Abstract
The asymmetric unit of the title compound, C16H16N4O2S, contains two molecules (A and B) with similar conformations: the benzene rings are oriented at dihedral angles of 80.94 (10)° and 84.54 (10)° with their adjacent 1H-benzimidazole groups. In the crystal, the molecules are connected by N—H⋯N hydrogen bonds, forming separate C(4) chains of both the A and B molecules along [010]. The A and B chains are cross-linked by several C—H⋯O interactions involving the benzene rings and the sulfonyl groups, which lead to R 2 1(5) loops in the linkage of the chains.
Related literature
For a related structure, see: Esparza-Ruiz et al. (2010 ▶).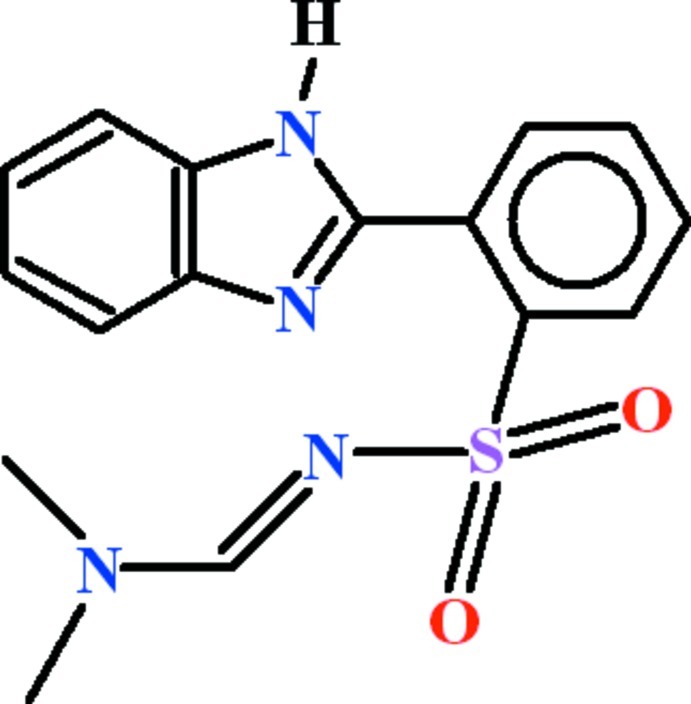
Experimental
Crystal data
C16H16N4O2S
M r = 328.39
Monoclinic,

a = 15.630 (5) Å
b = 10.003 (4) Å
c = 22.122 (5) Å
β = 110.657 (5)°
V = 3236.3 (18) Å3
Z = 8
Mo Kα radiation
μ = 0.22 mm−1
T = 296 K
0.28 × 0.20 × 0.18 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.930, T max = 0.952
26702 measured reflections
6347 independent reflections
3328 reflections with I > 2σ(I)
R int = 0.070
Refinement
R[F 2 > 2σ(F 2)] = 0.059
wR(F 2) = 0.159
S = 1.01
6347 reflections
419 parameters
H-atom parameters constrained
Δρmax = 0.34 e Å−3
Δρmin = −0.32 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and PLATON (Spek, 2009 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PLATON.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812025159/hb6832sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812025159/hb6832Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812025159/hb6832Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯N2i | 0.86 | 2.11 | 2.964 (3) | 174 |
| N5—H5A⋯N6ii | 0.86 | 2.10 | 2.955 (3) | 178 |
| C9—H9⋯O4i | 0.93 | 2.56 | 3.153 (4) | 122 |
| C10—H10⋯O4i | 0.93 | 2.57 | 3.153 (4) | 121 |
| C25—H25⋯O2ii | 0.93 | 2.47 | 3.114 (5) | 127 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
The authors acknowledge the provision of funds for the purchase of a diffractometer and encouragement by Dr Muhammad Akram Chaudhary, Vice Chancellor, University of Sargodha, Pakistan. The authors also acknowledge the technical support provided by Syed Muhammad Hussain Rizvi of Bana International, Karachi, Pakistan.
supplementary crystallographic information
Comment
The title compound (I), (Fig. 1) has been prepared in an attempt to attach benzenesulfonyl chloride with 2-(1H-benzimidazol-2-yl) benzenesulfonamide (Crystal structure has been determined) in the dimethylformamide.
The crystal structures of 2-(1H-benzimidazol-3-ium-2-yl)benzenesulfonate dimethylsulfoxide solvate (Esparza-Ruiz et al., 2010) has been published which is related to (I) upto some extent.
