Abstract
Trinucleotide repeat (TNR) expansions cause at least 17 heritable neurological diseases, including Huntington’s disease. Expansions are thought to arise from abnormal processing of TNR DNA by specific trans-acting proteins. For example, the DNA repair complex MutSβ (MSH2–MSH3 heterodimer) is required in mice for on-going expansions of long, disease-causing alleles. A distinctive feature of TNR expansions is a threshold effect, a narrow range of repeat units (∼30–40 in humans) at which mutation frequency rises dramatically and disease can initiate. The goal of this study was to identify factors that promote expansion of threshold-length CTG•CAG repeats in a human astrocytic cell line. siRNA knockdown of the MutSβ subunits MSH2 or MSH3 impeded expansions of threshold-length repeats, while knockdown of the MutSα subunit MSH6 had no effect. Chromatin immunoprecipitation experiments indicated that MutSβ, but not MutSα, was enriched at the TNR. These findings imply a direct role for MutSβ in promoting expansion of threshold-length CTG•CAG tracts. We identified the class II deacetylase HDAC5 as a novel promoting factor for expansions, joining the class I deacetylase HDAC3 that was previously identified. Double knockdowns were consistent with the possibility that MutSβ, HDAC3 and HDAC5 act through a common pathway to promote expansions of threshold-length TNRs.
INTRODUCTION
Expansions of trinucleotide repeats (TNRs) are the mutagenic cause of at least 17 hereditary neurological diseases, including Huntington’s disease (HD), myotonic dystrophy type 1 (DM1) and Friedreich’s Ataxia (FRDA) (1–4). A key characteristic of TNR instability is the presence of a threshold, which is a narrow range of uninterrupted repeat units that serves as a boundary between genetically stable (short) and unstable (long) alleles. In humans, the threshold is typically 30–40 triplet repeat units (2,5). Once the threshold number of repeat units is crossed, the probability of expansions increases dramatically and disease-causing alleles can result (2,4,6).
Expansions arise from incorrect processing of TNR DNA by the metabolic machinery, likely due to the ‘corruption’ of its normal biochemical activities by the TNR (2,4,7). In fact, it is currently believed that expansions mainly arise from the presence of key promoting factors, rather than the absence of protective proteins (4,7). A large body of evidence for promoting factors comes from a number of transgenic and knock-in mouse models for HD and DM1. Several of these studies used knockout animals lacking the DNA mismatch repair factors MSH2 or MSH3, which together form the MutSβ complex (8). It was found that long, disease-causing CTG•CAG repeat tracts were largely stabilized in knockout mice lacking either MSH2 or MSH3, both in somatic cells and during intergenerational transmission (9–13). These findings indicated that MSH2 and MSH3 are promoting factors for both intergenerational and somatic instability of CTG•CAG repeats. The role of the MutSα complex, formed by MSH2 and MSH6, in TNR instability is less clear, due to conflicting reports (11,14,15). Additionally, the DNA repair factors Pms2, Ogg1 and Xpa were identified as promoting proteins of somatic CTG•CAG repeat expansions in mice (16–18). Thus key DNA repair proteins act as promoting factors of CTG•CAG expansions in several mouse models.
The mice used in the studies described above carry long TNR tracts, from 84 to >300 repeat units in length (9–13,16–18). An unanswered question is whether there are mechanistic differences between expansions of long repeat tracts and short, threshold-length alleles, or whether the same factors promote expansions of both. Yeast studies indicate that some factors act differently during expansions of long and short repeat tracts. Specifically, the nucleases Sae2 and Mre11 promote expansions of short (CTG)20 alleles (19), while the same factors protect against expansion of longer (CTG)70 tracts (20). Additionally, while expansion rates of CTG runs from 13 to 25 repeats are largely unaffected in Δrad51 and Δrad52 backgrounds (21,22), expansion rates of (CTG)70 are increased in Δrad51 and Δrad52 mutants (20). Even though these short and long tracts yield overlapping expansion sizes of +5 to +30 repeats, the different genetic dependencies provide a precedent that triplet repeat length can help determine expansion mechanism. Biochemical studies using human cell extracts indicate that MutSβ is required for repair of short CTG slip-outs (23), while purified MutSβ also binds short (CAG)n and (CTG)n hairpins in vitro (24–26). Over-expression of MutSβ in humancells did not alter CTG/CAG instability at endogenous DM1 loci of 11 or 14 repeats (23). Thus, it is not known whether MutSβ binds to threshold-length CTG•CAG tracts and/or promotes their expansion in mammalian cells.
We recently reported that histone deacetylase complexes (HDACs) promote expansions in budding yeast and human cells (19). Mutation or chemical inhibition of the yeast HDACs Rpd3L and Hda1 suppress TNR expansions (19). The human class I deacetylase HDAC3, homologous to yeast Rpd3 (27), also promotes TNR expansions in human cells, based on RNA interference (RNAi) and small molecule inhibitor experiments (19). However, it was unknown whether human class II HDACs, which are homologous to Hda1 (27), also promote expansions. We therefore investigated a possible role for class II human histone deacetylases HDAC5 and HDAC9 in modulating TNR expansions in human cells. To do this, and to test potential roles for MutSα and MutSβ in facilitating expansion of threshold-length CTG•CAG repeats, we used a previously developed genetic assay for TNR expansions in an immortalized human astrocytic line called SVG-A (19,28). Evidence is presented that suggests MutSβ, HDAC3 and HDAC5 work through a common pathway that promotes expansions of threshold-length CTG•CAG repeats.
MATERIALS AND METHODS
Shuttle vector assays and molecular analysis of protein components
For genetic assays to assess expanded TNR alleles, a shuttle vector with a CAN1 reporter was used (19), which is summarized in Supplementary Figure S1. This shuttle vector has an SV40 ori that enables plasmid replication in SVG-A cells, which express replication competent SV40 large T antigen (29,30). Background levels of expansions were measured by directly transforming yeast with the stock shuttle vector, thereby bypassing the SVG-A cells (28). Background expansion values averaged 5% compared to expansion levels that arose after passage through the SVG-A cells (Supplementary Table S1).
