Summary
The kinetic properties of two classical inhibitors of aspartic proteases, pepstatin A and acetyl-pepstatin, were compared in their interactions with human immunodeficiency virus type-1 (HIV-1) and xenotropic murine leukemia virus-related virus (XMRV) proteases (PRs). Both compounds are substantially weaker inhibitors of XMRV PR than of HIV-1 PR. Previous kinetic and structural studies characterized HIV-1 PR/acetyl-pepstatin and XMRV PR/pepstain A complexes and suggested dramatically different binding modes. Interaction energies were calculated for the possible binding modes and suggested a strong preference for the one inhibitor-binding mode for HIV-1 PR, and two inhibitor-binding mode for XMRV PR-pepstatin A interaction. Comparison of the molecular models suggested that in the case of XMRV PR, the relatively unfavorable interactions at S3’ and the favorable interactions at S4 and S4’ sites with the statine residues may shift the ground state binding towards the two inhibitor-binding mode, whereas the single molecule ground state binding of statines to the HIV-1 PR appear to be more favorable. The preferred single molecular binding to HIV-1 PR allows the formation of the transition state complex, represented by substantially better binding constants. Intriguingly, crystal structure of the complex of acetyl-pepstatin with XMRV PR has shown a mixed type of binding: the unusual binding mode of two molecules of the inhibitor to the enzyme, in a mode very similar to the previously determined complex with pepstatin A, together with the classical binding mode found for HIV-1 PR. The structure is thus in good agreement with the very similar interaction energies calculated for the two types of binding.
Keywords: aspartic protease, enzyme inhibition, inhibitor binding, retrovirus, crystal structure
Introduction
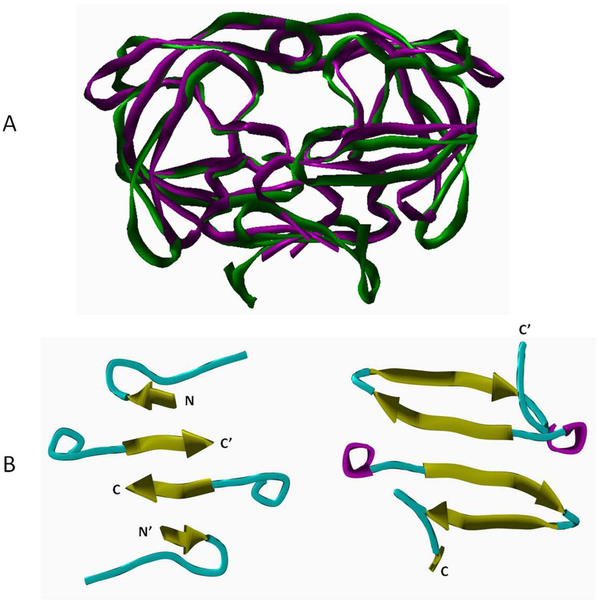
The proteolytic enzyme (PR) of the human immunodeficiency virus type-1 (HIV-1) has been recognized as a potential drug target almost a quarter century ago [1]. A number of its inhibitors have been synthesized, tested, and further developed, and currently 9 protease inhibitors out of 26 accepted antiretroviral agents are available in the clinical practice [2, 3]. Since the discovery of pathogenic human retroviruses, starting with human T-lymphotropic virus type-1 [4], several other retroviruses were also implicated as possible disease agents in humans. The most recent human retrovirus postulated to be associated with two distinct pathologies was the xenotropic murine leukemia virus-related virus (XMRV). Following extensive controversies (reviewed recently by Groom and Bishop) [5], it was recently reported that XMRV may have been created by passaging human tumors in mice [6] and it is no longer considered to be a causative agent of human disease. However, the genome of XMRV, including the segment encoding its protease, is highly similar to that of murine leukemia virus (MLV, Figure 1). Given the widespread use of MLV in gene transfer experiments as well as only limited amount of structural information on MLV PR, studies of XMRV and the proteins that it encodes are highly justified. Sequence identity between MLV and XMRV proteases is 98%, with 123 out of 125 amino acid residues being identical (Figure 1) and the differing two residues not involved in substrate binding. Multiple sequence alignment of retroviral proteases was performed previously by Eizert et al [7]. Sequence identity between HIV-1 and MLV PRs was found to be 24%, similarity was 41%, whereas residues involved in substrate binding showed 40% identity and 50% similarity [7] (Figure 1). While HIV-1 and XMRV PRs are structurally similar and their crystal structures superimpose well (Figure 2A), their mode of dimerization shows substantial differences. The dimer interface region of XMRV PR contains two hairpins which are formed by β-strands located near the C-terminal end of both monomers [8] (Figure 2B). The dimer interface of HIV-1 PR is also formed by four antiparalel β-strands, but the outer strands are located at the N-terminal ends of the monomers and the inner strands are formed by the C-terminal β-strands of the monomers [8] (Figure 2B). Based on the structure of the dimer interface region, XMRV PR shows higher similarity to the eukaryotic protein Ddi1 than to other retropepsins [8].
Figure 1. Structure-based sequence alignment of HIV-1, XMRV and MLV proteases.
Differing residues between XMRV and MLV proteases are indicated by arrows. Identical residues of the aligned sequences are indicated by asterisks and the binding site-forming residues are shown on gray background.
Figure 2. Comparison of the overall structures and dimer interfaces of HIV-1 and XMRV proteases.
(A) Superposition of overall structures of HIV-1 PR (magenta) and XMRV PR (green) using ribbon/tube representation. (B) Comparison of dimer interface regions of HIV-1 PR (left) and XMRV PR (right) using ribbon/tube representation (yellow: beta-sheet, cyan: loop, magenta: alpha-helix). The N- and C-terminal ends of the monomers are indicated.
