4R4 is the main complex formed by tubulin and R4. The T4R4 crystal structure further establishes the organization of tubulin in this complex.
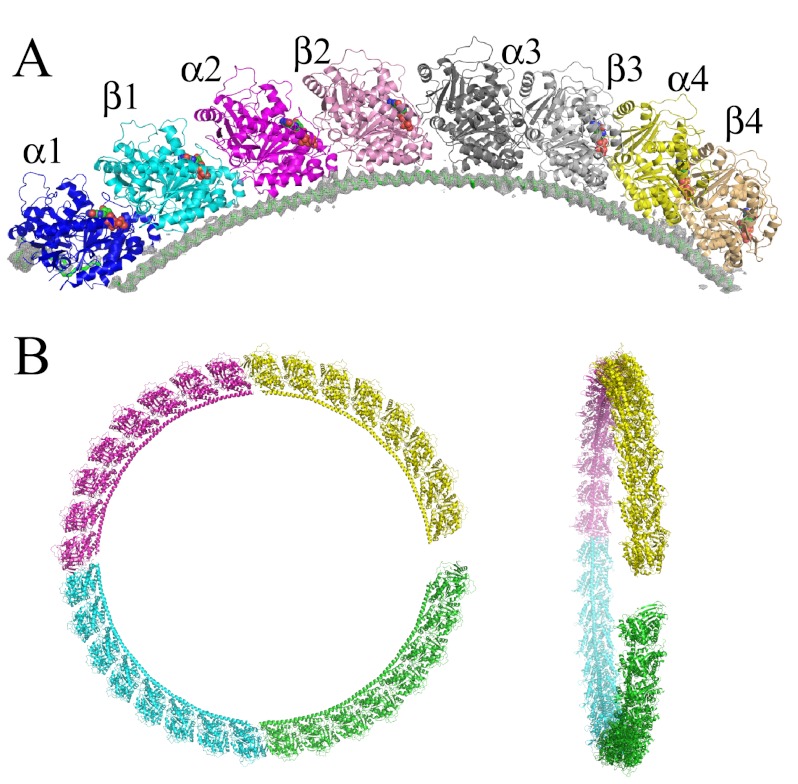
Tubulin in complex with R4 was crystallized, and the structure was determined by molecular replacement. There is one complex consisting of four tubulin heterodimers in the asymmetric unit. The elongated shape of the complex (Figs. 2C and 3A) is also reflected by its large hydrodynamic radius as determined by quasi-elastic light scattering in the SEC-MALLS experiment (Table 2). The R4 main chain was traced in the electron density maps, but consistent with the moderate resolution of the diffraction data set (4.2 Å, see Table 1), the signal for the side chains remained weak (Fig. 3A). As expected from T2R (16), the tubulin molecules are arranged head to tail as a curved protofilament-like assembly. Also similar to T2R (36), ∼11° rotations superimpose consecutive subunits, i.e. those within a tubulin molecule as well as the adjacent ones of consecutive heterodimers. The curvature of the complex is also pictured by a T4R4 helical superassembly (Fig. 3B). A comparison of the two T2R structures we have determined previously revealed that the intertubulin interface is variable (7) and gives rise to T2R helical superassemblies with pitches of opposite signs, depending on the crystal form considered. Superassembly of T4R4 in the crystal form described here yields a helix with a pitch that is close to zero, i.e. almost a ring (Fig. 3B), suggesting that a wide range of pitches within the limits initially found with T2R may indeed be observed.
FIGURE 3.
The T4R4 structure. A, overview of the complex in which each tubulin is colored differently. The α (β) subunits are in brighter (lighter) colors. The 4.2 Å resolution 2Fobs − Fcalc electron density map of the R4 molecule, contoured at the 1σ level, is displayed. B, the relative orientations of the tubulin subunits in T4R4 are close to those in a ring. The model resulting from the repetition of T4R4 was obtained by superimposing the β1 moiety of the (m+1)th complex onto the β3 moiety of the mth complex and by keeping in the final model the 1st, 3rd, 5th, and 7th complexes. Each T4R4 is colored differently. The resulting flat helix is viewed along its axis (left) and nearly perpendicularly to it (right).
