Abstract
Over the next decade, wheat grain production must increase to meet the demand of a fast growing human population. One strategy to meet this challenge is to raise wheat productivity by optimizing plant stature. The Reduced height 8 (Rht8) semi-dwarfing gene is one of the few, together with the Green Revolution genes, to reduce stature of wheat (Triticum aestivum L.), and improve lodging resistance, without compromising grain yield. Rht8 is widely used in dry environments such as Mediterranean countries where it increases plant adaptability. With recent climate change, its use could become increasingly important even in more northern latitudes. In the present study, the characterization of Rht8 was furthered. Morphological analyses show that the semi-dwarf phenotype of Rht8 lines is due to shorter internodal segments along the wheat culm, achieved through reduced cell elongation. Physiological experiments show that the reduced cell elongation is not due to defective gibberellin biosynthesis or signalling, but possibly to a reduced sensitivity to brassinosteroids. Using a fine-resolution mapping approach and screening 3104 F2 individuals of a newly developed mapping population, the Rht8 genetic interval was reduced from 20.5 cM to 1.29 cM. Comparative genomics with model genomes confined the Rht8 syntenic intervals to 3.3 Mb of the short arm of rice chromosome 4, and to 2 Mb of Brachypodium distachyon chromosome 5. The very high resolution potential of the plant material generated is crucial for the eventual cloning of Rht8.
Key words: Adaptation, brassinosteroid, height, Rht8, wheat
Introduction
Wheat genetics has the opportunity of understanding and improving a crop that has profound social and economic importance across countries and cultures. It is estimated that over the next decade, grain production must increase by 15% to meet the global demand and consumption of wheat as a result of a growing human population (Edgerton, 2009). One strategy to meet this challenge is to increase wheat productivity by optimizing plant architecture (defined by tillering, stature, and leaf and ear morphology). Plant architecture is of major agronomic importance as it determines the adaptability of a plant to cultivation, harvest index, and potential grain yield (Reinhardt and Kuhlemeier, 2002).
A decisive component of plant architecture is stature, mainly determined by stem elongation. Wheat (Triticum aestivum L.) is an annual crop with round, hollow, and jointed culms (stems). There are usually five elongated internodes in fully grown culms, with each internode progressively longer towards the ear. The last internode, the peduncle, is the longest. Numerous data from rice, barley, and Arabidopsis mutants indicate that internode elongation, which determines final plant height, is regulated by genes involved in brassinosteroid (BR) and gibberellin (GA) biosynthetic or signalling pathways (reviewed in Wang and Li, 2008).
Mutants defective in BR biosynthesis or signalling display characteristic growth-deficient phenotypes such as dwarfism. In rice, the Osdwarf4-1 mutant exhibits erect leaves and slight dwarfism without compromising grain yield (Sakamoto et al., 2006). This phenotype is due to loss of function of a cytochrome P450 (CYP90B2) involved in BR biosynthesis. In barley, the uzu semi-dwarfing allele of a gene encoding the putative BR receptor HvBRI1 is being introduced into all hull-less barley cultivars in Japan (Chono et al., 2003). These results provide a strategy for genetic improvement of crop production by modulation of BR biosynthesis and signal transduction. With a better understanding of the hormonal regulation of culm elongation, a similar strategy would also be possible in wheat, where to date there are no cloned genes involved in the BR pathway.
On the other hand, the deployment of genes influencing plant height through the GA pathway was a major factor in the success of the Green Revolution, which created high-yielding cultivars with shorter and sturdier culms (Khush, 2001). In contrast to the recessive semi-dwarf sd-1 Green Revolution allele in rice, that is a loss-of-function mutation in one of the major GA biosynthetic genes (GA 20-oxidase2 or GA20ox2) (Monna et al., 2002; Sasaki et al., 2002; Spielmeyer et al., 2002), the wheat reduced height Rht-B1b and Rht-D1b Green Revolution alleles are dominant gain-of-function mutations causing impaired GA signalling (Peng et al., 1999). Typically, mutants with decreased bioactive GA concentrations or impaired response are dwarf or semi-dwarf in stature, while elevated bioactive GA concentrations or increased signalling result in taller plants (Busov et al., 2008). The wheat Green Revolution genes are orthologues of the Arabidopsis gibberellic acid-insensitive (gai), the maize dwarf-8 (d8) (Peng et al., 1999), the rice gai (Ogawa et al., 2000) or slender1 (slr1) (Ikeda et al., 2001), and the barley slender1 (sln1) (Chandler et al., 2002) genes. The wild-type genes encode a DELLA protein that acts as a negative regulator of GA signalling (Harberd et al., 1998; Dill et al., 2001). Although Rht-B1 and Rht-D1 have influenced major advances in varietal performance (Gale and Youssefian, 1985), they are not universally beneficial, and in certain hot and dry climates they can reduce rather than promote crop performance (Worland and Law, 1986; Rebetzke et al., 2007). In addition to shortening plant stature, Rht-D1b and Rht-B1b also reduce coleoptile length and early vigour, which can reduce yield through poor seedling establishment when sown in unfavourable conditions (Rebetzke et al., 1999).
The Reduced height 8 (Rht8) semi-dwarfing gene is one of the few, together with the Green Revolution genes, to shorten wheat culms and improve lodging resistance without penalizing grain yield (Worland and Law, 1986). Rht8 is well adapted to southern and eastern European environments (Worland and Law, 1986) as it has no effect on coleoptile length or seedling vigour (Rebetzke et al., 1999; Rebetzke and Richards, 2000). It was introduced from the Japanese variety Akakomugi into European wheats in the 1930s by the Italian breeder Strampelli together with the photoperiod-insensitive, early flowering Ppd-D1a allele (Lorenzetti, 2000). Worland et al. (1990) described Rht8 as a weak allele of a gene for height promotion on the short arm of chromosome 2D. Successively, Korzun et al. (1998) detected a closely linked microsatellite marker, Xgwm261 that was mapped 0.6 cM distal to Rht8. The 192 bp allele of Xgwm261 corresponds to a height-reducing phenotype of Rht8 (Korzun et al., 1998). Varieties carrying the gwm261 192bp allele showed a height reduction of 7–8 cm (typically 8–10%) in England and the former Yugoslavia, without pleiotropic effects on other agronomic characters except for a slight increase in spikelet fertility (Worland et al., 1998). The gwm261 192bp allele was long proposed to be diagnostic for Rht8 (Chebotar et al., 2001; Ahmad and Sorrells, 2002; Bai et al., 2004; Schmidt et al., 2004; Ganeva et al., 2005; Liu et al., 2005; Zhang et al., 2006). However, the marker allele is indicative of Rht8 only for pedigrees derived from the original source variety Akakomugi (Ellis et al., 2007). In fact Norin10, a source variety of the Green Revolution semi-dwarfing genes, carries a gwm261 192bp allele that is independent of the height reduction effect of Rht8 (Ellis et al., 2007). Due to the wide spread of the Green Revolution genes and the gwm261 192bp allele independent of the height reduction effect of Rht8, studies assessing the distribution of height-reducing alleles at the Rht8 locus made by screening gwm261 alleles can mispredict the presence of the height-reducing allele of Rht8.
In the present study, the characterization of Rht8 was advanced by fine-resolution mapping and the identification of syntenic intervals in model genomes of Oryza sativa and Brachypodium distachyon. Furthermore, the relationships between Rht8 lines and the phytohormones involved in stature regulation, GA and BR, were investigated. It is clear that the genetic potential of optimizing wheat stature has not been fully exploited and the majority of genes and alleles affecting height remain uncharacterized. The work presented here addresses fundamental questions of the regulation of stature control in hexaploid wheat, and provides new tools for the more efficient deployment of Rht8 in international breeding programmes.
Materials and methods
Culm and total height measurements
Cappelle-Desprez, Cappelle-Desprez (Mara 2D) substitution line (SL), Mara, and the Cappelle-Desprez (Mara 2D) single chromosome recombinant inbred line (RIL) population of 89 lines (Triticum aestivum L.) were used to assess the effect of Rht8 on the wheat culm, and check the original height scores for Rht8 of Korzun et al. (1998). Seedlings were grown in a mixture of peat and sand and vernalized for 8 weeks under short days (10 h light/14 h dark). Vernalization growth chambers were maintained at 5 ºC with 70% relative humidity and were illuminated by tungsten lamps to provide a light intensity of 250±50 μmol m–2 s–1 at the top of the canopy. Subsequently, plants were transferred to an outdoor soil house, and hand dibbed. Two plants of each genotype were arranged in a randomized block design with five replicated plots (10 data points for each genotype). No plant growth regulators or triazoles were applied to the plots throughout development. Main tillers were tagged for each plant at the beginning of culm elongation. Final plant height, measured from the soil to the tip of the ear in fully grown and yellowing main tillers, was recorded for each plant. Individual internode lengths and ear lengths of the main tiller were also recorded. Two-way analysis of variance (ANOVA) followed by Tukey’s Honestly Significant Difference (HSD) were performed to determine height differences among height classes. Statistical analyses were performed in R 2.10.1 (R Development Core Team, 2005). The same analysis was done for ear, peduncle, and internode lengths.
Microscopic analyses
For transverse sections, main tillers of five wild-type RIL33 (Cappelle-Desprez tall rht8 allele) and five RIL4 (semi-dwarf Rht8 allele) lines were compared at ear emergence (growth stage 50–59; Zadoks et al., 1974). Sections were taken from the medial elongating zone of each culm segment (middle of peduncle, and middle of internode 1), and from the respective nodes. Cut tissues were immediately fixed (2.5% glutaraldehyde, 0.1 M phosphate buffer pH 7.0, 0.1% Triton X-100) and vacuum infiltrated overnight. Tissues were subsequently dehydrated in a graded ethanol series and embedded in Technovit 7100 resin (Kulzer-Technik, Germany) according to the manufacturer’s instructions. Embedded tissues were sectioned with a Leica Autocut microtome (RM 2055) to 5–10 μm thick slices, stained with toluidine blue, and visualized on a Nikon Microphot-SA microscope fitted with a Pixera Pro600ES DiRactor™ camera. All pictures were taken at the same magnification, and cell number per unit area was calculated using an arbitrary but constant area of 550×300 μm with ImageJ (Abramoff et al., 2004).
Scanning electron microscopy (SEM) was used to compare longitudinal sections of five Cappelle-Desprez (tall rht8 allele) and five RIL4 (semi-dwarf Rht8 allele) main tillers from fully elongated internode 1 and peduncle at straw stage (growth stage 90; Zadoks et al., 1974). Sections were taken from three different zones of each internode (basal, <1 cm after the basal node; medial, in the middle of the considered internode elongating zone; and distal, at >1 cm before the following node) under a dissecting microscope, and sputter coated with gold on an Agar, high resolution sputter-coater. Straw samples were observed directly using a FEI XL30 FEG scanning electron microscope, without the need of fixation and critical point drying. A minimum of 100 cells were measured for each sample using the instrument’s software.