In (I), two molecules (M1 and M2) in the asymmetric unit are present, which differ slightly from each other geometrically. In molecule M1, the group A (C1—C7/N1/N2) of 1H-benzimidazole, benzene ring B (C8—C13) and group C (N3/C14/N4/C15/C16) of N,N-dimethylimidoformamide moiety are planar with r. m. s. deviation of 0.0108 Å, 0.0046 Å and 0.0093 Å, respectively. The dihedral angle between A/B, A/C and B/C is 80.94 (10)°, 12.34 (4)° and 83.76 (18)°, respectively. The sulfonyl group D (O1/S1/O2) is of course planar. The dihedral angle between B/D and C/D is 70.86 (14)° and 53.88 (13)°, respectively. In second molecule M2, the similar groups E (C17—C23/N5/N6), F (C24—C29) and G (N7/C30/N8/C31/C32) are also planar with r. m. s. deviation of 0.0160 Å, 0.0054 Å and 0.0122 Å, respectively. The dihedral angle between E/F, E/G and F/G is 84.54 (10)°, 12.68 (8)° and 83.22 (20)°, respectively. In M2, dihedral angle between F/H and G/H is 69.47 (14)° and 54.53 (13)°, respectively where H (O3/S2/O4) is the sulfonyl group. Both molecules are interlinked with themselves with C (4) chains due to classical H–bonding of N—H···N type (Table 1, Fig. 2). These infinte one-dimensional chains exist along [010]. The polymeric chains are interlinked with each other through benzene ring and the sulfonyl groups due to H–bonding of C—H···O type in a different manner. There exist R21(5) ring motif in the linkage of polymeric chains.
Experimental
The 2-[o-(sulfamoyl)phenyl]benzimidazole (0.1 g, 0.37 mmol) in dimethylformamide (2 ml) was disolved to get a clear solution. Benzenesulfonyl chloride (0.065 g, 0.37 mmol) was added with catalytic amount of potassium carbonate to this solution and subjected to reflux for 2 h. The resulting solution was quenched in ice-cold distilled water (100 ml). Extracted the aqueous layer with ethyl acetate (3 × 25 ml) and dried the organic layer over anhydrous sodium sulfate to get light brown powder (0.11 g, 90.5%). The powder was recrystallized in methanol to get the light brown prisms of (I).
m.p. 593–594 K.
Refinement
The H-atoms were positioned geometrically at C—H = 0.93—0.96, nd N—H = 0.86 Å, respectively and included in the refinement as riding with Uiso(H) = xUeq(C, N), where x = 1.5 for metyl H-atoms and x = 1.2 for all other H-atoms.
Figures
Fig. 1.
View of the title compound with displecement ellipsoids drawn at the 50% probability level.
Fig. 2.
The partial packing (PLATON; Spek, 2009) which shows that molecules form C(4) chains and are interlinked. The H-atoms not involved in H-bondings are omitted for clarity.
Crystal data
| C16H16N4O2S | F(000) = 1376 |
| Mr = 328.39 | Dx = 1.348 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 3328 reflections |
| a = 15.630 (5) Å | θ = 2.0–26.0° |
| b = 10.003 (4) Å | µ = 0.22 mm−1 |
| c = 22.122 (5) Å | T = 296 K |
| β = 110.657 (5)° | Prism, light brown |
| V = 3236.3 (18) Å3 | 0.28 × 0.20 × 0.18 mm |
| Z = 8 |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 6347 independent reflections |
