Abstract
Approximately 15% of human genetic diseases are estimated to involve dysregulation of alternative pre-mRNA splicing. Antisense molecules designed to alter these and other splicing events typically target continuous linear sequences of the message. Here, we show that a structural feature in a pre-mRNA can be targeted by bipartite antisense molecules designed to hybridize with the discontinuous elements that flank the structure and thereby alter splicing. We targeted a hairpin structure at the boundary between exon 10 and intron 10 of the pre-mRNA of tau. Mutations in this region that are associated with certain forms of frontotemporal dementia, destabilize the hairpin to cause increased inclusion of exon 10. Via electrophoretic mobility shift and RNase protection assays, we demonstrate that bipartite antisense molecules designed to simultaneously interact with the available sequences that immediately flank the tau pre-mRNA hairpin do indeed bind to this structured region. Moreover, these agents inhibit exon 10 splicing and reverse the effect of destabilizing disease-causing mutations, in both in vitro splicing assays and cell culture. This general bipartite antisense strategy could be employed to modulate other splicing events that are regulated by RNA secondary structure.
INTRODUCTION
In general, alternative RNA splicing is a major source of proteomic diversity. Over 90% of human genes are alternatively spliced (1), with even more alternative splicing occurring in the human brain (2), and an estimated 15% of disease-causing mutations alter splicing (3). There is a growing awareness that structural features in the pre-mRNA can play an important role in determining alternative splicing events (4,5). The presence of such structures at or near splice sites provides opportunities for the design of agents that target these features and regulate specific splicing events. Such agents should help contribute both to the basic understanding of splice regulation in biology and to the development of practical therapeutics for diseases in which specific splicing events are dysregulated or where shunting splicing toward particular isoforms would be otherwise beneficial. The idea of developing agents to alter specific RNA splicing events by targeting structural features in the message is virtually unexplored.
As a proof of principle for this approach, we have chosen to target the pre-mRNA of the microtubule-associated protein tau. Assembly of the tau protein into intraneuronal filaments is a primary pathological feature of a spectrum of neurodegenerative diseases, including Alzheimer’s disease and frontotemporal dementia (FTD) (6). Some 40 mutations in the tau gene are associated with autosomal-dominant familial FTD, and many of these are silent or intronic mutations that shift the splicing of the tau pre-mRNA in favor of inclusion of exon 10 (7). This exon encodes for one of four possible repeated microtubule binding domains, with exclusion leading to a 3-repeat form (3R tau) and inclusion leading to the 4-repeat form (4R tau). A hairpin structure is located at the junction between exon 10 and intron 10. FTD mutations found in this region destabilize the hairpin and increase the proportion of splicing that leads to 4R tau (8–10). In contrast, artificial stabilizing mutations decrease exon 10 inclusion (11). Moreover, the degree of exon 10 exclusion correlates with the experimentally determined melting temperatures of hairpin oligonucleotide variants based on the exon 10/intron 10 junction (11).
To target this hairpin structure, we designed bipartite antisense oligonucleotides (ASOs) that would simultaneously interact with the 5′- and 3′-regions that immediately flank this structure. As the two parts of the ASO are held together in a single molecule, binding of the bipartite ASO should occur even in the presence of the structure and prevent exon 10 splicing. We show that such agents do bind simultaneously to both flanking sequences and specifically reduce inclusion of exon 10 of tau mRNA, both in vitro and in cells, with a tau minigene system, as well as endogenous tau.
MATERIALS AND METHODS
Electrophoretic mobility shift assay
RNA oligonucleotides were purchased from Dharmacon (part of Thermo Scientific). DNA oligonucleotides were purchased from Invitrogen. Prior to hybridization, RNA representing the tau hairpin and flanking sequences (sequence 5′-GGGAGGCGGCAGUGUGAGUACCUUCACACGUCCCAUGCGC-3′) or the mutated tau hairpin and flanking sequences (sequence 5′-GGGAGGCGGAAUCUCUUGUACCUUCACACGUCCCAUGCGC-3′) was renatured by heating to 80°C for 5 min and then cooling slowly to 25°C over 30 min. 0.5 µM hairpin-containing RNA was hybridized with 0.5 µM ASO in incubation buffer at 25°C for 30 min (50 mM TRIS–HCl, pH 7.5, 10 mM MgCl2, 10 mM NaCl, 0.1 mM EDTA). Novex hi-density Tris/borate/EDTA (TBE) sample buffer was added to the sample and hybridization was analyzed by 15% acrylamide gel electrophoresis in TRIS–borate–EDTA buffer (89 mM TRIS Base, 89 mM boric acid, 2 mM EDTA). The acrylamide gel and running buffer were pre-cooled to 4°C, and the samples were electrophoresed at 4°C. Bands were visualized by SYBR Gold staining (Invitrogen) under UV light, and images were captured with an AlphaImager system.
Synthesis of bipartite peptide nucleic acid oligomers
Peptide nucleic acid (PNA) oligomers were synthesized on Rink Amide PEGA resin (Novabiochem). Octa(L-lysine) peptide was covalently assembled on the PNA N-end as a cell penetrating peptide. PNA monomers (Fmoc/Bhoc protected) were commercially available (Link Technologies, UK). The monomer N-Fmoc-egl-OH (Sigma-Aldrich) was used to build the linker for bipartite PNA. Fmoc deprotection was carried out using 20% piperidine in dimethyl formamide (DMF). Amide coupling was performed using the coupling agent HATU. Final cleavage and deprotection were carried out with TFA/m-cresol solution. The crude PNA product cleaved from resin was confirmed to be pure by reverse-phase high pressure liquid chromatography (RP–HPLC), characterized by matrix-assisted laser desorption-time of flight (MALDI-TOF) mass spectrometry and used directly without further purification. The PNA aqueous solutions were prepared, and the concentrations were determined by the UV absorbance at 260 nm on a SPECTRAmax PLUS 384 spectrophotometer.
RNase A and T1 protection assays
5′-biotinylated hairpin-containing RNA (sequence as above) was renatured in structure buffer (10 mM TRIS pH 7.0, 100 mM KCl, 10 mM MgCl2) by heating to 80°C for 5 min and cooling to 25°C over 30 min. Streptavidin agarose beads (Sigma) were washed twice in structure buffer, and 0.8 µg of renatured biotinylated hairpin-containing RNA was bound to the beads for 30 min at 25°C on a rotator. An equimolar concentration of complementary DNA (9-2A-10) was added, where indicated and allowed to bind for 20 min at 25°C. RNase A or T1 was added, and the reaction was allowed to proceed for 10 min at 37°C. Final concentrations in the RNase reaction were 10 µM hairpin-containing RNA, 10 µM 9-2A-10, 1 µU/µl RNase A or 100 mU/µl RNase T1. Beads were washed twice in urea buffer at 50°C to remove cleaved 3′-fragments (20 mM sodium citrate pH 5.0, 1 mM EDTA, 7 M urea). 5′-biotinylated fragments were then eluted from the beads by heating for 5 min at 95°C in loading buffer II (Ambion). Samples were analyzed by denaturing acrylamide gel electrophoresis (20% acrylamide, 7M urea) in TBE buffer, alongside a ladder of all possible 5′-biotinylated cleavage fragments produced by alkaline hydrolysis. Cleavage fragments were visualized by SYBR Gold staining under UV light.
RNase H digestion
Complementary DNA oligonucleotides were hybridized to the hairpin-containing RNA (10 µM each) in structure buffer. The hybrids were then diluted to 1 µM in HEPES buffer (20 mM HEPES–KOH, pH 8.0, 50 mM KCl, 4 mM MgCl2, 1 mM DTT, 50 µg/ml BSA) containing RNase H (final concentrations of 1 µM hairpin-containing RNA, 1 µM complementary DNA, 1% vol/vol RNase H, Sigma). Samples were incubated at 37°C for 20 min to allow digestion of RNA in RNA:DNA hybrids by RNase H. Loading buffer II was added, and samples were heated for 5 min at 95°C before analyzing by denaturing acrylamide gel electrophoresis (20% acrylamide, 7 M urea). RNA and DNA bands were visualized by SYBR Gold staining under UV light.