Both transversal and longitudinal cell measurements were evaluated with Student’s t-test.
GA assays
The endogenous GA content of elongating main culms of Cappelle-Desprez (rht8) was compared with that of RIL4 (Rht8). Leaf sheathes covering the main culm were removed and developing ears discarded before collecting culm tissue from 6-week-old plantlets (35–45cm tall plants, flag leaf just visible, growth stage 37; Zadoks et al., 1974). Cut samples were immediately frozen in liquid nitrogen and freeze-dried. Each variety was replicated five times, and 4–5 main tillers were pooled per sample. GA extraction and analysis was performed as in Griffiths et al. (2006), and values were compared with the Student’s t-test.
To assess the culm response to exogenous GA3, Cappelle-Desprez (rht8), RIL4 (Rht8), and the Mercia Rht3 near isogenic line (NIL) were used. Prior to treatments, wheat seedlings were grown in a mixture of peat and sand and vernalized as before. Plantlets were then transferred to 1 litre pots containing cereal mix, randomized, and grown under long days (16 h light/8 h dark) at 18 ºC with 70% relative humidity with a light intensity of 350±50 μmol m–2 s–1 at the top of the canopy. GA3 (63492 Sigma) was dissolved in absolute ethanol at 1 mM, and treatments were prepared in fresh aqueous solutions. Test solutions (5 ml per seedling of 100 μM of GA3) were applied with spray bottles to the main culm surface every 3 d until anthesis. Control plants were sprayed with 100 μM ethanol. The experiment was repeated twice, with five plants for each treatment. Responses (final plant height) were compared with ANOVA followed by Tukey’s HSD test in R 2.10.1 (R Development Core Team, 2005).
BL assays
The root response of Cappelle-Desprez (rht8), and RIL6 (Rht8) plants to epi-brassinolide (epi-BL, E1641 Sigma) was measured and compared with an adapted method from Chono et al. (2003) and Hong et al. (2003). Seeds were surface sterilized with 70% ethanol for 30 s and 10% NaClO for 10 min, and germinated for 2 d in Petri dishes on wet filter paper at 24 ºC in the dark. Germinated seeds were transplanted to Magenta boxes containing 1% agar, half-strength Murashige and Skoog (MS) medium, and the appropriate epi-BL concentration (0, 0.01, 0.1, and 1 μM epi-BL). Samples were incubated at 23 ºC under long days (14 h light/8 h dark, 350±50 μmol m–2 s–1) for 4 d. The cotyledon and shoot length response of Cappelle-Desprez (rht8), and RIL6 (Rht8) plants to 1 μM epi-BL was measured and compared at 10 d post-germination. To determine root lengths, primary roots were stretched by forceps, photographed, and measured with ImageJ software (Abramoff et al., 2004). Root dry mass was measured with an analytical scale after all roots were removed from the plantlets and dried for 3 d at 60 °C. The experiment was repeated three times, with a minimum 10 plants for each treatment. Data were analysed with ANOVA followed by Tukey’s HSD test in R 2.10.1 (R Development Core Team, 2005).
The wheat leaf unrolling assay was performed according to Wada et al. (1985) on Cappelle-Desprez (rht8), RIL4 and RIL6 (Rht8), and selected homozygous F4 recombinants. After plants were grown in the dark for 6 d at 25 ºC, the first 1.5 cm long segment from the leaf tip was discarded, and the following 3×1.5 cm long leaf segments were excised and incubated in 2 ml of distilled water containing an appropriate epi-BL concentration. All excisions were conducted under a dim green safelight. Five epi-BL concentrations (0.0002, 0.002, 0.02, 0.2, and 2 μM) were tested against the control. A minimum of six leaf segments from six individual plants were incubated in each concentration, and each concentration was replicated three times. Leaf segments were allowed to unroll for 72 h in the dark at 25 ºC and photographed at 0, 24, 48, and 72 h after excision. Unrolling of leaf segments was determined by measuring their width using the software ImageJ (Abramoff et al., 2004). The unrolling percentage was calculated as described in Wada et al. (1985) with additional normalization for the initial leaf width and for the background (unrolling of the untreated control over time). Specifically, at a given time point (i) and treatment (j), ‘unrolling %’= [(Wij/W0j)–(Wi0–W00)], where Wij is the mean leaf width of the sample considered, W0j is the mean leaf width of the same sample at time ‘0’, Wi0 is the mean leaf width of the control at time ‘i’, and W00 is the mean leaf width of the control at time ‘0’. Statistical analysis (ANOVA and Tukey’s HSD test) was performed with R 2.10.1. (R Development Core Team, 2005). The experiment was repeated twice. Homozygous F4 recombinants were tested only at 0, 0.02, and 2 μM epi-BL.
Marker development
Gene-based markers were designed from wheat expressed sequence tags (ESTs) mapping to the chromosome 2DS1-0.33 deletion bin (Endo and Gill, 1996; http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi), from selected rice unspliced gene sequences (Rice MSU Rice Genome Annotation Project Database v6.1, ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_6.1/all.dir/; RAP database v5, http://rapdb.dna.affrc.go.jp/download/index.html), and from selected B. distachyon unspliced gene sequences (release 1.0, www.modelcrop.org; ftp://ftp.ensemblgenomes.org/pub/plants/release-3/fasta/brachypodium_distachyon/cdna/). Wheat assembled ESTs were downloaded from the TIGR Plant Transcript Assemblies database (ftp://ftp.tigr.org/pub/data/plantta/Triticum_aestivum/). For all data sets, a local BLAST database was produced using the command formatdb. A local similarity search (blastall –p blastn) (Altschul et al., 1990) was performed on wheat ESTs against the unspliced gene databases to identify the corresponding unspliced gene. Wheat EST sequences with a first-hit similarity score of >150 were aligned to the genomic sequence of the identified gene using the est2genome algorithm (EMBOSS). To reduce false positives, results were filtered for repetitive elements (Triticeae Repeat database: TREP http://wheat.pw.usda.gov/ITMI/Repeats), and for wheat ESTs that hit genes outside the desired genomic interval when re-BLASTed. Primers amplifying products of ~500–600 bp were designed on EST sequence-spanning predicted introns, and when possible on 3'-untranslated regions (UTRs), with Primer3 (http://frodo.wi.mit.edu/, Rozen and Skaletsky, 2000). Wheat ESTs from which markers were designed were also BLASTed against the SWISSPROT database for predicting the putative function (ftp://ftp.uniprot.org/pub/ databases/uniprot/knowledgebase/uniprot_sprot.fasta.gz).
Polymorphic markers (Table 1) between parents of the coarse mapping population (89 RILs; Korzun et al., 1998) were amplified from all RILs. Genomic DNA was extracted from 2-week-old seedlings with an adapted method of Pallotta et al. (2003). Amplification was conducted in a 20 μl volume containing 50 ng of genomic DNA, 1 μM of each primer, 1.25 mM dNTPs, 1× PCR buffer (Invitrogen), and 0.4 U of Taq DNA polymerase (Invitrogen). After an initial denaturation at 94 °C for 1 min, 35 amplification cycles were performed: 94 °C for 1 min, 58 °C for 1 min, and 72 °C for 1 min. Single-stranded amplification products of newly developed gene-based markers were separated according to their conformation by single strand conformation polymorphism (SSCP) analysis as described by Bertin et al. (2005). Linkage of molecular markers on chromosome 2DS was analysed with JoinMap version 3.0, with a logarithm (base 10) of odds (LOD) score >5.0, and the Haldane mapping function.
Table 1.
Polymorphic markers between parents of the Cappelle-Desprez (Mara 2D) RIL population developed in this study
| Marker | Primer_F | Primer_R | Ta (°C) | Wheat EST | Rice (MSU) Hit | Brachy Hit |
| DG025 | ACACGCACACATGAGCAAAT | ACGGGTTCAGGAAGATGTTG | 56 | CD490659 | LOC_Os04g02570 | Bradi0026s00210 |
| DG032 | AGGAGGCAGATGCAGAAGC | CCTGATCAAGACACCGTAAGC | 56 | CD897865 | LOC_Os04g02830 | Bradi5g01720 |
| DG035 | CATATGGCAGGAGCAGGAGT | TCCATCAGTCATAACCTCTTCTG | 56 | DN949138 | LOC_Os04g02870 | Bradi5g01430 |
| DG048 | GGAATGGCTTTTTCCCTGTT | TGGCGATAAGCCTTGAAAAT | 56 | CD906478 | LOC_Os04g04000 | Bradi3g06030 |
| DG057 | TGGACTCAACCATTGGAGAA | CGATCACTTGCTGTTGTTCA | 56 | TA27071_4565 | LOC_Os04g05010 | Bradi5g01180 |
| DG062 | GCAGGCATGGTTACTTCCAT | CCCTCTGACCTCCAGTTCC | 56 | TA24720_4565 | LOC_Os04g05050 | Bradi5g01130 |
| DG072 | CGTTCAATGTCTGGATCGAC | GGGTCACTGAGTTTCGCAAT | 58 | TA43575_4565 | LOC_Os04g11820 | Bradi5g02870 |
| DG086 | TCAATGGCCATATTAAGGCTCTA | AGCAATCTTTGTGTCCATATCAA | 58 | TA43575_4565 | LOC_Os04g11820 | Bradi5g02910 |
| DG087 | GATCTGCACTGCTCCATCAA | TCCACTGCGACATAAAACCA | 58 | CD900476 | LOC_Os04g12580 | Bradi5g02980 |
| DG118 | GCCTTCCGGAACAGGTACT | GCAGCTAGGACCCTCAAATG | 58 | BJ246143 | LOC_Os04g14760 | Bradi5g03700 |
| DG236 | CATCCAGACGCATGGATACT | CCATGCTTTCCAGTTCTTCC | 58 | BE489611 | LOC_Os12g16650 | Bradi5g02490 |
| DG241 | TCCCTGCAGGCGTAAGTAAC | GGGTCACTGAGTTTCGCAAT | 58 | TA43575_4565 | LOC_Os04g11820 | Bradi5g02870 |
| DG244 | GTTCAGATCAGGCGAGGAAG | GGAGGTCGTGATCGAGAAGA | 58 | TA11281_4565 | LOC_Os12g17910 | Bradi5g02890 |
| DG260 | ACCATTGGCTCCCTTCAGTA | TGGAGGCCTGATTCTGTTTC | 58 | CA629789 | LOC_Os07g04160 | Bradi5g02990 |
| DG273 | CTTGACGAGCTTGGAAATGG | GCAACAAGTGCTTCTGTCGT | 58 | CD877708 | LOC_Os04g12960 | Bradi5g03380 |
| DG274 | GGAGTCGCAGCCTTTGTTC | GCTCTCCATGTTAATTCCATGTACTC | 58 | TA41391_4565 | LOC_Os04g12720 | Bradi5g03390 |
| DG279 | TGCTCAAGGGAAAGACCATC | AAAGCCTGAGCCTGCTTCTA | 58 | TA44444_4565 | LOC_Os04g13210 | Bradi5g03460 |
| DG371 | CCACTTGACAAGCAAATTAAGA | ATCACGAGGCTGGTGTCG | 58 | BJ307036 | LOC_Os04g19140 | Bradi5g04710 |
Ta (°C) is the optimum annealing temperature. Wheat EST, wheat ESTs retrieved from GenBank or from the TIGR Plant Transcript Assemblies database (ftp://ftp.tigr.org/pub/data/plantta/Triticum_aestivum/). Rice (MSU) Hit, wheat EST BLASTN hit to Rice Genome Annotation MSU database version 6.1 (ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_6.1/all.dir/); Brachy Hit, wheat EST BLASTN hit to B. distachyon Genome Annotation Project database 1.0 (ftp://ftp.ensemblgenomes.org/pub/plants/release-/fasta/brachypodium_distachyon/cdna/).