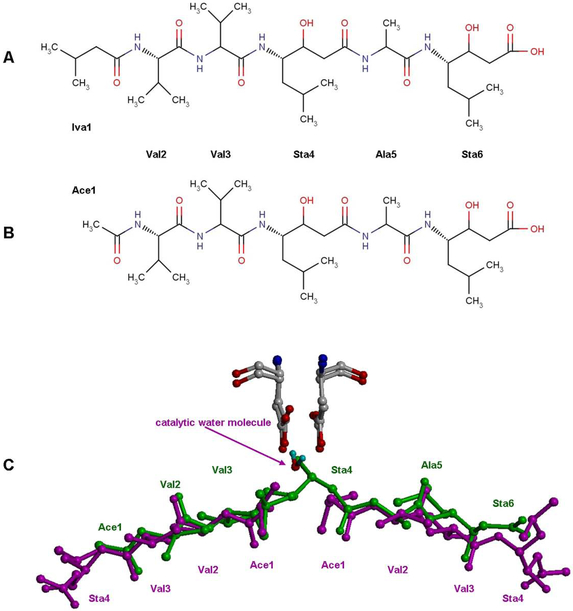
A number of known inhibitors of aspartic proteases were tested for their inhibitory properties against HIV-1 PR as soon as this enzyme was first isolated. One of the first ones was pepstatin A (Figure 3A), a general inhibitor of nearly all known aspartic proteases, which was found to be a rather weak inhibitor of the retroviral enzyme [9-11]. Another statine-containing compound, acetyl-pepstatin (Figure 3B), was subsequently described as a substantially more potent inhibitor of the HIV-1 PR, and the pH dependence of the binding was also demonstrated [12]. The crystal structure of HIV-1 PR complexed with acetyl-pepstatin [13] was published shortly after the publication of the first ever structure of HIV-1 PR with a bound inhibitor [14].
Figure 3. Chemical structures of the inhibitors and their mode of binding to retroviral proteases.
Chemical structures of (panel A) acetyl-pepstatin and (panel B) pepstatin A. Panel C: the binding mode of acetyl-pepstatin (green) to HIV-1 PR (PDB ID: 5HVP) [13] and of pepstatin A (magenta) to XMRV PR (PDB ID: 3SM1) [15].
Crystal structure of the unliganded XMRV PR was determined soon after XMRV was postulated to be a potentially harmful human retrovirus [8]. This structure was quickly followed by the crystal structures of several PR-inhibitor complexes, including a complex with pepstatin A [15]. Surprisingly, pepstatin A bound to the enzyme in a unique way that was not only very different from the way it bound to HIV-1 PR [13], but also distinct from the mode of binding of other inhibitors to XMRV PR [15]. The binding modes of the statine-containing inhibitors to XMRV and HIV-1 PRs are shown schematically in Figure 3C. The observed unique binding mode of pepstatin A to XMRV PR raised a question whether acetyl-pepstatin, a more potent inhibitor of retroviral enzymes, would bind in a typical way, or whether it would bind in the same way as pepstatin A, from which it differs only by the substitution of an isovarelyl group by an acetyl. In order to answer these questions we compared the kinetics of inhibition of HIV-1 and XMRV PRs by both pepstatin A and acetyl-pepstatin and determined the crystal structure of the complex of the latter inhibitor with XMRV PR. Another reported feature of XMRV PR which differentiated it from other known retroviral proteases was its unique dimer interface [8] which did not involve interpenetrating strands from both termini. In order to correlate the mode of dimerization with other properties of the enzyme, dimerization of XMRV PR was investigated here by kinetic methods.
Results
Inhibition of XMRV and HIV-1 proteases by pepstatin A and acetyl-pepstatin
Our previous study indicated that pepstatin A is only a very weak inhibitor of XMRV PR [15] (Table 1) and its apparent Ki value was in the range of the Km values determined for MLV PR [16]. As acetyl-pepstatin was found to be a more potent inhibitor of HIV-1 PR than pepstatin A [12], we compared the inhibition of XMRV PR by these two compounds and also determined the Ki values for HIV-1 PR (Table 1). Acetyl-pepstatin has also been described as a weak inhibitor of MLV PR, although its Ki was not determined [17]. Whereas acetyl-pepstatin was a stronger inhibitor for both PRs, the apparent Ki values were substantially lower for the HIV-1 PR than for XMRV PR. The inhibition of HIV-1 PR by both inhibitors in the nanomolar concentration range is in good agreement with their mode of action as transition state analogs. Such a mode of inhibition of HIV-1 PR was previously verified by the crystal structure with acetyl-pepstatin [13]. On the other hand, pepstatin A was earlier found to bind in a unique way to XMRV PR, with two inhibitor molecules binding to the enzyme, leaving the catalytic water in place [15]. Although acetyl-pepstatin is a better inhibitor of XMRV PR than pepstatin A, it is still a comparatively weak inhibitor and it is thus not surprising that the unusual mode of binding to the enzyme was also observed in this case (see below).
Table 1.
Inhibition and dimerization constants for XMRV and HIV-1 proteases
| Enzyme |
||
|---|---|---|
| Inhibitor | XMRV PR Ki (nM) |
HIV-1 PR Ki (nM) |
| Pepstatin A | 1442a± 123 | 22 ± 1,6 |
| Acetyl-pepstatin | 712 ±39 | 13 ±0,5 |
| Dimerization constant (K(dapp) | 115 nM | 1.0 nM |
| Urea dissociation (UC50) | 0.20 M | 1.47 M |
Ki values were determined by using an HPLC-based assay utilizing RSLLY↓PALTP and VSQNY↓PIVQ as substrates for XMRV and HIV-1 PRs, respectively, as described in the Materials and Methods.
Kdapp value for XMRV PR was determined using KARVNle↓F(NO2)EAL-NH2 substrate in 200 mM MES, 200 mM Tris 100 mM sodium acetate buffer (pH 5.0) in the presence of 2 M NaCl, using an HPLC assay.
This value was published previously [15].
