Abstract
There is an increasing call for the absolute quantification of time-resolved metabolite data. However, a number of technical issues exist, such as metabolites being modified/degraded either chemically or enzymatically during the extraction process. Additionally, capillary electrophoresis mass spectrometry (CE-MS) is incompatible with high salt concentrations often used in extraction protocols. In microbial systems, metabolite yield is influenced by the extraction protocol used and the cell disruption rate. Here we present a method that rapidly quenches metabolism using dry-ice ethanol bath and methanol N-ethylmaleimide solution (thus stabilising thiols), disrupts cells efficiently using bead-beating and avoids artefacts created by live-cell pelleting. Rapid sample processing minimised metabolite leaching. Cell weight, number and size distribution was used to calculate metabolites to an attomol/cell level. We apply this method to samples obtained from the respiratory oscillation that occurs when yeast are grown continuously.
Introduction
A comprehensive in vivo understanding of the underlying dynamics of metabolite reaction networks, enzyme kinetics and signalling requires the precise characterisation of intracellular metabolites at specific time points. Recent advances in high-throughput mass-spectrometry allow for the detailed metabolome-wide analysis with high accuracy [1]–[4]. However, development of metabolite extraction protocols has generally lagged behind detection methods. These protocols often suffer from complex experimental design (making time-series analysis difficult), metabolite leakage during processing, metabolite oxidation during sampling/extraction [5], [6], and metabolite specificity (acid-stable or alkali-stable metabolites). Furthermore, yields are influenced by the metabolic state (growth phase and rate) and properties of the species and/or strains used [7]–[10]. This leads to the paradoxical situation where the extraction protocol dictates the experimental conditions. An ideal metabolite extraction protocol should rapidly sample and quench the underlying metabolic processes, i.e., minimise degradation and modification of metabolites, and have a high and reproducible yield [7], [10], [11].
Turnover rates of metabolic intermediates change in the order of seconds and are highly sensitive to the changes in external conditions, thus rapid quenching is required [10], [12], [13]. Therefore, direct centrifugation or filtration of live cells prior to quenching should be avoided as they may alter the metabolite profile [7], [10]. However, the removal and subsequent analysis of the culture media is highly desirable and must be done rapidly to avoid interfering with intracellular metabolite concentrations [5], [7], [10]. Furthermore, the culture media may contain a sufficiently high concentration of salts that may suppress signals or interfere with the chromatography/electrophoresis stage.
There are several methods widely used for extracting metabolites from yeast cells such as freeze-thaw, sonication, hot water, boiling ethanol, permeabilisation using chloroform and treatment with extreme pH. However, these methods are mostly optimised and tested only on fast-growing low-density laboratory strains [9], [11]–[13] and the extraction buffers often use salts that are not compatible with capillary electrophoresis mass spectrometry (CE-MS). Moreover, non-laboratory strains are usually nutrient limited, slow-growing and recalcitrant to lysis. This resistance arises from changes in the cell wall structure [14], [15]. These physiological changes lead to differences in metabolite extraction efficiency and reproducibility, which are critical factors for the analysis of cultures grown in different conditions.
A combined chemical-mechanical disruption by bead-beating using zirconia/silica beads has a high and consistent cell disruption efficiency independent of respiratory state and the cell division cycle [16]. It has also been observed that the metabolic reactions of Saccharomyces cerevisiae can be efficiently quenched in methanol (final methanol concentration >50% v/v) at −40°C and having very low metabolite leaching when the media is removed [5], [12]. Moreover, extraction with chloroform-methanol has been used for the quantitative metabolite extraction of rapidly-growing, glucose-repressed laboratory strains of S. cerevisiae [9], [11], [12], [17]. The permeabilisation technique used was time consuming (45 min) and it is known that many laboratory strains have been selected for their ability to be easily disrupted. Any metabolite modifications happening during the extraction procedure can be avoided partially by keeping a low temperature (<−20°C) throughout the extraction process [10]. However, oxidation of metabolites remains an issue, for example, precise quantification of the redox sate of thiol groups are critical for understanding redox biochemistry in vivo [18]. However, oxidation of thiol groups during extraction usually hinders the accurate determination of redox state [6].
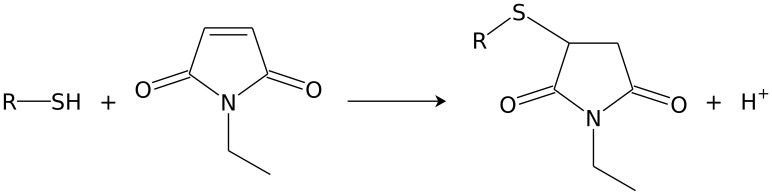
Previously we developed efficient methods for the disruption of budding yeasts for mRNA, proteins and DNA [16]. We optimised these methods for metabolite analyses that rapidly extract and fractionate the intracellular and extracellular metabolites. Our method was based on the rapid quenching of cell cultures with −80°C methanol and N-ethylmaleimide (NEM) solution. Here, NEM was used to protect thiols from oxidation by binding to -SH groups (figure 1) [19]. The quenching solution (which contains the extracellular metabolites) was rapidly removed and lyophilised. Cell pellets were bead-beated in chloroform/methanol/internal standard (IS) solution. Intracellular and extracellular metabolites were analysed using CE-MS. In parallel, we fixed a sample in ethanol to determine dry cell weight and cell number. We tested our method on continuously grown industrial S. cerevisiae cultures and it outperformed other tested extraction techniques (freeze-thaw and sonication), protected the redox state of the cell, has the potential to cover a large fraction of the known yeast metabolome, and gave comparable yields to those reported values for extracted metabolites from cells grown under similar conditions.
Figure 1. The general reaction scheme for N-ethylmaleimide on biological thiols.
The product formed is an N-ethylsuccinimido conjugate and the monoisotopic mass of the reactant is increased by 125.048. For example the glutathione (m/z 308.0911) conjugate is N-ethylsuccinimido-S-glutathione (m/z 433.1391).