In addition to R4, we designed another construct, termed R4a, which starts with modules M1 to M4, continues with twice a repeat of M3+M4, and ends with M5 (Fig. 1C). R4a differs from R4 in the stretch interacting with the β subunit of the third tubulin molecule in the T4R4 complex. R4a was produced and purified but because the initial characterization of its interaction with tubulin revealed no obvious difference compared with that of R4, it was not characterized any further. Using the same rational, we designed and produced the R3 construct (Fig. 1C), which also interacts with tubulin, likely giving rise to a T3R3 assembly (supplemental Fig. S1). The stoichiometries of the new complexes of SLDs with tubulin are as predefined. Unfortunately, the resolution of the structure we determined is not sufficient to demonstrate that in our designs, or at least in the complex crystallized, SLD residues interact with tubulin as expected, i.e. as in T2R. The design, based on the same principles, of an SLD (named R1, Fig. 1D) that binds one tubulin and the inspection of the structure of the resulting complex (TR1), has allowed us to validate the rational of the design of the three modules that constitute this SLD.
R1 Construct Makes a Tubulin-R1 1:1 High Affinity Complex
The R1 N-terminal part consists of an M1 module, the N-terminal end of RB3Q that interacts with a tubulin α subunit. Its C-terminal part comprises M4 and the capping motif M5, both regions constituting together the C-terminal region of RB3Q that interacts with a β subunit. In addition, we replaced the first residue of M4 by a lysine, to compensate for the negative charge of the residue that is four residue upstream (Glu58), i.e. on the same face of the SLD helix. A similar distribution of opposing charges is observed at the M1-M2 and M3-M4 boundaries of RB3SLD (Fig. 1).
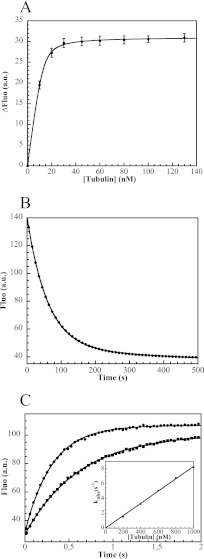
The interaction of R1 with tubulin was initially detected in a gel filtration assay. We observed a small shift upon addition of R1 to tubulin (slightly larger than 0.2 ml, see supplemental Fig. S1). Interestingly, a similar shift had been obtained upon addition to tubulin of a protein that makes a 1:1 complex with it and is slightly larger than R1 (113 versus 87 residues) (37). The tubulin-R1 interaction was quantified by fluorescence experiments using R1*, an acrylodan-labeled R1. The stepwise addition of tubulin to R1* leads to an increase of the acrylodan fluorescence signal (Fig. 4A). Fitting the data with a quadratic equation (Equation 1, see “Experimental Procedures”) yields the value of the equilibrium dissociation constant (Kd = 1 nm). This estimate reflects the high affinity of the TR1 complex, but it is probably not accurate, as the R1* concentration used in these experiments was ∼10-fold higher than the Kd. To confirm the tight interaction and characterize it further, we also determined the dissociation and association rate constants. A chase of R1* from tubulin by unlabeled R1 (Fig. 4B) yielded the dissociation rate constant koff, 0.016 s−1. The association rate constant was derived from association kinetics in pseudo-first-order conditions, yielding a kon of 8 × 106 m−1 s−1 (Fig. 4C). The Kd deduced (koff/kon = 2 nm) is consistent with the value determined by titrating tubulin with R1. To identify the interactions responsible for this tight complex, we determined its structure.
FIGURE 4.
