Background: Structural information concerning the phosphorylatable regulatory β subunit of phosphorylase kinase was lacking.
Results: Chemical, biochemical, biophysical, and computational approaches revealed secondary, tertiary, and quaternary structures for this subunit.
Conclusion: The β subunit is helical and forms the β4-bridged core in the (αβγδ)4 kinase complex.
Significance: These findings reveal the architecture of the complex, which provides an explanation for the conformational changes in its bridged core associated with activating β-phosphorylation.
Keywords: Computational Biology, Protein Complexes, Protein Cross-linking, Protein Domains, Protein Phosphorylation, Protein Structure, Phosphorylase Kinase, Threading, Top-down MS
Abstract
Phosphorylase kinase (PhK) is a hexadecameric (αβγδ)4 complex that regulates glycogenolysis in skeletal muscle. Activity of the catalytic γ subunit is regulated by allosteric activators targeting the regulatory α, β, and δ subunits. Three-dimensional EM reconstructions of PhK show it to be two large (αβγδ)2 lobes joined with D2 symmetry through interconnecting bridges. The subunit composition of these bridges was unknown, although indirect evidence suggested the β subunits may be involved in their formation. We have used biochemical, biophysical, and computational approaches to not only address the quaternary structure of the β subunits within the PhK complex, i.e. whether they compose the bridges, but also their secondary and tertiary structures. The secondary structure of β was determined to be predominantly helical by comparing the CD spectrum of an αγδ subcomplex with that of the native (αβγδ)4 complex. An atomic model displaying tertiary structure for the entire β subunit was constructed using chemical cross-linking, MS, threading, and ab initio approaches. Nearly all this model is covered by two templates corresponding to glycosyl hydrolase 15 family members and the A subunit of protein phosphatase 2A. Regarding the quaternary structure of the β subunits, they were directly determined to compose the four interconnecting bridges in the (αβγδ)4 kinase core, because a β4 subcomplex was observed through both chemical cross-linking and top-down MS of PhK. The predicted model of the β subunit was docked within the bridges of a cryoelectron microscopic density envelope of PhK utilizing known surface features of the subunit.
Introduction
Skeletal muscle phosphorylase kinase (PhK),2 a 1.3-MDa (αβγδ)4 complex, catalyzes the Ca2+-dependent phosphorylation of glycogen phosphorylase and thereby stimulates the breakdown of glycogen to form glucose 1-phosphate, leading to energy production to help sustain contraction (1). The catalytic γ subunit comprises an N-terminal catalytic core (residues 1–298) that has a typical protein kinase fold (2, 3) and a C-terminal regulatory domain (γCRD) that binds both the intrinsic calmodulin (δ) subunit (4, 5), as well as the regulatory α subunit (6). The activity of γ in the (αβγδ)4 complex is tightly regulated by the α (138.4 kDa), β (125.2 kDa), and δ (16.7 kDa) subunits, which together promote constraining quaternary (4°) interactions on γ (44.7 kDa) that are attenuated by signaling molecules from metabolic, hormonal, and neural pathways that target each of the three regulatory subunits (7, 8). Correspondingly, we have shown that the γCRD can be chemically cross-linked to α, β, and δ within the PhK complex (5–7), which led us to postulate that it is the allosteric activation switch through which the three regulatory subunits interact to control the kinase activity of γ (7).
Despite being the first protein kinase to be discovered (9), little is known about the structure of PhK. Reconstructions of images of PhK from EM reveal it to be a D2 symmetrical structure of two lobes (10–12), each composed of two αβγδ protomers packed head-to-head with the lobes associated through interconnecting bridges (13–16). The number of bridges reported for PhK from EM fields of the negatively stained complex on solid grids varies from a minimum of two to a maximum of four (13, 17), with reconstructions of the stained kinase revealing only two large bridges between the lobes (10, 12, 18). Direct evidence for four bridges was first obtained from a reconstruction of nonactivated PhK from particles of the complex frozen in vitreous ice (cryo-EM), demonstrating that the bridges are viable solution structures and not simply artifacts of staining (11). The cryo-EM results were corroborated by small angle x-ray scattering (SAXS) of nonactivated and Ca2+-activated PhK (19). SAXS modeling of both conformers demonstrated that only those models containing four, rather than two, bridge structures provided theoretical scattering profiles that accurately fit the experimental data. The divergent results reported above for the bridges in reconstructions of the negatively stained PhK complex may reflect either possible distortions imposed by the mechanical effects of drying and staining on a solid surface or a conformational change associated with the staining process (10). In support of the latter possibility, activation of PhK by Ca2+ has been shown by EM to perturb the structure of the bridges (10) and by SAXS to promote the approach of two bridges toward one another (19). These results suggest that in fully activated forms of PhK, two bridges may abut to form one large bridge, resulting in the overall visualization of only two bridges in low resolution structures of PhK (11, 19).
Attempts to determine the subunit composition of the bridges have proved difficult, because of a lack of structural information for the large homologous α and β subunits, which together constitute over 81% of the total mass of PhK. Although the primary structures are known for both α and β (20, 21), essentially all higher order secondary (2°) and tertiary (3°) structures reported for these subunits are based on predicted rather than experimental data (22–24). In contrast, high resolution structural information is available for the catalytic core of the γ subunit and for the δ subunit (3, 25); on the other hand, the small size and location of these subunits in the complex make them unlikely candidates for bridging the two (αβγδ)2 lobes (14, 16). The α subunit is theoretically large enough to span the distances calculated for the bridges (∼3 nm); however, its C terminus has been localized to the distal ends of the lobes, which would require the subunit to extend ∼90 Å in a straight line through one lobe before traversing the space to the adjacent lobe (10). Moreover, extensive digestion of this subunit in the complex has no effect on the bridges, whereas partial hydrolysis of both α and β disrupts the bridges, forming isolated lobes and smaller fragments in EM fields of the kinase (17). Proteolytic mapping of PhK also suggests α to be more peripherally located in the complex than β (26), and localization by immuno-EM of an epitope of the β subunit close to the bridges makes it a more likely candidate for composing the interlobal bridges joining the two (αβγδ)2 lobes of PhK (16). Nevertheless, no evidence has been previously obtained to indicate the direct involvement of any of the four subunits of PhK in bridge formation.