In vitro splicing assay
HeLa nuclear extract was prepared by a method adapted from Dignam et al. (12). In order to generate the splicing unit RNA, tau exon 10, the first 207 bases of intron 10, the last 213 bases of intron 10 and exon 11 were amplified from HEK-293 cell genomic DNA by In-Fusion PCR (Clontech), and cloned into pcDNA3.1/Zeo(+) (Invitrogen) between BamHI and XhoI sites. The insert was confirmed by sequencing. The vector was then linearized by digestion with XbaI, and the insert was transcribed into RNA in vitro using Ambion’s MEGAscript T7 kit. In vitro splicing reactions were performed for 3 h at 30°C in reaction volumes of 25 µl. Reactions contained 20 fmol splicing unit RNA, 0−1 µM RNA ASO, 10 µl HeLa nuclear extract, 20 mM phosphocreatine, 0.5 mM ATP, 3.2 mM MgCl2, 40 mU/µl creatine phosphokinase and 2.6% vol/vol polyvinyl alcohol in buffer D [20 mM HEPES–KOH, pH 8.0, 100 mM KCl, 0.2 mM EDTA, 20% (vol/vol) glycerol, 0.5 mM PMSF, 1 mM DTT]. In negative control reactions, HeLa nuclear extract was substituted with extra buffer D. Spliced products were purified by phenol/chloroform precipitation and treated with DNase. Spliced products were amplified by reverse-transcription PCR with primers specific for the splicing unit RNA (pcDNA3 f and tau E11 r), and analyzed by 8% acrylamide gel electrophoresis in TBE buffer with SYBR Gold staining under UV light.
Minigenes and cell transfections
The tau minigene used was as previously described (11), and comprised exons 9, 10 and 11 with shortened intervening introns cloned into the pcDNA3 vector (Invitrogen). The coding region for the photinus pyralis luciferase gene was ligated after exon 11 to provide a sequence for a reverse PCR primer unique to the minigene so as not to detect endogenous tau. The BACE1 minigene was comprised of exon 3, intron 3 and exon 4 truncated prior to the alternative 3′-splice site (13). The 5X stem mutant tau minigene was generated from the wild-type tau minigene using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer’s protocol. Primer sequences are shown in Supplementary Table S1 (nucleotide changes are italicized). Other minigenes were as previously described (14). HeLa, HEK-293 and SK-N-SH cells were grown in DMEM containing 4.5 mg/ml glucose, supplemented with 10% fetal bovine serum, 2 mM glutamine, 50 U/ml penicillin and 50 µg/ml streptomycin. For exon-trapping PCR experiments, SK-N-SH cells were seeded into 12-well plates and co-transfected at 80–90% confluence with 0.25 µg minigene plasmid DNA and 10 nM antisense RNA, using 4 µl lipofectamine per well. For real-time PCR experiments to investigate the effect of antisense RNA on the tau minigene, SK-N-SH cells were co-transfected at 80–90% confluence with 0.25 µg tau minigene, 0.05 µg pRL-TK vector (transfection control, Promega) and 0–100 nM antisense RNA, using 4 µl lipofectamine per well. For real-time PCR experiments to investigate the effect of antisense on the endogenous tau pre-mRNA, HEK-293 cells were seeded into plates coated with poly-D-lysine and transfected at 50% confluence with 0–1 µM RNA antisense, using 4.5 µl oligofectamine per well. The sequences of all antisense RNA oligonucleotides used are shown in Table 1.
Table 1.
Synthetic DNA, RNA and PNA antisense molecules used in this study
| 9-1A-10 DNA | 5′-GCGCATGGGAACCGCCTCCC-3′ |
| 9-2A-10 DNA | 5′-GCGCATGGGAAACCGCCTCCC-3′ |
| 9-3A-10 DNA | 5′-GCGCATGGGAAAACCGCCTCCC-3′ |
| Linear DNA | 5′-TGAAGGTACTCACACTGCCGC-3′ |
| Scrambled DNA | 5′-CATGTAATCCGGCCCAGTCGA-3′ |
| 5′-flank (9) DNA | 5′-CCGCCTCCC-3′ |
| 3′-flank (10) DNA | 5′-GCGCATGGGA-3′ |
| 7-2A-7 DNA | 5′-CATGGGAAACCGCCTC-3′ |
| 8-2A-0 DNA | 5′-AACCGCCTCC-3′ |
| 8-2A-1 DNA | 5′-AAACCGCCTCC-3′ |
| 8-2A-2 DNA | 5′-GAAACCGCCTCC-3′ |
| 8-2A-3 DNA | 5′-GGAAACCGCCTCC-3′ |
| 8-2A-4 DNA | 5′-GGGAAACCGCCTCC-3′ |
| 8-2A-5 DNA | 5′-TGGGAAACCGCCTCC-3′ |
| 8-2A-6 DNA | 5′-ATGGGAAACCGCCTCC-3′ |
| 8-2A-7 DNA | 5′-CATGGGAAACCGCCTCC-3′ |
| 8-2A-8 DNA | 5′-GCATGGGAAACCGCCTCC-3′ |
| Linear (40) DNA | 5′-GCGCATGGGACGTGTGAAGGTACTCACACTGCCGCCTCCC-3′ |
| 9-0A-10 RNA | 5′-GCGCAUGGGACCGCCUCCC-3′ |
| 9-1A-10 RNA | 5′-GCGCAUGGGAACCGCCUCCC-3′ |
| 9-2A-10 RNA | 5′-GCGCAUGGGAAACCGCCUCCC-3′ |
| 9-3A-10 RNA | 5′-GCGCAUGGGAAAACCGCCUCCC-3′ |
| E10B RNA | 5′-UGAAGGUACUCACACUGCCGC-3′ |
| Scrambled RNA | 5′-CACGGAGACCCGAUUCGUCAU-3′ |
| 5′-flank (9) RNA | 5′-CCGCCUCCC-3′ |
| 3′-flank (10) RNA | 5′-GCGCAUGGGA-3′ |
| lys-4-1egl-4-8lys | lysCGCCeglAGGG(lys)8 |
| lys-5-1egl-5-8lys | lysCCGCCeglAGGGT(lys)8 |
| lys-6-1egl-6-8lys | lysTCCGCCeglAGGGTA(lys)8 |
| lys-7-0egl-7-8lys | lysCTCCGCCAGGGTAC(lys)8 |
| lys-7-1egl-7-8lys | lysCTCCGCCeglAGGGTAC(lys)8 |
| lys-7-2egl-7-8lys | lysCTCCGCC(egl)2AGGGTAC(lys)8 |
| lys-7-3egl-7-8lys | lysCTCCGCC(egl)3AGGGTAC(lys)8 |
| lys-8-1egl-8-8lys | lysCCTCCGCCeglAGGGTACG(lys)8 |
| lys-9-1egl-9-8lys | lysCCCTCCGCCeglAGGGTACGC(lys)8 |
| BACE1 5′-splice site antisense RNA | 5′-GUUCCUCUCACCCUGGCAAUC-3′ |
| BACE1 3′-splice site antisense RNA | 5′-GGAGUCGUCAGGCUUUGGCAG-3′ |
RNA antisense molecules were modified with 2′-OMe groups and a phosphorothioate backbone.
Reverse transcription and exon-trapping PCR
RNA was isolated 24 h after transfection using Ambion’s RNAqueous™ kit according to the manufacturer’s protocol. RNA was quantified, and 1.5–2 µg was reverse transcribed into cDNA using Ambion’s RETROscript™ kit with oligo(dT) primer. Exon-trapping PCR was performed for 30 cycles using SuperTaq™ polymerase (Ambion) and 2.5 µl cDNA in a 25 µl reaction. For exon-trapping PCR from the tau minigene, a forward primer in exon 9 (tau E9 f) and a reverse primer in the luciferase coding region of the tau minigene (luciferase r) were used. For exon-trapping PCR from the BACE1 minigene, a forward primer in exon 3 (BACE E3 f) and a reverse primer in the bovine growth hormone sequence of the pcDNA vector (pcDNA3 BGH r) were used. Sequences of all primers are shown in Supplementary Table S1. PCR products were separated by 8% acrylamide gel electrophoresis alongside Bioline Hyperladder V, and visualized by SYBR Gold staining under UV light.