Fine mapping population development
To develop the fine-resolution mapping population for map-based cloning of Rht8, selected semi-dwarf lines of the 89 RIL population (Korzun et al., 1998) carrying Xgwm261 192bp, Rht8, and sensitive to photoperiod (Ppd-D1b), i.e. lines RIL4 and RIL6, were crossed with Cappelle-Desprez (Xgwm261 172bp, rht8, and Ppd-D1b). The resulting F1 plants were selfed to produce a segregating F2 population. F2 plants were screened using high-throughput fluorescent genotyping with Rht8-flanking SSR markers gwm261 and cfd53 to retain recombinants. Gwm261 and cfd53 forward primers were 5' labelled with four different dyes: 6-FAM, VIC, NED, and PET (Applied Biosystems), and their amplification multiplexed in a single reaction. Amplification was conducted in a 10 μl volume containing 50 ng of genomic DNA, 0.2 μM of each of the four primers, 1× Hot Star MasterMix (Qiagen), with the following conditions: denaturation at 94 °C for 15 min, 40 cycles of 94°C for 30 s, 58 °C for 30 s, 72 °C for 30 s, and final extension at 72 °C for 1 min. Following amplification, four different samples, each labelled with a different dye, were pooled in the first 1:50 (H2O) dilution at a ratio of 1:2:1:2 for 6-FAM:VIC:NED:PET. Each sample of the first dilution plate was further diluted 1:10 with 8.95 μl of denaturating agent HiDi Formamide (Applied Biosystems), and 0.05 μl of size standard LIZ500 (Applied Biosystems). Samples were separated by capillary electrophoresis on an ABI3730 (ABI PRISM 3730 DNA sequencer; Applied Biosystems), and semi-automated SSR allele sizing was performed using GeneMapper v.4 software (Applied Biosystems). This procedure allowed the analysis of eight data points per capillary run: two markers for each of the four DNA samples labelled in a different dye.
Identified recombinant F2 plants (i.e. homozygous for one SSR locus and heterozygous for the other one, or homozygous at both loci with a different genotype from the crossing parents) were further genotyped with gene-based markers within the interval (DG087, DG260, DG274, DG279, and DG371), phenotyped for height, and selfed to produce recombinant F3 families. The F3 families were genotyped as before to identify homozygous Rht8 recombinants (i.e. recombinants that have a different genotype from the crossing parents, and are homozygous at both loci of the flanking markers). Homozygous Rht8 F3 recombinants were selected, scored for height, and selfed to obtain homozygous F4 recombinant lines. The segregation fit of genotypic frequencies was estimated with the χ2 test, and the pair-wise recombination frequency (r) between Rh8 and the other seven loci (Xgwm261, DG260, DG087, DG274, DG279, DG371, and Xcfd53) in the F2 population was calculated as r = [(recombinant heterozygotes+2× recombinant homozygotes)/2× F2 plants] to determine marker order and genetic distances.
F2:3 height analysis
Height data from homozygous F2:3 recombinants were used to back-score the original F2 generation. F2:3 families deriving from recombinant F2 plants, each consisting of 16 individuals, were genotyped for Xgwm261 and Xcfd53 by high-throughput fluorescent genotyping, for the selection of homozygous Rht8 recombinants. Genotype frequencies were estimated with the χ2 test. Homozygous F3 recombinants were retained for each F2:3 family, and grown to maturity in glasshouse conditions in four randomized batches to produce homozygous F4 recombinants. Mean height values of F3 recombinants from each F2:3 family were used to determine a ‘b’ (short) or ‘a’ (tall) score, in a linear model expressed as the following equation: H=β0+β1V+β2E+e (‘H’ is the plant height; ‘β0’, ‘β1’, and ‘β2’ are the parameters evaluated by the model, ‘V’ is the variety, ‘E’ is the experiment, and ‘e’ is the error term). For each randomized batch, VCappelle (tall control) was imposed as 0, then β0 became the average heigh of Cappelle-Desprez, β1 the difference between Cappelle-Desprez and the other varieties, and β2 the correction factor for the experiment. The model evaluates if the mean height values of F3 recombinants from each F2:3 family have a statistically significant difference in height compared with Cappelle-Desprez (when β1 differs from 0). Analyses were done with R 2.10.1 (R Development Core Team, 2005).
Results
The effect of Rht8 on wheat culms
The most evident effect of the semi-dwarf Rht8 allele is to reduce wheat stature by shortening culm lengths (Fig. 1A, B). Worland et al. (1998) found that Rht8 causes a 7–8 cm reduction in plant height, resulting in plants that are typically 10% shorter than their wild-type counterparts. However, it was not known whether the reduction in culm length is due to fewer or shorter internodes, and whether the reduction in culm length is concentrated in a particular internode or spread along all internodes of the culm. It is also important to separate clearly the effects of Rht8 and Ppd-D1a which both influence culm length and are genetically linked. In varieties where Rht8 is used, it normally occurs together with the photoperiod-insensitive allele Ppd-D1a.
Fig. 1.
Phenotype of hexaploid wheat semi-dwarf Rht8 lines. (A) Gross plant morphology of wild-type (Cappelle-Desprez) and semi-dwarf Rht8 [RIL4, from Cappelle-Desprez (Mara 2D) RIL population] lines. (B) Culm morphology of wild-type (Cappelle-Desprez) and semi-dwarf Rht8 (RIL4) lines. (C) Schematic representation of internode elongation patterns in wild-type Cappelle-Desprez (Cappelle-D), semi-dwarf Cappelle-Desprez (Mara 2D) SL (substitution line), and the Cappelle-Desprez (Mara 2D) RIL population grouped in classes according to their Rht8, Ppd-D1, and Xgwm261 genotype: wild-type (wt), Ppd-D1a, Rht8, and Rht8+Ppd-D1a.
In a randomized and replicated experiment, individual internode lengths of main tillers in the Cappelle-Desprez (Mara 2D) RIL population (89 lines; Korzun et al., 1998) segregating for Rht8 and Ppd-D1, and parental lines Cappelle-Desprez (tall) and Cappelle-Desprez (Mara 2D) SL (short) were measured. By mapping Ppd-D1 using perfect markers developed by Beales et al. (2007), individual RILs were grouped into four classes according to their Rht8, Ppd-D1, and gwm261 genotype: wild-type Ppd-D1b+rht8 (33 lines, such as Cappelle-Desprez), Ppd-D1a+rht8 (five lines), Ppd-D1b+Rht8 (eight lines), and Ppd-D1a+Rht8 [43 lines, such as Cappelle-Desprez (Mara 2D) SL].
Wild-type RILs were of the same height as the tall Cappelle-Desprez parent (112±4.3 cm), while Rht8 RILs were 14 cm (13%) shorter than the tall parent. As previously reported (Borner et al., 1993; Worland et al., 1998), the Ppd-D1a allele also reduced the culm length of Ppd-D1a RILs by an average of 4 cm (3%). Under the long-day conditions used, an additive effect of Rht8 and Ppd-D1a alleles to shorten stature was not detected. In fact, the culm length of Rht8+Ppd-D1a RILs was not significantly different from that of Rht8 RILs or the short Cappelle-Desprez (Mara 2D) SL parent.
Analyses of internodal segments indicate that both Rht8 and Ppd-D1a alleles reduce plant height by acting throughout the culm, and not by decreasing the number of elongating internodes and nodes. In fact, all RIL classes have the same number of internodes (peduncle+5) and visible nodes (5) as the wild-type Cappelle-Desprez parent (Fig. 1C). As previously reported (Worland et al., 1998), the semi-dwarf Rht8 allele does not affect ear morphology in this population. The largest measurable difference in segment length between semi-dwarf Rht8 RILs and wild-type Cappelle-Desprez lines was observed in the peduncle (by an average of 15% length reduction) and internode 1 (by an average of 5% reduction), while other internodes were reduced proportionally. Internode 1 and peduncle are also the final two internodes to elongate, thus accounting for the majority of the final culm length.
Rht8 affects culm length by reducing cell elongation
In wheat, internode elongation is caused by cell division in the intercalary meristem, followed by cell elongation in the elongation zone. Shorter culms observed in Rht8 lines could be caused by changes in cell division, cell elongation, or both. To investigate these possibilities, the internal cellular morphology of the peduncle and internode 1 was examined.
First, transverse sections of culm tissues at ear emergence (wheat growth stage 50–59; Zadoks et al., 1974) in Rht8 (Ppd-D1b) and wild-type lines was examined. This developmental stage provided the opportunity to study both cell expansion and cell proliferation by examining expanded parenchyma cells in internode 1 and node 2, and proliferation activity in the intercalary meristem of node 1. In both wild-type and Rht8 lines, internode 1 was fully elongated and cell division at node 2 has ceased (Fig. 2I, J). The cellular structure of node 2 in both lines appeared very similar: tissues were fully differentiated with very thick cell walls, particularly the mechanically strong sclerenchyma cells surrounding vascular bundles. Transverse sections of medial elongating zones of fully elongated internode 1 of wild-type and Rht8 lines showed a similar anatomical organization of cell types (Fig. 2G, H). On the other hand, at the stage examined, the peduncle is still elongating and the meristem is active at node 1 until anthesis. A similar organization of cell types was also observed in medial elongating zones of the peduncle of wild-type and Rht8 lines (Fig. 2A, B). Cell divisions were still very pronounced in both wild-type and Rht8 lines (Fig. 2C–F). There was no apparent defect in cell proliferation in node 1 in the intercalary meristem of Rht8 lines. This suggests that intercalary meristems do develop in Rht8 mutants but perhaps cell elongation is reduced, resulting in shorter peduncles (and internodes).
Fig. 2.
Transverse culm sections of Rht8 lines at ear emergence. Transverse sections of the medial elongating zone of the peduncle (A, B) and internode 1 (G, H), of node 1 (C, D, E, F), and node 2 (I, J) in wild-type ([A, C, E, G, I, RIL33 from Cappelle-Desprez (Mara 2D) RIL population] and semi-dwarf Rht8 lines [B, D, F, H, J, RIL4 from Cappelle-Desprez (Mara 2D) RIL population]. Sections were stained with toluidine blue; red arrows indicate cell divisions. Bars = 100 μm. e, epidermis; ch, chlorenchyma; sc, sclerenchyma; vb, vascular bundle; p, parenchyma; a, aerenchymous centre.