Functionally, the β subunit places PhK at the interface of metabolic signaling pathways by being the subunit thought to bind the activator ADP (27). It is also the subunit that is predominantly responsible for the regulation of PhK activity by phosphorylation (7). The major regulatory phosphorylatable serine, Ser-26 (28), which is targeted in vivo by cAMP-dependent protein kinase (PKA) and is part of the β subunit's unique N-terminal phosphorylatable domain (NB1) (20), is also reportedly autophosphorylated, along with Ser-11, by the γ subunit within the complex (7, 20), triggering a conformational change in β that is detected by its cross-linking with 1,5-difluoro-2,4-dinitrobenzene (DFDNB) to form homodimers (29). This possible association of β subunits is consistent with phospho-mimetic S11E/S26E joint mutations that promote self-association of β chimeras in two-hybrid assays (7). We have shown that the NB1 domain is proximal to the γCRD and the C terminus of the β subunit by chemical cross-linking with N-[γ-maleimidobutyryloxy] succinimide ester (GMBS) (7). The sum of these data indicates that residues in the β C terminus, the NB1 domain, and γCRD are surface-accessible and proximal to one another within the (αβγδ)4 complex. Another likely surface-accessible region of β is Ser-700, which is reportedly autophosphorylated by PhK (20). We also report herein an epitope in the PhK complex for an anti-β-specific mAb that has been localized through β-truncation to within a region of this subunit from residues 704 to 815.
The regions of β discussed above represent the little that is experimentally known about its structure and approximate location in low resolution EM structures. A comparison of the sequences of the β subunit and its α subunit homolog indicated that both are multidomain structures, containing subunit-specific regulatory regions and large distinct sequence-similar domains, the latter of which suggested that the subunits are products of early gene duplication events (20). Since these earlier reports, both N-terminal glycosyl hydrolase (GH) clan 15 member-like (GHL) domains and C-terminal calcineurin-B like (CBL) domains have been predicted for α and β (22–24); however, attempts to model the entire structures were unsuccessful (22, 23). We have employed biochemical and biophysical methods in combination with predictive threading and ab initio approaches to model the full β subunit structure as an isolated protein and in the context of the (αβγδ)4 PhK complex. We report nearly full coverage of the subunit by threading with templates corresponding to GH-15 family members and the protein phosphatase 2A (PP2A) subunit PR65/A (PP2AA). We directly demonstrate for the intact complex by top-down MS and chemical cross-linking that the β subunits compose the central bridge region of PhK and that rigid-body docking of their theoretical counterparts in the individual bridges of the native PhK cryo-EM envelope correlates well with the known structural details for this subunit in the complex (7, 16, 30).
EXPERIMENTAL PROCEDURES
Proteins
PhK was purified from the psoas muscle of New Zealand White rabbits (31), dialyzed against 50 mm Hepes (pH 6.8), 0.2 mm EDTA, and 10% sucrose, and stored at −80 °C. Its concentration was determined by previously described methods (32). Autophosphorylated PhK was prepared as described previously (33). The mAbs against the α, β, and γ subunits of PhK were previously described (15, 16), and the anti-calmodulin mAb was from Zymed Laboratories Inc.. All other secondary conjugates were from Southern Biotechnology. Truncation mutants of the PhK β subunit were constructed as described previously (7).
CD
Far-UV CD spectra were collected for PhK and the αγδ subcomplex using previously described conditions (34). Secondary structure content was estimated using the Dichroweb software package (35), which permits analysis of secondary structure by CONTIN, SELCON, and CDSSTR (36, 37).
Cross-linking
PhK was cross-linked with DFDNB essentially as described (29), with cross-linking initiated by addition of DFDNB and carried out at 30 °C for 2.5 min at pH 8.2 in 50 mm Hepes, 0.2 mm EDTA. Final concentrations of PhK (αβγδ protomer) and GMBS in the reaction were 0.47 and 117 μm, respectively. The reaction was terminated by adding an equal volume of SDS buffer (0.125 m Tris (pH 6.8), 20% glycerol, 5% β-mercaptoethanol, 4% SDS), followed by brief vortexing. The PhK subunits were separated on 6–18% linear gradient polyacrylamide gels and stained with Coomassie Blue. Western blotting of the proteins was performed on PVDF membranes with subunit-specific mAbs as described previously (38). All cross-linking reactions were performed at least twice using different preparations of PhK.
To determine regions of cross-linking in the β monomer, the cross-linked PhK complex was resolved by preparative SDS-PAGE and stained with Coomassie Blue. The bands corresponding to the cross-linked and noncross-linked monomeric β subunits were excised from the gel, sectioned, and exchanged with three aliquots (each ∼5× the volume of the gel slice) of 50 mm ammonium bicarbonate, 50% acetonitrile to remove SDS. The proteins were then reduced in 10 mm dithiothreitol for 1 h at 55 °C, and carboxymethylated with 50 mm iodoacetic acid for 1 h in the dark. The gel pieces were washed as described above with 50 mm ammonium bicarbonate, followed by several exchanges with 50 mm ammonium bicarbonate, 50% acetonitrile. After removing the last wash, the gels were dried in a SpeedVac (Savant) and treated with trypsin (Promega; 12.5 ng/μl) for 24 h at 30 °C. Peptides were extracted from the gel pieces with 50% acetonitrile, 5% formic acid.
Bottom-up MS
All samples were concentrated on a CentriVac concentrator (Labconco) to a final volume of 20 μl and pressure-loaded onto a C18 reversed phase nano-column (75-μm inner diameter fused silica packed in-house with 9 cm of 100 Å, 5 μm, Magic C18 particles, Michrom Bioresources). Following a wash with 0.1% formic acid for 15 min at 0.5 μl/min, the column was mounted on the electrospray stage of a Fourier transform ion cyclotron resonance MS (LTQ FT, ThermoFinnigan, San Jose, CA), and peptides were eluted at an approximate flow rate of 0.3 μl/min over a 120-min period using a gradient of 0–90% acetonitrile (Buffer A = 0.1% formic acid; Buffer B = acetonitrile, 0.1% formic acid). The source was operated at 1.9 kV, with the ion transfer temperature set to 350 °C. LC MS data were obtained in a hybrid linear ion trap Fourier transform ion cyclotron resonance MS equipped with a 7 tesla magnet. The MS was controlled using an Xcalibur software package to continuously perform mass scan analysis on the FT, followed by MS/MS scans on the ion trap for the six most intense ions, with a dynamic exclusion of two repeat scans (30 s repeat duration and 90 s exclusion duration) of the same ion. Normalized collision energy for MS/MS was set to 35%.