Quantitative PCR and data analysis
RNA was isolated from transfected cells and reverse transcribed into cDNA as described above. Quantitative PCR was performed using an Applied Biosystems 7500 Real-Time PCR Fast System with SYBR® Green PCR mastermix (Invitrogen). PCR reactions were performed at 64°C for a combined annealing and extension. 3R tau from the minigene was amplified using tau E9-E11 f and luciferase r; 4R tau from the minigene was amplified using tau E9-E10 f and luciferase r; total tau from the minigene was amplified using luciferase f and luciferase r2. Endogenous 3R tau was amplified using tau E9-E11 f and tau E11 r; endogenous 4R tau was amplified using tau E9-E10 f and tau E11 r and total endogenous tau was amplified using total tau f and r (both in exon 9). Primer sequences are shown in Supplementary Table S1. Data were analyzed using Sequence Detection System software according to the ΔΔCt method, where the expression of 3R and 4R tau was normalized to pRL-TK (a transfection control for the tau minigene) or to total tau (endogenous tau experiments), and the expression of total tau was normalized to either pRL-TK (minigene experiments) or to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (endogenous tau experiments). Experiments were repeated on at least two separate occasions with replicates of three or more within each experiment. Graphs were drawn and statistical tests were carried out using GraphPad Prism; significant differences were determined using one-way ANOVA with the Bonferoni post-hoc test (*P < 0.05, **P < 0.01, ***P < 0.001).
RESULTS
Bipartite ASOs bind to a synthetic tau RNA oligonucleotide
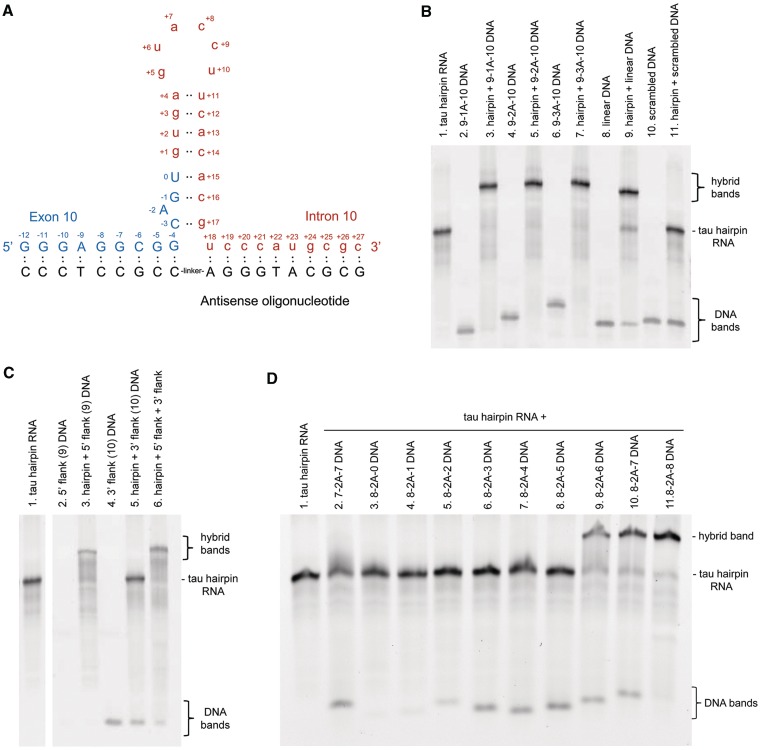
Our first designed bipartite ASOs were DNAs with up to 10 bases on either side, complementary to their respective partner regions that flank the stem–loop (or hairpin) structure at the tau pre-mRNA exon 10–intron 10 junction (Figure 1A). The DNA ASOs have 1, 2 or 3 adenosine spacers to bridge any potential gap between the flanking sequences (i.e. to ensure that both complementary parts can bind simultaneously). Adenosine was chosen because there are no complementary uridines at the base of the stem. To determine whether DNA bipartite ASOs could bind to the discontinuous region at the base of the tau hairpin, electrophoretic mobility shift assays (EMSA) were performed. An EMSA gel (Figure 1B) revealed that DNA with 9 bases complementary to the 5′-flank, a linker of one adenosine, and 10 bases complementary to the 3′-flank (termed 9-1A-10 DNA; Table 1 lists all ASOs used in this study) can hybridize virtually completely to a synthetic 40-nt tau hairpin-containing RNA at equimolar concentrations, as shown by a band of slower mobility (lane 3) compared with tau RNA alone (lane 1). Similar experiments in which the concentrations of DNA ASO were varied further supported a 1:1 binding ratio (Supplementary Figure S1). Linkers of two or three adenosines also allowed complete hybridization (lanes 5 and 7). In light of this, we tested the comparable DNA ASO with no linker adenosine and found that it hybridizes equally well (Supplementary Figure S2). A DNA ASO designed against the continuous, linear sequence at the exon 10–intron 10 junction (21 nts, from G−7 to A+13) also hybridized to the RNA (lane 9), although binding was not complete, with bands representing the tau RNA alone and the DNA ASO still observed. The complete binding of DNAs 9-1A-10, 9-2A-10 and 9-3A-10 to the discontinuous regions flanking the tau hairpin suggests that the equilibrium lies in favor of the hairpin structure over unfolded RNA under these assay conditions. DNA with a scrambled sequence did not bind the tau RNA (lane 11).
Figure 1.
Complementary bipartite ASOs can bind to the discontinuous sequences that flank the tau hairpin. (A) Diagram showing predicted RNA folding at the exon 10 (uppercase, blue): intron 10 (lowercase, red) boundary to form a hairpin secondary structure. The design of a bipartite ASO against the sequences flanking the hairpin is shown underneath in black. (B) EMSA to show the extent of hybridization of DNA ASOs to the tau hairpin-containing RNA sequence. ASOs were designed against the discontinuous sequences that flank the hairpin or against the unfolded linear sequence. A scrambled sequence was also tested. (C) EMSA showing the extent of hybridization of DNA ASOs designed separately against the 5′- and 3′-regions that flank the tau hairpin RNA. Note that unhybridized 5′-flank ASO is too small to be detected. (D) EMSA revealing the importance of binding of the 3′-flank. The hybridization of DNA ASOs with 7 or 8 bases complementary to the 5′-flank, and 0–8 bases complementary to the 3′-flank was examined.
Binding of ASOs directed to the 5′- or 3′-flank separately was also assessed using EMSA (Figure 1C). Addition of an ASO directed to the 5′-flank resulted in mobility shift (lane 3) but surprisingly, the 10 nucleotide ASO directed to the 3′-flank did not bind. When DNA ASOs against the 5′- and 3′-flanks were assessed together, the mobility of the hybrid is equivalent to that of the tau RNA bound to the 5′-flank DNA ASO alone. However, the 3′-flank-directed portion does play a role in binding of bipartite DNA ASO across the discontinuous region (Figure 1D). Bipartite DNA ASO 7-2A-7 does not hybridize (lane 2), nor does DNA 8-2A-0 (lane 3). By increasing the length of the region complementary to the 3′-flank in steps of one nucleotide, we see that 8-2A-1, 8-2A-2, 8-2A-3, 8-2A-4 and 8-2A-5 are not sufficient for binding (lanes 4–8). Interestingly, the DNA bands representing 8-2A-0 and 8-2A-1 (lanes 3 and 4) are not detected under these EMSA conditions due to their small size. However, they are not hybridized to the hairpin RNA since there is no mobility shift of the RNA in these lanes. In contrast, 8-2A-6, 8-2A-7 and 8-2A-8 bind effectively, resulting in a hybrid with slower electrophoretic mobility (lanes 9–11). This result indicates that the portion of the bipartite ASO directed to the 3′-flank is indeed involved in binding over this discontinuous region. Further evidence of binding to the 3′-flank is provided below using nuclease digestion.