Longitudinal sections of fully elongated peduncle and internode 1 at straw stage in wild-type and Rht8 lines were further examined to see if shorter culms in Rht8 lines are reflected in shorter parenchyma cells. Longitudinal sections were taken from three different zones of the internodes: basal, medial, and distal. In both wild-type Cappelle-Desprez and Rht8 lines, parenchyma cells in the peduncle and internode 1 were longitudinally elongated and organized in longitudinal files (Fig. 3A, B). However, throughout the sections, cells in the Rht8 lines were significantly shorter than those in Cappelle-Desprez plants (Fig. 3C, D, t-test P < 0.05). The observations show that the semi-dwarfing Rht8 allele affects culm cell elongation.
Fig. 3.
Longitudinal culm sections of Rht8 lines from straw. Scanning electron micrographs of the medial zone of fully elongated peduncle in (A) wild-type (Cappelle-Desprez) and (B) semi-dwarf Rht8 lines [RIL4 from Cappelle-Desprez (Mara 2D) RIL population]. Bars = 200 μm. Comparisons of parenchymatic cell lengths (μm) in basal (<1 cm after the basal node), medial (in the middle the considered segment), and distal (>1 cm before the following node) sections of the (C) peduncle and (D) internode 1 between wild-type and Rht8 lines. *t-test P < 0.05. Bars represent the positive value of the standard deviation.
Rht8 is not involved in gibberellin biosynthesis or signalling
Cell elongation in the culm is often regulated by gene products involved in the GA or BR pathways (reviewed in Wang and Li, 2008). Traditionally, Rht8 has been classified as a GA-sensitive gene because plants respond to the exogenous application of GAs (McIntosh et al., 2003), but its role, if any, in GA biosynthesis or signalling remains unknown. Ellis et al. (2004) found no difference in leaf elongation rate or responsiveness to GA using selected lines of the Vigour18 (tall)×Chuan-Mai 18 (semi-dwarf, Rht8) double haploid population. However, the response of semi-dwarf Rht8 plants to the exogenous application of GA or the quantification of endogenous GAs has never been analysed in elongating culms.
The endogenous GA content was characterized in rapidly expanding culm internodes of 6-week-old wild-type and semi-dwarf Rht8 lines (35–45 cm tall, flag leaf just visible, growth stage 37; Zadoks et al., 1974). GAs belonging to the early 13-hydroxylation GA pathway (GA1, GA8, GA19, GA20, GA29, and GA44), the main pathway occurring in wheat, were quantified by full-scan gas chromatography–mass spectrometry. GA53, GA3, and GA17, also synthesized via the early 13-hydroxylation GA pathway, were below the level of detection or too low to quantify. If Rht8 lines had a defect in GA biosynthesis, they would have lower levels of bioactive GA (GA1) compared with the wild type, while a defect in GA signal transduction would result in an accumulation of C19-GAs (Webb et al., 1998). However, no significant difference in the amount of endogenous GAs compared with wild-type plants was found (Table 2), at the developmental stage examined. The only significant difference was in the concentration of GA29, which is the inactivated product of GA20. As GA29 had the lowest concentration of all GAs measured, and levels of GA20 were the same in wild-type and Rht8 lines, an explanation for the observed difference is that GA29 was too low to quantify accurately. The result indicates that Rht8 is unlikely to be involved in GA biosynthesis or signalling.
Table 2.
Endogenous GA concentrations (ng g –1 dry weight) in uppermost expanding culm internodes of wild-type (wt, Cappelle-Desprez) and semi-dwarf Rht8 lines [RIL4 from from the Cappelle-Desprez (Mara 2D) RIL population]
| GA44 | GA19 | GA20 | GA29 | GA1 | GA8 | GA38 | |
| wt | 26.8±0.8 | 22.3±5.6 | 1.6±0.4 | 0.7±0.02 | 2.1±0.5 | 14.7±4.5 | 4.3±0.7 |
| Rht8 | 26.4±0.8 | 26±5.6 | 1.7±0.1 | 0.8±0.03 | 2.2±0.4 | 11.5±3.9 | 3.4±0.9 |
| t-test P-value | 0.509682 | 0.460928 | 0.683785 | 0.000146 | 0.644931 | 0.267896 | 0.119346 |
Values are given as the mean ±SE.
To confirm that semi-dwarf Rht8 lines do not differ from wild-type plants in GA content or signal transduction, wild-type and Rht8 lines were sprayed with 100 μM GA3 throughout development and their response was measured as final culm length. The Mercia Rht3 NIL containing the Rht-B1c GA-insensitive allele was used as a negative control. Both Cappelle-Desprez and Rht8 lines showed an increase in culm elongation following the treatments, whereas Mercia Rht3 NIL, as expected, did not (Supplementary Fig. S1 available at JXB online). Cappelle-Desprez and Rht8 lines responded with a very similar increase in plant height (Tukey’s HSD test P < 0.05): the wild type showed a 15% increase, whereas the semi-dwarf showed a 13% increase. Thus, Rht8 lines had an elongation response comparable with that of the wild type. The data confirm that Rht8 is unlikely to be directly involved in GA metabolism or signalling to reduce plant height.
Rht8 lines show an altered sensitivity to brassinolide
Typically, rice mutants with defects in BR biosynthesis or signalling are dwarfed, exhibit specific shortening of the second internode (equivalent to internode 1 of wheat), have very erect leaves, and show photomorphogenesis in the dark, also called the de-etiolated (DET) phenotype (Yamamuro et al., 2000). The altered phenotypes can be restored upon BR application in deficient, but not in insensitive BR mutants. In temperate cereals, the only characterized BR mutant is bri1, which encodes the brassinosteroid receptor HvBRI1, and was cloned using uzu barley (Chono et al., 2003). To the authors’ knowledge, it has not been reported whether uzu barley has an internode-specific reduction as in the case of rice BR mutants. To date, there are no characterized BR mutants in wheat, although precursors of the biologically active form BL are present in different parts of the wheat plant, and BRs are thought to influence wheat development at many levels (Takatsuto et al., 1999; Feng et al., 2007). Rht8 lines show a semi-dwarf phenotype, pronounced shortening of the peduncle and first internode, and no direct or indirect involvement in GA metabolism. These characters, in addition to the fact that BRs regulate cell elongation (Wang and Li, 2008), led to the decision to test the responsiveness of Cappelle-Desprez and isogenic Rht8 seedlings to BL in two distinct assays.
First, the root sensitivity to BL treatment was tested in 4-day-old wheat seedlings (wild-type Cappelle-Desprez and Rht8 RIL6 lines). In wild-type rice seedlings grown on agar media containing BL, coleoptiles elongate abnormally, resulting in a twisted shape, leaves grow poorly and do not break through the coleoptile, root elongation is inhibited, and the roots develop in a wavy form (Yamamuro et al., 2000). In contrast to rice, barley wild-type seedlings treated with BL show a clear response only in the roots, whereas BL effects on other tissues and body parts were inconclusive (Chono et al., 2003). Similarly, at 1 μM BL, no treatment effect was observed on coleoptiles and shoots of both wild-type and Rht8 lines (Supplementary Fig. S2 at JXB online). However, a differential root response after BL treatment was observed between the wild type and Rht8. In the wild type, the length of the primary root as well as the root dry mass decreased with increasing BL concentration (P < 0.05), while the semi-dwarf Rht8 line showed a slight but significant increase in root length at lower BL concentrations (0.01 μM and 0.1 μM, P < 0.05) but no response at a higher dose (1 μM, P > 0.05) (Fig. 4). Furthermore, there was no significant change in root dry mass upon treatment in the Rht8 line (Fig. 4, P > 0.05). Importantly, at the developmental stage examined, untreated wild-type and Rht8 lines did not differ in their shoot, coleoptile, or root length and root dry mass (Fig. 4, Supplementary Fig. S2, P > 0.05).
Fig. 4.
Seedling root response to different BL concentrations. Root elongation and root dry mass in response to different BL concentrations of wild-type Cappelle-Desprez and semi-dwarf Rht8 seedlings. Germinated seeds were transferred to media containing the indicated BL concentration and grown for 4 d. Bars indicate the standard deviation.
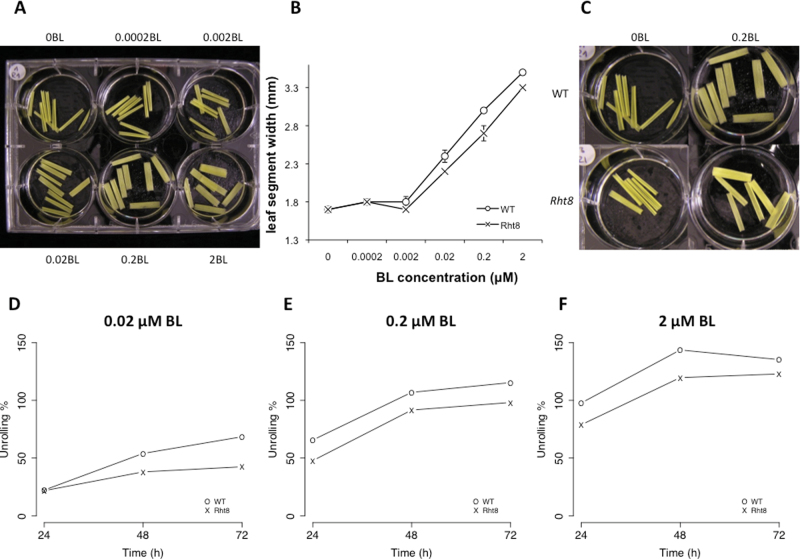
To better characterize the differential BR sensitivity observed between the wild-type and Rht8 seedlings, the established wheat leaf unrolling assay developed by Wada et al. (1985) was used. If Rht8 lines have a different sensitivity to BRs, the degree of leaf unrolling should be distinct from that of wild-type Cappelle-Desprez plants. Etiolated leaf segments were incubated in different concentrations of BL for 72 h and photographed every 24 h, as shown in Fig. 5A. Leaf segments of both wild-type and Rht8 lines did not respond to 0.0002 μM and 0.002 μM BL (Fig. 5B, test P > 0.05), but started to unroll at 0.02 μM BL (Fig. 5D), and the extent of unrolling (apparent width) increased over time at each concentration tested (Fig. 5D–F). For both lines the plateau was reached at 2 μM BL after 72 h (Fig. 5F), when leaf segments were totally flattened and of the same width. Although Cappelle-Desprez and Rht8 lines reached the same final leaf width (Fig. 5B, test P > 0.05), a significant difference was observed in unrolling percentage (leaf segment width) over time at the higher BL concentrations tested (0.02, 0.2, and 2 μM BL). The extent of leaf unrolling at each concentration was always lower in Rht8 lines (P < 0.05), suggesting that leaf sensitivity of Rht8 lines to BRs is less than that of wild-type Cappelle-Desprez plants. A hyposensitivity of Rht8 lines to BRs could explain the semi-dwarfed phenotype and the reduction in cell elongation.