Methods
Strain and Culture Conditions
Unless stated otherwise, all chemicals were supplied by Wako Chemicals, Japan or Fisher Chemicals, UK. In this study we used IFO 0233 diploid strain of Saccharomyces cerevisiae. The colonies were maintained at 4°C on yeast extract peptone dextrose (YEPD) agar plates, comprising 10 g/L yeast extract (Becton Dickinson, Japan/UK), 20 g/L glucose monohydrate, 20 g/L mycological peptone (Becton Dickinson, Japan/UK) and 30 g/L agar (Becton Dickinson, Japan/UK). Pre-cultures (10 mL; 0.5×YEPD broth) were inoculated with yeast colonies and incubated (30°C) in an orbital incubator (250 rpm) for 48 h. The basic medium used was composed of 5 g/L of (NH4)2SO4, 2 g/L of KH2PO4, 0.5 g/L of MgSO4.7H2O, 0.1 g/L of CaCl2.2H2O, 0.02 g/L of FeSO4.7H2O, 0.01 g/L of ZnSO4.7H2O, 0.005 g/L of CuSO4.5H2O, 0.001 g/L of MnCl2.4H2O, 0.5 mL/L of 70% H2SO4, 1 g/L of yeast extract (Becton Dickinson, Japan), 0.2 mL/L of Antifoam A (Sigma, USA) and 20 mL/L ethanol [20]. Cells were grown as described previously in a modified version of a MBF-250 bioreactor (Eyela, Japan), at 30°C (±0.02°C), pH 3.4 (±0.03), an aeration rate 0.150 L/min (±0.02 L/min), a working volume of 650 mL, 750 rpm (±3 rpm) agitation rate and the reactor pressure was maintained below 375 Pa, above atmospheric pressure [21]. Prior to initiating continuous culture mode, batch cultures were starved in fermentors for at least 6 h. Stable autonomous respiratory oscillation occurred 16 to 48 h after the initiation of the continuous culture (dilution rate at 0.087 h−1) and the respiration was monitored by measuring the dissolved oxygen (DO) content in the culture using an immersed polarographic electrode (InPro6800, Mettler Toledo, Japan/UK). As we could not measure a change in biomass or cell number during continuous growth (biomass was in steady state), this dilution rate equates to a cell doubling time of 8.1 h [22].
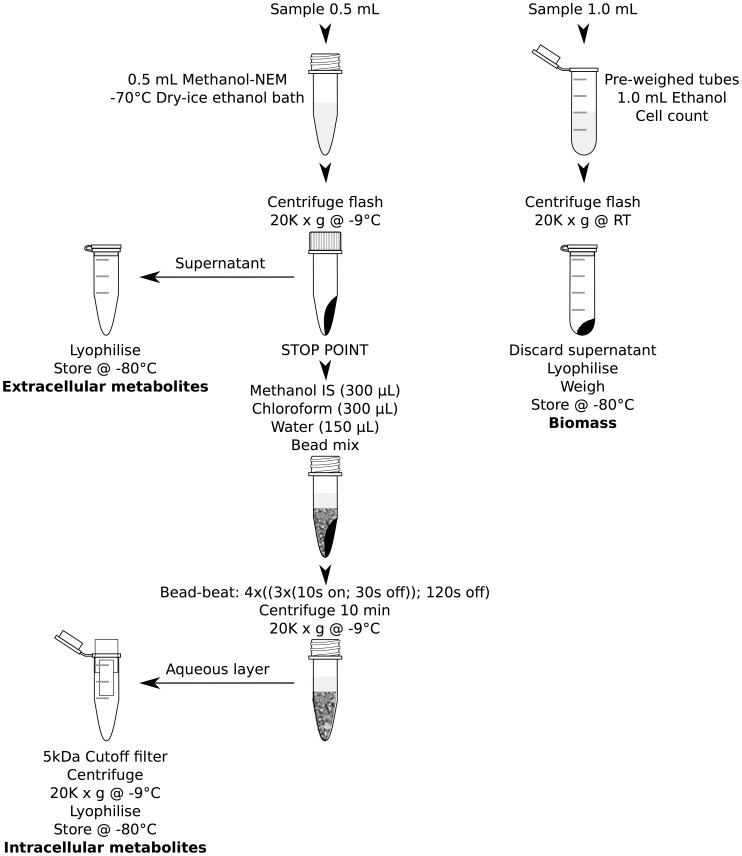
Metabolite Extraction (figure 2)
Figure 2. Extraction protocol flow diagram.
Sampling and quenching
To study the reproducibility (table 1) all samples were taken at the minimum dissolved oxygen concentration during the oscillation. Whereas, time-series samples were obtained at 4 min intervals over two oscillation cycles. To avoid any dead volume in the sampling needle 200 µL was removed 5 s prior to obtaining 0.7 mL of sample (gas and liquid) and the needle end of the syringe sealed. To rapidly degas the sample the plunger was removed rapidly and 500 µL of sample was rapidly pipetted into 1 volume of freshly prepared NEM methanol solution (4 mM) kept in screw cap tubes (1.5 mL; Simport, Canada) equilibrated to −70°C using dry ice/ethanol bath. The samples were then pelleted by flash centrifugation (20,000 g; −9°C) and the supernatant was transferred into fresh 1.5 mL tubes to measure the extracellular metabolites (appropriate standards added; lyophilised and store at −80°C prior to analysis). The sampling process to fixed samples took around 10 s. To monitor metabolite leakage, these samples were compared with lyophilised samples rapidly filtered through 0.2 µm pore size syringe filters 30 s later. After 30 s a further 1 mL of culture was sampled, degassed and added to 1 mL of ethanol in 2 mL pre-weighed centrifuge tubes. The samples were flash centrifuged (20,000 g; room temperature) and the supernatant removed. The cell pellet and extracellular metabolite samples were then lyophilised and cell pellet samples were weighed for the determination of biomass, although not determined here the biomass can be further digested to measure the biomass composition. A further 30 µL of culture was fixed with 70 µL ethanol (room temperature) for the determination of cell number and size.
Table 1. Metabolites (attomol/cell) extracted from samples taken during respiratory oscillation (minimum dissolved oxygen; figure 4), detected by CE-MS and the methods used to extract them. nd – not detected.