The tubulin-R1 interaction monitored by fluorescence spectroscopy. A, fluorescence variation of 13 nm R1* upon addition of tubulin. The curve is the fit of the experimental points with Equation 1, from which the Kd (1 nm) is extracted. Error bars correspond to the S.D. of the variation of fluorescence signal upon tubulin addition. B, dissociation of R1* from tubulin. The fluorescent TR1* complex was formed by mixing 30 nm R1* with 50 nm tubulin. The fluorescence decrease following addition of 4.2 μm R1 to this sample was monitored in a stopped-flow apparatus. The curve is the fit of the experimental points (5% of which are shown) with Equation 2. The same rate constant was obtained with two R1 concentrations (see “Experimental Procedures”) and is interpreted to be the dissociation rate constant of the complex (koff = 0.016 ± 0.003 s−1). C, determination of the association rate constant. Tubulin, at concentrations ranging from 200 nm to 1 μm, was added to a fixed concentration of R1* (30 nm). Fluorescence variations upon addition of 200 nm (square symbols) and 400 nm (dots) tubulin are shown (5% of the experimental points are displayed). The data were fitted according to Equation 3. The variation of kobs as a function of tubulin concentration is linear. kon is the slope (8 × 106 m−1 s−1) of that curve (inset). a.u., arbitrary units.
Structural Basis for Tubulin-R1 High Affinity
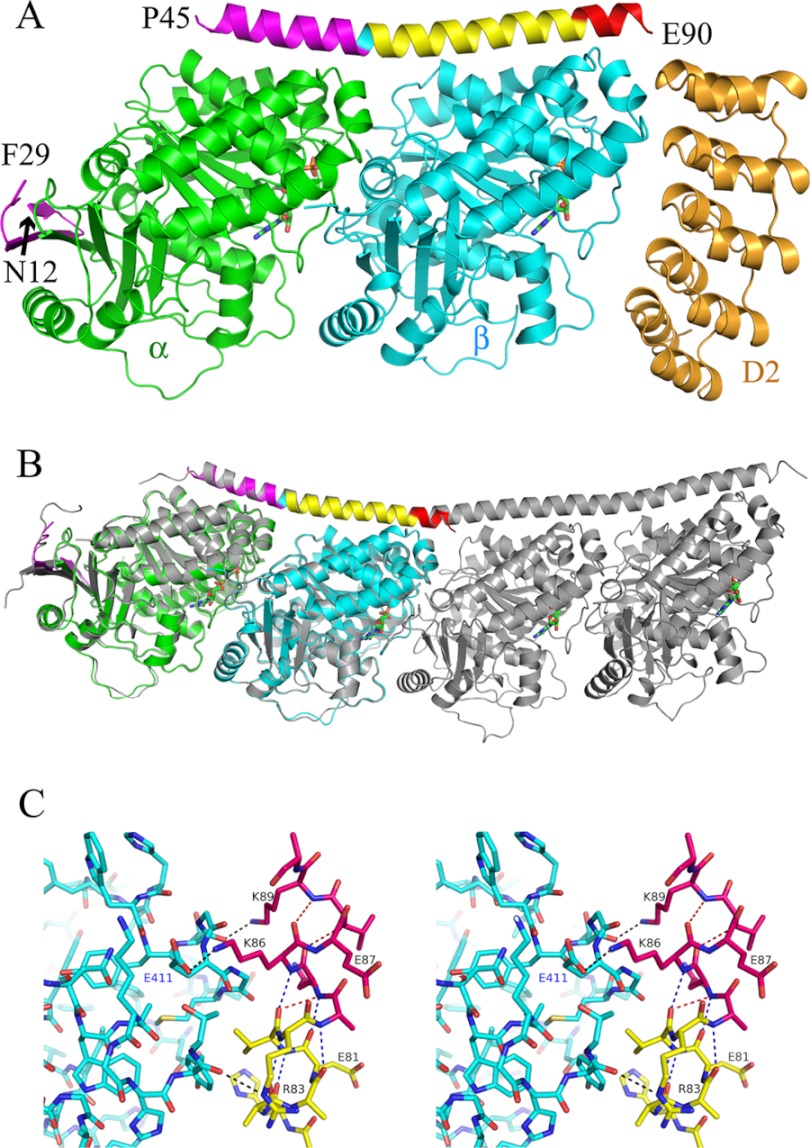
Crystals of TR1 were obtained as a ternary complex with the anti-tubulin DARPin D2 used as a crystallization chaperone. Data to 2.65 Å were collected from one crystal (Table 1), and the structure was solved by molecular replacement. D2 targets the β tubulin longitudinal interface. Hence, its binding site is distinct from that of R1, and the two proteins do not seem to interfere for tubulin binding (Fig. 5A). In the R1 structure, the C-terminal helix is ordered up to the end of the molecule, most of the side chains being defined in the electron density maps. Two R1 stretches could not be traced: the N-terminal half of the β hairpin first strand and the linker between this N-terminal β hairpin and the C-terminal helix. This loop is also mostly disordered in all the other tubulin-SLD structures determined so far (7, 36), probably reflecting its