For experiments investigating occupancy of MSH2, MSH3 and MSH6 at CTG•CAG repeats under normal conditions in SVG-A cells (i.e. without RNAi treatment), cells were seeded in 60 mm tissue culture dishes on Day 0. Cells were then transfected on Day 1 with 5 µg shuttle vector DNA containing either TNR or randomized control sequences, using Lipofectamine 2000 (Invitrogen Corporation). After 6 h, the DMEM transfection media was replaced by DMEM supplemented with 10% foetal bovine serum. On Day 3, cells were harvested and samples were then taken for chromatin immunoprecipitation (ChIP) assay, described later.
RNAi experiments were performed with minor variations and are summarized in Supplementary Figure S1. On Day 0, SVG-A astrocytes were seeded in 60 mm tissue culture dishes, and on Day 1 the cells were transfected with ON-TARGET plus or siGenome SMARTpool siRNAs (100 nM; all from Dharmacon) using DharmaFECT 1. siRNAs were against MSH2 (L-003909), MSH3 (L-019665), MSH6 (L-019287), CtIP (sequences from (31), Mre11 (M-009271), HDAC5 (M-003498; MU-003498), HDAC3 (L-003496, M-003496), HDAC9 (M-005241) or scrambled non-targeting siRNA (D-001810). On Day 3, cells were transfected with 7 µg of shuttle vector and also re-transfected with the relevant siRNA using Lipofectamine 2000 (Invitrogen Corporation). On Day 5, cells were harvested and samples were taken for expansion assay, assessment of knockdown by western blot analysis, reverse transcription polymerase chain reaction (PCR) or ChIP. To measure expansions, plasmid DNA was extracted and concentrated by using Hirt’s alkaline lysis (32) and Amicon Ultra 50 K centrifugal filter units (Millipore). Purified plasmid DNA was digested by DpnI (New England Biolabs) and then transformed into S. cerevisiae for measurement of canavanine (100 µg/ml) resistance or into E. coli for analysis of total plasmid numbers as measured by ampicillin-resistant colonies [Supplementary Figure S1; (19)]. Expansions were confirmed by PCR as described (19).
Western blot analysis
To assess knockdown of specific proteins following RNAi experiments, SVG-A cells were washed with ice-cold PBS (137 mM NaCl; 2.7 mM KCl; 4.3 mM Na2HPO4; 1.47 mM KH2PO4; pH 7.4) and re-suspended in RIPA buffer (150 mM NaCl, 10 mM Tris–HCl pH 7.5, 0.1% SDS, 0.1% Triton X, 1% sodium deoxycholate, 5 mM EDTA) at a concentration of 106/50 μl. Samples were incubated on ice for 30 min, followed by centrifugation at 4°C for 30 min. Supernatants containing whole-cell extracts were retained.
Whole-cell lysates were separated electrophoretically and transferred to PVDF membranes. Primary antibodies were against MSH2 (NA26, Calbiochem), MSH3 [kindly provided by Prof. Glenn Morris, Wolfson Centre for Inherited Neuromuscular Disease, UK; (33)], MSH6 (610919, BD Transduction Laboratories), CtIP [kindly provided by Prof. Richard Baer, Columbia University, USA; (34)], Mre11 (sc-5859, Santa Cruz Biotechnology), HDAC3 (sc-11417, Santa Cruz; ab16047, Abcam), HDAC5 (CH00150, Coriell Institute for Medical Research; ab1439, Abcam) and HDAC9 (CH00172, Coriell Institute for Medical Research; ab18970, Abcam). Actin (A2066, Sigma) and α-tubulin (T9026, Sigma) were used as loading controls.
Assessment of HDAC3 expression via western blot analysis resulted in two bands of ∼50 kDa, the predicted size of the protein, presumably representing the two reported isoforms of HDAC3 (35). Throughout all experiments, consistent knockdown of the top band was observed following HDAC3 siRNA treatment, however levels of the bottom band varied between experiments. Quantification of HDAC3 knockdown was performed by densitometric analysis of the top band only. Secondary antibodies conjugated to horseradish peroxidase were 711-035-152 (anti-rabbit), 705-035-147(anti-goat) and 115-035-003 (anti-mouse), all from Jackson ImmunoResearch Laboratories. Visualization was by chemiluminescence (Western Lightning Plus-ECL, PerkinElmer).
RNA isolation and reverse transcription PCR
RNA was isolated using a Qiagen RNeasy Kit. cDNA synthesis was performed using Precision nanoScript Reverse Transcription kit (Primer Design, Southampton, UK). For real-time PCR analysis of transcript levels, cDNA was analysed using SYBR GreenMaster Mix (Applied Biosystems) on the 7500 Fast Real-Time PCR system (Applied Biosystems). Each sample was assayed in triplicate for every run. Primer efficiencies were validated by performing a relative standard curve with a 2-fold serial dilution series for cDNA samples. Primer sequences and concentrations are listed in Table 1. Using the ΔΔCt method (36), normalization for cDNA quantity was performed with HPRT control primers for each template. Abundance values were expressed relative to scrambled siRNA, which was arbitrarily defined as 100%.
Table 1.