| Name | Sonication | Sonication -Q 1 | Freeze thaw | BB 2, 3 | BB 2 -NEM 4 before 5 | BB 2 -NEM 4 after 6 | BB 2 -NEM 4 -Q 1 |
| Pyruvate | 2.170±0.876 | 2.219±0.132 | 1.443±0.612 | 1.273 | 1.482±0.175 | 1.666±0.039 | 0.604±0.029 |
| Lactate | 1.653±1.187 | 2.495±0.069 | 0.381±0.124 | 1.339 | 0.949±0.064 | 1.261±0.107 | 1.437±0.088 |
| Fumarate | 1.607±0.617 | 2.180±0.144 | 1.096±0.657 | 0.683 | 0.629±0.040 | 0.626±0.061 | 0.473±0.002 |
| 2-Oxoisopentanoate | nd | nd | nd | 0.145 | 0.178±0.009 | 0.154±0.020 | 0.091±0.006 |
| Succinate | 1.451±0.306 | 5.073±0.356 | 1.680±0.423 | 1.373 | 1.137±0.036 | 1.709±0.262 | 8.124±0.172 |
| Malate | 2.231±1.592 | 6.874±0.560 | 2.150±1.509 | 2.585 | 2.292±0.074 | 2.221±0.151 | 2.036±0.036 |
| 2-Oxoglutarate | nd | nd | nd | 0.948 | 0.974±0.160 | nd | nd |
| PEP | nd | nd | nd | 0.220 | 0.429±0.079 | 0.363±0.002 | 0.193±0.017 |
| DHAP | nd | 0.984±0.210 | nd | 0.230 | 0.520±0.201 | 0.526±0.134 | 0.377±0.083 |
| Glycerophosphate | 1.269±0.275 | 2.175±0.321 | 1.390±0.989 | 0.629 | 0.613±0.021 | 0.672±0.125 | 0.719±0.062 |
| cis-Aconitate | 0.105±0.056 | nd | 0.141±0.068 | 0.521 | 0.365±0.007 | nd | nd |
| 3PG | 3.825±2.763 | nd | 0.873±1.010 | 4.432 | 4.317±0.343 | 4.969±0.225 | 0.703±0.045 |
| Ru5P | 0.264±0.113 | nd | nd | 0.254 | 0.473±0.120 | 0.453±0.040 | 0.172±0.038 |
| G1P | 0.757±0.353 | 0.699±0.030 | 0.011±0.004 | 0.504 | 0.539±0.060 | 0.656±0.119 | 0.631±0.020 |
| F6P | 0.498±0.232 | 0.716±0.028 | nd | nd | 0.451±0.048 | nd | nd |
| G6P | 0.641±0.873 | 2.399±0.110 | 0.010±0.003 | 5.110 | 7.325±0.247 | 5.432±0.333 | 1.661±0.046 |
| 2,3-DPG | nd | nd | nd | 0.032 | 0.045±0.011 | nd | 0.012±0.017 |
| 6-Phosphogluconate | 0.441±0.246 | 0.299±0.047 | nd | 0.889 | 1.165±0.350 | 0.933±0.066 | 0.122±0.019 |
| S7P | 0.817±0.431 | 0.386±0.011 | nd | 0.843 | 0.951±0.035 | 0.677±0.070 | 0.103±0.002 |
| F1,6P | 1.160±0.810 | 7.337±1.831 | 0.011±0.004 | 0.547 | 0.908±0.033 | 0.730±0.172 | 0.797±0.064 |
| Citrate | 2.470±0.457 | 3.825±0.270 | 1.035±1.119 | 2.174 | 2.316±0.153 | 2.040±0.250 | 1.650±0.037 |
| CMP | 0.217±0.040 | 0.223±0.024 | 0.106±0.066 | 0.178 | 0.123±0.007 | 0.211±0.042 | 0.620±0.010 |
| AMP | 3.668±0.599 | 2.264±0.222 | 1.760±1.211 | 2.239 | 1.278±0.099 | 2.505±0.562 | 6.824±0.225 |
| IMP | 0.540±0.086 | 0.483±0.035 | 0.268±0.168 | nd | 0.083±0.017 | nd | nd |
| GMP | 0.475±0.081 | 0.292±0.037 | 0.171±0.151 | 0.233 | 0.155±0.014 | 0.279±0.071 | 1.576±0.004 |
| NADPH | 0.003 | 0.280±0.010 | nd | nd | nd | nd | 0.198±0.002 |
| CoA | 0.094±0.027 | 0.125±0.008 | 0.018±0.023 | 0.020 | 0.160±0.017 | 0.139±0.036 | 0.228±0.014 |
| dTDP | 0.014±0.002 | 0.016±0.002 | nd | 0.046 | 0.035±0.004 | 0.041 | 0.039±0.001 |
| CDP | 0.135±0.017 | 0.151±0.027 | 0.034±0.044 | 0.297 | 0.235±0.019 | 0.315±0.009 | 0.221±0.004 |
| Acetyl CoA | 0.025±0.002 | nd | 0.010±0.012 | 0.068 | 0.095±0.005 | 0.085±0.014 | 0.084±0.004 |
| ADP | 1.918±0.274 | 1.979±0.326 | 0.506±0.699 | 3.597 | 3.110±0.080 | 3.716±0.333 | 3.309±0.025 |
| GDP | 0.264±0.004 | 0.273±0.052 | nd | 0.358 | 0.320±0.020 | 0.400±0.035 | 0.712±0.012 |
| dCTP | nd | nd | nd | 0.034 | 0.039±0.006 | 0.035±0.004 | nd |
| dTTP | 0.006±0.001 | 0.012±0.002 | nd | 0.062 | 0.076±0.004 | 0.063 | 0.026 |
| CTP | 0.054±0.008 | 0.127±0.046 | nd | 0.601 | 0.739±0.009 | 0.569±0.030 | 0.151±0.005 |
| UTP | 0.262±0.027 | 0.216±0.047 | 0.074±0.102 | 1.635 | 1.939±0.065 | 1.509±0.027 | 0.550±0.008 |
| dATP | 0.005±0.001 | 0.007±0.001 | nd | 0.051 | 0.058±0.012 | 0.046±0.010 | 0.026±0.006 |
| ATP | 1.167±0.218 | 2.271±0.490 | 0.361±0.506 | 8.814 | 10.520±0.588 | 7.605±0.088 | 3.082±0.037 |
| GTP | 0.165±0.016 | 0.240±0.071 | nd | 1.774 | 2.027±0.112 | 1.654±0.123 | 1.037±0.008 |
| NAD+ | 2.120±0.172 | 1.707±0.086 | nd | 3.504 | 3.711±0.107 | 3.639±0.013 | 3.358±0.082 |
| NADH | 0.025±0.003 | 2.421±0.313 | nd | nd | nd | nd | 0.964±0.149 |
| NADP+ | 0.306±0.023 | 0.183±0.013 | 0.068±0.093 | 0.520 | 0.520±0.013 | 0.504±0.032 | 0.382±0.011 |
| FAD | 0.052±0.007 | 0.072±0.007 | nd | nd | 0.184±0.012 | 0.144±0.087 | 0.087±0.027 |
| Gly | 1.938±0.171 | 2.006±0.120 | 1.895±0.056 | 7.184 | 7.270±0.745 | 8.590±0.027 | 16.670±5.088 |
| Ala | 2.018±0.080 | 2.012±0.031 | 2.060±0.054 | 190.603 | 198.371±16.626 | 228.475±3.048 | 302.253±0.606 |
| GABA | 2.000±0.085 | 1.975±0.198 | 1.974±0.106 | 1.021 | 1.097±0.113 | 1.259±0.068 | 1.377±0.037 |
| 2AB | 2.016±0.155 | 2.020±0.132 | 2.047±0.186 | 0.893 | 0.976±0.071 | 1.113±0.052 | 1.310±0.075 |
| Ser | 2.113±0.178 | 2.163±0.058 | 2.164±0.079 | 6.481 | 6.712±0.547 | 7.691±0.059 | 10.683±5.029 |
| Pro | 2.043±0.228 | 2.053±0.240 | 1.985±0.320 | 5.271 | 5.512±0.461 | 6.283±0.038 | 9.575±0.759 |
| Val | 1.871±0.315 | 2.087±0.123 | 2.059±0.097 | 38.476 | 43.287±3.415 | 48.741±1.008 | 55.543±1.200 |
| Homoserine | 2.362±1.049 | 1.714±0.121 | 1.742±0.134 | 1.691 | 1.971±0.168 | 2.127±0.082 | 1.410±0.042 |
| Thr | 2.500±1.057 | 2.567±0.899 | 2.854±1.111 | 12.228 | 13.525±1.159 | 14.918±0.192 | 16.956±0.692 |
| Cys | nd | nd | nd | nd | 0.310±0.049 | 0.343±0.022 | 0.444±0.008 |
| Ile | 2.042±0.119 | 2.045±0.162 | 2.072±0.032 | 4.053 | 4.592±0.456 | 5.340±0.084 | 6.518±0.618 |