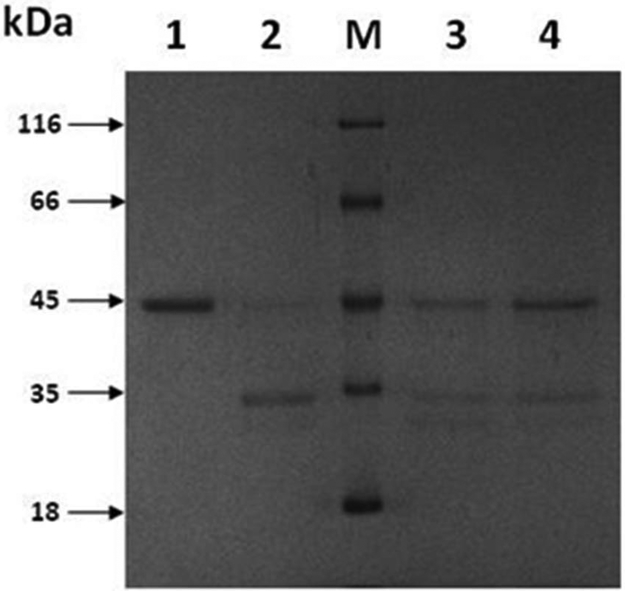
The relatively stronger inhibition of XMRV PR with acetyl-pepstatin compared to that with pepstatin A was also verified using a recombinant protein substrate. Compared to the high ionic-strength condition utilized for the HPLC assays, under this low ionic-strength condition 3 μM acetyl-pepstatin was required for 50% inhibition of protein cleavage compared to 28 μM pepstatin A (Figure 4).
Figure 4. Inhibition of the XMRV PR-mediated cleavage of a recombinant MLV Gag fragment by acetyl-pepstatin and pepstatin.
A. Cleavage of the recombinant MLV Gag fragment by XMRV protease. Recombinant MLV Gag fragment was incubated for 1 h alone (lane 1) or together with XMRV PR (30 nM) in the absence of any inhibitor (lane 2) as well as in the presence of acetyl-pepstatin (3.1 μM, lane 3) or pepstatin A (28 μM, lane 4). Reactions were stopped by the addition of loading buffer and subjected to SDS-PAGE followed by Coomassie staining. Molecular masses (kDa) of the protein markers (lane M, Fermentas, SM 0431) are indicated. Arrows indicate the uncleaved recombinant protein (Δp12-CA-NC) and its fragments.
Calculated interaction energies for the binding of acetyl-pepstatin and pepstatin A
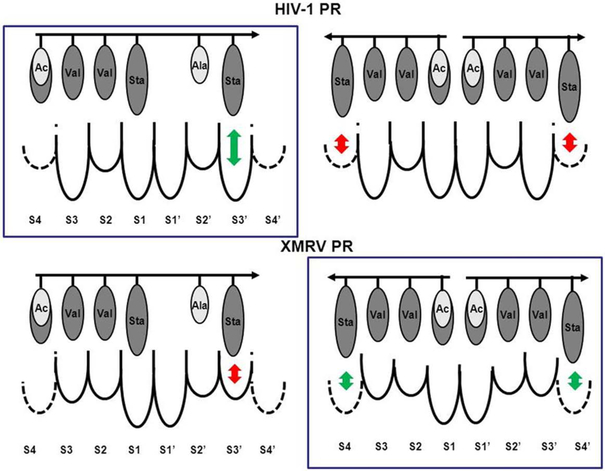
Energy calculations were performed in an attempt to evaluate the enzyme-inhibitor interactions and to explain the preferences for the binding of pepstatin A and acetyl-pepstatin to HIV-1 and XMRV PRs. Two binding modes were studied (Figure 5): in the first one a single inhibitor bound to the enzyme was modeled, as found in the HIV-1 PR/acetyl-pepstatin crystal structure [13], whereas in the second binding mode two molecules were simultaneously bound to the enzyme in a head-to-head manner, as found in the XMRV PR/pepstatin A crystal structure [15]. A single molecule of acetyl-pepstatin acts as a transition state analog in case of HIV-1 PR, where the hydroxyl group of the central statine (Sta) residue is placed between the catalytic aspartates, replacing the catalytic water molecule [13]. In the case of pepstatin A binding to XMRV PR, neither of the two inhibitor molecules bound to the enzyme could act as a transition state analog [15]. Pepstatin A and acetyl-pepstatin molecules differ only in their N-terminal residue (Figure 3). Interaction energies were calculated for both binding modes of these inhibitors to the enzymes (Table 2A). Calculated individual subsite interaction energies showed significant differences for the two inhibitors only at the subsites where the acetyl (Ace) or isovaleryl (Iva) groups were interacting (Table 2B). Our results showed that binding of a larger Iva group of pepstatin A resulted in higher interaction energy compared to the binding of a smaller Ace group of acetyl-pepstatin in all investigated binding modes (Table 2B), in good agreement with the generally hydrophobic nature of the enzyme-ligand interactions. In contrast, the enzyme kinetic studies showed a higher inhibitory potential of acetyl-pepstatin on both PRs, compared to that of pepstatin A. Therefore, the better binding of acetyl-pepstatin to the retroviral proteases compared to that of pepstatin A cannot be explained by considering only the interaction energies.
Figure 5. Schematic representation of the binding modes of pepstatin A and acetyl-pepstatin to HIV-1 and XMRV PRs.
Single inhibitor bound-states are presented on the left-hand side, while the two inhibitor bound-states are shown on the right-hand side. Preferred binding modes (based on the crystal structures) are shown in boxes. The arrows show the direction of the inhibitors from its N-terminal end to the C-terminal end, while the circles indicate the residues of the inhibitors. Shades of the residues approximate their hydrophobicity, darker higher and brighter lower. Pepstatin A and acetyl-pepstatin molecules differ only in their N-terminal residue, thus the brighter shade indicates the acetyl group at the N-terminal end of the inhibitor, while the isovaleryl group is indicated in the same N-terminal position with the darker shade. The size of the substrate binding pockets below the dotted lines complements the size of the most preferred residues [7], dashed lines for the S4 subsites indicate that these pockets are less defined than the other ones [7]. Relatively preferred side-chain-subsite interactions are indicated by green arrows and the non-preferred ones by red arrows.
Table 2.