In addition to DNA ASOs, RNA ASOs were tested for hybridization to the tau hairpin-containing RNA by EMSA. The RNA ASOs were modified with 2′-OMe groups and a phosphorothioate backbone to confer stability against nucleases, as these ASOs were also used to study splicing in cells in later experiments. Like their DNA counterparts, RNA ASOs 9-1A-10, 9-2A-10 and 9-3A-10 hybridized to the tau RNA, and linker length did not make a discernible difference (Figure 2A, lanes 2–4). E10B, an RNA ASO designed against the unfolded sequence at the exon 10–intron 10 junction (21 nts, from G−7 to A+13) also bound to the tau hairpin RNA, resulting in mobility shift (lane 5), whereas a scrambled RNA molecule did not hybridize (lane 6). A time course of binding showed a slower hybridization of E10B against the unfolded structure compared with RNA 9-2A-10 binding to the discontinuous region of the folded RNA, again suggesting that the equilibrium favors the hairpin structure under these assay conditions (Figure 2B and C). RNA ASO designed against only the 5′-flank was able to hybridize (Figure 2A, lane 7). Interestingly, in contrast to its DNA counterpart, RNA ASO directed only to the 3′-flank did show partial hybridization (lane 8), consistent with RNA:RNA hybrids having stronger binding affinity than RNA:DNA hybrids (15). Where these 5′- and 3′-directed RNA ASOs are included together in the hybridization reaction, both hybridize almost completely (lane 9), resulting in a band with slower mobility than the tau RNA hybridized to the 5′-directed or 3′-directed ASO alone. This result is further evidence that bipartite ASOs bind to the 3′-flank and suggests that the 3′-flank of the tau RNA becomes more available for ASO binding when the 5′-flank is also hybridized. Bipartite RNA ASOs can also bind to tau RNA with mutations that remove all complementarity in the hairpin (Supplementary Figure S3). This result suggests that while these ASOs bind to the discontinuous regions flanking the hairpin when the structure is present, they can also bind when the structure is absent, looping out the intervening non-complementary sequence as previously reported with other bipartite ASOs (16,17).
Figure 2.
RNA and PNA complementary ASOs can bind to the discontinuous sequences that flank the tau hairpin. (A) The extent of hybridization of RNA ASOs to the tau hairpin-containing RNA was tested by EMSA. Modified RNA does not stain with SYBR Gold, so bands representing unhybridized RNA ASOs are not seen on the gel. (B) EMSA showing the time-course of hybridization of 9-2A-10 and E10B RNA ASOs to the tau hairpin RNA. (C) Quantification of the intensity of the tau RNA band. Data are the mean ± S.E.M. from three independent experiments, normalized to the intensity of tau hairpin RNA alone. (D) EMSA showing the effect of increasing flank length of PNA ASOs on the extent of hybridization with the tau hairpin-containing RNA. PNA ASOs also contain lysine residues to aid cell penetration for experiments beyond the scope of the current study.
Bipartite PNA ASOs were also studied by EMSA (Figure 2D). These PNA ASOs contain linkers comprised of 0–3 ethylene glycol (egl) units, and one egl was found to be optimal for binding to the tau hairpin RNA (Supplementary Figure S4). PNA ASOs as short as 4 complementary bases on each side result in some hybridization to the tau RNA (lane 2), and the extent of hybridization increases with flank length. Since a DNA ASO composed of seven complementary nucleotides on each side could not hybridize, these RNA:PNA duplexes must have stronger binding affinity than RNA:DNA duplexes, consistent with what is known of RNA:PNA hybrids in general (15). Taken together, the EMSA results show that bipartite DNA, RNA and PNA ASOs can bind the tau hairpin-containing RNA.
Bipartite ASOs bind to both flanking regions of the tau hairpin RNA
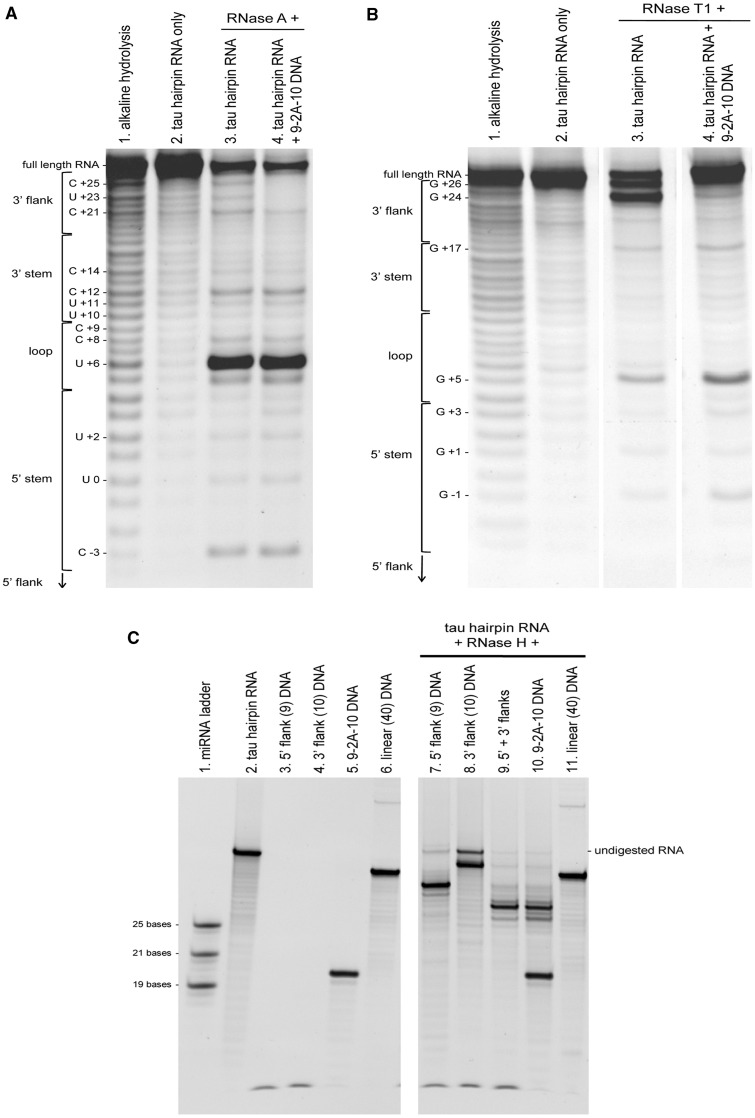
To show that these ASOs were binding where predicted, RNase protection assays were performed. In these experiments, a 5′-biotinylated version of the 40-nt synthetic tau RNA was tethered to streptavidin agarose beads and subjected to RNase A or T1 digestion in the absence or presence of DNA ASO 9-2A-10. 5′ biotinylated cleavage fragments were then analyzed by denaturing urea/acrylamide gel electrophoresis. The degradation profile of the tau RNA alone by RNase A, which cleaves 3′ of unpaired C and U residues, shows cleavage sites at C+25, U+23 and C+21 (in the 3′ flank), at C+12 (on the 3′ side of the stem) and at C-3 (on the 5′ side of the stem) (Figure 3A, lane 3). A predominant band is seen at U+6, which is found in the loop region. That the main cleavage event occurs at U+6 (and that this base must therefore be unpaired) lends further evidence for the hairpin structure, which has been described for a 25-nt RNA by nuclear magnetic resonance (NMR) spectroscopy (18) but is seen here for the first time in the presence of additional flanking sequences. In the presence of DNA ASO 9-2A-10, bands at C+25, U+23 and C+21 were reduced (lane 4), indicating that this region of RNA is protected from RNase A by hybridization to the bipartite DNA ASO. The degradation profile produced by RNase T1, which cleaves unpaired G residues, revealed predominant cleavage sites are at G+26 and G+24 (in the 3′-flank) and at G+5 (in the loop) (Figure 3B). Pre-treatment with DNA ASO 9-2A-10 results in a robust decrease of RNase T1 cleavage at G+24 and interestingly, promotes digestion within the loop region. Cleavage at G+26 may also be blocked, but the resolution between this band and the full-length RNA was not sufficient to determine this. These results are in good agreement with the RNase A footprinting results and with the proposed model of the RNA secondary structure (18). In addition, these results provide evidence that the 3′-flanking region is indeed bound by the bipartite DNA ASO.
Figure 3.
RNase digestion profiles of the 5′-biotinylated tau hairpin-containing RNA with bound DNA ASO. (A) RNase A 5′-digestion fragments of tau hairpin-containing RNA with and without protection by bipartite 9-2A-10 DNA. Digestion fragments were run alongside an alkaline hydrolysis ladder of all possible 5′-biotinylated fragments and alongside undigested RNA. Bands representing cleavage in the 5′-flank (below C−3) are too small to be resolved or detected on the gel. (B) RNase T1 digestion profile of the tau RNA as in panel A. (C) RNase H digestion of the tau RNA when bound to DNA ASOs. Digestion products were analyzed by denaturing urea/acrylamide gel electrophoresis and visualized by SYBR Gold staining under UV light.