MS Data Analyses
For data analyses, data files were created on Bioworks Browser version 3.2. The corresponding log file was used to generate a list of parent ions for which the corresponding charges and tandem mass spectra were obtained. A list of potential conjugates was generated from the resulting mass list ((M + H)+) by previously described methods (30), and the fragmentation patterns of all viable candidates were analyzed for consistency with the predicted chemistry of cross-linking (30).
The tandem mass spectrum of each conjugate was analyzed using a combination of programs that were subsequently verified by an “in-house” spreadsheet. A list of masses from the tandem mass spectrum of each candidate parent ion was up-loaded to MS2Links (39). Theoretical fragmentation of the conjugate in the positive mode was accomplished using the sequences for each peptide in the conjugate and the masses for each intervening chemical cross-link and its possible fragmentation products, using a mass error tolerance not exceeding 0.7 Da for each fragment ion. A modification table was generated for the DFDNB cross-linker based on the loss of NO2 observed for structural analogs of the reagent (40, 41). Potential matching ion assignments for each fragment mass generated by MS2Links were verified using a spreadsheet containing an array of masses predicted for the cross-linker (and fragments thereof) and also those generated for each peptide in the conjugate using MS Product, a fragmentation tool in Protein Prospector developed at the University of California, San Francisco (42). Final assignments were also checked by hand for verification and accuracy.
Top-down MS
A sample (130 μl) of autophosphorylated (1.85 and 0.90 mol of Pi incorporated per mol of α and β, respectively) PhK (∼1.5 mg/ml) was exchanged into 50 mm ammonium acetate (pH 6.8) using a 5K MWCO VivaSpin column (Vivascience) and then concentrated to a final volume of 30 μl. Nondenaturing nano-electrospray mass spectra were acquired on a Q-TOF 2 mass spectrometer (Micromass/Waters) that was modified for high mass detection (43). Optimal transmission of noncovalent protein complexes was carried out using a previously described protocol (44). Spectra were recorded in positive ion mode with the following settings: capillary voltage 1.7 kV; sample cone 194 V; extraction cone 5 V; collision energy 200 V; collision cell pressure 20 μbar; hexapole ion guide pressure 4.2 × 101 μbar; analyzer pressure 1.5 × 10−4 μbar; backing pressure 1.0 × 100 μbar, and TOF pressure 2.1 × 10−6 μbar.
Spectra were acquired and processed using MassLynx version 4.1 (Waters, Manchester, UK) and Analyst QS (MDS Sciex, Applied Biosystems). Errors are reported as ± 1 S.D. All spectra were calibrated externally using a standard (100 mg/ml) solution of cesium iodide, with minimal smoothing and without background subtraction. Subcomplex compositions were determined using the iterative search algorithm SUMMIT (45).
Threading and ab Initio Calculations
The theoretical atomic model of the β subunit was constructed using I-TASSER (46, 47). Using the rabbit muscle β subunit sequence (accession number = P12798) as query, multiple sequence-template alignments were initially generated by the meta-server LOMETS (48), with the domain boundaries decided based on unaligned regions of the threading alignments and the template structures. The β subunit sequence was divided into three domains. The first domain, corresponding to the GHL domain, matched well with several high scoring templates. The full-length model was constructed by I-TASSER, with spatial restraints (Cα distance map and side-chain contacts) extracted from the templates being used to guide replica-exchange Monte Carlo assembly simulation. The second domain, corresponding to the Huntingtin-elongation-A subunit-TOR (HEAT)-repeat like (HRL) domain, had only low scoring templates. The models were then constructed by I-TASSER ab initio structural assembly, which were guided with the sparse distance and contact restraints from short template fragments (46). The third domain, NB1, was also generated by the I-TASSER ab initio assembly simulations. Decoys generated during the structural assembly simulations were clustered by SPICKER (49), and the cluster centroid models were further refined by REMO to build the full-atomic model (50). The entire β subunit model was constructed by connecting all three domain models together with the domain orientation repacked based on the I-TASSER energy potential. Chemical cross-linking of residues Tyr-51 and Lys-53 from the GHL domain to Lys-1025 from the HRL domain was used as a constraint in packing the domains.
Rigid Body Docking
Docking of β atomic models was carried out using MVP-FIT,3 a program developed by the Zhang laboratory for flexible fitting of atomic models in EM density xmaps. The top 50 atomic models of PhK were docked in the bridge region of the cryo-EM envelope reconstructed from frozen solvated particles of nonactivated PhK (11), based on their localization to this region by top-down MS and intra-subunit chemical cross-linking of β within the PhK complex. The final model was chosen, based on best correspondence with known structural details of β in reconstructions of the PhK complex, as well as best fit in the cryo-EM envelope. All models were visualized using Chimera (51).
RESULTS
Determination of 2° Structure of the β Subunit within the PhK Complex
Predictions of 2° structure for the free β subunit suggest it to be a helical protein (52, 53), with helical content ranging from 41 to 70%. Within the (αβγδ)4 PhK complex, the structure of β is undoubtedly influenced by subunit interactions with itself and with the other three subunits, as evidenced by chemical cross-linking (7, 29, 30, 38, 54), EM three-dimensional reconstructions (10), and partial proteolysis (26). As opposed to the α, γ, and δ subunits, which have been isolated successfully either individually (8, 55, 56) or in subcomplexes of PhK (57, 58), the β subunit appears to be stable or isolable only in the context of the intact (αβγδ)4 complex. Although the β subunit undoubtedly influences the structure of α, γ, and δ in the PhK complex, we have shown that features of an established Ca2+-dependent communication network among the α, γ, and δ subunits are the same in both the (αβγδ)4 and αγδ complexes (57), suggesting a similar structural architecture for these subunits in each complex. To determine the 2° structure of β within the PhK complex, CD analyses of the intact (αβγδ)4 complex and an αγδ subcomplex, expressed and purified from insect cells (57), were carried out at 25 °C (Table 1). The percent 2° structural contents (helical, sheet, etc.) of the δ (endogenous calmodulin) subunit (25) and catalytic core of the γ subunit (3) were calculated from known crystal structures of the isolated proteins and were taken to be identical in both the αγδ and (αβγδ)4 complexes. Differences observed in the CD spectrum for each complex were assumed to reflect differences in contributions from the large homologous α and β subunits, which contribute 74% (α) and 87% (α and β) of the entire 2° structure content of PhK αγδ and (αβγδ)4, respectively. Based on 2° structural contributions predicted for α, γ, and δ in each complex using the CDSSTR algorithm (36, 37), the signal difference corresponding to β indicates that it is a helical (69%) protein, with minor amounts of sheet (16%), turn (8%), and unordered (7%) structure.