Calculated interaction energies
| A. Calculated total interaction energies (kcal/mol) between the enzymes and the inhibitor molecules: | ||||
|---|---|---|---|---|
| single inhibitor bound |
two inhibitors bound |
|||
| Pepstatin A | Ac-pepstatin | Pepstatin A | Ac-pepstatin | |
| HIV-1 PR | −114.9 | −112.6 | −111.5 | −97.9 |
| XMRV PR | −109.4 | −105.3 | −124.3 | −105.6 |
| B. Individual interaction energies (kcal/mol) contributing to the total interaction energies of the enzyme-ligand interactions: | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| SUBSITE: | S4 | S3 | S2 | S1 | S1’ | S2’ | S3’ | S4’ | |
| One inhibitor molecule | |||||||||
| HIV-1 | Iva1 | Val2 | Val3 | Sta4 | Ala5 | Sta6 | Total | ||
| Pep | −10.85 | −14.1 | −16.32 | −26.75 | −11.43 | −35.4 | −114.85 | ||
| Ace1 | Val2 | Val3 | Sta4 | Ala | Sta6 | Total | |||
| Ac-Pep | −7.06 | −13.87 | −16.63 | −26.44 | −12.05 | −36.51 | −112.56 | ||
| XMRV | Iva1 | Val2 | Val3 | Sta4 | Ala5 | Sta6 | Total | ||
| Pep | −10.9 | −15.41 | −15.14 | −26.56 | −11.1 | −30.2 | −109.35 | ||
| Ace1 | Val2 | Val3 | Sta4 | Ala5 | Sta6 | Total | |||
| Ac-Pep | −6.17 | −15.29 | −15.11 | −27.46 | −10.96 | −30.31 | −105.30 | ||
| Two inhibitor molecules | |||||||||
| HIV-1 | Sta4 | Val3 | Val2 | Iva1 | Iva1 | Val2 | Val3 | Sta4 | Total |
| Pep | −10.48 | −11.32 | −16.23 | −16.51 | −16.42 | −17.1 | −11.13 | −12.26 | −111.54 |
| Sta4 | Val3 | Val2 | Ace1 | Ace1 | Val2 | Val3 | Sta4 | Total | |
| Ac-Pep | −11.61 | −11.72 | −16.85 | −8.04 | −9.01 | −17.04 | −11.27 | −12.39 | −97.93 |
| XMRV | Sta4 | Val3 | Val2 | Iva1 | Iva1 | Val2 | Val3 | Sta4 | Total |
| Pep | −14.75 | −13.89 | −15.76 | −17.39 | −17.69 | −16.19 | −13.41 | −15.26 | −124.34 |
| Sta4 | Val3 | Val2 | Ace1 | Ace1 | Val2 | Val3 | Sta4 | Total | |
| Ac-Pep | −15.36 | −13.74 | −16.09 | −7.08 | −8.39 | −16.7 | −13.12 | −15.16 | −105.64 |
In contrast, simulation of binding of two pepstatin A molecules to XMRV PR resulted in substantially higher enzyme-inhibitor interaction energy compared to that of one molecule, whereas the preference for single inhibitor binding was observed for HIV-1 PR (Table 2A). A preference for two-inhibitor binding of XMRV PR was seen only in the case of pepstatin A, whereas simulation of binding of acetyl-pepstatin resulted in approximately the same interaction energies in the different binding modes (Table 2A). Individual inspection of the subsite residue interactions together with previous comparative specificity studies on HIV-1 and MLV PRs provided a rationale for the different preference of the two enzymes. It should be noted that, based on the high sequence identity of XMRV and MLV PRs and the identical substrate binding subsite compositions, the specificity of the two enzymes is expected to be identical. The subsite preferences were determined earlier for MLV PR in comparison with the HIV-1 (and other) PRs [7, 18, 19]. The mainly hydrophobic character of the substrate binding sites is preserved in both MLV (XMRV) and HIV-1 PRs. However, differences in substrate specificity were described, and are expected to be the consequence of different sub site-forming residues [8]. The SI subsite of both MLV and HIV-1 PRs is large, and showed a preference for aromatic residues [7]; therefore, together with SI’, it does not contribute to the differential binding mode of the inhibitors: a given residue at P1 or P1’ (Sta, Iva or Ace) provided similar interaction energies for both enzymes. However, HIV-1 PR showed a preference for small hydrophobic or even polar residues at the S2 subsite, whereas MLV preferred to bind large hydrophobic residues [19], in good agreement with its larger subsite [7]. In both inhibitor binding modes a Val residue occupied the S2 subsite, whereas either Ala or Val residue occupied subsite S2’, providing similar interaction energies for both enzymes (Table 2b). Unlike the S2 (and S2’ subsites), the S3/S3’ binding sites of MLV PR are smaller than the respective ones in HIV-1 PR and the average volume of the preferred P3 residues was also found to be larger for HIV-1 PR compared to that for MLV PR [7]. In both binding modes the S3 subsite is occupied by a Val residue, whereas the S3’ subsite is occupied either by a Val (two-inhibitor binding mode) or by a bulkier Sta residue. In good agreement with the pocket sizes, the Sta6-S3’ interaction is much more favorable in the case of the HIV-1 PR as compared to that of XMRV PR, favoring the single binding mode (Table 2). The S4/S4’ interactions also appear to substantially contribute to the different preferred binding mode of the inhibitors to the two proteases. The large hydrophobic S4/S4’ sites of MLV prefer to bind hydrophobic bulky residues [19] and the mean cavity volume of S4/S4' sites of MLV PR is substantially larger compared to those of HIV-1 PR [7] which allows for Sta4 residues to fill the S4/S4' sites in the case of simultaneous binding of pepstatin A or acetyl-pepstatin molecules. Therefore, the binding of the bulky Sta4 residues at the S4/S4' sites is relatively less favored for HIV-1 PR, with the difference confirmed by the lower calculated interaction energies at S4/S4' subsites compared to those with XMRV PR (Table 2). It is of interest to note that, whereas the two inhibitor-binding mode for XMRV PR-pepstatin A is clearly favored energetically, for XMRV PR-acetyl-pepstatin interaction simulation of the two binding modes provided very similar interaction energies, due to the substantially lower S1-P1 and S1’-P1’ interactions provided by the acetyl groups, compared to the isovaleryl ones of pepstatin A.