Fig. 5.
Leaf unrolling response of the wild type (WT) and Rht8 lines. (A) Example of the experimental outline: etiolated leaf segments of Cappelle-Desprez incubated in 0, 0.0002, 0.002, 0.02, 0.2, and 2 μM BL for 72 h in total darkness. (B) BL effect on the leaf unrolling of Cappelle-Desprez (WT) and Rht8 lines after 72 h. Bars represent the standard error. (C) Visual comparison of Cappelle-Desprez (WT) and Rht8 (RIL6) leaf unrolling after 72 h incubation in 0 and 0.2 μM BL. (D–F) Leaf ‘unrolling %’ at different BL concentrations over time of Cappelle-Desprez (WT) and Rht8 leaf segments. (D) 0.02 μM BL, (E) 0.2 μM BL, (F) 2 μM BL. At a given time point (i) and treatment (j), ‘unrolling %’ was calculated as =[(Wij/W0j)–(Wi0–W00)], where Wij is the mean leaf width of the sample considered, W0j is the mean leaf width of the same sample at time ‘0’, Wi0 is the mean leaf width of the control at time ‘i’, and W00 is the mean leaf width of the control at time ‘0’.
Comparative mapping of the Rht8 locus
Previously, Rht8 was mapped between the Xgwm261 (0.6 cM distal to Rht8) and the Xgwm484 SSR loci (19.9 cM proximal to Rht8) by Korzun et al. (1998). In this study, the same mapping population of 89 Cappelle-Desprez (Mara 2D) RILs was used for improving the 2DS map in this interval. Rht8 (plant height) was re-scored in the population according to the 95% confidence interval of the height controls: Cappelle-Desprez (Mara 2D) SL (short) or Cappelle-Desprez (tall). The 2DS map was also improved by mapping four additional SSR markers (wmc503, cfd53, wmc112, and gwm1418), and the Ppd-D1 perfect marker (Beales et al., 2007). Amplification conditions and primer sequences were obtained from GrainGenes, http://wheat.pw.usda.gov/cgi-bin/graingenes/browse.cgi?class=marker). To anchor collinear regions in model genomes, new gene-based markers were designed.
Initially, gene-based markers were designed from wheat ESTs mapping to the chromosome 2DS1-0.33 deletion bin. In fact, both gwm261 and gwm484 markers flanking Rht8 in the 89 Cappelle-Desprez (Mara 2D) RILs have been assigned to this chromosomal deletion bin (http://wheat.pw.usda.gov/cgi-bin/graingenes/browse.cgi?class=marker; Endo and Gill, 1996). In order to align the rice genomic sequence to the wheat Rht8 locus, sequence information of the gene-based marker COS2Q was used. COS2Q was originally developed for the WGIN project as a conserved orthologous sequence (COS), and has been previously mapped distal to Xgwm261 on 2DS using the Avalon×Cadenza double haploid population (http://www.wgin.org.uk). The wheat EST BE444541 (GenBank ID), from which it was developed, is orthologous to the rice gene LOC_Os04g01590 (Rice Genome Annotation project MSU 6.1). This marker was monomorphic between parents of the mapping population used, but LOC_Os04g01590 was taken as an anchoring point on the rice genome for marker development. Markers were designed every 10 kb in both directions from the anchoring point, screened for polymorphism, and, if polymorphic, mapped in wheat to establish the direction of the wheat–rice collinearity. After anchoring the rice genome with polymorphic markers, syntenic rice (MSU 6.1) and B. distachyon (release 1.0 at www.modelcrop.org) sequence information was used to target and saturate the Rht8 locus. Markers were named DG (for Dwarfing Gene) followed by three numbers. A total of 203 primer pairs were developed from collinearity with rice (from marker DG001 to marker DG203), and 228 from the collinearity with B. distachyon (from marker DG204 to marker DG431).
Following this approach, 18 newly developed gene-based markers (Table 1) were polymorphic and allowed mapping on the pre-existing genetic map (Korzun et al., 1998), identifying two separate linkage groups for part of 2DS (Fig. 6). A small linkage group of 11.7 cM was defined by one restriction fragment length polymorphism (RFLP) marker (Xpsr649) and two gene-based COS markers (DG048 and DG032). Four additional gene-based markers (DG025, DG035, DG057, and DG062) were tested on 24 lines of the mapping population, but were not genotyped on the remaining lines, as they map too distally from the target Rht8 locus. The order and orientation of the two linkage groups were established by analysing other publicly available wheat maps from GrainGenes (http://wheat.pw.usda.gov/GG2/index.shtml). The two linkage groups did not join because the genetic distance between them probably exceeds 30 cM. Nevertheless, the major objective of the project was to improve the map around the Rht8 locus.
Fig. 6.
Molecular linkage groups of part of wheat chromosome 2DS in the Cappelle-Desprez (Mara 2D) RIL population. Genetic distances are shown in cM at the left-hand side of each linkage group, while marker names are shown on the right-hand side. Newly developed gene-based markers are shown in bold and named DG followed by three numbers. Additional SSR and Ppd-D1 markers mapped on the population are also shown in bold. Rht8 was re-scored in the population as individual plant height in a replicated and randomized block design, and is marked with an asterisk.
A bigger linkage group of 48.1 cM, containing Rht8, comprises original SSR and RFLP markers mapped by Korzun et al. (1998). The mapping of Rht8 was furthered by identifying a genetic interval of 2.5 cM containing the locus: Rht8 is flanked 0.6 cM distally by two co-segregating SSR loci Xgwm261 and Xwmc503, and 1.9 cM proximally by two co-segregating SSR markers Xcfd53 and Xwmc112, and one co-segregating gene-based marker DG371. The Rht8 genetic interval contains five gene-based markers that co-segregate with Rht8 (DG087, DG260, DG273, DG274, and DG279). The closest gene-based markers that flank the Rht8 locus were developed from B. distachyon: DG244 is 1.8 cM distal and DG371 is 1.9 cM proximal to Rht8. DG244 corresponds to Bradi5g02890, and DG371 to Bradi5g04710, identifying a physical interval of 2.88 Mb in B. distachyon. The Rht8 proximal marker DG371 also corresponds to rice LOC_Os04g19140, and with DG086 (LOC_Os04g12480) defines a genetic interval of 5 cM containing Rht8 and coinciding with a 3.72 Mb physical interval in rice.
Fine-resolution mapping of the Rht8 locus
To delimit the Rht8 location further and resolve the order of the co-segregating markers, a new F2 fine mapping population was constructed. Selected short lines from the Cappelle-Desprez (Mara 2D) RIL population (Korzun et al., 1998) were crossed with Cappelle-Desprez (tall rht8 allele) to allow recombination. The selected short RILs (RIL4 and RIL6) and Cappelle-Desprez are polymorphic for Xgwm261 (192 bp and 174 bp) and Xcfd53 (274 bp and 254 bp), the SSR loci flanking Rht8, but carry the same photoperiod-sensitive Ppd-D1b allele. The short phenotype of F1 hybrids indicated that Rht8 is not inherited recessively, but that Rht8 exhibits incomplete dominance towards the short parent (Supplementary Fig. S3 at JXB online).
F2 seeds were collected from self-pollinated F1 plants, and 3104 F2 individuals were genotyped with SSR markers flanking Rht8: gwm261 and cfd53. Of the 3104 F2 plants, 712 had the parental short genotype (Xgwm261 192–192bp, Xcfd53 274–274bp), 738 had the parental tall genotype (Xgwm261 174–174bp, Xcfd53 254–254bp), and 1502 were heterozygous (Xgwm261 192–174bp, Xcfd53 274–254bp). The two co-dominant SSR markers showed the expected Mendelian segregation ratio of 1:2:1 (χ2 test value P=0.5), and their recombination frequency of 0.0248 is consistent with the genetic distance observed in the 89 RIL population of 2.5 cM (Fig. 6).
The screen identified 152 F2 recombinant plants between gwm261 and cfd53 that have a different genotype from the crossing parents (Supplementary Fig. S4 at JXB online). Recombinant F2 plants were retained and genotyped further with co-dominant gene-based markers mapping within the Xgwm261–Xcfd53 interval: DG087, DG260, DG274, DG279, and DG371 (DG273 does not exhibit a co-dominant amplification pattern and was therefore not used for further analyses). The recombinant plants were grouped in 14 recombinant classes according to their genotype at the seven marker loci (Supplementary Table S1 at JXB online; graphical genotypes of selected homozygous F2:3 recombinants). Segregation ratios for the five co-dominant markers were in agreement with the expected 1:2:1 ratios (P=0.72 for DG260, P=0.76 for DG087, P=0.72 for DG274, P=0.84 for DG279, and P=0.80 for DG371). The SSR and gene-based markers identify six genetic intervals of 0.69, 0.08, 0.18, 0.21, 1.29, and 0.04 cM in which Rht8 could be mapped (Fig.7).
To determine unequivocally the genotype at the Rht8 locus and map the gene in one of the identified genetic intervals, F2:3 phenotypic information was used. Sixteen individuals were genotyped with the two SSR markers for each segregating F2:3 family, and only the homozygous Rht8 recombinant lines (i.e. recombinants that have a different genotype from the crossing parents, and are homozygous at both loci of the flanking markers) were retained. Homozygous F3 recombinants were grown in three replicated and randomized experiments, and analysed for height with a linear model to back-score Rht8 in the F2 generation (Supplementary Table S1 at JXB online). By back-scoring F2 individuals with plant height observed in homozygous recombinant F2:3, it was also possible to discriminate homozygous and heterozygous individuals at the Rht8 locus. A segregation of 47 tall and 105 short plants was observed after back-scoring F2 individuals with F2:3 data. The phenotypic ratio fits the 1:3 segregation pattern (χ2 P=0.092), confirming F1 data that Rht8 exhibits incomplete dominance towards the short parent, and supporting the observation of Korzun et al. (1998) that the effect that Rht8 has on height is due to a major gene inherited in a Mendelian manner.
Rht8 was mapped within the 1.29 cM interval, between markers DG279 (0.79 cM distal) and DG371 (0.5 cM proximal). By increasing the size of the mapping population, a higher number of crossovers was observed between Xgwm261 and Rht8. In fact, the genetic distance between Xgwm261 and Rht8 was found to be larger (1.95 cM) than previously published (0.6 cM; Korzun et al., 1998). The gene-based markers identified the syntenic regions of the Rht8 locus within 2 Mb in B. distachyon chromosome 5 (Bradi5g03460-04710, Bd21 Genome Annotation 1.0) and 3.3 Mb in Oryza sativa chromosome 4 (LOC_Os04g13210-19140, MSU 6.1) (Fig. 7). The resolution potential of the F2 mapping population described in this work, is extremely high as there are still 48 and 31 F2 recombinants, respectively, on either side of the Rht8 locus.