mobility in the complex. Nonetheless, the accessible surface area buried by the ordered part of R1 is large, giving a rational for the high affinity of the TR1 complex (38). About 2250 Å2 are buried on the R1 side and a similar surface (2130 Å2) on tubulin. Two-thirds of the tubulin buried surface are on the α subunit, and one-third is on β; this difference reflects the contribution of the R1 N-terminal β hairpin that caps the α longitudinal interface. Tubulin in the TR1 complex is curved and retains the conformation it has in T2R (and in T4R4, see above) (Fig. 5B). Moreover, the footprint of R1 on tubulin is very similar to that of RB3SLD on the two distal tubulin subunits in T2R. Most important, the two R1 moieties make with tubulin in TR1 the exact same interactions as those established by the corresponding modules of RB3SLD in T2R. This fully validates the design principles used to define M1, M4, and M5 and strongly suggests that M2 and M3, which were similarly defined, also interact with tubulin as planned.
FIGURE 5.
The TR1 structure. A, overview of the TR1-D2 structure. The tubulin α and β subunits are in green and cyan, respectively. R1 is colored by modules as in Fig. 1. D2, the tubulin-binding DARPin with which crystals were obtained, is in orange. The nucleotides (GTP on α, GDP on β) are in green. The disordered eight N-terminal residues of R1 as well as the disordered linker between its N-terminal β hairpin and the C-terminal α helix (residues 30 to 44) are not displayed. B, comparison of TR1 with T2R. TR1 is colored as in A and superimposed on T2R (gray). The root mean square deviation after superposition (45) of Cαs of α, β, and R1 from TR1 and of α1, β1, and RB3SLD from T2R is 0.691 Å (872 atoms compared). C, stereo view of the interaction of the C-terminal end of R1 with tubulin in TR1. R1 and the tubulin β subunit are colored as in A. The hydrogen bonds between R1 and tubulin, including the Lys86–Gluβ411 salt bridge, are displayed as black dotted lines. The main chain hydrogen bonds in the R1 helix are displayed as red (310-helix) or blue (α-helix) dotted lines. R1 residues Glu80, Lys84, and Glu90, whose side chains are not defined, have been modeled as alanines. See supplemental Fig. S2 for the same view with R1 in its electron density map.