Crystal structure of the complex of XMRV PR and acetyl-pepstatin
Crystals of the complex of XMRV PR with acetyl-pepstatin are isomorphous with the previously described crystals of three other inhibitor complexes of this enzyme [15]. The crystals belong to the orthorhombic space group P212121 and contain a protein dimer in the asymmetric unit. The statistics of data collection and refinement at 2.0 Å resolution are listed in Table 3. The conformation of the protein is almost identical in the complexes of XMRV PR with acetyl-pepstatin and with pepstatin A, with the root-mean-square differences between all main chain coordinates of molecules A and B of 0.58 and 0.29 Å, respectively.
Table 3.
Data collection and structure refinement for a complex of XMRV PR with acetyl-pepstatin
| Data collection | |
| Space group | P212121 |
| Molecules/a.u. | |
| Unit cell a,=b,c {Å) | 46. 3, 65.2, 69.5 |
| Resolution (Å)* | 50.0-2.0 (2.03-2.0) |
| Rmerge† | 7.9 (28.4) |
| No. of reflections (measured/unique) | 77238/14051 |
| <I / σI> | 20.1 (3.3) |
| Completeness (%) | 93.0 (64.4) |
| Redundancy | 5.5 (2.1) |
| Refinement | |
| Resolution (Å) | 47.5-2.0 |
| No. of reflections (refinement/ Rfree) | 13292/719 |
| R / Rfree‡ | 0.196/0.238 |
| No. atoms | |
| Protein | 1723 |
| Ligand/ion | 113 |
| Water | 47 |
| B factors (Å2) | |
| Protein | 38.8 |
| Inhibitors | 27.1 |
| Water | 42.6 |
| R.m.s. deviations from ideality | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.3 |
| PDB code | 4EXH |
The highest resolution shell is shown in parentheses.
Rmerge = ΣhΣi∣Ii-⟨I⟩∣/ΣhΣiIi, where Ii is the observed intensity of the i-th measurement of reflection h, and ⟨I⟩ is the average intensity of that reflection obtained from multiple observations.
R = Σ∥Fo∣-∣Fc∥/Σ∣Fo∣, where Fo and Fc are the observed and calculated structure factors, respectively, calculated for all data. Rfree was defined in Brünger [31].
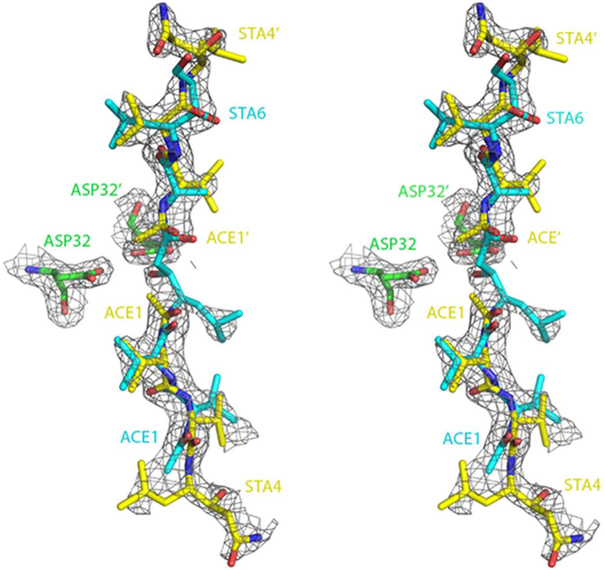
Modeling of the mode of binding of acetyl-pepstatin to XMRV PR was not straightforward. The presence of only very weak electron density near the catalytic aspartates and the length of the electron densities attributable to the inhibitors suggested that the mode of binding should be similar to that of pepstatin A, namely with two molecules binding head-to-head, with the outermost parts disordered. However, when this model was refined it resulted in significant residual electron density that could be explained by the “standard” mode of binding that would involve only a single molecule. The model that appears to agree best with the final electron density involves two molecules bound as in pepstatin A, with half occupancy each, overlaid by a third molecule in a “standard” conformation, also half occupied (Figure 5). The molecule that spans the active site appears to have a preferred orientation, although some small remaining density suggests a possibility of its slight disorder. The rms deviation of all the common atoms of pepstatin A and acetyl-pepstatin that are modeled in the same orientation is 0.21 Å for inhibitor J and 0.35 Å for those in inhibitor M. These differences are comparable to the errors in the atomic positions expected in the structures at comparable resolution.
Dimerization of XMRV and HIV-1 PRs
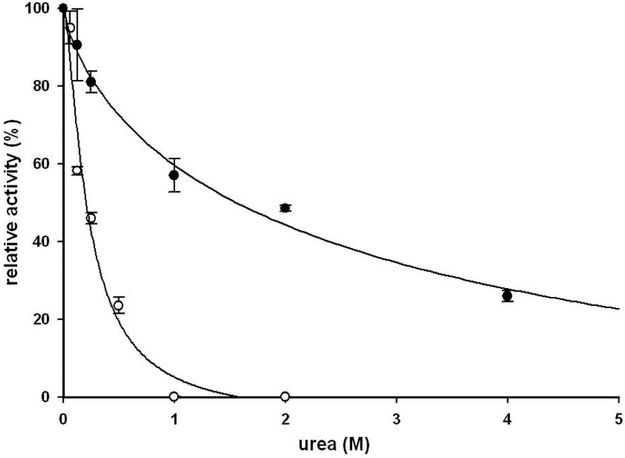
The apparent Kd value for the XMRV PR was found to be 115 nM (Table 1), whereas that of HIV-1 PR determined using the same substrate and the HPLC method was found to be dramatically lower at 1.0 nM. This value for HIV-1 PR is in good agreement with the photometric assay value, where it was found to be below the detection limit of the assay (< 5 nM [20] or <0.37 nM [21], with different substrates and detection methods). It is of interest to note that another PR having extended terminal regions, such as HTLV-1 PR, also showed substantially higher Kd (491 nM) [16]. Stability of the dimer was also assessed by measuring urea denaturation, in a manner previously applied for several HIV-1 PR mutants [22]. While the UD50 value for HIV-1 PR is similar to the value previously determined using photometric [22] or HPLC [21] detection, XMRV PR appeared to be substantially more sensitive to urea concentrations, in good agreement with its higher Kd value (Table 1, Figure 7).