| Radiation source: fine-focus sealed tube | 3328 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.070 |
| Detector resolution: 8.00 pixels mm-1 | θmax = 26.0°, θmin = 2.0° |
| ω scans | h = −19→19 |
| Absorption correction: multi-scan (SADABS; Bruker, 2005) | k = −12→12 |
| Tmin = 0.930, Tmax = 0.952 | l = −27→22 |
| 26702 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.059 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.159 | H-atom parameters constrained |
| S = 1.01 | w = 1/[σ2(Fo2) + (0.0638P)2 + 0.4292P] where P = (Fo2 + 2Fc2)/3 |
| 6347 reflections | (Δ/σ)max < 0.001 |
| 419 parameters | Δρmax = 0.34 e Å−3 |
| 0 restraints | Δρmin = −0.32 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.37734 (6) | 0.20738 (11) | 0.16157 (4) | 0.0540 (4) | |
| O1 | 0.38444 (16) | 0.3454 (2) | 0.18274 (13) | 0.0766 (11) | |
| O2 | 0.41107 (17) | 0.1781 (4) | 0.11124 (13) | 0.1060 (16) | |
| N1 | 0.25436 (18) | 0.0481 (2) | 0.27589 (11) | 0.0378 (9) | |
| N2 | 0.25592 (18) | 0.2709 (2) | 0.27357 (12) | 0.0369 (9) | |
| N3 | 0.42077 (18) | 0.1030 (3) | 0.21989 (13) | 0.0507 (10) | |
| N4 | 0.49756 (19) | 0.0855 (3) | 0.32963 (14) | 0.0548 (11) | |
| C1 | 0.2819 (2) | 0.2276 (3) | 0.33732 (15) | 0.0362 (11) | |
| C2 | 0.3058 (3) | 0.2996 (3) | 0.39450 (15) | 0.0529 (14) | |
| C3 | 0.3299 (3) | 0.2312 (4) | 0.45146 (16) | 0.0563 (14) | |
| C4 | 0.3314 (3) | 0.0922 (3) | 0.45294 (15) | 0.0541 (16) | |
| C5 | 0.3069 (3) | 0.0181 (3) | 0.39697 (15) | 0.0510 (14) | |
| C6 | 0.2813 (2) | 0.0883 (3) | 0.33941 (14) | 0.0341 (11) | |
| C7 | 0.24058 (19) | 0.1606 (3) | 0.23952 (14) | 0.0293 (10) | |
| C8 | 0.20535 (19) | 0.1538 (3) | 0.16793 (13) | 0.0278 (9) | |
| C9 | 0.1123 (2) | 0.1313 (3) | 0.13747 (15) | 0.0398 (11) | |
| C10 | 0.0741 (2) | 0.1237 (3) | 0.07082 (16) | 0.0479 (11) | |
| C11 | 0.1275 (2) | 0.1359 (3) | 0.03388 (15) | 0.0429 (11) | |
| C12 | 0.2199 (2) | 0.1569 (3) | 0.06293 (15) | 0.0388 (11) | |
| C13 | 0.25898 (19) | 0.1670 (3) | 0.12983 (14) | 0.0304 (10) | |
| C14 | 0.4588 (2) | 0.1557 (3) | 0.27730 (16) | 0.0472 (11) | |
| C15 | 0.4997 (3) | −0.0594 (4) | 0.3281 (2) | 0.1017 (19) | |
| C16 | 0.5413 (3) | 0.1492 (4) | 0.39179 (17) | 0.0774 (16) | |
| S2 | 0.62238 (5) | 0.50313 (10) | 0.33544 (4) | 0.0466 (3) | |
| O3 | 0.62214 (15) | 0.3695 (2) | 0.31038 (12) | 0.0628 (9) | |
| O4 | 0.58453 (16) | 0.5169 (3) | 0.38516 (12) | 0.0784 (13) | |
| N5 | 0.73966 (18) | 0.6823 (2) | 0.22561 (11) | 0.0399 (9) | |
| N6 | 0.75783 (18) | 0.4613 (2) | 0.22988 (12) | 0.0400 (9) | |
| N7 | 0.57612 (18) | 0.6125 (3) | 0.28025 (13) | 0.0495 (10) | |
| N8 | 0.5008 (2) | 0.6440 (4) | 0.17119 (14) | 0.0673 (13) | |
| C17 | 0.7302 (2) | 0.5032 (3) | 0.16597 (15) | 0.0412 (11) | |
| C18 | 0.7158 (3) | 0.4298 (4) | 0.10993 (16) | 0.0602 (14) | |
| C19 | 0.6876 (3) | 0.4976 (4) | 0.05251 (17) | 0.0652 (16) | |
| C20 | 0.6720 (3) | 0.6343 (4) | 0.04889 (16) | 0.0612 (16) | |
| C21 | 0.6871 (3) | 0.7089 (3) | 0.10395 (16) | 0.0539 (14) | |
| C22 | 0.7180 (2) | 0.6412 (3) | 0.16255 (14) | 0.0382 (11) | |