TABLE 1.
Predicted percent secondary structure content of the PhK αγδ-trimer and the PhK hexadecamer from CD analysis at 25 °C and the contributions from individual subunits
| Secondary structure type |
||||
|---|---|---|---|---|
| Helix | Sheet | Turn | Unordered | |
| Nonactivated trimera | 65 ± 2 | 14 ± 2 | 5 ± 1 | 16 ± 1 |
| Nonactivated hexadecamera | 64 ± 5 | 16 ± 5 | 6 ± 4 | 14 ± 5 |
| Trimer subunit | Individual subunit percent secondary structure | |||
| PhK α | 67 | 16 | 6 | 11 |
| PhK γa | 40 | 18 | 4 | 38 |
| PhK δb | 59 | 12 | 0 | 29 |
| Trimer subunit | Calculated contribution to percent secondary structure | |||
| PhK α | 53 | 10 | 4 | 7 |
| PhK γb | 7 | 3 | 1 | 6 |
| PhK δc | 5 | 1 | 0 | 3 |
| Hexadecamer subunit | Individual subunit percent secondary structure | |||
| PhK α | 67 | 16 | 6 | 11 |
| PhK β | 69 | 16 | 8 | 7 |
| PhK γb | 40 | 18 | 4 | 38 |
| PhK δc | 59 | 12 | 0 | 29 |
| Hexadecamer subunit | Calculated contribution to percent secondary structure | |||
| PhK α | 29 | 7 | 2 | 5 |
| PhK β | 27 | 6 | 3 | 3 |
| PhK γb | 5 | 2 | 1 | 5 |
| PhK δc | 3 | 1 | 0 | 1 |
a The percent secondary structure of the PhK holoenzyme and the αγδ -trimer CD spectra was predicted using the CDSSTR algorithm, found in the DICHROWEB software suite (35).
b The percent secondary structure of the PhK γ subunit was calculated from the 2.1 Å crystal structure of the PhK γ peptide substrate complex (PDB code 2PHK).
c The percent secondary structure of the PhK δ subunit was calculated from the 1.7 Å crystal structure of calmodulin (PDB code 1CLL).
Modeling of the 3° Structure of the β Subunit
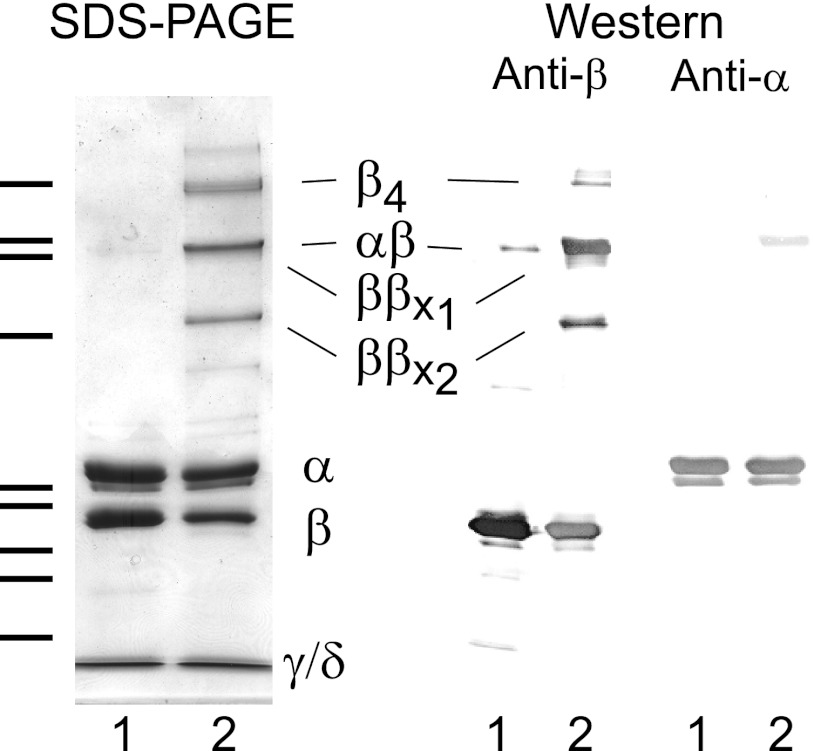
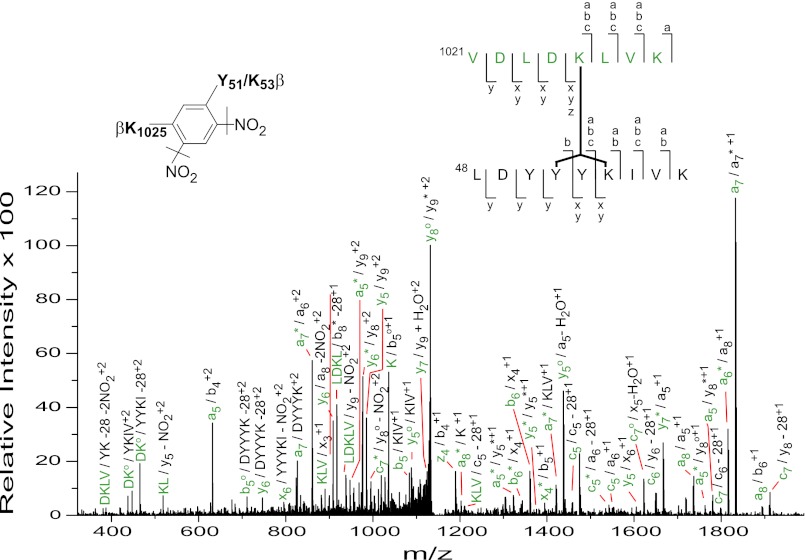
As was the case with its 2° structure, little was previously known about the 3° structure of β, whether free or as a component of the PhK complex, a lack that we addressed through modeling. To obtain a physical constraint that could be used in the structural assembly of theoretical three-dimensional models, we employed chemical cross-linking. This technique has been used successfully to reveal intermediate resolution 3° structural information for proteins that are refractive to crystallographic and/or NMR methods (59). In an initial attempt to probe the 3° structure of β within the PhK complex, we had used the cross-linker GMBS and deduced that the polypeptide backbone of β folds back on itself in such a way that the N and C termini of the subunit approach each other (7). GMBS, however, has a spacer arm that is both long (∼12 Å) and highly flexible and thus would not provide a reliable distance constraint. We turned instead to DFDNB, which has a rigid spacer arm and reportedly cross-links side chains separated by 3–5 Å. Autophosphorylated PhK (1.85 and 0.90 mol of Pi incorporated per mol of α and β, respectively) was treated with DFDNB (Fig. 1), which has previously been shown to produce intermolecular and intramolecular cross-linked forms of the β subunit with this form of the enzyme (29). Four major high molecular weight β-containing conjugates were identified by their apparent mass and cross-reactivity against subunit-specific mAbs (15, 16). With the exception of an αβ dimer (massExp = 263.6 kDa, 4.0% error), all the remaining conjugates cross-reacted only with an anti-β subunit-specific mAb and were consistent by mass with a β4 tetramer (massExp = 500.8 kDa, 4.1% error) and two β2 dimers (massTheo = 250.2 kDa, 5.0% error (ββx1); 23.3% error (ββx2)). The faster migrating DFDNB ββx2 cross-linked dimer has been reported previously and was suggested to differ from ββx1 by alternative intermolecular cross-linking of the β subunits in the PhK complex and/or intramolecular cross-linking of one or both of the β subunits that hinders unfolding of the protein in SDS, leading to an anomalously fast migration (29). To screen for potential intramolecular conjugates, the β monomeric band remaining after treatment with DFDNB was digested in-gel with trypsin, and the resultant peptides were resolved and measured by Fourier transform ion cyclotron resonance MS. The mass list for the digest was then evaluated using the CrossSearch method (30). After culling experimental masses corresponding to those predicted either for nonmodified peptides or for side products of chemical cross-linking, a mass (m/zTheor = 2297.207) predicted for a peptide corresponding to cross-linking between