Figure 7. Urea dissociation curve of XMRV and HIV-1 PRs.
The activity of the HIV-1 PR (solid circles) and XMRV PR (open circles) were measured at increasing urea concentration, using an HPLC detection of the substrate cleavage as described in the Materials and Methods.
Discussion
Acetyl-pepstatin was found to be a stronger inhibitor than pepstatin A for both HIV-1 and XMRV PRs, but, in general, the apparent Ki values were substantially lower for the HIV-1 PR than for XMRV PR. Although acetyl-pepstatin is a better inhibitor of XMRV PR than pepstatin A, it is still a comparatively weak inhibitor, as also indicated by the mixed mode of its binding. The relatively stronger inhibition of XMRV PR with acetyl-pepstatin compared to that with pepstatin A was also verified using a recombinant protein substrate. Under the low ionic-strength conditions 3 μM acetyl-pepstatin was required for 50% inhibition of protein cleavage compared to 28 μM pepstatin A.
In spite of the substantially higher calculated interaction energy of the real two inhibitor bound acetyl-pepstatin/XMRV PR complex (−124.3 kcal/mol) compared to that of single inhibitor-bound HIV-1 PR (−112.6 kcal/mol), the inhibitor is substantially more potent against HIV-1 PR (Table 1). The energy calculation can account for only the enzyme-ligand interactions, while the transition state binding mode of HIV-1 PR complex utilizes the energy of the transition state.
The relatively unfavorable Sta6-S3’ interactions between the statines and XMRV PR and the favorable Sta4-S4 and Sta4-S4’ interactions may shift the ground state binding towards the two-inhibitor-binding mode, whereas the single molecule ground state binding of statines to the HIV-1 PR appears to be more favorable. The preferred binding mode of a single inhibitor molecule to HIV-1 PR allows formation of the transition state complex, represented by the substantially better binding constants. The relatively small S4 binding site of HIV-1 PR has already been suggested as the reason for better binding of acetyl-pepstatin as compared to pepstatin A [13].
The mode of binding of two inhibitor molecules resembles the previously reported case of binding of the inhibitor SB203386 to SIV PR [23]. Binding modes are quite similar in the case of SIV and XMRV PRs, with the catalytic water molecules still present between the catalytic aspartates. However, in the case of SIV PR the binding stoichiometry was found to be one inhibitor molecule/protease dimer using titration calorimetry, and significantly higher B factors were obtained for the inhibitors compared to the protein. In the case of pepstatin A and acetyl-pepstatin binding to XMRV PR the inhibitor B factors are comparable (or lower) than for the protein (Table 3), indirectly suggesting the presence of more than one molecule bound in this mode to each dimer of the enzyme.
The stability of the dimer was also assessed by measuring urea denaturation, as well as by measuring the Kd value for dimer stability. Both the UD50 and the Kd values for XMRV PR suggested a substantially weaker dimer stability as compared to that of HIV-1 protease. These results, together with previous findings, suggest unique characteristics for the HIV-1 (and other primate lentiviral) PRs: as a consequence of rapid, massive evolution due to the high number of progeny virus production and replication through RNA intermediate these viral proteases evolved to acquire not only unique specificity among the retroviral proteases [7] but also exceptionally high dimer stability.
Materials and Methods
Determination of the inhibition constants
XMRV PR used in kinetic studies and crystal structure determination was expressed and purified following the previously described procedures [8, 24]. Recombinant XMRV PR engineered with an N-terminal, non-cleavable 6-His purification tag was expressed in E. coli and purified on a nickel affinity column. The resulting polypeptide consisted of 132 amino acids (initial Met, His6, and the complete 125 residue-long PR). Purified HIV-1 PR, containing stabilizing mutations (Q7K, L33I, L63I, C67A and C95A) was prepared as described previously [24]. PR stock solutions were diluted with 20 mM Pipes, pH 7.0, containing 100 mM NaCl, 10 % glycerol and 0.5% NP40. Activity of XMRV PR was measured with an HPLC-based assay as described previously [15]. For the determination of Ki values, the PR assays were initiated by mixing 5 μl of PR, 10 μl buffer (0.5 M potassium phosphate, pH 5.6, containing 10% glycerol, 10 mM DTT, 4 M NaCl), 4.8 μl 0.17 mM (final concentration) RSLLY↓PALTP (XMRV PR) or 0.27 mM (final concentration) VSQNY↓PIVQ (HIV-1 PR) and 0.2 μl inhibitor in DMSO or DMSO alone. The reaction mixture was incubated at 37 °C for 1 hour and terminated by the addition of 180 μl 1% trifluoroacetic acid (TFA). Enzyme concentration in the assay was selected to cause less than 20% substrate hydrolysis. Separation of cleavage products with reversed-phase chromatography was performed as described previously [15]. The Ki values were obtained from the IC50 values determined from the inhibitor dose-response curves using the equation Ki = (IC50 - [E]/2) / (1 + [S]/Km), where [E] and [S] are the protease and substrate concentrations, respectively. The exact amount of active PRs in the preparations used for kinetic measurements was determined by active center titration with amprenavir, using the HPLC method. Kinetic parameters were determined by fitting the data to the Michaelis-Menten equation using Enzyme Kinetics Module 1.1 of SigmaPlot 8.0 (Systat Software Inc).