R1 makes a stronger complex with tubulin than previous constructs of similar length (e.g. see Refs. 19 and 20). These consisted in a continuous N-terminal region of an SLD, i.e. they were based on constructs including the M1 and M2 modules. The tighter interaction established by R1 may be attributed to two main reasons. First, RB3SLD makes fewer contacts with β1 in T2R than R1 with β tubulin in TR1 (or than RB3SLD with the β2 subunit in T2R). In particular, two salt bridges are mediated by the M4–M5 stretch (between Asp76 of R1 and Lysβ156 of tubulin and, as shown in Fig. 5C, between Lys86 and Gluβ411) whereas only one (mediated by Arg76 of RB3SLD and Gluβ159) is contributed by the M2 module (tubulin residue numbering is as in Ref. 12). Secondly, the R1 C-terminal end is derived from an RB3SLD ending at position 141. Glu141 is the residue at the boundary between the helix and the downstream non-helical region (7). In TR1, the R1 helix is not capped in the usual sense (39), but it is stabilized at its C-terminal end by interactions with tubulin of the side chains of two residues: Lys86, as mentioned above, and Lys89 (respectively equivalent to Lys137 and Lys140 in RB3SLD). In addition, 310 helix main chain hydrogen bonds are made in the last turn, by the carbonyls of residues 85 and 86 with the amide NH of residues 88 and 89, respectively (Fig. 5C). Constructs with an untimely helix termination probably lacked these interactions and have resulted in proteins that make less stable complexes with tubulin.
DISCUSSION
We have presented the design and construction of artificial SLD proteins that efficiently bind tubulin in a range of predefined stoichiometries. This may serve at least two purposes. The first purpose is to provide alternative options for tubulin crystallization. The second purpose is the design of homogenous and stable tubulin assemblies larger than T2R for cryo-electron microscopy. Both objectives have been reached. First, new crystals of a single sequestered tubulin have been characterized. Second, as a proof of principle, we designed SLDs that form a T4R4 complex. Both EM (Fig. 2C) and SEC-MALLS analyses (Fig. 2A) indicate that the designed stoichiometry has been obtained. The stability of the T4R4 complex, higher than those obtained with SLDs from invertebrates (18), together with its crystal structure showing the longitudinal arrangements of the tubulins in a curved protofilament-like assembly, make it a well characterized entity for EM studies. In addition to defining the limits in RB3SLD sequence of tubulin subunit binding modules, our study identifies one important feature of strong RB3SLD-based tubulin binders: the presence at their C-terminal end of a capping module, termed here as M5 (Fig. 1). M5 ending at residue 141 confers strong binding to the SLDs we designed, whereas an SLD ending at residue 138 is a weaker binder. Residues up to Lys89 in R1 (Lys140 in RB3SLD) make important contributions to the stability of the SLD helix both through side chain interactions with tubulin and main chain intramolecular hydrogen bonds (see above). Therefore, Glu141 is probably close to the C-terminal end of the shortest M5 module for strong tubulin binding.
In addition to the new SLD constructs described above, our work provides new information on the tubulin-SLD interaction. Most studies of this interaction have been centered on ternary complexes of an SLD with two tubulin heterodimers. It is not straightforward to distinguish biochemically the binding of the two tubulins in these complexes. This has led to discrepancies of the estimates of their affinities and of their binding cooperativity (reviewed in Ref. 40). R1, as it associates with tubulin in a 1:1 complex, represents a simplified starting point for such studies. Both the kinetics and the strength of the association are of interest. We have found that the tubulin-R1 association is fast, its rate constant being close to that estimated from Brownian dynamics simulations (41). By comparison, the kon in T2R is ∼400 times slower (42). The suggestion has been made that the slow association kinetic in T2R may be related to structural rearrangements within the complex (42). As the association rate constants of SLDs other than RB3SLD are significantly faster (14), the slow association rate of T2R may be due to adjustments of the RB3SLD structure rather than to the tubulin-tubulin association. The question then arises of whether the RB3SLD region where these adjustments take place may be narrowed down. Because the association of R1 with tubulin is fast, it is tempting to suggest that they take place in the RB3SLD modules that are not included in R1, i.e. in the modules that interact with the β1 and α2 tubulin subunits.