PCR primers
| Region/gene | Forward primer | Reverse primer | Conc. (nM) | Reference |
|---|---|---|---|---|
| HPRT (RT–PCR) | TGACACTGGCAAAACAATGCA | GGTCCTTTTCACCAGCAAGCT | 300 | (39) |
| HDAC3 (RT–PCR) | CTGGCTTCTGCTATGTCAAC | ACATATTCAACGCATTCCCCA | 300 | (40) |
| HDAC5 (RT–PCR) | GTCTCGGCTCTGCTCAGTGTAGA | GGCCACTGCGTTGATGTTG | 300 | (41) |
| HDAC9 (RT–PCR) | CAGGCTGCTTTTATGCAACA | TTTCTTGCAGTCGTGACCAG | 300 | (42) |
| Adh1 promoter (ChIP) | ACTTCTTTCTCTCTCGCTTTCCT | CAAGTAACATCGGCAAAGCA | 50 | This study |
| MSH2 (RT–PCR) | GGCTTCTCCTGGCAATCTCT | CCCAACTCCAACCTGTCTCT | 300 | PrimerDesign, Ltd. (Southampton, UK) |
Chromatin immunoprecipitation
ChIP assays were performed as described by Koch et al. (37), with some modifications. Cell pellets from 1 × 106 SVG-A cells transfected with shuttle vector were re-suspended in ∼1.5× pellet volume of Cell Lysis Buffer (10 mM Tris–HCl, pH 8.0; 10 mM NaCl; 0.2% NP-40; 10 mM sodium butyrate; 50 μg/ml PMSF; 1 μg/ml leupeptin). Following incubation on ice for 10 min, cell nuclei were collected by centrifugation at 4°C. Nuclei were then re-suspended in 300 µl Nuclear Lysis Buffer (50 mM Tris–HCl, pH 8.0; 10 mM EDTA; 1% SDS; 10 mM sodium butyrate; 50 μg/ml PMSF; 1 μg/ml leupeptin) and were incubated on ice for 10 min. Following lysis of nuclei, 180 µl IP Dilution Buffer (20 mM Tris–HCl, pH 8.0; 150 mM NaCl; 2 mM EDTA; 1% Triton X-100; 0.01% SDS; 10 mM sodium butyrate; 50 μg/ml PMSF; 1 μg/ml leupeptin) was added. Chromatin was sonicated using a Bioruptor UCD-200 Sonication System (Diagenode) to generate fragments of ∼200 to ∼500 bp. Following dilution of chromatin with IP Dilution Buffer to 1.5 ml, 300 µl of this chromatin was used to set up each ChIP reaction. IP reactions were set up with 5 µg MSH2 antibody (ab16833, Abcam), 5 µg MSH3 antibody (ab74607) and 2.5 µg MSH6 antibody (sc-10798, Santa Cruz), while a FLAG antibody (A8592, Sigma) was used as a negative control to assess levels of background. Following overnight antibody incubation, 30 µl Protein G Magnetic Beads (S1430S, NEB) were added and incubated with rotation for 4 h at 4°C. After the wash steps, immune complexes were eluted with 100 µl Elution Buffer 1 (50 mM Tris–HCl, pH 8.0; 10 mM EDTA; 1% SDS) and incubated at 65°C for 10 min. The samples were vortexed briefly and the supernatants were transferred into a new tube. The beads were then re-suspended in 100 µl Elution Buffer 2 (50 mM Tris–HCl, pH 8.0; 10 mM EDTA; 0.67% SDS) and vortexed briefly. The supernatant from this second elution step was added to that of the first elution. Following reversal of cross-links by incubation at 65°C overnight, proteinase K (03115879001, Roche) digests were performed at 37°C for 2 h. After phenol:chloroform extraction and precipitation, samples were re-suspended in 10 μl dH2O. Real-time PCR analysis was carried out using 0.125 ng of each ChIP sample as template.
ChIP data are averaged over five independent experiments with real-time PCR performed in triplicate for each. Signals were calculated as described by Pfaffl (38) using the following formula: fold increase = EΔCt(input−IP)/EΔCt(input−background). Primer sequences and concentrations used for the ChIP PCRs are listed in Table 1. The position of the real-time PCR primers and ChIP amplicon relative to the (CTG)22 tract or randomized (C,T,G)22 sequence is shown in the schematic in Figure 2B.
Figure 2.
(A) MSH2 and MSH3, but not MSH6, are enriched at the (CTG)22 repeat tract in human SVG-A cells. Cells were transfected with a shuttle vector containing either a (CTG)22 repeat tract (red bars) or a corresponding randomized (C,T,G)22 sequence as a control (white bars). ChIP reactions were performed subsequently with antibodies specific for MSH2, MSH3 or MSH6, while background signals were assessed using a FLAG tag antibody. Real-time PCR signals for occupancy of MSH2, MSH3 and MSH6 are presented as fold enrichment over background signals. Error bars, ±SEM; *P < 0.05 compared to randomized (C,T,G)22 control; n = 5. (B) Schematic showing position of real-time PCR primers for ChIP experiments, relative to the (CTG)22 tract or randomized (C,T,G)22 sequence. The ChIP amplicon is 89 bp, with 69 bp of flanking sequence between the amplicon and the repeat tract or control sequence.
Statistical analyses
All P-values were determined by two-tailed Student’s t-tests, except for the ChIP experiments in Figure 2. In this case, one-tailed Student’s t-tests were used to assess enrichment of specific factors at the repeat tract. P- and n-values for each data set are specified in each of the figure legends.
RESULTS
Rationale
This study focuses on expansions of repeats near the crucial threshold length of ∼30–40 repeats. The goal is to understand better the genesis of these disease-initiating mutations in human cells. The approach takes advantage of a unique genetic assay for expansions in a human astrocytic cell line, SVG-A [Supplementary Figure S1; (19)]. The assay quantitatively monitors changes from a starting (CTG)22 tract to final lengths of 26–46 repeats. SVG-A cells are amenable to siRNA manipulation to investigate the functional consequences of knockdowns on expansions, and ChIP is used to measure the physical presence of relevant proteins at the TNR (Supplementary Figure S1). Together, these aspects allowed us to test candidate proteins, identified in other studies, for their potential mechanistic roles in driving threshold-length expansions. In several cases, genetic interactions were also examined to identify whether these proteins function together to promote expansions.
The mismatch repair proteins MSH2 and MSH3 promote expansions and are enriched at the TNR
In several HD and DM1 mouse models, MSH2 and MSH3 promote both intergenerational and somatic instability of long CTG•CAG repeat tracts (9–13,15). Biochemical studies using human cell extracts indicate that MutSβ is also required for repair of short CTG slip-outs in vitro (23). Therefore, in the current study, we investigated whether MutSβ promotes expansions of short, threshold-length TNR tracts in human cells. We also investigated MSH6, for which there are conflicting reports regarding its role in TNR instability (11,14,15).
If MutSβ promotes expansions in SVG-A cells, then siRNA knockdown of either MSH2 or MSH3 should result in lower expansion frequencies. This result was observed. Knockdown of MSH2 and MSH3 resulted in 66% and 62% reductions in expansion frequency, respectively, while knockdown of MSH6 did not change the expansion phenotype (Figure 1A). Knockdown efficiencies were similar for all three proteins (68–79%; Figure 1B and Supplementary Figure S2A). Knockdown of MSH6 to a similar level in a human fibroblast line led to suppression of MMR-induced double-strand breaks following chromium treatment (43), indicating that the absence of a phenotype in our system is unlikely to be due to residual expression of MSH6. The expansion data (Figure 1A) indicate that MutSβ, and not MutSα, promotes expansions of threshold-length CTG•CAG repeats in SVG-A cells. Analysis of the spectra of expansion sizes in MSH2 and MSH3 knockdown cells revealed an overlap with that of scrambled siRNA control cells (range +4 to +21) (Supplementary Figure S3), indicating that loss of MutSβ affected expansions throughout the entire size range.