regions of the N terminus (residues 48–56, LDYYYKIVK) and C terminus of β (residues 1021–1028, VDLDKLVK) matched that of a peptide mass (m/zExp = 2297.197; 4.37 ppm error) measured experimentally. The match was verified by MS/MS analysis (Fig. 2), demonstrating the presence of a pool of conjugates with the same peptide partners, but with Lys-1025 in the C-terminal peptide stretch cross-linked to either Tyr-51 or Lys-53 in the N-terminal region of β. No direct evidence for cross-linking between Lys-1025 and Tyr-52 was observed in the MS/MS spectrum. The observed cross-linking between Lys-1025 and both Tyr-51 and Lys-53 within the N-terminal peptide stretch may indicate that these regions may be conformationally dynamic, which would be consistent with the fact that the conformation of β correlates with its phosphorylation and with the activation state of PhK (16, 26, 29). These results with the shorter, rigid cross-linker DFDNB corroborate our previous findings that the N and C termini of β are proximal and provide a viable physical constraint to be used in the three-dimensional modeling of this regulatory subunit.
FIGURE 1.
Cross-linking of PhK with DFDNB. Left panel, PhK (lane 1) was cross-linked with DFDNB (lane 2) and resolved by SDS-PAGE. Right panel, parallel samples were transferred to PVDF membranes and probed with mAbs against all of the subunits. All major conjugates cross-reacted only with anti-β and anti-α mAbs and not with anti-γ or anti-δ (δ = integral calmodulin subunit) mAbs. Cross-linking of PhK by DFDNB resulted in the formation of four major β-containing conjugates that corresponded by mass and cross-reactivity to a β4 tetramer (501 kDa), αβ dimer (264 kDa), intermolecularly cross-linked ββx1 (250 kDa), and intramolecularly cross-linked ββx2 (250 kDa) dimers. As discussed under “Results,” the ββx2 dimer migrates faster than its theoretical mass of 250 kDa would predict. Progressing from top to bottom, hatchmarks to the left of column 1 correspond to molecular mass markers with masses (kDa) of 500, 279, 251, 164, 121, 117, 77, 64, and 52.
FIGURE 2.
MS/MS analysis of the signal at m/z 1149.102 identifying a conjugate comprising residues 48–56 and 1021–1028 of the regulatory β subunit of PhK. The composition of the ions identifying the cross-linked peptide and chemical structure of the cross-link are shown. Lowercase letters denote ions arising from amide cleavages of the peptide backbone and are color-coded for each peptide in the conjugate pair (black for residues 48–56 and green for 1021–1028). Intact covalent links formed between peptide fragments are indicated by a forward slash (/). For singly charged ions, it should be noted that one of the two peptide ions covalently attached to either position of the ring is a neutral product of the indicated backbone amide cleavage. Lines bisecting the bonds between the ring carbons and NO2 nitrogens in the cross-link structure represent the loss of nitro groups, indicated in several fragment ions (40, 41). Heavy black bars between each peptide indicate residues cross-linked and are consistent with a pool of peptides, containing either Lys-1025/Tyr-51 or Lys-1025/Lys-53 cross-linked side chains.
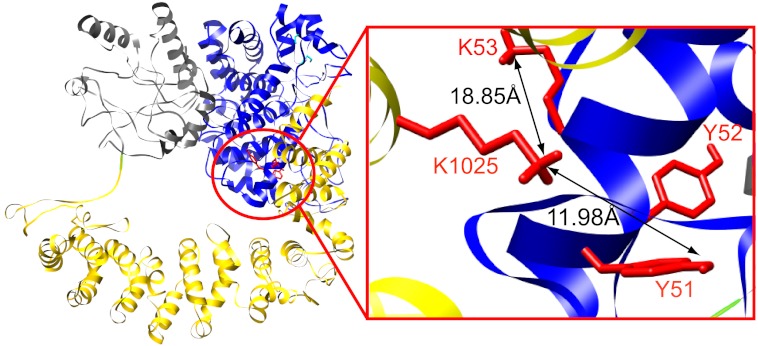
Three-dimensional models for the β subunit were generated using the I-Tasser hierarchical structural modeling approach (46). Multiple threading alignments of the β sequence were carried out to identify template structures from the Protein Data Bank (PDB) library (48), followed by structural assembly and refinement steps, with subsequent reconstruction of atomic models (50). As shown for the best fit structure (Fig. 3), two major domains comprising residues 41–670 and 671–1092 were predicted and modeled, respectively, using as templates members of the GH family 15 (blue and gray traces) and PP2AA (yellow trace). The remainder of the molecule, the small N-terminal phosphorylatable domain of β (NB1, residues 1–40), is apparently unique and was therefore modeled ab initio (Fig. 3, blue trace) (60). The models were constrained using the DFDNB cross-linking results by forcing the closest possible approach of the α-carbons of Lys-1025 and Tyr-52. In the best fit model, this constraint automatically oriented without further manipulation the side chains of Lys-1025 and Tyr-51/Lys-53 in positions that would allow their observed cross-linking (Fig. 3).