We have also found that the tubulin:R1 affinity is large (Kd in the low nanomolar range). Because no natural tubulin-SLD 1:1 complex has been identified so far, it might have been thought that SLD-based sequesterers are inherently unable to bind one tubulin molecule efficiently. R1 proves that it is not the case as it binds one heterodimer tightly. Therefore, the failure to identify such a natural assembly is not due to an impossibility to produce it using SLD-based sequences. There must be another explanation. In the balance between mechanisms in which SLDs favor either microtubule assembly or disassembly depending on their phosphorylation state, our results give weight to the hypothesis that a main function of SLDs is to store tubulin at specific localizations in the cell (43) to deliver it for microtubule polymerization when required. In this scheme, tubulin becomes available for assembly upon SLD phosphorylation. Obviously, an SLD that releases more than one tubulin heterodimer at a time fulfills this function more efficiently than R1, at least from the point of view of the quantity of kinases required to release a number of tubulin molecules. The question then arises of whether higher stoichiometry complexes would be even more efficient. It seems that in such complexes the tubulin binding cooperativity is less pronounced. We observed this with R4 which, in certain conditions, gives rise to unsaturated assemblies (T2R4 and T3R4) (Fig. 2) and this was also observed with Drosophila SLDs (18). SLDs binding two heterodimers may therefore be an optimal choice for efficient sequestration and phosphorylation-inducible tubulin release.
Supplementary Material
Acknowledgments
Diffraction data were collected at the following synchrotron beam lines: ID14-1, ID23-1, and ID29 at the European Synchrotron Radiation Facility (Grenoble, France) and Proxima 1 at SOLEIL (Saint-Aubin, France). We are most grateful to the machine and beam line groups for making these experiments possible. We thank J. Massonneau and D. Mauchand (Unité Commune d'Expérimentation Animale, Institut National de la Recherche Agronomique) for providing us with the material from which tubulin was purified, Drs. B. Collinet and N. Lazar (Institut de Biochimie et Biophysique Moléculaire et Cellulaire, Orsay) for a preliminary SEC-MALLS characterization, and Drs. M. Argentini and D. Cornu (SiCaps, Imagif, Gif-sur-Yvette) for the mass spectrometry analysis.
This work was supported by the Centre National de la Recherche Scientifique, the Agence Nationale de la Recherche Grant ANR-09-BLAN-0071 and the Fondation pour la Recherche Médicale Grant DEQ20081213979.

This article contains supplemental “Results,” Figs. S1 and S2, and additional references.
The atomic coordinates and structure factors (codes 4F61 and 4F6R) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
B. Gigant, L. Pecqueur, B. Dreier, A. Plückthun, and M. Knossow, manuscript in preparation.
- TEM
- transmission electron microscopy
- DARPin
- designed ankyrin repeat protein
- SEC-MALLS
- size exclusion chromatography coupled to multi-angle laser light scattering
- SLD
- stathmin-like domain.
REFERENCES
- 1. Desai A., Mitchison T. J. (1997) Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117 [DOI] [PubMed] [Google Scholar]
- 2. Amos L. A., Schlieper D. (2005) Microtubules and maps. Adv. Protein Chem. 71, 257–298 [DOI] [PubMed] [Google Scholar]
- 3. Brouhard G. J., Stear J. H., Noetzel T. L., Al-Bassam J., Kinoshita K., Harrison S. C., Howard J., Hyman A. A. (2008) XMAP215 is a processive microtubule polymerase. Cell 132, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Howard J., Hyman A. A. (2007) Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 19, 31–35 [DOI] [PubMed] [Google Scholar]
- 5. Jourdain L., Curmi P., Sobel A., Pantaloni D., Carlier M. F. (1997) Stathmin: A tubulin-sequestering protein, which forms a ternary T2S complex with two tubulin molecules. Biochemistry 36, 10817–10821 [DOI] [PubMed] [Google Scholar]