| C23 | 0.76194 (19) | 0.5718 (3) | 0.26304 (14) | 0.0309 (10) | |
| C24 | 0.7940 (2) | 0.5793 (3) | 0.33451 (14) | 0.0326 (10) | |
| C25 | 0.8858 (2) | 0.6104 (3) | 0.36720 (16) | 0.0478 (11) | |
| C26 | 0.9208 (2) | 0.6157 (4) | 0.43360 (17) | 0.0591 (14) | |
| C27 | 0.8658 (3) | 0.5928 (3) | 0.46862 (17) | 0.0539 (12) | |
| C28 | 0.7741 (2) | 0.5633 (3) | 0.43755 (15) | 0.0415 (11) | |
| C29 | 0.7384 (2) | 0.5552 (3) | 0.37065 (14) | 0.0320 (10) | |
| C30 | 0.5412 (2) | 0.5667 (4) | 0.22150 (17) | 0.0555 (16) | |
| C31 | 0.4949 (4) | 0.7876 (5) | 0.1769 (2) | 0.122 (3) | |
| C32 | 0.4600 (3) | 0.5844 (5) | 0.10749 (19) | 0.108 (2) | |
| H1 | 0.24759 | −0.03292 | 0.26196 | 0.0453* | |
| H2 | 0.30546 | 0.39255 | 0.39403 | 0.0639* | |
| H3 | 0.34568 | 0.27844 | 0.49005 | 0.0673* | |
| H4 | 0.34927 | 0.04845 | 0.49255 | 0.0651* | |
| H5 | 0.30761 | −0.07483 | 0.39780 | 0.0608* | |
| H9 | 0.07525 | 0.12129 | 0.16212 | 0.0479* | |
| H10 | 0.01148 | 0.11022 | 0.05105 | 0.0573* | |
| H11 | 0.10149 | 0.12994 | −0.01087 | 0.0516* | |
| H12 | 0.25644 | 0.16446 | 0.03780 | 0.0467* | |
| H14 | 0.45815 | 0.24824 | 0.28106 | 0.0564* | |
| H15A | 0.45358 | −0.09097 | 0.28921 | 0.1522* | |
| H15B | 0.48820 | −0.09457 | 0.36489 | 0.1522* | |
| H15C | 0.55872 | −0.08863 | 0.32916 | 0.1522* | |
| H16A | 0.53553 | 0.24449 | 0.38678 | 0.1162* | |
| H16B | 0.60491 | 0.12542 | 0.40845 | 0.1162* | |
| H16C | 0.51255 | 0.11996 | 0.42130 | 0.1162* | |
| H5A | 0.73910 | 0.76310 | 0.23871 | 0.0478* | |
| H18 | 0.72508 | 0.33784 | 0.11139 | 0.0719* | |
| H19 | 0.67852 | 0.45024 | 0.01457 | 0.0784* | |
| H20 | 0.65107 | 0.67604 | 0.00876 | 0.0736* | |
| H21 | 0.67706 | 0.80070 | 0.10202 | 0.0649* | |
| H25 | 0.92372 | 0.62779 | 0.34384 | 0.0575* | |
| H26 | 0.98240 | 0.63487 | 0.45476 | 0.0711* | |
| H27 | 0.88990 | 0.59712 | 0.51348 | 0.0645* | |
| H28 | 0.73658 | 0.54898 | 0.46149 | 0.0496* | |
| H30 | 0.54482 | 0.47545 | 0.21474 | 0.0667* | |
| H31A | 0.53722 | 0.81604 | 0.21800 | 0.1837* | |
| H31B | 0.43392 | 0.81161 | 0.17345 | 0.1837* | |
| H31C | 0.50972 | 0.83024 | 0.14296 | 0.1837* | |
| H32A | 0.48518 | 0.62559 | 0.07828 | 0.1617* | |
| H32B | 0.39504 | 0.59798 | 0.09207 | 0.1617* | |
| H32C | 0.47291 | 0.49031 | 0.11006 | 0.1617* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0310 (5) | 0.0841 (8) | 0.0447 (6) | −0.0017 (5) | 0.0108 (4) | 0.0174 (5) |
| O1 | 0.0589 (17) | 0.0562 (18) | 0.087 (2) | −0.0297 (13) | −0.0085 (15) | 0.0247 (14) |
| O2 | 0.0421 (16) | 0.230 (4) | 0.0547 (19) | 0.022 (2) | 0.0280 (14) | 0.026 (2) |
| N1 | 0.0621 (18) | 0.0204 (13) | 0.0280 (15) | −0.0069 (12) | 0.0123 (13) | −0.0025 (11) |
| N2 | 0.0582 (18) | 0.0215 (14) | 0.0280 (15) | −0.0024 (12) | 0.0116 (13) | −0.0009 (11) |
| N3 | 0.0426 (16) | 0.0581 (19) | 0.0441 (19) | 0.0099 (14) | 0.0063 (14) | 0.0020 (15) |
| N4 | 0.0478 (17) | 0.058 (2) | 0.0480 (19) | −0.0014 (15) | 0.0037 (15) | 0.0097 (16) |
| C1 | 0.0462 (19) | 0.0310 (18) | 0.0311 (19) | −0.0014 (14) | 0.0133 (15) | −0.0018 (14) |