Figure 1.
Expansions are suppressed following siRNA knockdown of MSH2 and MSH3, but not by knockdown of MSH6, CtIP or Mre11. (A) Expansion frequencies subsequent to treatment with MSH2 siRNA, MSH3 siRNA, MSH6 siRNA, CtIP individual siRNAs (denoted #1 and #2) and Mre11 siRNA. All frequencies are normalized to scrambled siRNA, denoted as ‘Scr’ (white bars). Error bars denote ±1 SEM; *P < 0.05 compared to scrambled siRNA control; n = 4 for MSH2 and MSH6 siRNA, n = 3 for MSH3, CtIP and Mre11 siRNA. (B) Quantification of protein levels following knockdown, normalized to actin and to the scrambled siRNA control. Error bars denote ±1 SEM; n = 3.
MutSβ is thought to exert its influence on expansions through direct interaction with the TNR, since the pure protein binds CAG and CTG hairpins in vitro (24–26). Additionally, ChIP experiments indicated that both MSH2 and MSH3, but not MSH6, were enriched immediately downstream of a long GAA repeat tract in induced pluripotent stem cells (iPSCs) derived from FRDA fibroblasts compared to control iPSCs derived from normal fibroblasts (44). In this experiment, the length of the repeat tract varied between the two cell lines. To address whether MutSβ and/or MutSα is enriched at short threshold-length CTG•CAG repeats in human cells, ChIP experiments were performed. Parallel cultures were transfected with either the shuttle vector containing a (CTG)22 repeat tract or a control plasmid that was identical except for a randomized order of nucleotides, denoted (C,T,G)22. ChIP was performed to assess occupancy of MSH2, MSH3 and MSH6 at the repeat and the randomized sequence (Figure 2B). Both MSH2 and MSH3 were specifically enriched at a (CTG)22• (CAG)22 tract compared to the randomized control plasmid (Figure 2A). In contrast, there was no enrichment for MSH6. The ChIP data are consistent with the expansion frequency results (Figure 1). Together, they imply a direct role for MutSβ, but not MutSα, in promoting expansions of threshold-length CTG•CAG repeats in SVG-A cells.
The DNA repair nucleases CtIP and Mre11 were also assessed for potential roles in promoting expansions. Recent findings with S. cerevisiae indicated that expansion frequencies were suppressed in sae2 and mre11 mutants (19), the yeast homologs of CtIP and Mre11. If CtIP and Mre11 also promote expansions, one would predict that knockdown of these proteins would suppress expansions. Contrary to this hypothesis, knockdown of either CtIP (using two customized individual siRNA oligonucleotides) or Mre11 gave no detectable changes in expansion frequency (Figure 1A). The knockdown efficiencies were 65–77% (Figure 1B; Supplementary Figure S2B and C). Similar levels of knockdown led to measurable phenotypes in other studies. For example, knockdown of Mre11 to similar levels sensitized human adenocarcinoma cells to ionizing radiation (45). Additionally, knockdown of CtIP to similar levels reduced the ability of its interacting protein AdE1A to transactivate a luciferase reporter (46). In light of these reports, it seems less likely that residual protein levels are masking any expansion phenotype in our system. Therefore, we did not find any evidence to suggest that either CtIP or Mre11 drive CTG•CAG expansions in SVG-A cells.
MSH2 acts in the same pathway as HDAC3 in promoting CTG•CAG repeat expansions
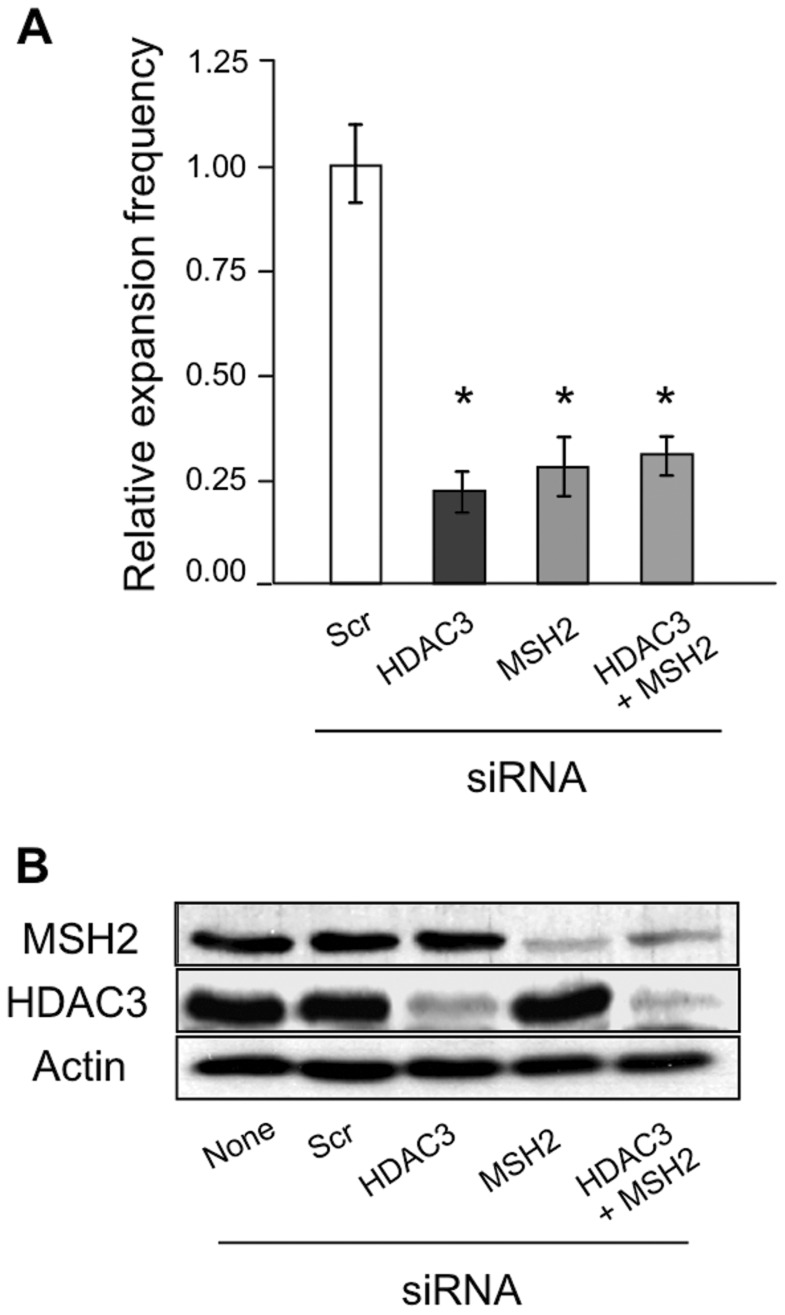
We investigated whether HDAC3, which was previously identified as a promoting factor of CTG•CAG expansions (19), might act in the same pathway as MSH2 to promote CTG•CAG instability. To test this, expansions were measured in SVG-A cells in which both MSH2 and HDAC3 were knocked down simultaneously. Double knockdown of MSH2 and HDAC3 led to an expansion phenotype that was indistinguishable from knockdown of either MSH2 or HDAC3 alone (Figure 3A). Efficiencies for the single and double knockdowns, measured as residual MSH2 and/or HDAC3 protein levels, were 17–21% of control levels (Figure 3B and Supplementary Figure S2D). About 75% of expansions were suppressed in both the single and double knockdown situations, consistent with previous work on HDAC3 (19). Since the background in the expansion assay is 5% (Supplementary Table S1), we infer that the dynamic range of the assay allows measurement of up to 95% suppression. Thus the double knockdowns at 75% suppression are consistent with epistasis, not due to an artificial limit of the assay. The finding of epistasis implies that HDAC3 and MutSβ act in a common pathway that promotes expansions of CTG•CAG repeats in SVG-A cells.