FIGURE 3.
Theoretical three-dimensional model of the PhK β subunit. Hierarchical protein structural modeling of the β subunit was carried out using I-TASSER (46). X-ray crystal structures of Aspergillus awamori glucoamylase (blue and gray ribbon traces) and human PP2A PR65/A subunit (yellow trace) were used to thread, respectively, residues 41–670 and 671–1092 of the multidomain β subunit primary sequence (61, 65). The remaining N-terminal residues (1–40, blue trace) were modeled ab initio using QUARK (60). Models were constrained using the DFDNB cross-linking results, forcing the approach of the Lys-1025 and Tyr-52 α-carbons. Side chains of the DFDNB cross-linked residues (red) and the distances between the Lys-1025/Lys-51 and Lys-1025/Lys-53 cross-linked pairs (arrows) are indicated in a magnified view of the cross-link site linking the N and C termini of β.
Our multidomain model derived from threading is consistent with previous structural predictions for β that suggested it to be a multidomain structure with an N-terminal GHL domain (22, 24). The GH-15 thread coverage of β is indicated in a two-color scheme, with blue and gray representing, respectively, the (α/α)6-barrel fold for the catalytic domain (A) and a mixed 2° structure corresponding to the B and C domains found in bacterial and archaeal glucoamylases and glucodextranases (61). The protein structurally closest to the best model of this region is Anthrobacter globiformus glucodextranase (PDB code 1ULV) (61), with a template modeling score of 0.8462 and root mean square deviation of 1.92 Å, indicating a good topographical match (62). The crystal structure of the glucodextranase template revealed the protein in complex with the pseudo tetrasaccharide acarbose, a potent transition state inhibitor of glucoamylases (63). Correspondingly, we recently showed that PhK binds acarbose, which in turn promotes a conformational change in the β subunit (64). The large C-terminal domain of the β model was a relatively good topographical match (template modeling score = 0.6272; root mean square deviation = 5.41) with PP2AA (PDB code 1B3U), and the predominantly helical HEAT repeat structure corresponded well with the helical content estimated for β by CD measurements (65), revealing an overall helical topology for β.
Determination of the 4° Structure of the β Subunits within the PhK Complex
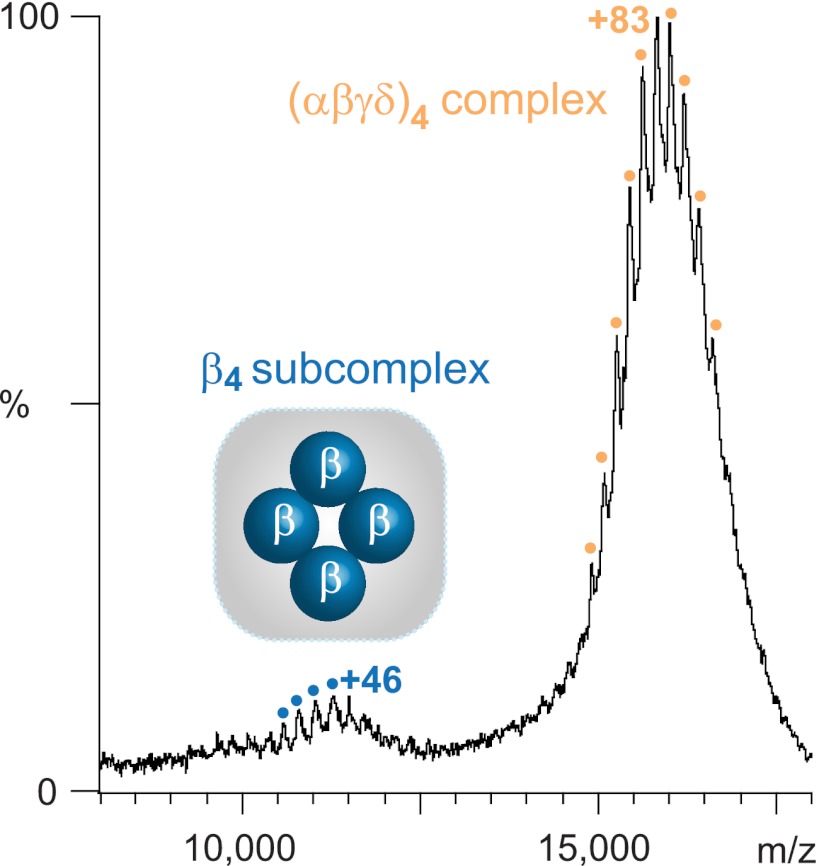
Having analyzed the 2° and 3° structures of PhKβ, we set out to determine the arrangement of these subunits in the 4° structure of PhK. Both cryo-EM and SAXS analyses of the native (αβγδ)4 PhK complex show it to be a large bilobal structure, in which the lobes associate with each other in D2 symmetry through four interconnecting bridges (11, 19). Previous evidence from our laboratory indicated that an unknown epitope on the β subunit is centrally located on the lobes near the bridges (16) and suggested that at least two β subunits may self-associate (7, 29). If the β subunits actually composed the bridges and self-association were more extensive, we asked whether direct evidence could be obtained for a β4 complex that would by definition represent the bridged central core of PhK upon which the stable αγδ trimers might be arrayed. To investigate this possibility, we first screened the (αβγδ)4 PhK complex with cross-linkers that preferentially targeted the β subunits and analyzed for cross-linked β4 complexes. Several reagents formed conjugates that corresponded to a β4 conjugate by apparent mass and cross-reactivity against subunit-specific mAbs (results not shown). The best resolved species was formed by cross-linking phosphorylated PhK with DFDNB (Fig. 1); however, despite the fact that an anti-α mAb did not detect any α subunit within the β4 band, it did contain minor amounts of co-migrating α/β heteromeric conjugates, because bottom-up MS indicated very small amounts of α peptides in the β4 band. As an alternative corroborating approach, we employed top-down MS to analyze large partially solvated complexes under near-native conditions (66). With the phosphorylated, but not the nonactivated, PhK complex, we observed one subcomplex by nondenaturing nano-electrospray MS that corresponded by mass only to a β4 tetramer (Fig. 4; massTheo = 500. 3 kDa; 1.3% difference), with water from the solvation sphere accounting primarily for the small mass addition observed for the tetramer (67). Given the known D2 symmetrical head-to-head packing of αβγδ protomers in each of the two octameric lobes of PhK (15), the only possible arrangement for the β subunits in the complex that can account for these results and our previous findings is a centrally located core, with the four β subunits occupying a region comprising the bridges and internal lobe-faces of PhK.