| Leu | 2.103±0.143 | 2.139±0.197 | 2.112±0.228 | 1.567 | 1.589±0.114 | 1.913±0.045 | 3.025±0.766 |
| Asn | 2.009±0.124 | 2.034±0.095 | 2.050±0.134 | 3.447 | 3.920±0.304 | 4.161±0.124 | 5.038±0.168 |
| Ornithine | 1.924±0.087 | 1.933±0.071 | 1.865±0.061 | 9.902 | 8.199±0.726 | 8.850±0.796 | 10.901±2.848 |
| Asp | 2.004±0.090 | 1.955±0.121 | 1.969±0.050 | 13.221 | 55.773±1.133 | 56.621±0.508 | 55.545±2.242 |
| Homocysteine | nd | nd | nd | 0.132 | 0.709±0.097 | 0.737±0.006 | 0.795±0.036 |
| Adenine | nd | nd | nd | 0.151 | 0.172±0.007 | 0.155±0.007 | 0.139±0.003 |
| Gln | 2.008±0.113 | 2.046±0.143 | 2.013±0.155 | 104.444 | 223.521±10.435 | 228.527±0.257 | 225.379±0.076 |
| Lys | 2.003±0.136 | 2.009±0.095 | 2.036±0.144 | 12.354 | 34.010±1.118 | 35.580±0.021 | 35.572±0.776 |
| Glu | 1.980±0.129 | 1.977±0.115 | 1.997±0.121 | 835.237 | 864.465±19.172 | 887.669±10.922 | 886.352±2.001 |
| Met | nd | nd | nd | 0.454 | 0.487±0.061 | 0.554±0.012 | 1.003±0.033 |
| Guanine | nd | nd | nd | 0.068 | 0.071±0.001 | 0.066±0.006 | nd |
| His | 1.948±0.094 | 1.980±0.060 | 1.944±0.069 | 12.022 | 13.150±1.267 | 14.154±0.263 | 14.336±1.289 |
| Phe | 2.038±0.099 | 2.006±0.090 | 2.012±0.045 | 1.113 | 1.254±0.073 | 1.330±0.014 | 1.710±0.387 |
| Arg | 2.020±0.107 | 1.964±0.065 | 2.002±0.080 | 93.775 | 218.231±10.111 | 225.316±4.592 | 214.317±2.327 |
| Citrulline | 2.034±0.084 | 2.007±0.082 | 2.022±0.083 | 7.458 | 8.877±0.960 | 9.170±0.372 | 8.951±0.139 |
| Tyr | 2.114±1.093 | 2.076±0.968 | 2.374±1.191 | 1.227 | 1.400±0.090 | 1.509±0.017 | 1.843±0.584 |
| SAH | 1.932±0.257 | 1.910±0.223 | 1.987±0.283 | 0.318 | 0.330±0.020 | 0.407±0.002 | 0.696±0.007 |
| SAM+ | nd | nd | nd | 0.530 | 0.549±0.028 | 0.553±0.079 | 0.532±0.008 |
| Trp | 2.060±0.099 | 2.053±0.069 | 2.058±0.124 | 0.254 | 0.287±0.015 | 0.287±0.028 | 0.365±0.073 |
| Cystathionine | 1.981±0.119 | 1.957±0.119 | 1.976±0.135 | 5.837 | 7.127±0.603 | 7.281±0.039 | 6.559±0.062 |
| γ-Glu-2AB | nd | nd | nd | 0.255 | 0.282±0.026 | 0.313±0.001 | 0.279±0.004 |
| γ-Glu-Cys | nd | nd | nd | 1.436 | 7.796±0.500 | 8.822±0.415 | 8.220±0.438 |
| Adenosine | 2.009±0.152 | 1.970±0.124 | 1.949±0.190 | 0.281 | 0.444±0.064 | 0.421±0.156 | 0.408±0.016 |
| Guanosine | nd | nd | nd | 0.108 | 0.125±0.005 | 0.131±0.022 | 0.090±0.008 |
| Glutathione(ox) | 2.103±0.279 | 2.078±0.243 | 2.162±0.308 | 13.105 | 14.840±0.114 | 15.060±0.199 | 14.559±0.086 |
| Glutathione(red) | 2.004±0.148 | 2.015±0.150 | 1.923±0.125 | 36.580 | 143.608±12.226 | 147.043±5.378 | 129.318±1.290 |
-Q not quenched.
BB bead-beating.
All other samples were performed in triplicate, except this one.
NEM N-ethylmaleimide.
NEM added pre bead-beating.
NEM added post bead-beating.
Determination of dry cell weight and cell number
Dry cell weight was calculated by weighing the tube and lyophilised pellet from the tube weight using an accurate balance (+/−0.1 mg, HR-202i, AND, Japan). Cell number and volume distributions were measured using a particle counter (50 µm orifice, CDA-500, Sysmex, Japan) by vortexing the ethanol fixed samples for 30 s and pipetting 2.5 µL into 19.9975 mL of Manoresh™ (Sysmex, Japan). The cell number and volume was used to calculate the average metabolite concentration (attomol/cell) for each sample.
N-Ethylmaleimide bead-beating method
The cell pellets were resuspended in internal standards-methanol solution (300 µL; −70°C). The internal standards comprised of 2-morpholinoethanesulfonic acid (MES; 10 nmol), methionine sulfone (10 nmol) and D-camphor-10-sulfonic acid (CSA; 10 nmol). As the analysis volume was 50 µL, the final amount added of each standard was adjusted to a 200 µM solution on rehydration prior to analysis. Chloroform and deionised water (final ratio 1∶1∶0.5) was then added. A mixture of chilled acid washed zirconia/silica beads (∼300 µL; 0.1 mm and 0.5 mm; 1∶1; Tomy Seiko Co., Ltd., Japan) were then added and the samples were disrupted in a multi-tube bead-beater (5500 rpm; Micro Smash™ MS-100, Tomy, Japan), using 12 cycles of 10 s beat, 30 s rest (for cooling) and a further 120 s rest after every third cycle, with inbuilt refrigeration unit turned on. The samples were then centrifuged (12,000 g; 10 min; −9°C), and the aqueous phase (if a clear phase separation was not obtained more water can be added and the samples mixed then re-centrifuged) transferred to 5 kDa cut-off filter tubes (Ultrafree®-MC, Millipore, USA) and centrifuged (20,000 g; −9°C until the supernatant had passed through the filter). The filtration step removes many proteins and the filtrate was then lyophilised and stored at −80°C prior to CE-MS analyses. For comparison we also have extracted metabolites using different cell disruption methods and conditions: (1) sonication, 20 min, with and without quenching (2) freeze-thaw using liquid nitrogen, thaw at −30°C, 10 times (3) bead-beating without NEM treatment (4) NEM treatment after bead-beating, with and without quenching. Prior to analysis intracellular metabolite samples were rehydrated in 50 µL and extracellular samples were rehydrated in 500 µL of Milli-Q water, containing 200 µM 1,3,5-benzenetricarboxylate (anion) and 3-aminopyrrolidine dihydrochloride (cation).