RNase H is a nuclease which digests RNA only where it is hybridized with DNA, leaving the DNA and unhybridized regions of RNA intact. After digestion of the tau RNA:5′-flank-directed DNA ASO hybrid by RNase H (Figure 3C, lane 7), the remaining RNA is shorter than in its full length form (lane 2), indicating that the region bound by the 5′-flank-directed DNA has been degraded. When the RNA is hybridized to the 3′-flank-directed DNA, RNase H digestion occurs (lane 8), although the remaining fragment is not as short as expected, suggesting that the 3′-flank is incompletely bound, possibly due to involvement in further secondary structure. Additionally, some undigested full-length RNA remains, which may indicate that this reaction does not proceed as quickly as the digestion of the RNA:5′-flank-directed DNA hybrid. When both 5′- and 3′-flank-directed DNA ASOs are bound to the tau RNA before RNase H digestion (lane 9), the remaining RNA is shorter than the fragment left after digestion with 5′- or 3′-flank-directed DNA alone, and this RNA fragment is the same size as that seen after digestion of the hairpin RNA:9-2A-10 hybrid (lane 10). This result again suggests that ASO binding to the 5′-flanking region makes the 3′-flanking region more accessible and gives further support to the hypothesis that when the tau RNA is hybridized with 9-2A-10, both 5′- and 3′-regions flanking the hairpin are bound to the bipartite ASO.
Bipartite ASOs alter tau pre-mRNA in vitro splicing
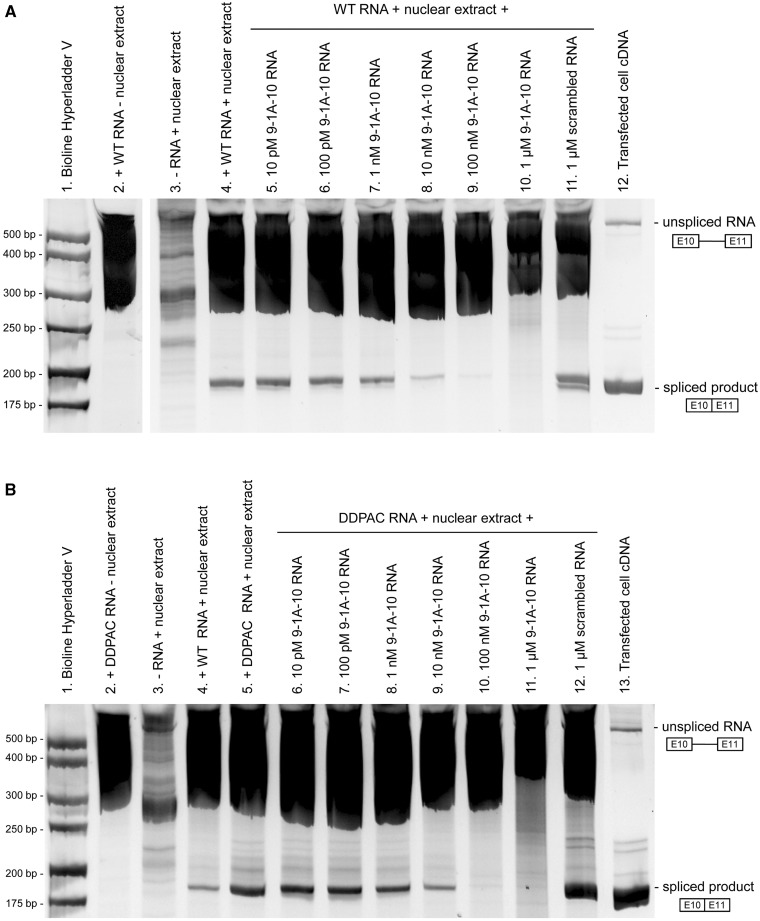
We next sought to address whether the binding of bipartite ASOs across this discontinuous region would stabilize the secondary structure sufficiently to inhibit exon 10 splicing. To this end, we designed an in vitro splicing assay to assess exon 10 splicing in a cell-free environment. The splicing unit RNA comprised exon 10, a shortened intron 10 and exon 11. The splicing unit RNA was incubated with nuclear extract from HeLa cells to allow the splicing machinery to recognize and remove intron 10, and products were detected by RT–PCR. As shown in Figure 4A, reactions containing both tau splicing unit RNA and HeLa nuclear extract lead to a band near the expected 179 bp of the correctly spliced product (lane 4), whereas tau RNA or HeLa nuclear extract alone do not (lanes 2 and 3, respectively). The splicing efficiency of our system is apparently quite low, as RT–PCR is needed to detect the spliced product, and large smears for unspliced products are seen in all lanes with splicing unit RNA, regardless of the presence of nuclear extract. Nevertheless, the presence of the spliced product is clear, and the ability of ASOs to block its formation can be assessed. Upon inclusion of 9-1A-10 RNA antisense, this band becomes less intense in a concentration-dependent manner (lanes 5–10). This antisense effect is sequence-specific since a scrambled RNA at the highest concentration tested (1 µM) had no inhibitory activity on exon 10 splicing (lane 11). cDNA reverse transcribed from RNA from SK-N-SH cells transfected with the splicing unit vector served as a positive control for the correctly spliced product (lane 12). Because antisense oligonucleotides can inhibit reverse transcription (19–22), a control experiment was performed during which 9-1A-10 RNA antisense (1 nM–1 µM) was included in the reverse transcription step. There was no block of reverse transcription of the spliced product (unsurprising, as half of this bipartite ASO is targeted to an intronic sequence) (Supplementary Figure S5), indicating that the effect of 9-1A-10 RNA antisense is due to inhibition of splicing.
Figure 4.
In vitro splicing of a tau splicing unit comprising exon 10, a shortened intron 10 and exon 11 in the absence or presence of a bipartite RNA ASO. Products from the in vitro splicing reactions were purified by phenol/chloroform precipitation and amplified by reverse transcription PCR. Unspliced RNA and spliced products were visualized by 8% acrylamide gel electrophoresis and SYBR Gold staining under UV light. In vitro splicing of a wild-type tau splicing unit (A) and a tau splicing unit containing the FTDP-17-associated DDPAC mutation (B) was inhibited by 9-1A-10 RNA in a concentration-dependent manner.
Since an aberrant increase in exon 10 splicing is a feature of certain FTD-causing tau mutations, we examined whether this bipartite ASO strategy would also be effective in inhibiting exon 10 splicing from a hairpin that is destabilized by such a mutation. The DDPAC mutation (a C to T mutation at position +14 after the exon 10–intron 10 boundary) was introduced into the splicing unit RNA. A comparison of the wild-type and DDPAC splicing units (Figure 4B, lanes 4 and 5) revealed increased exon 10 splicing for the DDPAC splicing unit, in agreement with previous reports (8,10,11,23). Exon 10 splicing from the DDPAC splicing unit RNA could also be inhibited by inclusion of RNA ASO 9-1A-10 in the splicing reaction (lanes 6–11), but not by a scrambled RNA (lane 12). Collectively, these results provide evidence that bipartite ASOs can bind to the discontinuous regions that flank the tau mRNA hairpin and inhibit exon 10 splicing in a cell-free system.