| C2 | 0.086 (3) | 0.0307 (19) | 0.036 (2) | −0.0013 (19) | 0.014 (2) | −0.0063 (16) |
| C3 | 0.082 (3) | 0.052 (2) | 0.033 (2) | −0.001 (2) | 0.018 (2) | −0.0100 (17) |
| C4 | 0.074 (3) | 0.056 (3) | 0.028 (2) | −0.002 (2) | 0.0125 (18) | 0.0081 (17) |
| C5 | 0.081 (3) | 0.0332 (19) | 0.035 (2) | −0.0088 (18) | 0.0156 (19) | 0.0047 (16) |
| C6 | 0.0458 (19) | 0.0259 (18) | 0.0288 (18) | −0.0067 (14) | 0.0109 (15) | −0.0004 (14) |
| C7 | 0.0359 (17) | 0.0244 (16) | 0.0289 (17) | −0.0020 (14) | 0.0132 (14) | 0.0021 (14) |
| C8 | 0.0324 (16) | 0.0200 (16) | 0.0293 (17) | −0.0010 (13) | 0.0089 (14) | −0.0007 (12) |
| C9 | 0.0370 (18) | 0.045 (2) | 0.039 (2) | −0.0078 (15) | 0.0154 (16) | −0.0052 (15) |
| C10 | 0.0317 (18) | 0.058 (2) | 0.041 (2) | −0.0067 (16) | −0.0034 (16) | −0.0018 (17) |
| C11 | 0.050 (2) | 0.043 (2) | 0.0280 (19) | −0.0027 (16) | 0.0042 (17) | 0.0017 (15) |
| C12 | 0.049 (2) | 0.0368 (19) | 0.0322 (19) | −0.0013 (16) | 0.0162 (16) | 0.0034 (14) |
| C13 | 0.0319 (16) | 0.0262 (16) | 0.0317 (18) | 0.0009 (13) | 0.0095 (14) | 0.0019 (13) |
| C14 | 0.0338 (18) | 0.053 (2) | 0.050 (2) | −0.0061 (16) | 0.0089 (17) | 0.0087 (18) |
| C15 | 0.126 (4) | 0.066 (3) | 0.082 (3) | 0.028 (3) | −0.002 (3) | 0.010 (2) |
| C16 | 0.077 (3) | 0.087 (3) | 0.046 (2) | −0.024 (2) | −0.006 (2) | 0.009 (2) |
| S2 | 0.0311 (4) | 0.0672 (7) | 0.0420 (5) | 0.0038 (4) | 0.0134 (4) | 0.0173 (5) |
| O3 | 0.0529 (15) | 0.0439 (15) | 0.0805 (19) | −0.0146 (12) | 0.0099 (14) | 0.0084 (13) |
| O4 | 0.0459 (15) | 0.146 (3) | 0.0526 (17) | 0.0202 (16) | 0.0291 (14) | 0.0309 (16) |
| N5 | 0.0627 (18) | 0.0266 (14) | 0.0311 (15) | −0.0061 (13) | 0.0176 (13) | 0.0016 (12) |
| N6 | 0.0562 (18) | 0.0334 (15) | 0.0306 (15) | −0.0038 (13) | 0.0155 (13) | −0.0043 (12) |
| N7 | 0.0475 (17) | 0.0610 (19) | 0.0343 (17) | 0.0147 (14) | 0.0072 (14) | 0.0086 (14) |
| N8 | 0.0533 (19) | 0.099 (3) | 0.0369 (19) | −0.0115 (19) | 0.0002 (15) | 0.0157 (18) |
| C17 | 0.057 (2) | 0.0347 (19) | 0.0308 (19) | −0.0103 (16) | 0.0141 (16) | −0.0033 (15) |
| C18 | 0.091 (3) | 0.047 (2) | 0.040 (2) | −0.010 (2) | 0.020 (2) | −0.0105 (18) |
| C19 | 0.092 (3) | 0.068 (3) | 0.033 (2) | −0.021 (2) | 0.019 (2) | −0.015 (2) |
| C20 | 0.082 (3) | 0.070 (3) | 0.028 (2) | −0.025 (2) | 0.015 (2) | 0.0067 (18) |
| C21 | 0.075 (3) | 0.046 (2) | 0.037 (2) | −0.0132 (19) | 0.015 (2) | 0.0075 (17) |
| C22 | 0.051 (2) | 0.037 (2) | 0.0264 (18) | −0.0136 (15) | 0.0134 (15) | −0.0023 (14) |
| C23 | 0.0309 (16) | 0.0298 (17) | 0.0320 (18) | −0.0047 (14) | 0.0112 (14) | −0.0010 (14) |
| C24 | 0.0362 (17) | 0.0317 (18) | 0.0284 (18) | −0.0015 (14) | 0.0097 (14) | 0.0006 (14) |
| C25 | 0.0394 (19) | 0.059 (2) | 0.042 (2) | −0.0086 (17) | 0.0107 (17) | −0.0008 (17) |
| C26 | 0.041 (2) | 0.079 (3) | 0.046 (2) | −0.0135 (19) | 0.0015 (19) | −0.003 (2) |
| C27 | 0.059 (2) | 0.059 (2) | 0.033 (2) | 0.002 (2) | 0.0029 (19) | −0.0040 (17) |
| C28 | 0.052 (2) | 0.047 (2) | 0.0290 (19) | 0.0068 (16) | 0.0185 (16) | 0.0038 (15) |