Capillary Electrophoresis Analytical Methods
In all CE-TOFMS experiments we used the Agilent CE capillary electrophoresis system (Agilent Technologies, Waldbronn, Germany), the Agilent G3250AA LC/MSD TOF system (Agilent Technologies, Palo Alto, CA), the Agilent 1100 series binary HPLC pump, the G1603A Agilent CE-MS adapter, and the G1607A Agilent CE-ESI-MS sprayer kit. Data acquisition was with G2201AA Agilent ChemStation software for CE and Analyst QS software for Agilent TOFMS (Agilent technologies, Japan). For anionic metabolite profiling, the original Agilent stainless steel ESI needle was replaced with the Agilent G7100–60041 platinum needle [23].
CE-TOFMS conditions for cationic metabolite profiling
Metabolites were separated in a fused silica capillary (50 µm i.d. x 100 cm) filled with 1 M formic acid as the electrolyte [4]. Prior to first use, a new capillary was flushed with the running electrolyte (1 M formic acid) for 20 min. A sample solution was injected at 5 kPa for 3 s (3 nL), and 30 kV voltage was applied. The capillary temperature and the sample tray were set at 20°C and below 5°C, respectively. Methanol:water (50% v/v) containing 0.1 µM hexakis (2,2-difluoroethoxy) phosphazene was delivered as the sheath liquid at 10 µL/min. ESI-TOFMS was in the positive ion mode, and the capillary voltage was set at 4 kV. The flow rate of heated dry nitrogen gas (heater temperature 300°C) was maintained at 170 kPa. At TOFMS, the fragmentor, skimmer and Oct RFV voltages were set at 75, 50, and 125 V, respectively. Automatic recalibration of each acquired spectrum was with reference masses of reference standards ([13C isotopic ion of protonated methanol dimer (2MeOH+H)]+; m/z 66.0632) and ([hexakis (2,2-difluoroethoxy) phosphazene+H]+; m/z 622.0290). Exact mass data were acquired at a rate of 1.5 spectra/s over a 50–1,000 m/z range [24].
CE-TOFMS conditions for anionic metabolite profiling
A cationic polymer-coated COSMO(+) capillary (50 µm i.d. x 110 cm) (Nacalai Tesque, Kyoto, Japan) was used as the separation capillary. A 50 mM ammonium acetate solution (pH 8.5) was used as the electrolyte for the capillary separation. Prior to first use, a new capillary was flushed successively with the running electrolyte, 50 mM acetic acid (pH 3.4), and then the electrolyte again for 20 min each. Before each injection, the capillary was equilibrated for 2 min by flushing with 50 mM acetic acid (pH 3.4), and then for 5 min with the running electrolyte. A sample solution (30 nL) was injected at 5000 kPa for 30 s, and −30 kV of voltage applied. 5 mM ammonium acetate in 50% (v/v) methanol-water containing 0.1 µM hexakis (2,2-difluoroethoxy) phosphazene was delivered as the sheath liquid at 10 µL/min. ESI-TOFMS was conducted in the negative ionization; the capillary voltage was set at 3,500 V. At TOFMS, the fragmenter, skimmer, and Oct RFV voltages were set at 100 V, 50 V, and 200 V, respectively. Automatic recalibration of each acquired spectrum was performed using reference masses of reference standards ([13C isotopic ion of deprotonated acetic acid dimer (2CH3COOH-H)]-, m/z 120.03841), and ([hexakis (2,2-difluoroethoxy) phosphazene + deprotonated acetic acid (CH3COOH-H)]-, m/z 680.03554). Other conditions were identical to those used in cationic metabolite analysis [23].
Data analysis
We first visualised the raw data in Analyst QS to check for any problems in the CE-MS. The data were then converted into mzXML format using mzWiff [25] for Agilent “.wiff” and trapper [26] for Agilent “.d” formats, using the following parameters:
mzWiff.exe -z -c1 -v –mzXML <filename>
trapper.exe -c -v –mzXML <filename>
Data was then loaded into the xcms package [27] of Bioconductor for further processing. CE-MS spectra tend to drift significantly as the column properties change with time [28]. This makes the inbuilt alignment algorithms in XCMS perform poorly. Therefore, peaks were pre-aligned using a regression analysis of the internal standards. The alignment was then carried out using XCMS for the datasets, where we used the matchedFilter method and set the following parameters, bw = 8, mzwid = 0.25, fwhm = 6, snthresh = 5 and step = 0.1. The concentrations were calculated using external standards that were prepared and lyophilised weekly, then reconstituted freshly prior to each run. This freshly prepared high quality set was a subset (61 anionic and 59 cationic) of the whole standard set (272 anionic and 318 cationic; prepared and lyophilised monthly) which was also run (data not shown).
Results and Discussion
Metabolome
The overall internal standard recovery rate (average for all the samples taken) for 2-(N-morpholino)ethanesulfonate (anionic) was 89.2%±2.32% and methionine sulfone (cationic) was 93.8%±3.8%. These standards were added prior to bead-beating therefore give an idea of the recovery from the extraction protocol. We used three methods to extract metabolites from yeast cells. Out of these, bead-beating performed the best (table 1) with a higher yield of most metabolites. Freeze thaw, although limiting the number of handling steps produced very low, variable yields. A striking example of this was the yield of ATP and amino acids, which were at least a magnitude higher using bead-beating. Quenching with methanol prior to extraction is known to cause metabolite leaching [8], however removing the cells from the media using centrifugation and to a lesser extent filtration causes a rapid changes in physiology [29]. Therefore, any quenching method is a trade-off. We examined the effect of quenching and found that the metabolites that increased between the filtered samples and the methanol-NEM supernatant were glutamate (5.03%), citrate (2.85%), succinate (8.34%) and pyruvate (12.34%) and these were within the error of the residual levels of the filtered samples. More significantly, flash centrifugation (10 s process) of the cultures caused very rapid metabolome-wide changes (table 1). For example [ATP] decreased from 10.5 to 3.1 attomol/cell, [AMP] showed a reverse trend increasing from 1.3 to 6.8 attomol/cell, suggesting ATP is rapidly utilised when the samples are centrifuged. There were also significant changes in TCA cycle intermediates where succinate increased, citrate decreased, and NADH became detectable when quenched and non-quenched samples were compared. This indicates that live cell pelleting may inhibit the respiratory chain, perhaps by limiting oxygen transfer.