Fig.7.
Rht8 fine map. Wheat 2DS genetic map around the Rht8 locus, and comparative mapping with B. distachyon (Bd21 Genome Annotation 1.0) and O. sativa (MSU 6.1) using gene-based markers developed for this project.
Altered sensitivity to brassinolide co-segregates with Rht8
To determine further the relationship between Rht8 and the altered BR response observed in semi-dwarf Rht8 lines and to test whether the BR response co-segregates specifically with the 1.29 cM Rht8 interval, leaf unrolling tests were performed on selected homozygous F4 recombinant lines segregating for Rht8. At 0.02 μM BL, it was possible to group the tall and short F4 recombinant lines into two groups of BL sensitivity, according to the parental controls (Fig. 8A).
Fig. 8.
Leaf unrolling response of parents of the fine-resolution mapping population, Cappelle-Desprez (WT) and RIL6 (Rht8), and selected homozygous F4 recombinant lines around the Rht8 locus. (A and B) BL effect on the leaf unrolling perecntage over time in Cappelle-Desprez (WT), RIL6 (Rht8), and selected tall (dotted black lines) and short (dotted red lines) homozygous F4 recombinant lines, at (A) 0.02 μM BL and (B) 2 μM BL. (C) Graphical genotypes of homozygous F4 recombinant lines grouped in recombinant classes, and comparison of the leaf unrolling response of tall (unshaded, unrolling like tall wild-type Cappelle-Desprez) and short (shaded in grey, unrolling less than normal like the semi-dwarf parent RIL6) F4 lines. The leaf unrolling response co-segregates with the height phenotype (Rht8).
The semi-dwarf homozygous F4 lines (Rht8 allele) were less sensitive to BL, showing a minor leaf unrolling response when compared with the tall F4 group (rht8 allele) at 0.02 μM BL. The group of tall F4 plants clearly unrolled more readily and grouped with the tall Cappelle-Desprez control. The leaf unrolling of all plants plateaued at 2 μM BL after 72 h (Fig. 8B), and the final leaf width was the same, demonstrating that the difference in BL sensitivity is not due to a difference in leaf width. The differential unrolling response at 0.02 μM BL in the F4 lines tested co-segregates with the Rht8 locus (Fig. 8C), indicating that the Rht8 locus is possibly involved in modulating the BR response of the wheat plant.
Rht8 candidates in the rice chromosome 4 and B. distachyon chromosome 5 syntenic regions
The gene content in the physical intervals of the two model genomes was analysed to consider possible candidates for Rht8, as there was no genomic sequence information available for hexaploid wheat at this time. The identified rice interval (LOC_Os04g13210-19140, MSU 6.1) contains 584 loci. According to rice MSU 6.1 annotation, 379 of the total 584 loci are repetitive elements (65%). Similarity searches of the remaining 205 loci against the TREP and the rice annotation Rice Annotation Project database (RAP-DB, 5) revealed that 73 additional loci are similar to repetitive elements, leaving 132 non-transposable element (non-TE) loci. In the rice genomic region considered, there is a sequence assembly gap of 120 kb according to the RAP database (Chr4:9277690..9387689 missing data). The same genomic portion (LOC_Os04g16712-16896) is presumably misannotated in the MSU database. In fact, there are 57 fragmented predicted genes in a 123 kb genomic interval, mainly involved in photosynthesis (photosystem I and II proteins). Excluding the annotation/assembly gap, the Rht8 equivalent region of rice contains 75 non-TE genic loci (Supplementary Table S2, worksheet 1 at JXB online). The same procedure was used to filter repetitive elements in the identified region of B. distachyon (2 Mb defined by Bradi5g03460-04710, Bd21 Genome Annotation 1.0). A high content of repetitive elements was also observed in B. distachyon, leaving 62 non-TE genic loci (Supplementary Table S2, worksheet 2). Moreover, many predicted genes have unknown functions and have not been characterized.
Good conservation of gene content and order is observed between the two species (Supplementary Fig. S5 at JXB online). The only exception in gene order is between LOC_Os04g14510 and Bradi5g04580. The complexity, size, and gene content in the two model organisms represent a limiting factor for the description and analysis of functional candidate genes. However, the physiological assays indicate that a possible cause of the impaired cell elongation in Rht8 lines is an altered BR sensitivity or response. Similarly, Rht8 gene candidates could be involved in cell elongation and have a direct or indirect role in BR signalling. In the two intervals, loci with possible roles in cell elongation include: LOC_Os04g14510 and Bradi5g04580, LOC_Os04g16970 and Bradi5g04540, LOC_Os04g14110, LOC_Os04g 14190 (putative zinc-finger domain-containing proteins); LOC_Os04g15840 and Bradi5g04120 (putative expansin EXPA1). A gene cluster, present in the intervals of both model genomes, consisting of several adjacent protein kinases (PKs) was also identified. In rice, the PK cluster consists of a leucine-rich repeat (LRR)-RLK (LOC_Os04g15560), a Ser/Thr PK (LOC_Os04g15580), and three additional LRR-RLKs (LOC_Os04g15630-Xa21, LOC_Os04g15650, and LOC_Os04g15660). A similar gene content was also observed in the B. distachyon PK cluster, which consists of three Ser/Thr PKs containing N-terminal domains of unknown function (DUF) (Bradi5g03900, Bradi5g03930, and Bradi5g03940), two LRR-RLKs (Bradi5g03960 and Bradi5g04000), and an additional LRR-receptor like protein lacking the cytoplasmic kinase domain (Bradi5g03980). The LRR-RLKs present in the physical intervals of the two model genomes are particularly interesting putative Rht8 candidates as BRI1, the BR receptor (Li and Chory, 1997), was shown to be an LRR-RLK.
Nevertheless, all of the genes present in the syntenic intervals need to be considered as Rht8 putative candidates based on comparative genomics studies. A total of 152 primer pairs (from 47 different rice and B. distachyon loci) were developed to target and saturate the DG279–DG371 interval, and refine the map location of Rht8. However, due to lack of polymorphism between parents of the fine-resolution mapping population it was not possible to map Rht8 to a smaller genetic interval.
Discussion
The reduced stature of varieties with the semi-dwarf Rht8 allele confers a substantial agronomic impact in environments where the use of other semi-dwarfing genes is not advantageous (Law and Worland, 1985; Worland and Law, 1986; Rebetzke et al., 1999; Worland et al., 2001). In southern European and Russian areas, a selective advantage for preserving the linkage between Ppd-D1a and Rht8 has been observed (Worland et al., 1998). However, the use of Ppd-D1a is restricted to those environments conducive to earlier flowering and a shorter life cycle. Most North European and American areas are associated with extended life cycles, so Ppd-D1a cannot be used to increase grain yield (Worland et al., 1997). Probably, by selecting away from Ppd-D1a and thereby also losing Rht8, breeders in northern countries have never had the opportunity to test Rht8 semi-dwarfing activity independently. Therefore, the identification of the gene underlying the semi-dwarf phenotype will not only broaden the knowledge of stature control in wheat, but will also facilitate the effective deployment of favourable Rht8 alleles into modern breeding varieties in conjunction with other appropriate genes and their alleles.
The physiological analyses carried out show that the semi-dwarfed stature of Rht8 plants is a consequence of shorter internodes throughout the culm, and the largest measurable difference lies in a shorter peduncle and the first internode (Fig. 1). Peduncle and internode 1 undergo the fastest and largest elongation process of all internodes in the culm (Kirby and Appleyard, 1981), thus accounting for the majority of the length reduction in the semi-dwarf line. Microscopic analyses revealed that the Rht8 difference in internode length is due to impaired cell elongation in culm segments. Rht8 lines have shorter parenchymatic cells in elongated peduncle and internode 1 (Fig. 3). No apparent defect in meristematic activity in intercalary meristems below elongating internodes was observed (Fig. 2). The extent of elongation depends on the cell type and is often regulated by environmental cues and endogenous hormones with crucial roles in cell expansion and elongation in culms and stems (Wang and Li, 2008).
Auxins have long been known to play a central role in orchestrating plant architecture during development, such as maintenance of apical dominance (Evans, 1985; Qi et al., 2008; McSteen, 2009). Although auxins are involved in cell elongation (Wada et al., 1968), no agriculturally beneficial rice, barley, or wheat auxin mutants are known to date, probably due to major pleiotropic effects other than plant stature. On the other hand, agriculturally beneficial mutants shortening plant stature are often involved in the biosynthesis or perception of GAs and BRs (Wang and Li, 2008). The semi-dwarf stature of Rht8 lines is unlikely to be due to an altered GA biosynthesis or signalling as wild-type rht8 and semi-dwarf Rht8 lines responded similarly to the exogenous application of GA3 (Supplementary Fig. S1 at JXB online), as well as containing the same amount of endogenous GAs (Table 2). Instead, a differential response to applied BL was observed between wild-type rht8 and semi-dwarf Rht8 lines. The whole plant response of wild-type wheat seedlings grown on media containing BL was similar to that of barley (Chono et al., 2003) as opposed to that of rice: root elongation was inhibited, while aerial parts remained unaffected (Fig. 4; Supplementary Fig. S2). In contrast to wheat and barley, rice seedlings grown on BL media, show clear responses in both aerial and submerged parts of the plant (Yamamuro et al., 2000). The observations strengthen the hypothesis that temperate cereals have a different BR uptake or transport mechanism when compared with rice (Chono et al., 2003), and question the possibility of using rice as a physiological model to understand the BR pathway in wheat. A differential response in root length and root dry mass was also observed between semi-dwarf Rht8 lines compared with the wild-type. While the wild-type line showed a decrease in primary root length and root dry mass with increasing BL concentrations, the semi-dwarf Rht8 line showed a slight increase in root length at lower BL concentrations (0.01 μM and 0.1 μM) but no response at a higher dose (1 μM; Fig. 4). The altered BR sensitivity in the semi-dwarf Rht8 line could be due to a decreased BR sensitivity. To examine further the differential BR response between wild-type and Rht8 seedlings and quantify the BR sensitivity, a more sensitive method established for temperate cereals (Wada et al., 1985; Chono et al., 2003) was used. The leaf unrolling response (Wada et al., 1985) showed that although wild-type and semi-dwarf lines did not differ in their final leaf width, semi-dwarf Rht8 lines showed a reduced rate of leaf unrolling (Fig. 5). The differential leaf unrolling response is likely to be a consequence of reduced BL sensitivity.