| C29 | 0.0338 (17) | 0.0313 (17) | 0.0294 (18) | 0.0046 (13) | 0.0093 (14) | 0.0053 (13) |
| C30 | 0.038 (2) | 0.073 (3) | 0.050 (3) | −0.0114 (18) | 0.0088 (18) | 0.012 (2) |
| C31 | 0.155 (6) | 0.104 (4) | 0.078 (4) | 0.053 (4) | 0.004 (4) | 0.029 (3) |
| C32 | 0.093 (4) | 0.164 (5) | 0.043 (3) | −0.056 (3) | −0.005 (2) | 0.011 (3) |
Geometric parameters (Å, º)
| S1—O1 | 1.450 (2) | C4—H4 | 0.9300 |
| S1—O2 | 1.420 (3) | C5—H5 | 0.9300 |
| S1—N3 | 1.612 (3) | C9—H9 | 0.9300 |
| S1—C13 | 1.778 (3) | C10—H10 | 0.9300 |
| S2—O4 | 1.428 (3) | C11—H11 | 0.9300 |
| S2—N7 | 1.609 (3) | C12—H12 | 0.9300 |
| S2—C29 | 1.780 (3) | C14—H14 | 0.9300 |
| S2—O3 | 1.447 (2) | C15—H15C | 0.9600 |
| N1—C6 | 1.377 (4) | C15—H15A | 0.9600 |
| N1—C7 | 1.356 (4) | C15—H15B | 0.9600 |
| N2—C7 | 1.310 (4) | C16—H16C | 0.9600 |
| N2—C1 | 1.392 (4) | C16—H16B | 0.9600 |
| N3—C14 | 1.308 (4) | C16—H16A | 0.9600 |
| N4—C14 | 1.306 (4) | C17—C22 | 1.392 (4) |
| N4—C16 | 1.450 (5) | C17—C18 | 1.389 (5) |
| N4—C15 | 1.451 (5) | C18—C19 | 1.369 (5) |
| N1—H1 | 0.8600 | C19—C20 | 1.386 (6) |
| N5—C22 | 1.377 (4) | C20—C21 | 1.376 (5) |
| N5—C23 | 1.351 (4) | C21—C22 | 1.390 (4) |
| N6—C17 | 1.389 (4) | C23—C24 | 1.482 (4) |
| N6—C23 | 1.316 (4) | C24—C29 | 1.394 (5) |
| N7—C30 | 1.303 (5) | C24—C25 | 1.395 (5) |
| N8—C32 | 1.454 (5) | C25—C26 | 1.376 (5) |
| N8—C30 | 1.319 (5) | C26—C27 | 1.364 (6) |
| N8—C31 | 1.448 (6) | C27—C28 | 1.385 (6) |
| N5—H5A | 0.8600 | C28—C29 | 1.388 (4) |
| C1—C2 | 1.387 (4) | C18—H18 | 0.9300 |
| C1—C6 | 1.394 (4) | C19—H19 | 0.9300 |
| C2—C3 | 1.365 (5) | C20—H20 | 0.9300 |
| C3—C4 | 1.391 (5) | C21—H21 | 0.9300 |
| C4—C5 | 1.376 (4) | C25—H25 | 0.9300 |
| C5—C6 | 1.384 (4) | C26—H26 | 0.9300 |
| C7—C8 | 1.484 (4) | C27—H27 | 0.9300 |
| C8—C9 | 1.389 (5) | C28—H28 | 0.9300 |
| C8—C13 | 1.389 (4) | C30—H30 | 0.9300 |
| C9—C10 | 1.384 (5) | C31—H31A | 0.9600 |
| C10—C11 | 1.364 (5) | C31—H31B | 0.9600 |
| C11—C12 | 1.374 (5) | C31—H31C | 0.9600 |
| C12—C13 | 1.391 (4) | C32—H32A | 0.9600 |
| C2—H2 | 0.9300 | C32—H32B | 0.9600 |
| C3—H3 | 0.9300 | C32—H32C | 0.9600 |
| O1—S1—O2 | 116.3 (2) | N3—C14—H14 | 118.00 |
| O1—S1—N3 | 113.32 (16) | N4—C14—H14 | 118.00 |
| O1—S1—C13 | 107.12 (16) | N4—C15—H15B | 109.00 |
| O2—S1—N3 | 109.02 (19) | N4—C15—H15A | 109.00 |
| O2—S1—C13 | 105.41 (16) | N4—C15—H15C | 109.00 |
| N3—S1—C13 | 104.75 (15) | H15B—C15—H15C | 109.00 |
| O3—S2—N7 | 113.45 (15) | H15A—C15—H15C | 109.00 |
| O3—S2—C29 | 107.61 (15) | H15A—C15—H15B | 109.00 |
| O4—S2—N7 | 108.77 (16) | H16A—C16—H16B | 109.00 |
| O4—S2—C29 | 105.65 (15) | H16B—C16—H16C | 109.00 |
| N7—S2—C29 | 104.41 (15) | N4—C16—H16B | 109.00 |
| O3—S2—O4 | 116.02 (17) | H16A—C16—H16C | 109.00 |
| C6—N1—C7 | 106.9 (2) | N4—C16—H16A | 109.00 |
| C1—N2—C7 | 104.4 (2) | N4—C16—H16C | 110.00 |
| S1—N3—C14 | 115.8 (2) | N6—C17—C22 | 110.0 (3) |
| C14—N4—C16 | 121.4 (3) | N6—C17—C18 | 130.0 (3) |
| C14—N4—C15 | 121.6 (3) | C18—C17—C22 | 120.0 (3) |
| C15—N4—C16 | 117.0 (3) | C17—C18—C19 | 117.7 (4) |