- 6. Dorléans A., Knossow M., Gigant B. (2007) Studying drug-tubulin interactions by x-ray crystallography. Methods Mol. Med. 137, 235–243 [DOI] [PubMed] [Google Scholar]
- 7. Nawrotek A., Knossow M., Gigant B. (2011) The determinants that govern microtubule assembly from the atomic structure of GTP-tubulin. J. Mol. Biol. 412, 35–42 [DOI] [PubMed] [Google Scholar]
- 8. Sui H., Downing K. H. (2010) Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure 18, 1022–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sindelar C. V., Downing K. H. (2010) An atomic level mechanism for activation of the kinesin molecular motors. Proc. Natl. Acad. Sci. U.S.A. 107, 4111–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fourniol F. J., Sindelar C. V., Amigues B., Clare D. K., Thomas G., Perderiset M., Francis F., Houdusse A., Moores C. A. (2010) Template-free 13-protofilament microtubule-MAP assembly visualized at 8 Å resolution. J. Cell Biol. 191, 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maurer S. P., Fourniol F. J., Bohner G., Moores C. A., Surrey T. (2012) EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell 149, 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Löwe J., Li H., Downing K. H., Nogales E. (2001) Refined structure of αβ-tubulin at 3.5 Å resolution. J. Mol. Biol. 313, 1045–1057 [DOI] [PubMed] [Google Scholar]
- 13. Henderson R. (1995) The potential and limitations of neutrons, electrons, and x-rays for atomic resolution microscopy of unstained biological molecules. Q Rev. Biophys. 28, 171–193 [DOI] [PubMed] [Google Scholar]
- 14. Charbaut E., Curmi P. A., Ozon S., Lachkar S., Redeker V., Sobel A. (2001) Stathmin family proteins display specific molecular and tubulin binding properties. J. Biol. Chem. 276, 16146–16154 [DOI] [PubMed] [Google Scholar]
- 15. Steinmetz M. O., Kammerer R. A., Jahnke W., Goldie K. N., Lustig A., van Oostrum J. (2000) Op18/stathmin caps a kinked protofilament-like tubulin tetramer. EMBO J. 19, 572–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gigant B., Curmi P. A., Martin-Barbey C., Charbaut E., Lachkar S., Lebeau L., Siavoshian S., Sobel A., Knossow M. (2000) The 4 Å x-ray structure of a tubulin:stathmin-like domain complex. Cell 102, 809–816 [DOI] [PubMed] [Google Scholar]
- 17. Frank J. (2006) Three-dimensional Electron Microscopy of Macromolecular Assemblies, 2nd Ed., pp. 139–140, Oxford University Press, New York [Google Scholar]
- 18. Lachkar S., Lebois M., Steinmetz M. O., Guichet A., Lal N., Curmi P. A., Sobel A., Ozon S. (2010) Drosophila stathmins bind tubulin heterodimers with high and variable stoichiometries. J. Biol. Chem. 285, 11667–11680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jourdain I., Lachkar S., Charbaut E., Gigant B., Knossow M., Sobel A., Curmi P. A. (2004) A synergistic relationship between three regions of stathmin family proteins is required for the formation of a stable complex with tubulin. Biochem. J. 378, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Segerman B., Larsson N., Holmfeldt P., Gullberg M. (2000) Mutational analysis of Op18/stathmin-tubulin-interacting surfaces. Binding cooperativity controls tubulin GTP hydrolysis in the ternary complex. J. Biol. Chem. 275, 35759–35766 [DOI] [PubMed] [Google Scholar]
- 21. Fauquant C., Redeker V., Landrieu I., Wieruszeski J. M., Verdegem D., Laprévote O., Lippens G., Gigant B., Knossow M. (2011) Systematic identification of tubulin-interacting fragments of the microtubule-associated protein Tau leads to a highly efficient promoter of microtubule assembly. J. Biol. Chem. 286, 33358–33368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stemmer W. P., Crameri A., Ha K. D., Brennan T. M., Heyneker H. L. (1995) Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164, 49–53 [DOI] [PubMed] [Google Scholar]