Thiol Analysis
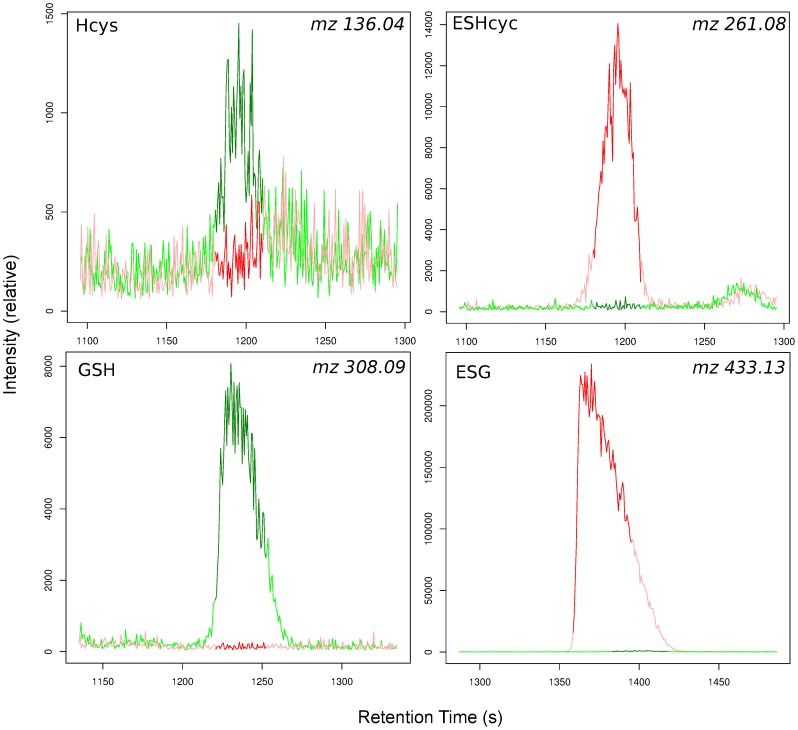
Thiols are critical for the understanding of redox biochemistry and all the tested methods were not able to measure thiols in a consistent way, and produced much lower concentrations than expected. Therefore, we used NEM which binds to the -SH group of thiols, thus protecting it from oxidation (figure 1; cysteine, homocysteine, L-γ-glutamyl-L-cysteine and reduced glutathione) [19]. The spectra were compared to identify peaks that corresponded to NEM-conjugated thiols (+125.048 mass; figure 3). All of the thiol compounds were measurable with low error (table 1), in addition the peaks also tended to migrate slower. Cysteine and L-γ-glutamyl-L-cysteine could be detected and homocysteine and glutathione were present at ∼70 fold higher concentrations compared to either sonicated samples or non-NEM treated samples. NEM is not charged therefore excess NEM does not influence the CE-MS run. Samples were oxidised after extraction as thiol metabolite concentrations were similar for the addition of NEM pre or post bead-beating (table 1).
Figure 3. Electrophoretograms of glutathione (GSH) and homocysteine (Hcys) and their NEM derivatives (ESG and ESHcys) in representative samples with (red line) and without (green line) the addition of 2 mM NEM.
Time-series Analysis
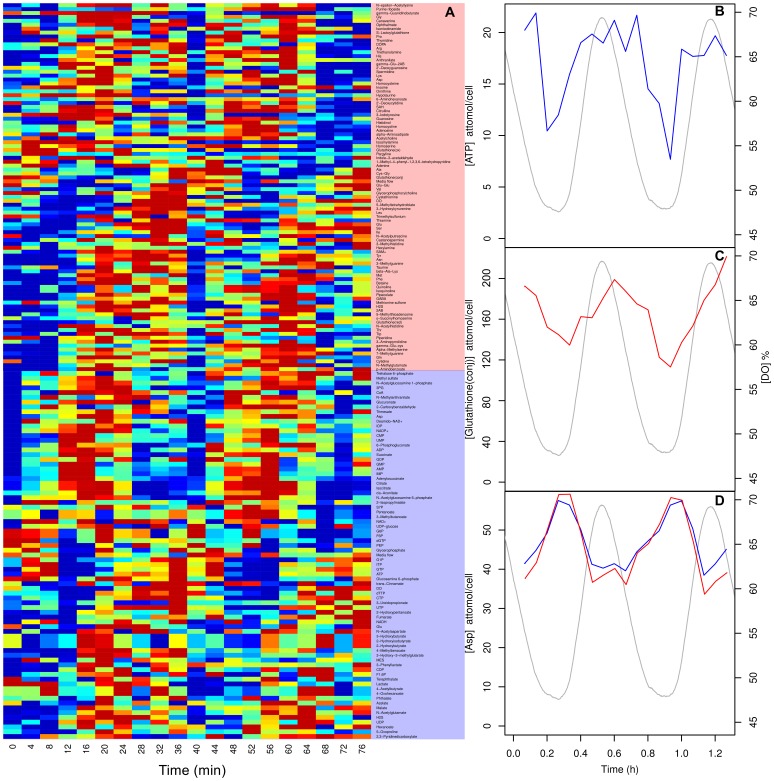
Next we tested our extraction methods during experimental conditions. When grown continuously, budding yeast synchronise their respiratory activity to form a robust respiratory oscillation [20], [30] whose period ranges from 35 min to 8 h [31]. We measured the intracellular metabolites for two cycles for S. cerevisiae (figure 4). We highlight three metabolites, ATP, glutathione and aspartate whose concentrations had previously been determined by other methods in S. cerevisiae under similar conditions [20], [32], [33]. Our extraction protocol produced reproducible time-series for all calibrated metabolites (figure 4A) many with a robust oscillation. ATP oscillated (figure 4B) with an amplitude of 7–20 attomol/cell (0.6–1.5 mM) which was in the range (1–2 mM) of recent online measurements using online nanosensors [34]. Glutathione-NEM conjugate oscillated (figure 4C) with an amplidude of 120–205 attomol/cell (8.4–14.2 mM) which was an order of magnitude higher than previously reported values for GSH (0.6–1.2 mM) extracted with perchloric acid during the oscillation [35]. We argue that this increase in yield was due to enhanced disruption, rapid quenching and thiol protection during the extraction process. Aspartate is an interesting example of a cross-platform replicate as it is an acidic amino acid therefore can be detected in CE-MS anionic and cationic detection modes (figure 4D). It oscillated in the range of 37–56 attomol/cell (2–4 mM), which was double previously reported values for hot water extraction of 1–2 mM [33], [36]. Perhaps this increase in yield was due to avoiding live cell centrifugation steps.