| C6—N1—H1 | 127.00 | C18—C19—C20 | 122.3 (3) |
| C7—N1—H1 | 127.00 | C19—C20—C21 | 120.8 (3) |
| C22—N5—C23 | 107.1 (2) | C20—C21—C22 | 117.1 (3) |
| C17—N6—C23 | 104.5 (2) | N5—C22—C21 | 132.8 (3) |
| S2—N7—C30 | 115.9 (3) | N5—C22—C17 | 105.1 (2) |
| C30—N8—C32 | 119.6 (4) | C17—C22—C21 | 122.0 (3) |
| C31—N8—C32 | 118.2 (3) | N6—C23—C24 | 124.7 (3) |
| C30—N8—C31 | 122.3 (3) | N5—C23—C24 | 122.0 (3) |
| C23—N5—H5A | 126.00 | N5—C23—N6 | 113.2 (3) |
| C22—N5—H5A | 126.00 | C23—C24—C25 | 117.6 (3) |
| N2—C1—C6 | 110.0 (3) | C23—C24—C29 | 124.0 (3) |
| C2—C1—C6 | 119.4 (3) | C25—C24—C29 | 118.5 (3) |
| N2—C1—C2 | 130.6 (3) | C24—C25—C26 | 120.7 (3) |
| C1—C2—C3 | 118.6 (3) | C25—C26—C27 | 120.5 (3) |
| C2—C3—C4 | 121.3 (3) | C26—C27—C28 | 120.2 (3) |
| C3—C4—C5 | 121.3 (3) | C27—C28—C29 | 119.9 (3) |
| C4—C5—C6 | 116.9 (3) | S2—C29—C24 | 123.0 (2) |
| N1—C6—C5 | 132.5 (3) | S2—C29—C28 | 116.6 (2) |
| C1—C6—C5 | 122.3 (3) | C24—C29—C28 | 120.3 (3) |
| N1—C6—C1 | 105.1 (2) | N7—C30—N8 | 122.9 (4) |
| N1—C7—N2 | 113.6 (3) | C17—C18—H18 | 121.00 |
| N2—C7—C8 | 125.2 (3) | C19—C18—H18 | 121.00 |
| N1—C7—C8 | 121.1 (3) | C18—C19—H19 | 119.00 |
| C9—C8—C13 | 118.3 (3) | C20—C19—H19 | 119.00 |
| C7—C8—C13 | 124.6 (3) | C19—C20—H20 | 120.00 |
| C7—C8—C9 | 117.1 (3) | C21—C20—H20 | 120.00 |
| C8—C9—C10 | 120.7 (3) | C20—C21—H21 | 121.00 |
| C9—C10—C11 | 120.5 (3) | C22—C21—H21 | 121.00 |
| C10—C11—C12 | 119.9 (3) | C24—C25—H25 | 120.00 |
| C11—C12—C13 | 120.2 (3) | C26—C25—H25 | 120.00 |
| S1—C13—C12 | 116.2 (2) | C25—C26—H26 | 120.00 |
| C8—C13—C12 | 120.4 (3) | C27—C26—H26 | 120.00 |
| S1—C13—C8 | 123.3 (2) | C26—C27—H27 | 120.00 |
| N3—C14—N4 | 123.6 (3) | C28—C27—H27 | 120.00 |
| C3—C2—H2 | 121.00 | C27—C28—H28 | 120.00 |
| C1—C2—H2 | 121.00 | C29—C28—H28 | 120.00 |
| C2—C3—H3 | 119.00 | N7—C30—H30 | 119.00 |
| C4—C3—H3 | 119.00 | N8—C30—H30 | 119.00 |
| C3—C4—H4 | 119.00 | N8—C31—H31A | 109.00 |
| C5—C4—H4 | 119.00 | N8—C31—H31B | 109.00 |
| C4—C5—H5 | 122.00 | N8—C31—H31C | 109.00 |
| C6—C5—H5 | 122.00 | H31A—C31—H31B | 110.00 |
| C10—C9—H9 | 120.00 | H31A—C31—H31C | 109.00 |
| C8—C9—H9 | 120.00 | H31B—C31—H31C | 109.00 |
| C9—C10—H10 | 120.00 | N8—C32—H32A | 109.00 |
| C11—C10—H10 | 120.00 | N8—C32—H32B | 109.00 |
| C12—C11—H11 | 120.00 | N8—C32—H32C | 109.00 |
| C10—C11—H11 | 120.00 | H32A—C32—H32B | 109.00 |
| C11—C12—H12 | 120.00 | H32A—C32—H32C | 109.00 |
| C13—C12—H12 | 120.00 | H32B—C32—H32C | 109.00 |
| O1—S1—N3—C14 | −2.2 (3) | C1—C2—C3—C4 | 0.4 (7) |
| O2—S1—N3—C14 | 129.0 (3) | C2—C3—C4—C5 | −1.3 (8) |
| C13—S1—N3—C14 | −118.6 (3) | C3—C4—C5—C6 | 0.4 (7) |
| O1—S1—C13—C8 | −70.0 (3) | C4—C5—C6—N1 | 179.1 (4) |
| O1—S1—C13—C12 | 106.1 (3) | C4—C5—C6—C1 | 1.5 (6) |
| O2—S1—C13—C8 | 165.6 (3) | N2—C7—C8—C13 | 82.7 (4) |
| O2—S1—C13—C12 | −18.3 (3) | N1—C7—C8—C9 | 77.7 (4) |
| N3—S1—C13—C8 | 50.6 (3) | N1—C7—C8—C13 | −101.6 (4) |
| N3—S1—C13—C12 | −133.3 (2) | N2—C7—C8—C9 | −98.0 (4) |
| O4—S2—N7—C30 | −127.7 (3) | C7—C8—C13—S1 | −5.2 (4) |
| C29—S2—N7—C30 | 119.9 (3) | C7—C8—C13—C12 | 178.9 (3) |