Determination of the apparent Kd value and the urea dissociation constant (UD50) of XMRV and HIV-1 PRs
The apparent Kd values for the dimers were studied essentially as described previously [25]. Specific activity values were measured in duplicate as a function of dimeric enzyme concentration in 100 mM sodium-acetate buffer pH 5.0, containing 100 mM MES, 200 mM Tris, 2M NaCl, using KARVNle↓F(NO2)EAL-NH2 chromogenic substrate (0.3 mM final concentration). Samples were incubated for 20 minute at 37 °C and subjected to HPLC analysis as described above. The dimerization constant (Kdapp) value was obtained by plotting the relative specific activity values against the PR concentrations and fitting a curve using “Hyperbola single rectangular 2 parameters equation” setting of the SigmaPlot 8.0 software. To assay the urea denaturation, PR activity was measured with increasing concentration of urea (0–4.0 M) at a final concentration of 34 nM XMRV or 5.5 nM HIV-1 PR, and 0.3 mM substrate concentration. The UC50 values for half-maximal velocity were obtained by plotting the relative specific activity values against the urea concentrations and fitting a curve using “Hill 3 parameters equation” setting of the SigmaPlot 8.0 software (Figure 7).
Cleavage of recombinant MLV Gag fragment by XMRV PR
Recombinant MLV Gag fragment (0.9 μM MLVGagΔ2) [26] was incubated in 75 mM phosphate buffer, pH 5.6, 0.5 mM EDTA, for 1 h at 37 °C without XMRV PR, or with XMRV PR (30 nM) in the absence and presence of acetyl-pepstatin (3.0 μM) or pepstatin A (28.8 μM). Reactions were stopped by the addition of loading buffer and subjected to SDS-PAGE, followed by staining with Coomassie Brillant Blue. Protein ladder (Fermentas, SM 0431) was used to determine the molecular mass of protein fragments.
Molecular modeling
Structure-based multiple sequence alignment of HIV-1, MLV and XMRV PR sequences was performed as described previously [18]. Crystal structures of HIV-1 PR complexed with acetyl-pepstatin (PDB ID: 5HVP) [13] and XMRV PR complexed with pepstatin A (PDB ID: 3SM1) [15] were used for building up the 3D structures of HIV-1 and XMRV PRs complexed with one or two pepstatin A and acetyl-pepstatin inhibitor molecules, using the SYBYL program package (Tripos Inc., St. Louis, MO, USA). Initial structures of enzyme–inhibitor complexes were generated by merging the inhibitor molecule(s) into the active site. Water molecules present in the crystal structures were involved in the calculations, only water molecules bumping into the modeled inhibitors were removed. In case of binding of a single inhibitor molecule the catalytic water molecule was removed and replaced by the hydroxyl group of the central statine residue. The catalytic water was left in place in case of binding of two inhibitors molecules. Inhibitor positions were refined by a short minimization procedure using SYBYL (500 Powell iterations, dielectric constant 4, AMBER7_FF99 force field). The enzyme–inhibitor complexes were further energy minimized without any fixed atoms by SYBYL (AMBER7_FF99 force field, non-bonded cutoff 8 Å, 500 Powell iterations, and dielectric constant 4). Interaction energies between the enzyme and the residues of inhibitor molecules were calculated for each substrate binding site. Calculations and molecular visualizations were performed on Silicon Graphics Fuel workstations (Silicon Graphics International, Fremont, CA, USA).
Determination of the crystal structure of XMRV PR complexed with acetyl-pepstatin
Before addition of the inhibitor for crystallization, the XMRV PR sample buffer was exchanged to 20 mM Na-citrate, pH 5.5, also containing 0.2 M NaCl, and the protein was concentrated to 6 mg/ml. Acetyl-pepstatin was added at the 4:1 PR (monomer) to inhibitor molar ratio. Crystallization was carried out using the hanging drop, vapor diffusion method. Each drop contained 4 μl of the complex sample mixed with 2 μl of well solution and was equilibrated with 500 μl of the latter. The well solution contained 3.5 M Na-formate, pH 5.5. The crystals grew slowly, taking over a month to reach the size of 0.05×0.05×0.2 mm. Diffraction data extending to 2.0 Å resolution were collected on the SER-CAT beamline 22-ID at the APS using a MAR300CCD detector. The crystal was cryoprotected before rapid freezing and diffraction intensities were measured at 100 K in a single pass at 2 seconds/degree. Data were indexed, integrated, and scaled with the HKL2000 package [27]. The structure was solved by molecular replacement with PHASER [28] using a monomer of XMRV PR (3NR6) as a search model. The structure was refined with REFMAC5 [29] and rebuilt with COOT [30]. The final coordinates have been submitted to the Protein Data Bank (PDB ID: 4EXH).
Figure 6. Catalytic residues of XMRV PR and bound acetyl-pepstatin.
The 2Fo-Fc electron density map was contoured at 1.0 σ. Partially occupied molecules are shown in cyan for the “standard” orientation, and in yellow for the orientation resembling the mode of binding of pepstatin A. The figure was prepared with PyMol [32].
Acknowledgements
We acknowledge the use of beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT), located at the Advanced Photon Source, Argonne National Laboratory. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No.W-31-109-Eng-38. The work was supported in part by the TÁMOP 4.2.1./B-09/1/KONV-2010-0007 project and by the Hungarian Science and Research Fund (OTKA K68288, OTKA 101591). This work was also supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U. S. Government.
Abbreviations:
- MLV
murine leukemia virus
- PR
protease
- XMRV
xenotropic murine leukemia virus-related virus
Footnotes
Authors declared no conflict of interest.
Database section: The final coordinates of the crystal structure of XMRV protease complexed with acetyl-pepstatin are available in the Protein Data Bank (PDB) under the accession number: 4EXH.