FIGURE 4.
β4 subcomplex revealed by nano-ESMS of autophosphorylated PhK. Nondenaturing MS of intact phosphorylated PhK yielded two charge state series centered at 12,500 and 16,000 m/z, the latter of which corresponds to the intact hexadecameric PhK complex (discussed in detail in Ref. 72). The former charge state series (blue circles), with charges ranging from +46 to +50, yielded a measured mass of 507,062 ± 55 Da that was in close agreement by mass with only one combination of subunits, namely a β4 tetramer (massTheo = 500,338 Da).
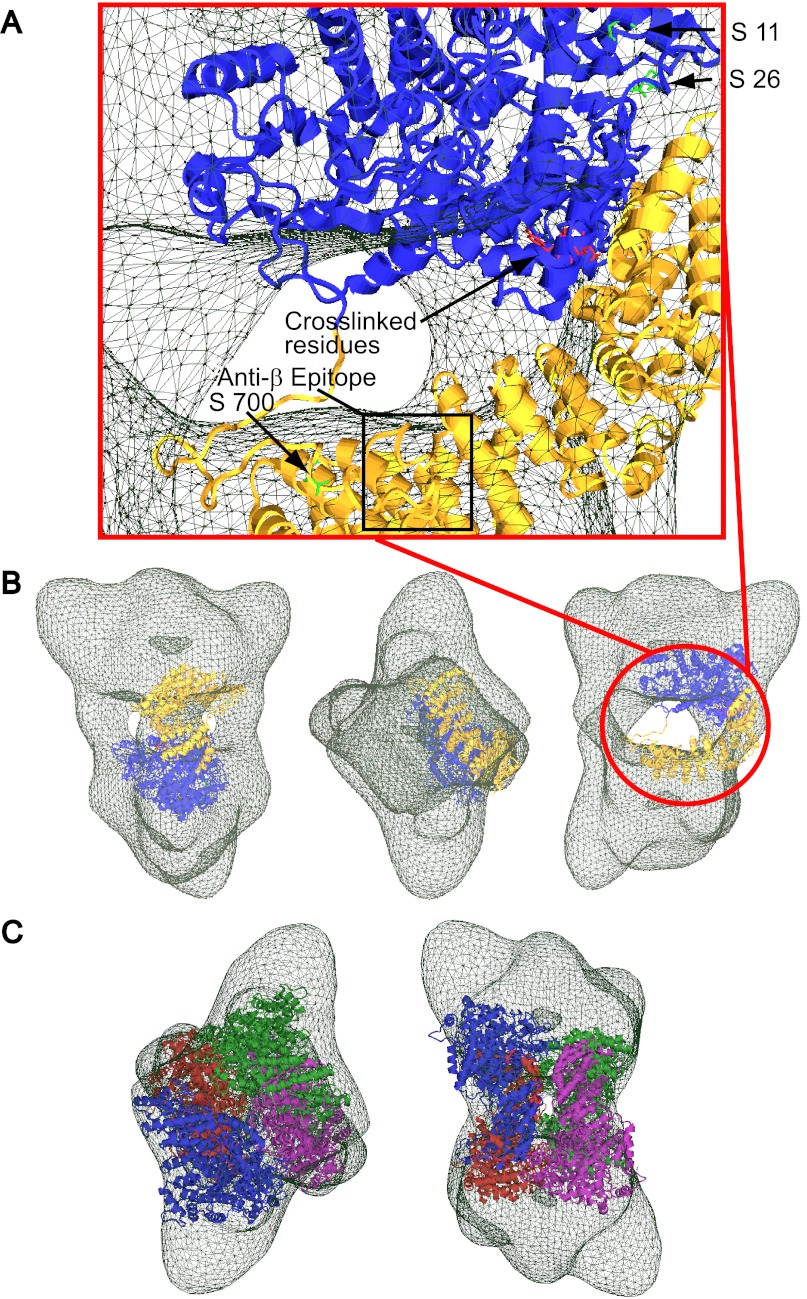
Having a three-dimensional cryo-EM envelope for PhK (11), knowledge of the location of the β subunits in its 4° structure, and a model for the β subunit in it 3° structure, we asked whether the modeled β subunits could be docked within the four central bridges of the cryo-EM envelope. To facilitate docking, we reasoned that all known solvent-accessible structures in the modeled β structure, including phosphorylatable serines 11, 26, and 700 (green side chains) and DFDNB-cross-linked residues (red side chains), should be placed on the periphery of the EM structure (Fig. 5A). We also knew the solvent-accessible binding location for the anti-β mAb that was revealed by immuno-EM of PhK in complex with this mAb (16), but we did not know the actual epitope recognized by the mAb. Consequently, we set out to map the epitope by determining the cross-reactivity of the mAb against expressed engineered GST fusion constructs of the complete β subunit and seven previously used truncation mutants as follows: 1–285, 1–350, 1–520, 1–703, 1–815, 1–916, and 33–1092 (7). After these constructs were expressed, purified, resolved on SDS-polyacrylamide gels, transferred to PDF membranes, and probed with the anti-β mAb, only the full-length β and the last three truncation mutants cross-reacted with the mAb (data not shown). Thus, the epitope most likely occurs somewhere in the stretch between residues 704 and 815, and this information was also used in docking the β structure in the PhK EM envelope (Fig. 5A). Only in the orientation shown in Fig. 5B does the shape of the β structure match well with the bridge structure and regions of the lobes, suggesting a β core structure in the complex that encompasses all four bridges. All four subunits were then docked as rigid bodies in the cryo-EM envelope, forming a central core comprising the four bridges and the interior faces of both lobes (Fig. 5C).