| O3—S2—C29—C24 | 67.4 (3) | C13—C8—C9—C10 | −0.7 (4) |
| O3—S2—C29—C28 | −108.7 (3) | C7—C8—C9—C10 | 180.0 (3) |
| O4—S2—C29—C24 | −168.1 (3) | C9—C8—C13—C12 | −0.4 (4) |
| O4—S2—C29—C28 | 15.9 (3) | C9—C8—C13—S1 | 175.5 (2) |
| O3—S2—N7—C30 | 3.0 (3) | C8—C9—C10—C11 | 1.1 (5) |
| N7—S2—C29—C28 | 130.5 (2) | C9—C10—C11—C12 | −0.5 (5) |
| N7—S2—C29—C24 | −53.4 (3) | C10—C11—C12—C13 | −0.6 (5) |
| C7—N1—C6—C5 | −177.8 (4) | C11—C12—C13—C8 | 1.1 (5) |
| C6—N1—C7—N2 | −0.1 (4) | C11—C12—C13—S1 | −175.2 (2) |
| C7—N1—C6—C1 | 0.2 (4) | N6—C17—C18—C19 | 179.5 (4) |
| C6—N1—C7—C8 | −176.2 (3) | C22—C17—C18—C19 | −1.5 (6) |
| C7—N2—C1—C6 | 0.0 (4) | N6—C17—C22—N5 | 0.6 (4) |
| C7—N2—C1—C2 | −179.5 (4) | N6—C17—C22—C21 | −177.6 (4) |
| C1—N2—C7—N1 | 0.1 (4) | C18—C17—C22—N5 | −178.6 (3) |
| C1—N2—C7—C8 | 176.0 (3) | C18—C17—C22—C21 | 3.3 (6) |
| S1—N3—C14—N4 | −179.6 (3) | C17—C18—C19—C20 | −1.0 (7) |
| C15—N4—C14—N3 | −1.3 (6) | C18—C19—C20—C21 | 2.0 (8) |
| C16—N4—C14—N3 | 178.0 (4) | C19—C20—C21—C22 | −0.3 (7) |
| C23—N5—C22—C21 | 177.1 (4) | C20—C21—C22—N5 | −179.9 (4) |
| C22—N5—C23—N6 | 0.8 (4) | C20—C21—C22—C17 | −2.3 (6) |
| C23—N5—C22—C17 | −0.8 (4) | N5—C23—C24—C25 | −81.3 (4) |
| C22—N5—C23—C24 | 176.6 (3) | N5—C23—C24—C29 | 99.8 (4) |
| C23—N6—C17—C22 | −0.2 (4) | N6—C23—C24—C25 | 94.1 (4) |
| C17—N6—C23—N5 | −0.4 (4) | N6—C23—C24—C29 | −84.9 (4) |
| C23—N6—C17—C18 | 178.9 (4) | C23—C24—C25—C26 | −178.3 (3) |
| C17—N6—C23—C24 | −176.1 (3) | C29—C24—C25—C26 | 0.7 (5) |
| S2—N7—C30—N8 | 179.8 (3) | C23—C24—C29—S2 | 3.5 (4) |
| C31—N8—C30—N7 | 2.3 (6) | C23—C24—C29—C28 | 179.5 (3) |
| C32—N8—C30—N7 | −177.3 (4) | C25—C24—C29—S2 | −175.5 (2) |
| C6—C1—C2—C3 | 1.3 (6) | C25—C24—C29—C28 | 0.5 (4) |
| N2—C1—C6—N1 | −0.1 (4) | C24—C25—C26—C27 | −1.1 (5) |
| N2—C1—C2—C3 | −179.2 (4) | C25—C26—C27—C28 | 0.3 (5) |
| N2—C1—C6—C5 | 178.1 (4) | C26—C27—C28—C29 | 0.9 (5) |
| C2—C1—C6—N1 | 179.5 (3) | C27—C28—C29—S2 | 174.9 (2) |
| C2—C1—C6—C5 | −2.3 (6) | C27—C28—C29—C24 | −1.3 (5) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···N2i | 0.86 | 2.11 | 2.964 (3) | 174 |
| N5—H5A···N6ii | 0.86 | 2.10 | 2.955 (3) | 178 |
| C9—H9···O4i | 0.93 | 2.56 | 3.153 (4) | 122 |
| C10—H10···O4i | 0.93 | 2.57 | 3.153 (4) | 121 |
| C25—H25···O2ii | 0.93 | 2.47 | 3.114 (5) | 127 |
Symmetry codes: (i) −x+1/2, y−1/2, −z+1/2; (ii) −x+3/2, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB6832).
References
- Bruker (2005). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2009). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Esparza-Ruiz, A., Gonzalez-Gomez, G., Mijangos, E., Pena-Hueso, A., Lopez-Sandoval, H., Flores-Parra, A., Contreras, R. & Barba-Behrens, N. (2010). Dalton Trans. 39, 6302–6309. [DOI] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812025159/hb6832sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812025159/hb6832Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812025159/hb6832Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