Figure 4. A heatmap of the calibrated intracellular metabolite time-series during the respiratory oscillation (A) and time-series for ATP (B), glutathione (C), and aspartate (D) in S. cerevisiae.
Cationic and anionic data are shown in red and blue (lines or shaded areas), respectively. The corresponding oscillation in dissolved oxygen (DO) is represented by a grey line.
It is clear from the time-series data (figure 4) that sample-to-sample error is very low between extractions and CE-MS detection methods. Indeed, none of the data presented here was normalised to the CE-MS internal standards (these were only used to align chromatograms). When this normalisation is done, paradoxically, the data becomes slightly noisier, perhaps because of the errors introduced during small volume pipetting operations or integration.
Conclusions
Here we present an extraction protocol for yeast metabolites that rapidly quenches metabolism using methanol-NEM at sub −40°C temperatures from a small volume of culture (0.5 mL). NEM was shown to protect biological thiols from oxidation in subsequent sample processing steps. The method was tested on samples taken during the respiratory oscillation and was found to be reproducible and give equivalent or higher yields than previous data, which used different extraction protocols and analytical techniques. No one analysis platform can cover the entire scope of the metabolome, but our method removes ion contamination from the media (making it compatible with CE-MS) and, although not tested, our method has the potential to be expanded to analyse biomass composition (acid hydrolysis of biomass followed by LC/GC-MS) and the non-polar metabolites (in the chloroform phase of the extraction by LC/GC-MS). We envisage our methods can be readily applied for the reliable extraction of metabolites from other microbial systems.
Acknowledgments
We are grateful to Martin Robert (Keio University) and Ken Haynes (Exeter University) for their input into the metabolite extraction and data analyses. We are also indebted to Kazutaka Ikeda, Maki Sugawara and Hiroko Ueda for their technical assistance and advice.
Funding Statement
The authors are grateful to Tsuruoka city and Yamagata prefecture for supporting this work. In part this work was funded by a joint JST-BBSRC grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ban E, Park SH, Kang M-J, Lee H-J, Song EJ, et al. (2012) Growing trend of CE at the omics level: The frontier of systems biology - An update. Electrophoresis 33: 2–13. Available:http://www.ncbi.nlm.nih.gov/pubmed/22139583. Accessed 12 January 2012. [DOI] [PubMed]
- 2.Buchholz A, Takors R, Wandrey C (2001) Quantification of intracellular metabolites in Escherichia coli K12 using liquid chromatographic-electrospray ionization tandem mass spectrometric techniques. Analytical biochemistry 295: 129–137. Available:http://www.ncbi.nlm.nih.gov/pubmed/11488613. Accessed 12 January 2012. [DOI] [PubMed]
- 3.Ohta D, Kanaya S, Suzuki H (2010) Application of Fourier-transform ion cyclotron resonance mass spectrometry to metabolic profiling and metabolite identification. Current opinion in biotechnology 21: 35–44. Available:http://www.ncbi.nlm.nih.gov/pubmed/20171870. Accessed 12 January 2012. [DOI] [PubMed]
- 4.Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, et al. (2003) Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. Journal of proteome research 2: 488–494. Available:http://www.ncbi.nlm.nih.gov/pubmed/14582645. Accessed 3 October 2011. [DOI] [PubMed]
- 5.Canelas AB, Ras C, Pierick A, Dam JC, Heijnen JJ, et al. (2008) Leakage-free rapid quenching technique for yeast metabolomics. Metabolomics 4: 226–239. Available:http://www.springerlink.com/content/n4w4117132644470/. Accessed 18 June 2011.
- 6.Sporty JL, Kabir MM, Turteltaub KW, Ognibene T, Lin S-J, et al. (2008) Single sample extraction protocol for the quantification of NAD and NADH redox states in Saccharomyces cerevisiae Journal of separation science 31: 3202–3211. Available:http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2640230&tool=pmcentrez&rendertype=abstract. Accessed 13 January 2012. [DOI] [PMC free article] [PubMed]
- 7.Gonzalez B, François J, Renaud M (1997) A rapid and reliable method for metabolite extraction in yeast using boiling buffered ethanol. Yeast (Chichester, England) 13: 1347–1355. Available:http://www.ncbi.nlm.nih.gov/pubmed/9392079. Accessed 5 September 2011. [DOI] [PubMed]
- 8.Villas-Bôas SG, Højer-Pedersen J, Akesson M, Smedsgaard J, Nielsen J (2005) Global metabolite analysis of yeast: evaluation of sample preparation methods. Yeast (Chichester, England) 22: 1155–1169. Available:http://www.ncbi.nlm.nih.gov/pubmed/16240456. Accessed 29 November 2011. [DOI] [PubMed]
- 9.Bolten CJ, Kiefer P, Letisse F, Portais J-C, Wittmann C (2007) Sampling for metabolome analysis of microorganisms. Analytical chemistry 79: 3843–3849. Available:http://www.ncbi.nlm.nih.gov/pubmed/17411014. Accessed 26 July 2011. [DOI] [PubMed]
- 10.de Koning W, van Dam K (1992) A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Analytical biochemistry 204: 118–123. Available:http://www.ncbi.nlm.nih.gov/pubmed/1514678. Accessed 13 January 2012. [DOI] [PubMed]
- 11.Canelas AB, ten Pierick A, Ras C, Seifar RM, van Dam JC, et al. (2009) Quantitative evaluation of intracellular metabolite extraction techniques for yeast metabolomics. Analytical chemistry 81: 7379–7389. Available:http://www.ncbi.nlm.nih.gov/pubmed/19653633. Accessed 13 January 2012. [DOI] [PubMed]
- 12.Faijes M, Mars AE, Smid EJ (2007) Comparison of quenching and extraction methodologies for metabolome analysis of Lactobacillus plantarum. Microbial cell factories 6: 27. Available:http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2031893&tool=pmcentrez&rendertype=abstract. Accessed 13 January 2012. [DOI] [PMC free article] [PubMed]
- 13.Theobald U, Mailinger W, Baltes M, Rizzi M, Reuss M (1997) In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae: I. Experimental observations. Biotechnology and bioengineering 55: 305–316. Available:http://www.ncbi.nlm.nih.gov/pubmed/18636489. Accessed 13 January 2012. [DOI] [PubMed]