Bipartite ASOs alter tau pre-mRNA splicing in cells
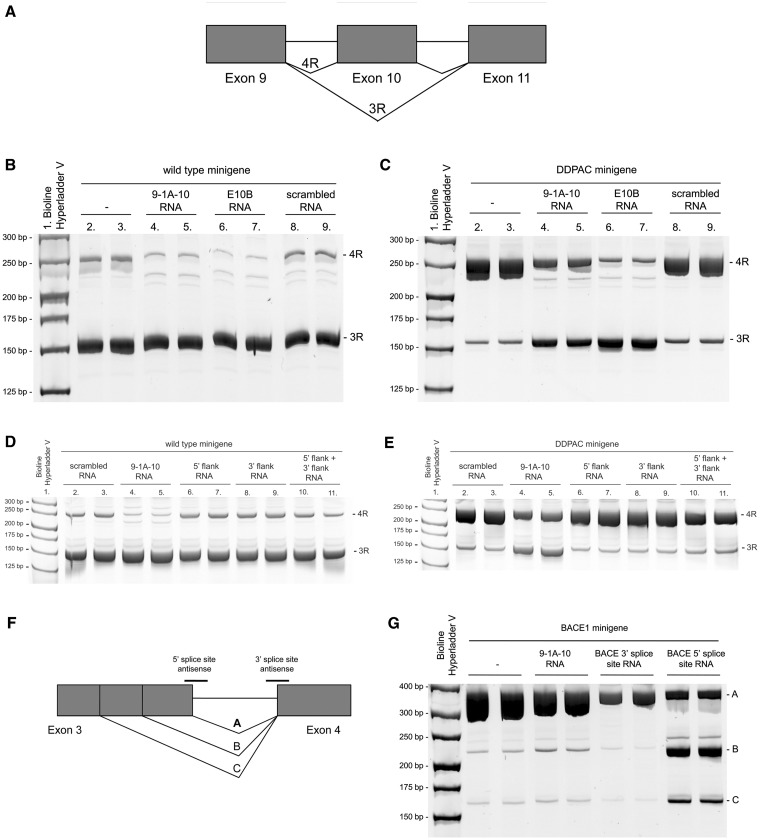
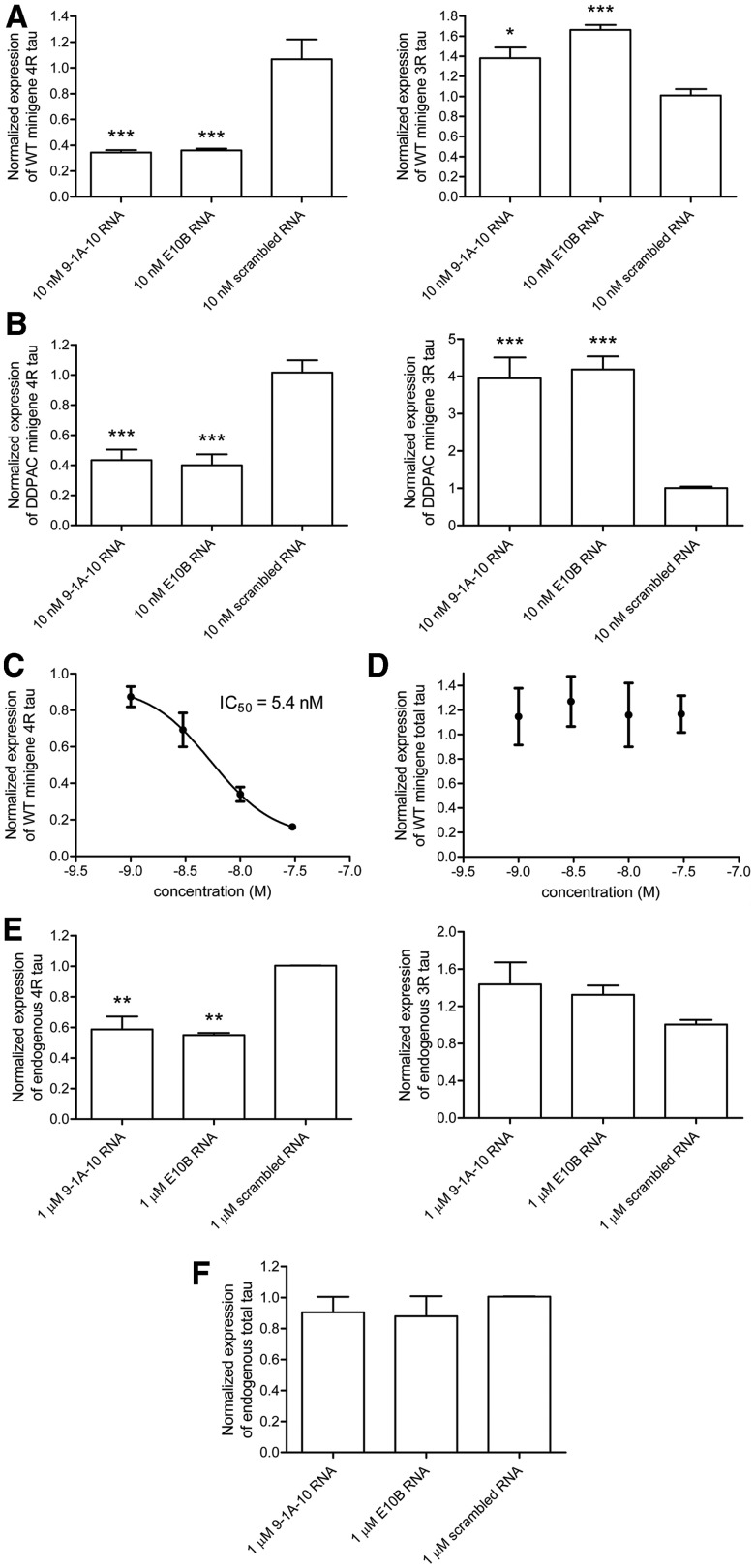
We next determined whether these RNA ASOs could inhibit exon 10 splicing from a tau minigene (11,23) in SK-N-SH neuroblastoma cells. The splicing pathway for this minigene is depicted in Figure 5A. The minigene was transfected alone or co-transfected with 9-1A-10, E10B or a scrambled RNA ASO, and exon 10 splicing was assessed by exon-trapping PCR. E10B is an antisense RNA complementary to the linear sequence of the splice site of the unfolded exon10–intron 10 boundary and has been previously shown to be effective in decreasing exon 10 splicing from a tau minigene (11,24). In cells transfected with the minigene alone, exon 10 was predominantly excluded, and the PCR product representing 3R tau was more intense than the PCR product representing 4R tau (Figure 5B, lanes 2 and 3). This deviation from the expected 1:1 ratio of 3R:4R tau that is seen in healthy adult brain may be due to a number of reasons (e.g. different 5′- and 3′-UTRs, shortened introns, absence of other exons, heterologous overexpression in undifferentiated neuroblastoma cells). Upon co-transfection with 10 nM RNA ASO 9-1A-10, the expression of the isoform representing 4R tau (in which exon 10 is included) was decreased (lanes 4 and 5). Faint additional bands at ∼220 bp are also seen which, according to in silico predictions, likely represent splicing from a cryptic donor splice site in intron 9. Co-transfection of the wild-type minigene with RNA ASO E10B resulted in a similar decrease of 4R tau (lanes 6 and 7). Co-transfection with a scrambled antisense did not alter the pattern of splicing compared with the tau minigene transfected alone (lanes 8 and 9).
Figure 5.
The effect of RNA ASOs on alternative exon splicing from a tau minigene and from a BACE1 minigene expressed in SK-N-SH cells. (A) Diagram of alternative splicing pathways of a minigene encoding tau exons 9, 10 and 11. (B) Exon-trapping PCR from the wild-type minigene co-expressed with 9-1A-10, E10B, scrambled RNA antisense or no antisense in SK-N-SH cells. PCR products were analyzed by 8% acrylamide gel electrophoresis and SYBR Gold staining under UV light. (C) Exon-trapping PCR from the DDPAC minigene expressed in SK-N-SH cells. (D) RNA ASOs against the 5′- or 3′-flank of the tau hairpin (alone or in combination) have no effect on splicing from the wild-type or DDPAC (E) minigene. (F) Diagram of alternative splicing pathways of a minigene encoding BACE1 exons 3 and 4. (G) Exon-trapping PCR from the BACE1 minigene.