Figure 3.
Double knockdown of HDAC3 and MSH2 leads to an expansion phenotype that is indistinguishable from knockdown of either protein alone. (A) Expansion frequencies following separate siRNA treatments against HDAC3 and MSH2 and a simultaneous HDAC3+MSH2 siRNA treatment. All frequencies were normalized to scrambled siRNA (‘Scr’, white bar). Error bars denote ±1 SEM; *P < 0.05 compared to scrambled control; n = 3. (B) Representative western blot confirming knockdown of HDAC3 and MSH2 expression following siRNA treatments. Actin was used as a loading control.
Two potential mechanisms were tested. First, it was hypothesized that HDAC3 might control access of MutSβ to the repeat tract, for example through chromatin modification. If so, knockdown of HDAC3 should reduce MutSβ occupancy at the TNR. ChIP data; however, showed no detectable difference in MutSβ binding to the (CTG)22 tract upon HDAC3 knockdown (Figure 4A), with the MutSα subunit MSH6 acting as a negative control. The second potential mechanism is that HDAC3 ablation alters transcription or protein stability of MSH2 or MSH3. Previous microarray studies showed that shRNA knockdown of HDAC3 in a colon cancer cell line led to 50% and 37% reductions in transcript levels for MSH2 and MSH3, respectively (47). Immunoblot analysis of SVG-A cells following HDAC3 depletion showed no apparent change (<2%) in steady-state levels of either MSH2 or MSH3 compared to treatment with scrambled siRNA (Figure 4B). In summary, we could find no compelling evidence in SVG-A cells to suggest that HDAC3 promotes CTG•CAG expansions by controlling access of MutSβ to the repeat tract or by regulating MSH2 or MSH3 protein levels.
Figure 4.
RNAi knockdown of HDAC3 does not affect occupancy of MSH2, MSH3 or MSH6 at (CTG)22 repeat tracts, or protein expression of MSH2 or MSH3, in SVG-A cells. (A) SVG-A cells were transiently transfected with a (CTG)22-repeat containing shuttle vector, in the presence of scrambled siRNA (white bars) or HDAC3 siRNA (navy bars). ChIP reactions were performed subsequently with antibodies specific for MSH2, MSH3 or MSH6, while background signals were assessed using a FLAG tag antibody. Signals for occupancy of MSH2, MSH3 and MSH6 are presented as fold enrichment over background signals. Error bars, ±SEM; *P < 0.05 compared to control; n = 5. (B) Representative immunoblots for MSH2 and MSH3 protein expression in SVG-A cells following transfection with scrambled siRNA or HDAC3 siRNA. For assessment of MSH2 and MSH3 protein expression, α-tubulin was used as a loading control. For assessment of HDAC3 protein expression, actin was used as a loading control.
HDAC5, a class II HDAC, also promotes expansions
In yeast, mutations in subunits of Rpd3L suppress expansions (19). Human HDAC3 is a class I enzyme, homologous to yeast Rpd3 (27). Similarly, mutation of the yeast Hda1 complex also suppresses expansions (19), so we tested human class II HDACs, which are homologous to Hda1 (27). Knockdown of the class II enzyme HDAC5 reduced expansion frequencies by 62% using a combined pool of four siRNAs, and by 76% for two individual siRNAs denoted #5 and #7 (Figure 5A). Knockdown efficiency, measured as residual HDAC5 transcript level, was 38–48% of control (Figure 5B). There was no detectable difference in expansion size range, weighted mean or median between HDAC5 knockdowns and control values (Supplementary Figure S3B). The activity of HDAC5 on expansions was specific, since knockdown of another class II enzyme HDAC9 actually increased expansion levels (Supplementary Figure S4).
Figure 5.
Single and double knockdowns of HDAC3 and HDAC5 lead to similar reductions in expansions. (A) Expansion frequencies subsequent to treatment with HDAC5 SMARTpool siRNA (labelled HDAC5 in the graph; n = 6), individual HDAC5 siRNAs (labelled #5 and #7; n = 3), HDAC3 siRNA (n = 3), HDAC5 SMARTpool siRNA (n = 3) and simultaneous HDAC3 and HDAC5 siRNA treatments (n = 3). Error bars denote ± 1 SEM; *P < 0.05 compared to scrambled siRNA control. (B) Expression levels of HDAC5 and HDAC3 determined by real-time RT–PCR normalized to scrambled siRNA and HPRT levels following siRNA treatment with HDAC5 SMARTpool (n = 3), HDAC5 siRNAs #5 and HDAC #7 (n = 2), HDAC3 SMARTpool (n = 2), separate siRNA treatments against HDAC3 and HDAC5 (n = 2) and a simultaneous HDAC3+HDAC5 siRNA treatment (n = 2). Error bars denote range.