As well as providing novel physiological evidence for the mechanism of Rht8 action, the present work has achieved fine mapping for the eventual map-based cloning of the gene and the provision of superior molecular markers for immediate use. Using gene-based markers co-segregating with Rht8 in the coarse mapping population (Korzun et al., 1998) and screening 3104 F2 individuals of the fine-resolution mapping population developed for this project, it was possible to reduce the Rht8 genetic interval from 20.5 cM (Korzun et al., 1998) to 1.29 cM (Fig. 6). the wheat Rht8 syntenic region was identified and limited to 3.3 Mb of the short arm of rice chromosome 4 (from LOC_Os04g13210 to LOC_Os04g19140, MSU v6.1.), and to 2 Mb of B. distachyon chromosome 5 (from Bradi5g03460 to Bradi5g04710, genome assembly v1.) (Fig. 7). Moreover, following the observation of an altered BR sensitivity in parental lines of the fine-resolution mapping population, analyses of the leaf unrolling response in homozygous F4 recombinant lines indicate a co-segregation between BR hyposensitivity and the height reduction of the Rht8 locus (Fig. 8). Due to the compensatory dosage effect of the three wheat genomes, BR-deficient lines would be difficult to obtain, except in gain-of-function mutations. The semi-dwarfed phenotype and reduced culm length observed in Rht8 lines could be explained by a reduced sensitivity to BRs and—unlikely—to a reduced endogenous BR concentration.
Although an obvious gene candidate involved in the regulation of the BR response was not found in the genomic interval of the two model genomes, and the large gene content makes it difficult to make assumptions, the intervals contain several candidates with possible roles in cell elongation.
Altered activity of zinc-finger (Zn-finger) transcription factors can influence a wide range of processes during plant growth and development, including reduced plant stature of A. thaliana (AtSHI; Fridborg et al., 2001) and rice (OsLOL2; Xu and He, 2007). Several genes encoding putative Zn-finger proteins were found in the Rht8 syntenic intervals of rice and B. distachyon (LOC_Os04g14510 and Bradi5g04580, LOC_Os04g16970 and Bradi5g04540, LOC_Os04g14110, and LOC_Os04g 14190).
Expansins (EXPs) are cell wall proteins that induce wall extension (Cosgrove, 2000). Among EXP subfamilies (http://www.bio.psu.edu/expansins), EXPA and EXPB are known to induce cell wall loosening for the expansion of plant cells (Cosgrove et al., 2002). The EXPA subfamily is composed of 26 genes in Arabidopsis and 34 genes in rice (Choi et al., 2006). The diverse and specific expression patterns of the different EXPA transcripts suggest their distinct roles in plant growth and development (Choi et al., 2006). The collinear genic loci LOC_Os04g15840/Bradi5g04120 encode a putative EXPA1 protein. Shin et al. (2005) showed that rice EXPA1 is specifically expressed in leaf blades, whereas transcript levels of EXPA7, 14, 15, 18, 21, and 29 were greater in stems. Recently, Park et al. (2010) found that AtEXPA5 is a growth-regulating gene whose expression is controlled by BR signalling downstream of BZR1 in A. thaliana.
The cytochrome P450 family is a large family of enzymes with important roles in lipid metabolism, metabolism of phytohormones including BRs, and defence responses (Nelson, 2006). None of the CYPs found in the physical intervals appears to be related to BR metabolism.
The LRR-RLKs present in the physical intervals or the two model genomes are particularly interesting putative Rht8 candidates as several LRR-RLKs have been shown to play critical roles in development. Characterized LRR-RLKs include ERECTA that regulates organ shape (Torii et al., 1996), CLAVATA1 which controls cell differentiation at the shoot meristem (Clark et al., 1997), HAESA which regulates the floral abscission process (Jinn et al., 2000), and BRI1 the BR receptor (Li and Chory, 1997). LRR-RLKs also play a role in disease resistance (XA21; Song et al., 1995). LRR-RLKs comprise the largest class of plant RLKs, with 216 members in A. thaliana (Dievart and Clark, 2004). These have been divided into 14 subfamilies (LRR I–LRR XIV) classified on the basis of the organization of LRRs in the extracellular domain and the phylogenetic relationship between the kinase domains of subfamily members (Shiu and Bleecker, 2003). According to this classification BRI1, BRL1, BRL2, and BRL3 belong to the LRRX clade, whereas XA21 belongs to the XII clade (Morillo and Tax, 2006). To examine the phylogenetic relationships of the LRR-RLK present in the regions of interest with classified LRR-RLKs, a cladogram tree was constructed using the Neighbor–Joining method including at least one representative member of each LRR-RLK clade (data not shown). The five LRR-RLKs present in the region of interest (LOC_Os04g15560, LOC_Os04g15650, LOC_Os04g15660, Bradi5g03960, and Bradi04000) cluster to the LRR XII clade, comprising members involved in pathogen response such as XA21, FLS2, and EFR (Morillo and Tax, 2006), and not to the LRRX clade involved in developmental processes such as BR signalling and vascular differentiation. Therefore, both LRR-RLK clusters present in rice and B. distachyon seem to be involved in disease resistance.
Sequence information from rice and B. distachyon was used to saturate further the Rht8 genetic interval, defined by DG279 (0.79 cM distal) and DG371 (0.5 cM proximal). However, all 152 primer pairs (from 47 different loci) were monomorphic between parents of the fine-resolution mapping population, and it was not possible to map Rht8 to a smaller genetic interval. The low level of diversity found in the D-genome is expected from the evolutionary history of hexaploid wheat (Bossolini et al., 2006; Dubcovsky and Dvorak, 2007). Nevertheless, the resolution potential of the F2 mapping population is very high as there are 48 and 31 F2 recombinants, respectively, either side of Rht8, and the population can be exploited further once polymorphic markers are identified (Fig. 7).
In addition to the fundamental scientific interest of the Rht8 locus showing altered BR sensitivity, the work described presents exciting opportunities for wheat breeding and improved crop agronomy. The new gene-based Rht8 flanking markers (DG279 and DG371) can be used to generate a diagnostic haplotype for sources of Rht8 where gwm261 has been shown to be uninformative. The molecular marker-based prediction that a line carries Rht8 can then be validated by the application of the BL leaf unrolling assay. This combined assay will arm breeders with an equivalent tool to the GA sensitivity test (McIntosh et al., 1998) that has allowed the discrimination of GA-sensitive and GA-insensitive wheat varieties, marking the genotype for Rht-B1b and Rht-D1b types. The precise selection of Rht8 in the same way will allow it to be efficiently and immediately deployed in the water-limited target environments where it has very high potential impact.
Supplementary Material
Acknowledgements
We are very grateful to Fan Gong (PH lab) for his technical assistance in GA quantification, to Andrew Davis (JIC) for taking the wheat photographs, and to David Laurie (JIC) and John Snape (JIC) for critical reading of the manuscript. We thank Kim Findlay (JIC) for assistance on SEM, and the JIC horticultural staff for their valuable and skilled help with general plant husbandry. This work was supported by the Biotechnology and Biological Sciences Research Council, UK. DG was supported by an NIAB PhD studentship.
Footnotes
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Exogenous GA treatment of semi-dwarf Rht8 lines.
Figure S2. BL treatment.
Figure S3. The mode of inheritance of Rht8.
Figure S4. Electropherograms of 6-FAM-labelled Xgwm261 and Xcfd53 amplification products from parental, heterozygous, and recombinant F2 genotypes.
Figure S5. Graphical comparison of genomic segments of rice chromosome 4 with B. distachyon chromosome 5 around the wheat Rht8 locus.
Table S1. Graphical genotypes of homozygous F3 recombinant lines grouped in recombinant classes according to their genotype and Rht8 phenotype.
Table S2. Worksheet 1—rice gene content. Worksheet 2—Bd gene content.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Ahmad M, Sorrells ME. Distribution of microsatellite alleles linked to Rht8 dwarfing gene in wheat. Euphytica. 2002;123:235–240. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215,:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bai GH, Das MK, Carver BF, Xu XY, Krenzer EG. Covariation for microsatellite marker alleles associated with Rht8 and coleoptile length in winter wheat. Crop Science. 2004;44:1187–1194. [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.) Theoretical and Applied Genetics. 2007;115:721–733. doi: 10.1007/s00122-007-0603-4. [DOI] [PubMed] [Google Scholar]
- Bertin I, Zhu JH, Gale MD. SSCP-SNP in pearl millet—a new marker system for comparative genetics. Theoretical and Applied Genetics. 2005;110L:1467–1472. doi: 10.1007/s00122-005-1981-0. [DOI] [PubMed] [Google Scholar]
- Borner A, Worland AJ, Plaschke J, Schumann E, Law CN. Pleiotropic effects of genes for reduced height (Rht) and day-length insensitivity (Ppd) on yield and its components for wheat grown in middle Europe. Plant Breeding. 1993;111:204–216. [Google Scholar]
- Bossolini E, Krattinger SG, Keller B. Development of simple sequence repeat markers specific for the Lr34 resistance region of wheat using sequence information from rice and Aegilops tauschii. Theoretical and Applied Genetics. 2006;113:1049–1062. doi: 10.1007/s00122-006-0364-5. [DOI] [PubMed] [Google Scholar]
- Busov VB, Brunner AM, Strauss SH. Genes for control of plant stature and form. New Phytologist. 2008;177:589–607. doi: 10.1111/j.1469-8137.2007.02324.x. [DOI] [PubMed] [Google Scholar]
- Chandler PM, Marion-Poll A, Ellis M, Gubler F. Mutants at the Slender1 locus of barley cv Himalaya: molecular and physiological characterization. Plant Physiology. 2002;129:181–190. doi: 10.1104/pp.010917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebotar SV, Korzun VN, Sivolap YM. Allele distribution at locus WMS261 marking the dwarfing gene Rht8 in common wheat cultivars of Southern Ukraine. Russian Journal of Genetics. 2001;37:894–898. [PubMed] [Google Scholar]
- Choi D, Cho H-T, Lee Y. Expansins: expanding importance in plant growth and development. Physiologia Plantarum. 2006;126:511–518. [Google Scholar]
- Chono M, Honda I, Zeniya H, Yoneyama K, Saisho D, Takeda K, Takatsuto S, Hoshino T, Watanabe Y. A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiology. 2003;133,:1209–1219. doi: 10.1104/pp.103.026195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis . Cell. 1997;89,:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D. The growing world of expansins. Plant and Cell Physiology. 2002;43:1436–1444. doi: 10.1093/pcp/pcf180. [DOI] [PubMed] [Google Scholar]
- Dievart A, Clark SE. LRR-containing receptors regulating plant development and defense. Development. 2004;131:251–261. doi: 10.1242/dev.00998. [DOI] [PubMed] [Google Scholar]