- 23. Wurch T., Lestienne F., Pauwels P. J. (1998) A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnology Techniques 12, 653–657 [Google Scholar]
- 24. Barbier P., Dorléans A., Devred F., Sanz L., Allegro D., Alfonso C., Knossow M., Peyrot V., Andreu J. M. (2010) Stathmin and interfacial microtubule inhibitors recognize a naturally curved conformation of tubulin dimers. J. Biol. Chem. 285, 31672–31681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Charbaut E., Redeker V., Rossier J., Sobel A. (2002) N-terminal acetylation of ectopic recombinant proteins in Escherichia coli. FEBS Lett. 529, 341–345 [DOI] [PubMed] [Google Scholar]
- 26. Castoldi M., Popov A. V. (2003) Purification of brain tubulin through two cycles of polymerization depolymerization in a high-molarity buffer. Protein Expr. Purif. 32, 83–88 [DOI] [PubMed] [Google Scholar]
- 27. Pecqueur L., Duellberg C., Dreier B., Jiang Q., Wang C., Plückthun A., Surrey T., Gigant B., Knossow M. (2012) A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end. Proc. Natl. Acad. Sci. U.S.A. 109, 12011–12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mastronarde D. N. (1997) Dual-axis tomography: An approach with alignment methods that preserve resolution. J. Struct. Biol. 120, 343–352 [DOI] [PubMed] [Google Scholar]
- 29. Kabsch W. (2010) XDS, Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Navaza J. (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 57, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 31. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O. S., Vonrhein C., Womack T. O. (2011) BUSTER, version 2.8.0, Global Phasing, Ltd., Cambridge, UK [Google Scholar]
- 32. Knipling L., Hwang J., Wolff J. (1999) Preparation and properties of pure tubulin S. Cell Motil. Cytoskeleton 43, 63–71 [DOI] [PubMed] [Google Scholar]
- 33. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maucuer A., Doye V., Sobel A. (1990) A single amino acid difference distinguishes the human and the rat sequences of stathmin, a ubiquitous intracellular phosphoprotein associated with cell regulations. FEBS Lett. 264, 275–278 [DOI] [PubMed] [Google Scholar]
- 36. Ravelli R. B., Gigant B., Curmi P. A., Jourdain I., Lachkar S., Sobel A., Knossow M. (2004) Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 428, 198–202 [DOI] [PubMed] [Google Scholar]
- 37. Cormier A., Clément M. J., Knossow M., Lachkar S., Savarin P., Toma F., Sobel A., Gigant B., Curmi P. A. (2009) The PN2–3 domain of centrosomal P4.1-associated protein implements a novel mechanism for tubulin sequestration. J. Biol. Chem. 284, 6909–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kastritis P. L., Moal I. H., Hwang H., Weng Z., Bates P. A., Bonvin A. M., Janin J. (2011) A structure-based benchmark for protein-protein binding affinity. Protein Sci. 20, 482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aurora R., Rose G. D. (1998) Helix capping. Protein Sci. 7, 21–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Steinmetz M. O. (2007) Structure and thermodynamics of the tubulin-stathmin interaction. J. Struct. Biol. 158, 137–147 [DOI] [PubMed] [Google Scholar]
- 41. Northrup S. H., Erickson H. P. (1992) Kinetics of protein-protein association explained by Brownian dynamics computer simulation. Proc. Natl. Acad. Sci. U.S.A. 89, 3338–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krouglova T., Amayed P., Engelborghs Y., Carlier M. F. (2003) Fluorescence correlation spectroscopy analysis of the dynamics of tubulin interaction with RB3, a stathmin family protein. FEBS Lett. 546, 365–368 [DOI] [PubMed] [Google Scholar]
- 43. Levy A. D., Devignot V., Fukata Y., Fukata M., Sobel A., Chauvin S. (2011) Subcellular Golgi localization of stathmin family proteins is promoted by a specific set of DHHC palmitoyl transferases. Mol. Biol. Cell 22, 1930–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 45. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol. Crystallogr 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.