Since both HDAC3 and HDAC5 promote expansions in our system, we investigated whether they did so through a common pathway. Expansions were measured in SVG-A cells in which both HDAC3 and HDAC5 proteins were knocked down simultaneously. Simultaneous knockdown of HDAC3 and HDAC5 resulted in a significant, 67% decrease in expansions that was comparable to the phenotype observed following single knockdowns for HDAC3 and HDAC5 (56–76% decreases in expansions; Figure 5A). Knockdown efficiencies were similar for the single and double siRNA treatment (Figure 5B). Together, these findings imply that the class I enzyme HDAC3 and the class II deacetylase HDAC5 promote CTG•CAG expansions through a common pathway. Similarly, HDAC5 was tested for epistasis with MutSβ by double knockdowns. Treatment with siRNA against either HDAC5 or MSH2 resulted in reductions in expansion frequency of 79% and 77%, respectively, similar to our earlier results. The HDAC5 MSH2 double knockdown suppressed expansions by about the same extent, 68% (Figure 6A). Expression levels of HDAC5 and MSH2 were similar whether single or double siRNA was utilized (Figure 6B). In summary, the double knockdown experiments in Figures 3A, 5A and 6A can be explained most easily as genetic interactions within a common pathway of MutSβ, HDAC3 and HDAC5 (Figure 6C).
Figure 6.
Double knockdown of HDAC5 and MSH2 leads to an expansion phenotype that is indistinguishable from knockdown of either protein alone. (A) Expansion frequencies following separate siRNA treatments against HDAC5 or MSH2 or a simultaneous HDAC5+MSH2 siRNA treatment. All frequencies were normalized to scrambled siRNA. Error bars denote ±1 SEM; *P < 0.05 compared to scrambled control; n = 3–4. (B) Expression levels of HDAC5 and MSH2 determined by real-time RT–PCR. Expression was normalized to scrambled siRNA and HPRT levels following siRNA treatment with individual HDAC5 or MSH2 SMARTpool siRNAs or a simultaneous HDAC5+MSH2 siRNA treatment (n = 2). Error bars denote range. (C) Schematic diagram of the genetic relationship between HDAC3, HDAC5 and MutSβ in regard to TNR expansions. Double-headed arrows signify genetic epistasis, as inferred from siRNA double knockdowns.
DISCUSSION
This study revealed that the mismatch repair protein MutSβ promotes expansions of threshold length TNR tracts in human astrocytes. Knockdown of either MSH2 or MSH3 with siRNA led to as much as a 66% reduction in expansion frequency, whereas knockdown of the MutSα subunit MSH6 did not lead to any detectable effects. We addressed the question as to whether the MutSβ or MutSα complex is enriched at short CTG•CAG repeats, since previous experiments indicated that MSH2 and MSH3, but not MSH6, were enriched downstream of long GAA repeat tracts in FRDA iPSCs (44). In our study, we used parallel cultures of SVG-A cells transfected with either a shuttle vector containing a single CTG•CAG repeat tract or a control plasmid that was identical except for a randomized control sequence. We found that MutSβ, but not MutSα, is specifically enriched at short CTG•CAG repeats compared to the control sequence. These findings indicate that MutSβ facilitates the majority of the critical initiating expansions that cross the TNR threshold and lead to enhanced instability. Our results are consistent with a large body of evidence from several mouse models indicating a role for MutSβ in promoting expansions of long disease-causing alleles (9–13). The current study also identified the class II deacetylase HDAC5 as a novel promoting factor of CTG•CAG expansions, adding to the recent finding that the class I enzyme HDAC3 facilitates expansions (19). Additionally, double knockdowns in all three combinations were consistent with the possibility that MutSβ, HDAC3 and HDAC5 act through a common pathway to promote expansions of threshold TNR tracts in human cells (Figure 6C). The mechanistic and therapeutic implications of these findings are considered below.
A recent study identified HDAC3 as a novel promoting factor of TNR expansions, while it appears that other class I candidates HDAC1 and HDAC2 are unlikely to play a role in expansion of threshold-length CTG•CAG repeats (19). HDAC3 has been shown to be highly expressed in the striatum (48) where expansion is particularly frequent in HD patients (49). Furthermore, HDAC3 is present in striatal tissue from R6/2 HD mice at early and terminal disease stages (50). The class II deacetylase HDAC5 is also highly expressed in neuronal nuclei of the striatum and cortex in human HD brains (51). Furthermore, in yeast both HDACs Rpd3L and Hda1, prototypes of human class I and class II enzymes, respectively, promote expansions (19). Therefore, in the current study, we investigated whether HDAC5 also plays a role in TNR expansions. We found that depletion of HDAC5 led to a significant reduction in expansions. Additionally, double knockdown of HDAC3 and HDAC5 led to an expansion phenotype that was similar to those following knockdown of either HDAC3 or HDAC5 alone (Figure 5A). These findings indicate a mechanistic link between the two HDACs in facilitating expansions. Since HDAC3 and HDAC5 are reported to interact physically (52), we suggest the possibility that they may physically and functionally cooperate to promote expansions of CTG•CAG repeats. To investigate if other HDACs also promote expansions, we investigated another class II deacetylase HDAC9. Knockdown of HDAC9 increased expansions in our system. This indicates that HDAC9 normally plays a protective role in preventing against expansions, suggesting that specific members of the HDAC family regulate expansions in a differential manner.