References:
- 1.Kohl NE, Emini EA, Schleif WA, Davis LJ, Heimbach JC, Dixon RA, Scolnick EM & Sigal IS (1988) Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 85, 4686–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wlodawer A & Vondrasek J (1998) Inhibitors of HIV-1 protease: A major success of structure-assisted drug design. Annu. Rev. Biophys. Biomol. Struct 27, 249–284. [DOI] [PubMed] [Google Scholar]
- 3.Scourfield A, Waters L, Nelson M (2011) Drug combinations for HIV: what's new? Expert Rev Anti Infect Ther 9: 1001–1011. [DOI] [PubMed] [Google Scholar]
- 4.Poiesz BJ, Ruscetti FW, Reitz MS, Kalyanaraman VS & Gallo RC (1981) Isolation of a new type C retrovirus (HTLV) in primary uncultured cells of a patient with Sezary T-cell leukaemia. Nature 294, 268–271. [DOI] [PubMed] [Google Scholar]
- 5.Groom HC & Bishop KN (2012) The tale of xenotropic murine leukemia virus-related virus. J. Gen. Virol 93, 915–924. [DOI] [PubMed] [Google Scholar]
- 6.Paprotka T, Delviks-Frankenberry KA, Cingöz O, Martinez A, Kung HJ, Tepper CG, Hu WS, Fivash MJ Jr., Coffin JM & Pathak VK (2011) Recombinant origin of the retrovirus XMRV. Science 333, 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eizert H, Bander P, Bagossi P, Sperka T, Miklóssy G, Boross P, Weber IT & Tőzsér J (2008) Amino acid preferences of retroviral proteases for amino-terminal positions in a type 1 cleavage site. J. Virol 82, 10111–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Dimaio F, Zhou D, Gustchina A, Lubkowski J, Dauter Z, Baker D & Wlodawer A (2011a) Crystal structure of XMRV protease differs from the structures of other retropepsins. Nat. Struct. Mol. Biol 18, 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seelmeier S, Schmidt H, Turk V & von der Helm K (1988) Human immunodeficiency virus has an aspartic-type protease that can be inhibited by pepstatin A. Proc. Natl. Acad. Sci. USA 85, 6612–6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giam CZ & Boros I (1988) In vivo and in vitro autoprocessing of human immunodeficiency virus protease expressed in Escherichia coli. J. Biol. Chem 263, 14617–14620. [PubMed] [Google Scholar]
- 11.Hansen J, Billich S, Schulze T, Sukrow S & Moelling K (1988) Partial purification and substrate analysis of bacterially expressed HIV protease by means of monoclonal antibody. EMBO J 7, 1785–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards AD, Roberts R, Dunn BM, Graves MC & Kay J (1989) Effective blocking of HIV-1 proteinase activity by characteristic inhibitors of aspartic proteinases. FEBS Lett 247, 113–117. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald PM, McKeever BM, Van Middlesworth JF, Springer JP, Heimbach JC, Leu CT, Herber WK, Dixon RA & Darke PL (1990) Crystallographic analysis of a complex between human immunodeficiency virus type 1 protease and acetyl-pepstatin at 2.0-A resolution. J. Biol. Chem 265, 14209–14219. [PubMed] [Google Scholar]
- 14.Miller M, Schneider J, Sathyanarayana BK, Toth MV, Marshall GR, Clawson L, Selk L, Kent SB & Wlodawer A (1989) Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution. Science 246, 1149–1152. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Gustchina A, Matúz K, Tőzsér J, Namwong S, Goldfarb NE, Dunn BM & Wlodawer A (2011b) Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease. FEBS J 278, 4413–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kádas J, Boross P, Weber IT, Bagossi P, Matúz K & Tőzsér J (2008) C-terminal residues of mature human T-lymphotropic virus type 1 protease are critical for efficient dimerization and for catalytic activity. Biochem. J 416, 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menendez-Arias L, Gotte D & Oroszlan S (1993) Moloney murine leukemia virus protease: bacterial expression and characterization of the purified enzyme. Virology, 196, 557–563. [DOI] [PubMed] [Google Scholar]
- 18.Bagossi P, Sperka T, Fehér A, Kádas J, Zahuczky G, Miklóssy G, Boross P & Tőzsér J (2005) Amino acid preferences for a critical substrate binding subsite of retroviral proteases in type 1 cleavage sites. J. Virol 79, 4213–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menéndez-Arias L, Weber IT, Soss J, Harrison RW, Gotte D & Oroszlan S (1994) Kinetic and modeling studies of subsites S4-S3' of Moloney murine leukemia virus protease. J. Biol. Chem 269, 16795–16801. [PubMed] [Google Scholar]
- 20.Wondrak EM & Louis JM (1996) Influence of flanking sequences on the dimer stability of human immunodeficiency virus type 1 protease. Biochemistry 35, 12957–12962. [DOI] [PubMed] [Google Scholar]
- 21.Olivares I, Mulky A, Boross PI, Tőzsér J, Kappes JC, López-Galíndez C, Menéndez-Arias LJ (2007) HIV-1 protease dimer interface mutations that compensate for viral reverse transcriptase instability in infectious virions. J Mol Biol 372: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu F, Boross PI, Wang YF, Tőzsér J, Louis JM, Harrison RW & Weber IT (2005) Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S. J. Mol. Biol 354, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoog SS, Towler EM, Zhao B, Doyle ML, Debouck C, Abdel-Meguid SS (1996) Human immunodeficiency virus protease ligand specificity conferred by residues outside of the active site cavity. Biochemistry 35:10279–86. [DOI] [PubMed] [Google Scholar]
- 24.Gillette WK, Esposito D, Taylor TE, Hopkins RF, Bagni RK & Hartley JL (2010) Purify First: Rapid expression and purification of proteins from XMRV. Protein Expr. Purif 76, 238–247. [DOI] [PubMed] [Google Scholar]
- 25.Mahalingam B, Louis JM, Hung J, Harrison RW & Weber IT (2001) Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins 43, 455–464. [DOI] [PubMed] [Google Scholar]
- 26.Fehér A, Boross P, Sperka T, Miklóssy G, Kadas J, Bagossi P, Oroszlan S, Weber IT & Tőzsér J (2006) Characterization of the murine leukemia virus protease and its comparison with the human immunodeficiency virus type 1 protease. J. Gen. Virol 87, 1321–1330. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z & Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC & Read RJ (2007) Phaser crystallographic software. J. Appl. Cryst 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F & Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D67, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P & Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 31.Brünger AT (1992) The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–474. [DOI] [PubMed] [Google Scholar]
- 32.DeLano WL (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA. [Google Scholar]