- Dill A, Jung H-S, Sun T-p. The DELLA motif is essential for gibberellin induced degradation of RGA. Proceedings of the National Academy of Sciences, USA. 2001;98:14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgerton MD. Increasing crop productivity to meet global needs for feed, food, and fuel. Plant Physiology. 2009;149:7–13. doi: 10.1104/pp.108.130195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MH, Bonnett DG, Rebetzke GJ. A 192bp allele at the Xgwm261 locus is not always associated with the Rht8 dwarfing gene in wheat (Triticum aestivum L.) Euphytica. 2007;157:209–214. [Google Scholar]
- Ellis MH, Rebetzke GJ, Chandler P, Bonnett D, Spielmeyer W, Richards RA. The effect of different height reducing genes on early growth characteristics of wheat. Functional Plant Biology. 2004;31:583–589. doi: 10.1071/FP03207. [DOI] [PubMed] [Google Scholar]
- Endo TR, Gill BS. The deletion stocks of common wheat. Journal of Heredity. 1996;87:295–307. [Google Scholar]
- Evans ML. The action of auxin on plant-cell elongation. Critical Reviews in Plant Sciences. 1985;2,:317–365. doi: 10.1080/07352688509382200. [DOI] [PubMed] [Google Scholar]
- Feng X, Jiang D, Shan Y, Dai TB, Jing Q, Dong YF, Cao WX. Identification of steroids in wheat bran. Asian Journal of Chemistry. 2007;19,:87–94. [Google Scholar]
- Fridborg I, Kuusk S, Robertson M, Sundberg E. The Arabidopsis protein SHI represses gibberellin responses in Arabidopsis and barley. Plant Physiology. 2001;127:937–948. [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Youssefian S. Dwarfing genes in wheat. In: Russed GE, editor. Progress in plant breeding. London: Butterworths; 1985. pp. 1–35. [Google Scholar]
- Ganeva G, Korzun V, Landjeva S, Tsenov N, Atanasova M. Identification, distribution and effects on agronomic traits of the semi-dwarfing Rht alleles in Bulgarian common wheat cultivars. Euphytica. 2005;145:305–315. [Google Scholar]
- Griffiths J, Murase K, Rieu I, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. The Plant Cell. 2006;18:3399–414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd NP, King KE, Carol P, Cowling RJ, Peng J, Richards DE. Gibberellin: inhibitor of an inhibitor of…? BioEssays. 1998;20:1001–1008. doi: 10.1002/(SICI)1521-1878(199812)20:12<1001::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. The Plant Cell. 2003;15:2900–2910. doi: 10.1105/tpc.014712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. Slender rice, a constitutive gibberellin response mutant is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. The Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes and Development. 2000;14:108–117. [PMC free article] [PubMed] [Google Scholar]
- Khripach V, Zhabinski V, De Groot A. Twenty years of brassinosteroids: steroidal plant hormones warrant better crops for the XXI century. Annals of Botany. 2000;86:441–447. [Google Scholar]
- Khush GS. Green revolution: the way forward. Nature Reviews Genetics. 2001;2:15–822. doi: 10.1038/35093585. [DOI] [PubMed] [Google Scholar]
- Kirby EJM, Appleyard M. Cereal development guide. Stoneleigh, UK: Cereal Unit, National Agricultural Centre; 1981. [Google Scholar]
- Korzun V, Roder MS, Ganal MW, Worland AJ, Law CN. Genetic analysis of the dwarfing gene (Rht8) in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat. (Triticum aestivum L.) Theoretical and Applied Genetics. 1998;96,:1104–1109. [Google Scholar]
- Law CN, Worland AJ. Annual Report of the Plant Breeding Institute 1984. Cambridge, UK: Plant Breeding Institute; 1985. An effect of temperature on the fertility of wheats containing dwarfing genes Rht1, Rht2 and Rht3; pp. 69–71. [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90,:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu DC, Zhang HY, Wang J, Sun JZ, Guo XL, Zhang AM. Allelic variation, sequence determination and microsatellite screening at the XGWM261 locus in Chinese hexaploid wheat (Triticum aestivum) varieties. Euphytica. 2005;145,:103–112. [Google Scholar]
- Lorenzetti R. Wheat science. The green revolution of Nazareno Strampelli. Journal of Genetic Breeding. 2000:40. [Google Scholar]
- McIntosh RA, Hart GE, Devos KM, Gale MD, Rogers WJ. Catalogue of gene symbols for wheat. In: Slinkard AE, editor. Proceedings of the 9th International Wheat Genetics Symposium. University of Saskatchewan, Canada: University Extension Press; 1998. [Google Scholar]
- McIntosh RA, Yamazaki Y, Devos KM, Dubcovski J, Rogers WJ, Apples R. Catalogue of gene symbols in wheat 2003.
- McSteen P. Hormonal regulation of branching in grasses. Plant Physiology. 2009;149:46–55. doi: 10.1104/pp.108.129056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y. Positional cloning of rice semi-dwarfing gene, sd-1: rice ‘green revolution gene’ encodes a mutant enzyme involved in gibberellin synthesis. DNA Research. 2002;9,:11–17. doi: 10.1093/dnares/9.1.11. [DOI] [PubMed] [Google Scholar]
- Morillo SA, Tax FE. Functional analysis of receptor-like kinases in monocots and dicots. Current Opinion in Plant Biology. 2006;9:460–469. doi: 10.1016/j.pbi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Nelson DR. Plant cytochrome P450s from moss to poplar. Phytochemistry Reviews. 2006;5:193–204. [Google Scholar]
- Ogawa M, Kusanoa T, Katsumib M, Sano H. Rice gibberellin-insensitive gene homolog, OsGAI, encodes a nuclear-localized protein capable of gene activation at transcriptional level. Gene. 2000;245:21–29. doi: 10.1016/s0378-1119(00)00018-4. [DOI] [PubMed] [Google Scholar]
- Pallotta MA, Warner P, Fox RL, Kuchel H, Jefferies SP, Langridge P. 10th International Wheat Genetics Symposium. Paestum, Italy: 2003. Marker assisted wheat breeding in the southern region of Australia; pp. 789–791. [Google Scholar]
- Park CH, Kim TW, Son SH, Hwang JY, Lee SC, Chang SC, Kim SH, Kim SW, Kim SK. Brassinosteroids control AtEXPA5 gene expression in Arabidopsis thaliana. Phytochemistry. 2010;71:380–387. doi: 10.1016/j.phytochem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, et al. ‘Green Revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400,:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Qi J, Qian Q, Bu QY, et al. Mutation of the rice narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiology. 2008;147:1947–1959. doi: 10.1104/pp.108.118778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. [Google Scholar]
- Rebetzke GJ, Richards RA. Gibberellic acid-sensitive dwarfing genes reduce plant height to increase kernel number and grain yield of wheat. Australian Journal of Agricultural Research. 2000;52,:1221–1234. [Google Scholar]
- Rebetzke GJ, Richards RA, Fettell NA, Long M, Condon AG, Forrester RI, Botwright TL. Genotypic increases in coleoptile length improves stand establishment, vigour and grain yield of deep-sown wheat. Field Crops Research. 2007;100,:10–23. [Google Scholar]
- Rebetzke GJ, Richards RA, Fisher VM, Mickelson BJ. Breeding long coleoptile reduced height wheats. Euphytica. 1999;106:1159–1168. [Google Scholar]
- Reinhardt D, Kuhlemeier C. Plant architecture. EMBO Reports. 2002;3:846–851. doi: 10.1093/embo-reports/kvf177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods in Molecular Biology. 2000;32,:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nature Biotechnology. 2006;24:105–109. doi: 10.1038/nbt1173. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, et al. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- Schmidt AL, Gale KR, Ellis MH, Giffard PM. Sequence variation at a microsatellite locus (XGWM261) in hexaploid wheat (Triticum aestivum) varieties. Euphytica. 2004;135:239–246. [Google Scholar]
- Shin JH, Jeong DH, Park MC, An G. Characterization and transcriptional expression of the alpha-expansin gene family in rice. Molecules and Cells. 2005;20,:210–218. [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proceedings of the National Academy of Sciences, USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM. Semidwarf, (sd-1), ‘green revolution’ rice, contains a defective gibberellin 20-oxidase gene. Proceedings of the National Academy of Sciences, USA. 2002;99:9043–9048. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuto S, Kosuga N, Abe B, Noguchi T, Fujioka S, Yokota T. Occurrence of potential brassinosteroid precursor steroids in seeds of wheat and foxtail millet. Journal of Plant Research. 1999;112:27–33. [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. The Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Kondo H, Marumo S. A simple bioassay for brassinosteroids: a wheat leaf-unrolling test. Agricultural and Biological Chemistry. 1985;49:2249–2251. [Google Scholar]
- Wada S, Tanimoto E, Masuda Y. Cell elongation and metabolic turnover of cell wall as affected by auxin and cell wall degrading enzymes. Plant and Cell Physiology. 1968;9:369–376. [Google Scholar]
- Wang Y, Li J. Molecular basis of plant architecture. Annual Reviews of Plant Biology. 2008;59:253–79. doi: 10.1146/annurev.arplant.59.032607.092902. [DOI] [PubMed] [Google Scholar]
- Webb SE, Appleford NE, Gaskin P, Lenton JR. Gibberellins in internodes and ears of wheat containing different dwarfing alleles. Phytochemistry. 1998;47:671–677. [Google Scholar]
- Worland A J, Borner A, Korzun V, Li WM, Petrovic S, Sayers EJ. Wheat: prospects for global improvement. Ankara, Turkey: 1997. The influence of photoperiod genes on the adaptability of European winter wheats; pp. 10–14.pp. 517–526. [Google Scholar]
- Worland AJ, Korzun V, Roder MS, Ganal MW, Law CN. Genetic analysis of the dwarfing gene Rht8 in wheat. Part II. The distribution and adaptive significance of allelic variants at the Rht8 locus of wheat as revealed by microsatellite screening. Theoretical and Applied Genetics. 1998;96:1110–1120. [Google Scholar]
- Worland AJ, Law CN. Genetic analysis of chromosome 2D of wheat. 1. The location of genes effecting height, daylength insensitivity, hybrid dwarfism and yellow-rust resistance. Zeitschrift für Pflanzenzüchtung. 1986;96,:331–345. [Google Scholar]
- Worland AJ, Law CN, Petrovic S. Height reducing genes and their importance to Yugoslavian winter wheat varieties. Savremena Poljoprivreda. 1990;38:245–258. [Google Scholar]
- Worland AJ, Sayers EJ, Korzun V. Allelic variation at the dwarfing gene Rht8 locus and its significance in international breeding programmes. Euphytica. 2001;119,:155–159. [Google Scholar]
- Xu C, He C. The rice OsLOL2 gene encodes a zinc finger protein involved in rice growth and disease resistance. Molecular Genetics and Genomics. 2007;278:85–94. doi: 10.1007/s00438-007-0232-2. [DOI] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. The Plant Cell. 2000;12:1591–606. doi: 10.1105/tpc.12.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF. A decimal code for the growth stages of cereals. Weed Research. 1974;14:415–42. [Google Scholar]
- Zhang X, Yang S, Zhou Y, He Z, Xia X. Distribution of the Rht-B1b, Rht-D1b and Rht8 reduced height genes in autumn-sown Chinese wheats detected by molecular markers. Euphytica. 2006;152:109–116. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.