As with the in vitro splicing experiments, we next sought to determine whether bipartite ASOs could inhibit exon 10 splicing from a tau minigene containing a hairpin-destabilizing FTD mutation (Figure 5C). In cells transfected with the DDPAC minigene alone (lanes 2 and 3), the ratio of 3R:4R was shifted in favor of the 4R isoform in contrast to the wild-type minigene, as predicted and previously demonstrated (8,10,11,23). Co-transfection of bipartite RNA ASO 9-1A-10 (lanes 4 and 5), targeted to the regions flanking the hairpin, or E10B RNA (lanes 6 and 7), targeted to the exon–intron junction, both resulted in correction of the splicing defect by decreasing the 4R and increasing the 3R isoforms. Co-transfection of the DDPAC minigene with a scrambled RNA (lanes 8 and 9) resulted in a splicing profile similar to that seen from the minigene transfected alone. RNA ASOs targeted to only the 5′-flanking region or only the 3′-flanking region did not alter the splicing profile of the wild-type or DDPAC tau minigene, and neither did the combination of these two ASOs (Figure 5D and E). This result reveals the importance of connecting the two ASOs into a single bipartite ASO, in order to simultaneously bind the discontinuous regions that flank the hairpin and thereby inhibit exon 10 splicing. However, the presence of the hairpin is not necessary for the bipartite RNA ASOs to alter splicing, as simultaneously disrupting five hairpin base pairs with FTD-associated mutations still allow these ASOs to shift splicing from 4R to 3R tau (Supplementary Figure S6). These results are consistent with the EMSA experiments described earlier, showing that disruption of the hairpin complementarity still allowed RNA ASO binding. The specificity of the bipartite ASOs for tau exon 10 was confirmed using a minigene encoding exons 3 and 4 of β-site APP cleaving enzyme (BACE1, Figure 5F) (13). Upon transfection of this minigene alone, variant A is the predominant isoform (Figure 5G, lanes 2 and 3); with much lower expression of variants B and C. This splicing profile is largely unchanged upon co-transfection with RNA ASO 9-1A-10 (lanes 4 and 5) with the exception of a small increase in isoform B. However, co-transfection with either of two different RNA ASOs complementary to the BACE1 minigene 3′-splice site (lanes 6 and 7) or 5′-splice site (lanes 8 and 9) resulted in the expected substantial alterations of the splicing profiles. RNA ASO 9-1A-10 was also found to have no effect on splicing from minigenes for SMN1, SMN2, ApoA-II and Casp3 (Supplementary Figure S7A, B).
The effect of the RNA ASO 9-1A-10 was further examined by quantitative real-time PCR. In order to compare the approaches of targeting the exon–intron junction with RNA ASO E10B, and targeting the regions flanking the hairpin with RNA ASO 9-1A-10, we quantified the expression of 4R and 3R transcripts from tau minigenes co-expressed in SK-N-SH cells with these antisense molecules (Figure 6). 4R tau was amplified using a forward primer across the exon 9–exon 10 boundary, whereas 3R tau was amplified by using a forward primer across the exon 9–exon 11 boundary. Compared with a scrambled RNA ASO, 10 nM 9-1 A-10 or E10B significantly reduced expression of 4 R tau from the wild-type minigene by 68 and 66%, respectively. In addition, 10 nM 9-1 A-10 and E10B significantly increased expression of 3R tau by 37 and 65%, respectively (one-way ANOVA with post-hoc Bonferroni’s test, P < 0.05) (Figure 6A). Similar results were obtained for the DDPAC tau minigene; co-transfection of the DDPAC minigene with 10 nM 9-1A-10 or E10B significantly reduced expression of 4R tau by 57 and 61%, respectively with concomitant increase in 3R tau expression (a 4-fold increase in both cases) (Figure 6B). These results are in good agreement with the exon trapping PCR data. The effect of 9-1A-10 was also shown to be concentration-dependent (Figure 6C). At these bipartite ASO concentrations, the total tau was not altered (Figure 6D).
Figure 6.
Bipartite RNA ASO inhibits exon 10 splicing from a tau minigene expressed in SK-N-SH cells and from the endogenous tau pre-mRNA in HEK-293 cells, as quantified by real-time PCR. 10 nM 9-1A-10 RNA or E10B RNA caused a significant reduction in exon 10 splicing from the wild-type minigene (A), and from the DDPAC minigene (B), with a concomitant increase in the 3R isoform compared with 10 nM scrambled control RNA. (C) The inhibition of exon 10 splicing by 9-1A-10 RNA was concentration-dependent, with an IC50 of 5.4 nM. (D) 9-1A-10 RNA ASO does not alter total tau expression from the tau minigene in SK-N-SH cells. Total tau expression from the minigene in transfected cells was normalized to the expression of pRL-TK, which was used as a transfection control. (E) Transfection of HEK-293 cells with 1 µM 9-1A-10 or E10B antisense RNA significantly inhibits exon 10 splicing from the endogenous tau pre-mRNA in comparison with transfection with a scrambled control. (F) Total endogenous tau is unaltered in cells transfected with 1 µM 9-1A-10, E10B or scrambled RNAs. Expression of total tau was normalized to expression of GAPDH.
Finally, we sought to inhibit exon 10 splicing from endogenous tau pre-mRNA. Due to the low endogenous expression of tau mRNA in SK-N-SH neuroblastoma cells, HEK-293 cells, which have higher tau expression, were used for these experiments. Cells were transfected with 0–1 µM antisense RNA, and the expression of total tau, 3R and 4R tau was analyzed after 24 h by real-time PCR. Transfection of cells with RNA ASOs 9-1A-10 or E10B resulted in a significant reduction in 4R tau (42 and 45% reductions, respectively) compared with cells transfected with a scrambled RNA ASO (Figure 6E). The concomitant increase in 3R tau was not statistically significant, possibly due to high expression of the 3R isoform compared with the 4R isoform in these cells (23). There was no change in the levels of total tau mRNA when normalized to the expression of GAPDH for any of the transfections, indicating that this was a specific effect on exon 10 splicing (Figure 6F). The bipartite RNA ASO showed a maximum effect of a 42% reduction of exon 10 splicing at 1 µM (Figure 6E); the inhibition is not complete even at this high concentration of RNA ASO, likely due to imperfect transfection efficiency. This ASO is specific for tau since it had no effect on splicing of the endogenous IG-20 gene in SK-N-SH cells (Supplementary Figure S7C). Taken together, these results demonstrate that a bipartite ASO designed to target the regions flanking the hairpin structure at the exon 10–intron 10 boundary is effective in specifically inhibiting exon 10 splicing from a tau minigene and from the endogenous tau pre-mRNA in cell culture.
DISCUSSION
Antisense technology is an important alternative to small molecules and proteins for manipulating biological systems (15), and in contrast to RNA interference, antisense approaches allow more varied effects than simply knocking down the target of interest. Among these effects is modulation of RNA splicing, which allows skipping of exons with premature stop codons, changing the proportions of protein isoforms, or shunting toward isoforms that undergo nonsense-mediated decay. Typical antisense design involves directly blocking splice sites or regulatory elements with an ASO complementary to the linear mRNA sequence (25). It is generally thought that targeting secondary structures should be avoided, as structures such as hairpins can interfere with ASO binding (26). However, structural features in mRNA can have important functions, including the regulation of alternative splicing (4), and strategies for targeting these structured regions should be developed. Here, we have focused on a hairpin motif in the pre-mRNA of the microtubule-associated protein tau as a proof of principle that targeting such structures with ASOs to promote alternative splicing is possible.
The ratio of 3R to 4R isoforms of tau, resulting from the exclusion or inclusion of exon 10, respectively, is approximately 1:1 in the adult human brain. Altering the balance of these isoforms can cause disease, as silent or intronic mutations in the tau gene that are associated with frontotemporal dementia increase the 4R-to-3R ratio from the normal 1:1 balance (8,9). Such alteration of tau splicing is sufficient for the development of tau pathology in the brain and dementia in mid-life. The inclusion or exclusion of exon 10 is controlled in part by a hairpin structure at the boundary between exon 10 and intron 10. Self-complementarity in this region was first noted upon initial discovery of genetic mutations in tau associated with FTD (8,9). A number of these disease-causing mutations disrupt the hairpin self-complementarity and thereby increase the proportion of 4R tau (7). In contrast, designed mutations that increase self-complementarity in this region, stabilizing the hairpin structure, increase the proportion of 3R tau (11). Indeed, the exclusion of exon 10 correlates with the stability of the hairpin structure (11). Further evidence for the hairpin structure is provided in this study, in which we show that the stem region of the tau hairpin is protected from RNase degradation, with cleavage occurring in the single-stranded loop region.
To target the hairpin of the tau pre-mRNA, we designed bipartite ASOs to interact simultaneously with the regions that flank either side of this structure. The two antisense stretches were connected in a single ASO, so that simultaneous binding to the 5′- and 3′-flanking regions could take place in the presence of the hairpin structure. A similar strategy had been reported from the laboratory of Alanna Schepartz (27–29), in which ‘tethered oligonucleotide probes’, linking two oligonucleotides through a polyethylene glycol spacer, were designed to interact with sequences flanking RNA structural motifs; i.e. these tethered oligonucleotides were targeted to structure, as well as sequence. Another report describes DNA aptamers that bind an engineered RNA hairpin and thereby inhibit in vitro translation when this artificial hairpin is inserted into a 5′-untranslated region (30). In addition, a recent report describes the design of oligodeoxyribonucleotides against discontinuous regions of RNA to induce bulges in order to study the action of ribonuclease mimics (16,17). However, the current work is the first demonstration to our knowledge of the use of a bipartite ASO to target a naturally occurring RNA secondary structure in order to modulate alternative splicing.