How might HDACs promote expansions? We tested three possibilities. The first idea was based on recent findings in S. cerevisiae indicating that the nuclease Sae2 is deacetylated and thereby stabilized in an Rpd3 and Hda1 dependent manner (53) and that expansions were suppressed in sae2 and mre11 mutants (19). Therefore, we investigated the human homologs of these DNA repair nucleases, CtIP and Mre11, respectively. Knockdown of either CtIP or Mre11 did not suppress expansions in SVG-A cells. A probable explanation for this outcome is that CtIP is deacetylated by the class III HDAC, SIRT6, with no reported involvement for class I or II HDACs (54). The other two hypotheses derived from double knockdowns of MSH2 and HDAC3, which suggested that these factors operate in a common pathway. The second possibility was that HDAC3 controls access of MutSβ to the repeat tract, possibly through chromatin modification. If this were the case, knockdown of HDAC3 should reduce MutSβ occupancy at the TNR. ChIP experiments, however, failed to indicate a change in MutSβ binding to the (CTG)22 tract following HDAC3 knockdown. The final hypothesis we tested is that HDAC3 depletion might alter transcription or protein turnover of MSH2 or MSH3. Previous microarray studies using a colon cancer cell line SW480 showed that shRNA knockdown of HDAC3 led to moderately decreased mRNA levels for both MSH2 and MSH3 (47). In SVG-A cells, however, HDAC3 depletion did not lead to any appreciable changes in steady-state protein levels of either MSH2 or MSH3 (Figure 4B). Due to the absence of any reduction in MSH2 or MSH3 levels or changes in occupancy of MutSβ at the repeat tract, other possibilities need to be considered. One explanation yet to be investigated is that HDAC3 depletion might lead to increased acetylation of MSH2, thereby regulating MutSβ function without changing its localization or expression. Mass spectrometry identified putative MSH2 acetylation sites at lysine 555 and lysine 635, which are within its MSH3/MSH6-binding domain (55). A final possibility is that HDAC3 depletion affects the expression, TNR occupancy or function of another unidentified factor that functionally interacts with MutSβ to promote expansions.
The current study presents new evidence to suggest that both HDAC3 and HDAC5 are relevant pharmacological targets for TNR expansion diseases due to their role as promoters of CTG•CAG instability. HDAC inhibitors are currently being evaluated for their therapeutic value in treating the transcriptional defects in several TNR expansion diseases (56,57). As well as relieving disease phenotype and transcriptional abnormalities, our work implies that specific HDAC inhibitors may have the added benefit of suppressing somatic expansions that lead to disease progression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
Science Foundation Ireland [10/IN.1/B2973 to R.S.L.]; NUI Galway’s Millenium Fund (to R.S.L.); the Irish Research Council for Science, Engineering and Technology (to A-M.M.G. and A.F.); the Thomas Crawford Hayes Fund (to A.F.); the National University of Ireland, Galway (to E.H.). Funding for open access charge: Science Foundation Ireland [10/IN.1/B2973].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 3.Kovtun IV, McMurray CT. Features of trinucleotide repeat instability in vivo. Cell Res. 2008;18:198–213. doi: 10.1038/cr.2008.5. [DOI] [PubMed] [Google Scholar]
- 4.Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell. Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 5.Paulson HL, Fischbeck KH. Trinucleotide repeats in neurogenetic disorders. Annu. Rev. Neurosci. 1996;19:79–107. doi: 10.1146/annurev.ne.19.030196.000455. [DOI] [PubMed] [Google Scholar]
- 6.Leeflang EP, Zhang L, Tavare S, Hubert R, Srinidhi J, MacDonald ME, Myers RH, de Young M, Wexler NS, Gusella JF, et al. Single sperm analysis of the trinucleotide repeats in the Huntington's disease gene: quantification of the mutation frequency spectrum. Hum. Mol. Genet. 1995;4:1519–1526. doi: 10.1093/hmg/4.9.1519. [DOI] [PubMed] [Google Scholar]
- 7.McMurray CT. Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease. DNA Repair. 2008;7:1121–1134. doi: 10.1016/j.dnarep.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peltomaki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum. Mol. Genet. 2001;7:735–740. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- 9.Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 10.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 11.van den Broek WJAA, Nelen MR, Wansink DG, Coerwinkel MM, te Riele H, Groenen PJTA, Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 12.Savouret C, Brisson E, Essers J, Kanaar R, Pastink A, te Riele H, Junien C, Gourdon G. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler VC, Lebel L-A, Vrbanac V, Teed A, te Riele H, MacDonald ME. Mismatch repair gene Msh2 modifies the timing of early disease in HdhQ111 striatum. Hum. Mol. Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- 14.Foiry L, Dong L, Savouret C, Hubert L, te Riele H, Junien C, Gourdon G. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum. Genet. 2006;119:520–526. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- 15.Dragileva E, Hendricks A, Teed A, Gillis T, Lopez ET, Friedberg EC, Kucherlapati R, Edelmann W, Lunetta KL, MacDonald ME, et al. Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiol. Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomes-Pereira M, Fortune MT, Ingram L, McAbney JP, Monckton DG. Pms2 is a genetic enhancer of trinucleotide CAG•CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum. Mol. Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 17.Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubert L, Jr, Lin Y, Dion V, Wilson JH. Xpa deficiency reduces CAG trinucleotide repeat instability in neuronal tissues in a mouse model of SCA1. Hum. Mol. Genet. 2011;20:4822–4830. doi: 10.1093/hmg/ddr421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debacker K, Frizzell A, Gleeson O, Kirkham-McCarthy L, Mertz T, Lahue RS. Histone deacetylase complexes promote trinucleotide repeat expansions. PLoS Biol. 2012;10:e1001257. doi: 10.1371/journal.pbio.1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sundararajan R, Gellon L, Zunder RM, Freudenreich CH. Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics. 2010;184:65–77. doi: 10.1534/genetics.109.111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miret JJ, Pessoa-Brandao L, Lahue RS. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharyya S, Lahue RS. Yeast Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Mol. Cell. Biol. 2004;24:7324–7330. doi: 10.1128/MCB.24.17.7324-7330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panigrahi GB, Slean MM, Simard JP, Gileadi O, Pearson CE. Isolated short CTG/CAG slip-outs are repaired efficiently by hMutSbeta, but clustered slip-outs are poorly repaired. Proc. Natl Acad. Sci. USA. 2010;107:12593–12598. doi: 10.1073/pnas.0909087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owen BAL, Yang Z, Lai M, Gajek M, Badger JD, II, Hayes JJ, Edelman W, Kucherlapati R, Wilson TM, McMurray CT. (CAG)n-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 25.Lang WH, Coats JE, Majka J, Hura GL, Lin Y, Rasnik I, McMurray CT. Conformational trapping of mismatch recognition complex MSH2/MSH3 on repair-resistant DNA loops. Proc. Natl Acad. Sci. USA. 2011;108:E837–E844. doi: 10.1073/pnas.1105461108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian L, Hou C, Tian K, Holcomb NC, Gu L, Li G-M. Mismatch recognition protein MutSβ does not hijack (CAG)n hairpin repair in vitro. J. Biol. Chem. 2009;284:20452–20456. doi: 10.1074/jbc.C109.014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X-J, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell. Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claassen DA, Lahue RS. Expansions of CAG•CTG repeats in immortalized human astrocytes. Hum. Mol. Genet. 2007;16:3088–3096. doi: 10.1093/hmg/ddm270. [DOI] [PubMed] [Google Scholar]
- 29.Major EO, Miller AE, Mourrain P, Traub RG, de Widt E, Sever J. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc. Natl Acad. Sci. USA. 1985;82:1257–1261. doi: 10.1073/pnas.82.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gee GV, Manley K, Atwood WJ. Derivation of a JC virus-resistant human glial cell line: implications for the identification of host cell factors that determine viral tropism. Virology. 2003;314:101–109. doi: 10.1016/s0042-6822(03)00389-1. [DOI] [PubMed] [Google Scholar]
- 31.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 33.Holt I, Lam LT, Tome S, Wansink DG, te Riele H, Gourdon G, Morris GE. The mouse mismatch repair protein, MSH3, is a nucleoplasmic protein that aggregates into denser nuclear bodies under conditions of stress. J. Cell. Biochem. 2011;112:1612–1621. doi: 10.1002/jcb.23075. [DOI] [PubMed] [Google Scholar]
- 34.Yu X, Baer R. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 2000;275:18541–18549. doi: 10.1074/jbc.M909494199. [DOI] [PubMed] [Google Scholar]
- 35.Yang W-M, Yao Y-L, Sun J-M, Davie JR, Seto E. Isolation and characterization of cDNAs corresonding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baelde HJ, Eikmans M, Lappin DWP, Doran PP, Hohenadel D, Brinkkoetter P-T, van der Woude FJ, Rabelink TJ, de Heer E, Bruijn JA. Reduction of VEGF-A and CTGF expression in diabetic nephropathy is associated with podocyte loss. Kidney Int. 2007;71:637–645. doi: 10.1038/sj.ki.5002101. [DOI] [PubMed] [Google Scholar]
- 40.Atsumi A, Tomita A, Kiyoi H, Naoe T. Histone deacetylase 3 (HDAC3) is recruited to target promoters by PML-RARα as a component of the N0CoR co-repressor complex to repress transcription in vivo Biochem. Biophys. Res. Comm. 2006;345:1471–1480. doi: 10.1016/j.bbrc.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 41.Milde T, Oehme I, Korshunov A, Kopp-Schneider A, Remke M, Northcott P, Deubzer HE, Lodrini M, Taylor MD, von Deimling A, et al. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin. Cancer Res. 2010;16:3240–3252. doi: 10.1158/1078-0432.CCR-10-0395. [DOI] [PubMed] [Google Scholar]
- 42.Choi D, Kang S. identification and characterization of RNF2 response elements in human kidney cells. Mol. Cell. 2011;31:247–253. doi: 10.1007/s10059-011-0033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zecevic A, Menard H, Gurel V, Hagan E, DeCaro R, Zhitkovich a. WRN helicase promotes repair of DNA double-strand breaks caused by aberrant mismatch repair of chromium-DNA adducts. Cell Cycle. 2009;8:2769–2778. doi: 10.4161/cc.8.17.9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ku S, Soragni E, Campau E, Thomas EA, Altun G, Laurent LC, Loring JF, Napierala M, Gottesfeld JM. Friedreich's ataxia induced pluripotent stem cells model intergenerational GAA•TTC triplet repeat instability. Cell Stem Cell. 2010;7:631–637. doi: 10.1016/j.stem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu M, Myerson RJ, Hunt C, Kumar S, Moros EG, Straube WL, Roti Roti JL. Transfection of human tumour cels with Mre11 siRNA and the increase in radiation sensitivity and the reduction in heat-induced radiosensitization. Int. J. Hyperthermia. 2004;20:157–162. doi: 10.1080/02656730310001625986. [DOI] [PubMed] [Google Scholar]
- 46.Bruton RK, Rasti M, Mapp KL, Young N, Carter RZ, Abramowicz IA, Sedgwick GG, Onion DF, Shuen M, Mymryk JS, et al. C-terminal-binding protein interacting protein binds directly to adenovirus early region 1A through its N-terminal region and conserved region 3. Oncogene. 2007;26:7467–7479. doi: 10.1038/sj.onc.1210551. [DOI] [PubMed] [Google Scholar]
- 47.Godman CA, Joshi R, Tierney BR, Greenspan E, Rasmussen TP, Wang H-w, Shin D-G, Rosenberg DW, Giardina C. HDAC3 impacts multiple oncogenic pathways in colon cancer cells with effects on Wnt and vitamin D signaling. Cancer Biol. Ther. 2008;7:1570–1580. doi: 10.4161/cbt.7.10.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1-11 in the rat brain. J. Mol. Neurosci. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- 49.Gonitel R, Moffitt H, Sathasivam K, Woodman B, Detloff PJ, Faull RLM, Bates GP. DNA instability in postmitotic neuros. Proc. Natl Acad. Sci. USA. 2008;105:3467–3472. doi: 10.1073/pnas.0800048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinti L, Chopra V, Rotili D, Valente S, Amore A, Franci G, Meade S, Valenza M, Altucci L, Maxwell MM, et al. Evaluation of histone deacetylases as drug targets in Huntington's disease models. Study of HDACs in brain tissues from R6/2 and CAG140 knock-in HD mouse models and human patients and in a neuronal HD cell model. PLoS Curr. 2010;2 doi: 10.1371/currents.RRN1172. pii: RRN1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoshino M, Tagawa K, Okuda T, Murata M, Oyanagi K, Arai N, Mizutani T, Kanazawa I, Wanker EE, Okazawa H. Histone deacetylase activity is retained in primary neuros expressing mutant huntington protein. J. Neurochem. 2003;87:257–267. doi: 10.1046/j.1471-4159.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- 52.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 53.Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Botrugno OA, Parazzoli D, Oldani A, Minucci S, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–79. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulated major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 56.Butler R, Bates GP. Histone deacetylase inhibitors as therapeutics for polyglutamine disorders. Nat. Rev. Neurosci. 2006;7:784–796. doi: 10.1038/nrn1989. [DOI] [PubMed] [Google Scholar]
- 57.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat. Rev. Drug Disc. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.