- 14.Elliott B, Futcher B (1993) Stress resistance of yeast cells is largely independent of cell cycle phase. Yeast (Chichester, England) 9: 33–42. Available:http://www.ncbi.nlm.nih.gov/pubmed/8442385. Accessed 22 December 2011. [DOI] [PubMed]
- 15.Klosinska MM, Crutchfield CA, Bradley PH, Rabinowitz JD, Broach JR (2011) Yeast cells can access distinct quiescent states. Genes & development 25: 336–349. Available:http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3042157&tool=pmcentrez&rendertype=abstract. Accessed 27 November 2011. [DOI] [PMC free article] [PubMed]
- 16.Sasidharan K, Amariei C, Tomita M, Murray DB (2012) Rapid DNA, RNA and protein extraction protocols optimised for slow continuously growing yeast cultures. Yeast. Available:http://onlinelibrary.wiley.com/doi/10.1002/yea.2911/abstract. Accessed 20 June 2012. [DOI] [PubMed]
- 17.Bolten CJ, Wittmann C (2008) Appropriate sampling for intracellular amino acid analysis in five phylogenetically different yeasts. Biotechnology letters 30: 1993–2000. Available:http://www.ncbi.nlm.nih.gov/pubmed/18604477. Accessed 13 January 2012. [DOI] [PubMed]
- 18.Murray DB, Haynes K, Tomita M (2011) Redox regulation in respiring Saccharomyces cerevisiae Biochimica et biophysica acta 1810: 945–958. Available:http://www.ncbi.nlm.nih.gov/pubmed/21549177. Accessed 25 July 2011. [DOI] [PubMed]
- 19.D’Agostino LA, Lam KP, Lee R, Britz-McKibbin P (2011) Comprehensive plasma thiol redox status determination for metabolomics. Journal of proteome research 10: 592–603. Available:http://www.ncbi.nlm.nih.gov/pubmed/21053925. Accessed 22 December 2011. [DOI] [PubMed]
- 20.Satroutdinov AD, Kuriyama H, Kobayashi H (1992) Oscillatory metabolism of Saccharomyces cerevisiae in continuous culture. FEMS microbiology letters 77: 261–267. Available:http://www.ncbi.nlm.nih.gov/pubmed/1334018. Accessed 9 November 2011. [DOI] [PubMed]
- 21.Murray DB, Beckmann M, Kitano H (2007) Regulation of yeast oscillatory dynamics. Proceedings of the National Academy of Sciences of the United States of America 104: 2241–2246. Available:http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1794218&tool=pmcentrez&rendertype=abstract. Accessed 9 November 2011. [DOI] [PMC free article] [PubMed]
- 22.Monod J (1950) The technique of continuous culture. Ann Inst Pasteur 79: 390–410. Available:http://www.cabdirect.org/abstracts/19522202986.html. Accessed 27 July 2012.
- 23.Soga T, Igarashi K, Ito C, Mizobuchi K, Zimmermann H-P, et al. (2009) Metabolomic profiling of anionic metabolites by capillary electrophoresis mass spectrometry. Analytical chemistry 81: 6165–6174. Available:http://www.ncbi.nlm.nih.gov/pubmed/19522513. Accessed 21 June 2012. [DOI] [PubMed]
- 24.Soga T, Baran R, Suematsu M, Ueno Y, Ikeda S, et al. (2006) Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. The Journal of biological chemistry 281: 16768–16776. Available:http://www.ncbi.nlm.nih.gov/pubmed/16608839. Accessed 9 March 2012. [DOI] [PubMed]
- 25.Software:mzWiff - SPCTools. Available:http://tools.proteomecenter.org/wiki/index.php?title=Software:mzWiff. Accessed 21 June 2012.
- 26.Software:trapper - SPCTools. Available:http://tools.proteomecenter.org/wiki/index.php?title=Software:trapper. Accessed 21 June 2012.
- 27.Tautenhahn R, Böttcher C, Neumann S (2008) Highly sensitive feature detection for high resolution LC/MS. BMC bioinformatics 9: 504. Available:http://www.biomedcentral.com/1471-2105/9/504. Accessed 15 March 2012. [DOI] [PMC free article] [PubMed]
- 28.Baran R, Kochi H, Saito N, Suematsu M, Soga T, et al. (2006) MathDAMP: a package for differential analysis of metabolite profiles. BMC bioinformatics 7: 530. Available:http://www.biomedcentral.com/1471-2105/7/530. Accessed 2 March 2012. [DOI] [PMC free article] [PubMed]
- 29.Barri T, Holmer-Jensen J, Hermansen K, Dragsted LO (2012) Metabolic fingerprinting of high-fat plasma samples processed by centrifugation- and filtration-based protein precipitation delineates significant differences in metabolite information coverage. Analytica chimica acta 718: 47–57. Available:http://www.ncbi.nlm.nih.gov/pubmed/22305897. Accessed 21 June 2012. [DOI] [PubMed]
- 30.Murray DB, Engelen F, Lloyd D, Kuriyama H (1999) Involvement of glutathione in the regulation of respiratory oscillation during a continuous culture of Saccharomyces cerevisiae Microbiology 145: 2739–2745. Available:http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10537195. [DOI] [PubMed]
- 31.Murray DB, Lloyd D (2006) A tuneable attractor underlies yeast respiratory dynamics. Bio Systems 90: 287–294. Available:http://www.ncbi.nlm.nih.gov/pubmed/17074432. [DOI] [PubMed]
- 32.Machné R, Murray DB (2012) The Yin and Yang of Yeast Transcription: Elements of a Global Feedback System between Metabolism and Chromatin. PLoS ONE 7: e37906. Available:http://dx.plos.org/10.1371/journal.pone.0037906. Accessed 8 June 2012. [DOI] [PMC free article] [PubMed]
- 33.Sohn HY, Kum EJ, Kwon GS, Jin I, Kuriyama H (2005) Regulation of branched-chain, and sulfur-containing amino acid metabolism by glutathione during ultradian metabolic oscillation of Saccharomyces cerevisiae J Microbiol 43: 375–380. Available:http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16145554. [PubMed]
- 34.Ozalp VC, Pedersen TR, Nielsen LJ, Olsen LF (2010) Time-resolved measurements of intracellular ATP in the yeast Saccharomyces cerevisiae using a new type of nanobiosensor. The Journal of biological chemistry 285: 37579–37588. Available:http://www.jbc.org/cgi/content/abstract/285/48/37579. Accessed 13 March 2012. [DOI] [PMC free article] [PubMed]
- 35.Sohn H-Y, Kum E-J, Kwon G-S, Jin I, Adams CA, et al. (2005) GLR1 plays an essential role in the homeodynamics of glutathione and the regulation of H2S production during respiratory oscillation of Saccharomyces cerevisiae Bioscience, biotechnology, and biochemistry 69: 2450–2454. Available:http://www.ncbi.nlm.nih.gov/pubmed/16377908. [DOI] [PubMed]
- 36.Sohn HY, Kuriyama H (2001) The role of amino acids in the regulation of hydrogen sulfide production during ultradian respiratory oscillation of Saccharomyces cerevisiae. Archives of Microbiology 176: 69–78. Available:<Go to ISI>://000170363300009. [DOI] [PubMed]