EMSAs demonstrated that bipartite ASOs do indeed bind to an RNA oligonucleotide representing the normal human tau hairpin motif and flanking sequences, that this binding requires both parts of the designed ASO and 1:1 stoichiometry. RNase digestion experiments confirmed effective binding to the 3′-flanking region, as this region was protected in the presence of a bipartite DNA ASO. Bipartite ASOs also decreased the splicing of exon 10, both in vitro and in cells, with wild-type and FTD-mutant tau minigenes, and with endogenous tau as well. Notably, the bipartite ASO does not inhibit the correct splicing of exon 9 to exon 11 (to generate the 3R tau isoform), nor does it inhibit normal or alternative splicing of control minigenes. The bipartite ASO also effectively reduced the level of the endogenous 4R tau isoform without altering total endogenous tau mRNA.
Taken together, these results indicate that the designed bipartite ASOs block tau exon 10 splicing by simultaneous binding to the 5′- and 3′-regions that flank the hairpin structure at the boundary between exon 10 and intron 10. Neither half of the bipartite ASOs binds as effectively as the bipartite ASOs themselves, and both halves of the bipartite sequence are required for inhibition of exon 10 splicing. By virtue of their bipartite nature, these ASOs can bind to tau pre-mRNA in the presence of the hairpin structure.
We believe this is the first demonstration that bipartite ASOs can target a structural motif in a pre-mRNA and thereby inhibit splicing. We have shown this strategy to be as effective for inhibition of splicing as directly blocking the linear sequence at the splice site. Targeting secondary structure to modulate splicing rather than binding directly to splice sites or regulatory sequences may be a useful strategy to improve specificity, given the degree of consensus of splice site and regulatory sequences. Recently, we reported a small molecule, mitoxantrone, that binds to and stabilizes the tau hairpin structure (31). The site of mitoxantrone interaction in the hairpin is located near the base of the stem, proximal to where the bipartite ASOs bind (32). Thus, it should be possible to connect this compound to bipartite ASOs through the linker region, to form tripartite agents that simultaneously bind the 5′-flanking region, the 3′-flanking region, and the stem of the hairpin with high potency and specificity. Such studies are presently underway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–7.
FUNDING
Zenith Fellows Award [ZEN-08-91633 to M.S.W.]; Alzheimer’s Association. Alzheimer Drug Discovery Foundation. Funding for open access charge: Alzheimer's Association.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Robin Reed, Eric Folco and Jeanne Hsu (Harvard Medical School) for providing an improved method for preparing nuclear extract from HeLa cells suitable for cell-free splicing experiments prior to publication. We also thank Jean-François Fisette for providing the modified BACE1 minigene and Dr. Ravindra Singh (Iowa State University) for SMN1, SMN2, ApoA and Caspase3 minigenes.
REFERENCES
- 1.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 4.Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 2009;35:169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Curr. Opin. Genet. Dev. 2011;21:373–379. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat. Rev. Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- 7.Wolfe MS. Tau mutations in neurodegenerative diseases. J. Biol. Chem. 2009;284:6021–6025. doi: 10.1074/jbc.R800013200. [DOI] [PubMed] [Google Scholar]
- 8.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 9.Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl Acad. Sci. USA. 1998;95:7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grover A, Houlden H, Baker M, Adamson J, Lewis J, Prihar G, Pickering-Brown S, Duff K, Hutton M. 5′ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J. Biol. Chem. 1999;274:15134–15143. doi: 10.1074/jbc.274.21.15134. [DOI] [PubMed] [Google Scholar]
- 11.Donahue CP, Muratore C, Wu JY, Kosik KS, Wolfe MS. Stabilization of the tau exon 10 stem loop alters pre-mRNA splicing. J. Biol. Chem. 2006;281:23302–23306. doi: 10.1074/jbc.C600143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 13.Fisette JF, Montagna DR, Mihailescu MR, Wolfe MS. A G-rich element forms a G-quadruplex and regulates bace1 mRNA alternative splicing. J. Neurochem. 2012;121:763–773. doi: 10.1111/j.1471-4159.2012.07680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 16.Kuznetsova IL, Zenkova MA, Gross HJ, Vlassov VV. Enhanced RNA cleavage within bulge-loops by an artificial ribonuclease. Nucleic Acids Res. 2005;33:1201–1212. doi: 10.1093/nar/gki264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baughan TD, Dickson A, Osman EY, Lorson CL. Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy. Hum. Mol. Genet. 2009;18:1600–1611. doi: 10.1093/hmg/ddp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varani L, Hasegawa M, Spillantini MG, Smith MJ, Murrell JR, Ghetti B, Klug A, Goedert M, Varani G. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc. Natl Acad. Sci. USA. 1999;96:8229–8234. doi: 10.1073/pnas.96.14.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boiziau C, Thuong NT, Toulme JJ. Mechanisms of the inhibition of reverse transcription by antisense oligonucleotides. Proc. Natl Acad. Sci. USA. 1992;89:768–772. doi: 10.1073/pnas.89.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boiziau C, Moreau S, Toulme JJ. A phosphorothioate oligonucleotide blocks reverse transcription via an antisense mechanism. FEBS Lett. 1994;340:236–240. doi: 10.1016/0014-5793(94)80145-2. [DOI] [PubMed] [Google Scholar]
- 21.Boiziau C, Larrouy B, Sproat BS, Toulme JJ. Antisense 2′-O-alkyl oligoribonucleotides are efficient inhibitors of reverse transcription. Nucleic Acids Res. 1995;23:64–71. doi: 10.1093/nar/23.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatta T, Kim SG, Nakashima H, Yamamoto N, Sakamoto K, Yokoyama S, Takaku H. Mechanisms of the inhibition of reverse transcription by unmodified and modified antisense oligonucleotides. FEBS Lett. 1993;330:161–164. doi: 10.1016/0014-5793(93)80264-u. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Z, Cote J, Kwon JM, Goate AM, Wu JY. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol. Cell Biol. 2000;20:4036–4048. doi: 10.1128/mcb.20.11.4036-4048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalbfuss B, Mabon SA, Misteli T. Correction of alternative splicing of tau in frontotemporal dementia and parkinsonism linked to chromosome 17. J. Biol. Chem. 2001;276:42986–42993. doi: 10.1074/jbc.M105113200. [DOI] [PubMed] [Google Scholar]
- 25.Du L, Gatti RA. Progress toward therapy with antisense-mediated splicing modulation. Curr. Opin. Mol. Ther. 2009;11:116–123. [PMC free article] [PubMed] [Google Scholar]
- 26.Vickers TA, Wyatt JR, Freier SM. Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res. 2000;28:1340–1347. doi: 10.1093/nar/28.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richarson PL, Schepartz A. Tethered oligonucleotide probes (TOPs): a strategy for the recognition of structured RNA. J. Am. Chem. Soc. 1991;113:5109–5111. [Google Scholar]
- 28.Cload ST, Schepartz A. Polyether tethered oligonucleotide probes. J. Am. Chem. Soc. 1991;113:6324–6326. [Google Scholar]
- 29.Cload ST, Richardson PL, Huang YH, Schepartz A. Kinetic and thermodynamic analysis of RNA binding by tethered oligonucleotide probes: alternative structures and conformational changes. J. Am. Chem. Soc. 1993;115:5005–5014. [Google Scholar]
- 30.Le Tinevez R, Mishra RK, Toulme JJ. Selective inhibition of cell-free translation by oligonucleotides targeted to a mRNA hairpin structure. Nucleic Acids Res. 1998;26:2273–2278. doi: 10.1093/nar/26.10.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Peacey E, Dickson J, Donahue CP, Zheng S, Varani G, Wolfe MS. Mitoxantrone analogues as ligands for a stem-loop structure of tau pre-mRNA. J. Med. Chem. 2009;52:6523–6526. doi: 10.1021/jm9013407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng S, Chen Y, Donahue CP, Wolfe MS, Varani G. Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by novantrone (mitoxantrone) Chem. Biol. 2009;16:557–566. doi: 10.1016/j.chembiol.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.