FIGURE 5.
Docking the β model in the PhK cryo-EM envelope. A, magnified view of the β subunit docked in the three-dimensional cryo-EM envelope (green mesh) solved for the native PhK complex (11). The β model is shown positioned in the EM envelope, proximal to the interconnecting bridge region of PhK (11), using the known location of an anti-β subunit-specific mAb epitope (mapped to residues 703–815) as a marker (16). All known solvent-accessible structures (phosphorylatable serines 11, 26, and 700; cross-linked residues Lys-1025, Tyr-51, and Lys-53 and the anti-β epitope) are shown located at the periphery of the EM envelope. B, three predominant orientations of PhK are shown, revealing an optimal fit for the β model in the bridge region of PhK. C, β core in PhK, modeled by docking a single color-coded β structure in each of the four bridges of the EM envelope with overall D2 symmetry.
DISCUSSION
Previous analyses of the primary structures of α and β noted that the two subunits were homologous and putatively products of gene duplication, but each was unique as a complete protein (20). Predictions of 2° structure for both subunits suggest them to be predominantly α-helical, and our CD measurements of the αγδ and (αβγδ)4 complexes are in agreement, showing α and β to have similar 2° structure content. Some minor differences in 2° structure were observed between α and β, however, and these likely reflect the presence of several nonhomologous stretches, some of which include phosphorylatable regulatory domains specific to each subunit (20, 21). The α subunit has a large phosphorylatable C-terminal regulatory domain with multiple phosphorylation sites (1), whereas the β subunit contains two known phosphorylatable regions, Ser-700 plus Ser-11/Ser-26 in the N-terminal phosphorylatable domain (NB1, residues 1–40) (7, 28). NB1 was the first of three functional domains predicted in our analysis of the β subunit 2° and 3° structures.
Several previous reports suggest that the small NB1 domain, which by sequence similarity is unique to β, is a distinct functional moiety (7, 20). By two-hybrid analyses, this domain is an inhibitor of β-β dimerization (7), with only mutants of β either lacking the entire NB1 domain or containing the phospho-mimetic point mutations S11E or S26E being capable of self-interaction. The self-association of β subunits has been associated with activation in the intact PhK complex (27, 29). NB1 contains the major regulatory phosphorylation site, Ser-26, targeted by PKA in vivo (28), and there have also been several reports that within the PhK complex Ser-11 and Ser-26 are autophosphorylated by the γ subunit (7, 33), leading to activation (28, 33). NB1 is also a potential interaction site for the intrinsic calmodulin (δ) subunit, as a synthetic peptide corresponding to residues 5–28 of NB1 inhibited active forms of the PhK complex and had nanomolar affinity for free calmodulin (68). We also previously demonstrated by cross-linking that Arg-18 of NB1 is proximal (≤12 Å) to the γCRD (7), which binds the δ subunit within the PhK complex (4, 5). Based on its high affinity for calmodulin, this region of NB1 (residues 5–28) was suggested to have the characteristic amphipathic α-helical structure of many known calmodulin targets (68), an architecture that is also observed in our current ab initio model of this domain. The proximity of NB1 to γ in the PhK complex indicated by cross-linking and by its phosphorylation mandated that this region be surface-accessible and proximal to the position of γ in the EM structure of the (αβγδ)4 complex. The localization of γ was achieved by mapping two-dimensional EM images of PhK in complex with an anti-γ mAb directed against the C-terminal lobe of γ (16). An optimal fit of the β subunit model in the bridge region occurred when satisfying the above conditions.
Aside from the small NB1 domain, the remaining 93% of the 1092-residue β subunit structure was covered by templates from two distinct protein families. The first of these, the GH-15 family of glycosyl hydrolases, provided several good template matches that extended from residues 41 to 670. These results differed from two previous reports that predicted only the GH-15 (α/α)6 catalytic core to be present in β (22, 24), extending approximately through its first 480 residues (22). Just C-terminal to the GH-15 catalytic core, Carriere et al. (23) predicted a small loop followed by an additional domain (termed B), which extended from residues 492 to 676; however, they were unable to achieve fold recognition for this domain. Our GH fold recognition extended throughout this region and corresponded to the β GH-15 template archaeal glucodextranase B and C domains, which show homology with proteins containing the immunoglobulin folds and carbohydrate-binding domains of several glycosidases, respectively (61). An independent analysis of the primary structure of β using the Pfam database also predicted a GHL fold for this region of the subunit (69). Our predicted domain structures for residues 40–670 of the β subunit mirrored that of the shorter domain predicted by Carriere et al. (23) and are topologically similar to several domains observed in the template glucodextranase crystal structure (61), comprising an (α/α)6 catalytic core connected to the B and C domains by a short loop (61).
In addition to threading, we used the constraint from intramolecular cross-linking with DFDNB to influence the structure of the β model by positioning the GHL domain proximal to the β C terminus (Fig. 3). In the absence of this constraint, the top 100 models generated for the β subunit did not complement well the shape of the PhK cryo-EM envelope when positioned in all possible orientations in the central bridge region of the molecule. We assume that cross-linking of the β subunit in the intact complex captures a conformation of this subunit that is induced by its interactions with all subunits, including itself, in the 4° structure of PhK. In further support of the use of this cross-linking constraint, we recently demonstrated that PhK binds the glucoamylase inhibitor acarbose and that this binding perturbs intramolecular cross-linking of the β subunit by GMBS (64), which cross-links the GHL domain to the C terminus of the β subunit (30). The direct binding of acarbose by the β GHL domains would allow a potential mechanism for inducing a conformational change in this subunit through altering potential interactions between the predicted β GHL and the HRL domains, consistent with results from the intramolecular cross-linking of β by DFDNB. Acarbose also stimulates the protein kinase activity of PhK γ (64), and functionally it mimics glycogen, which also binds to PhK, activating the catalytic γ subunit (9). It must be cautioned, however, that because the homologous α and β subunits are both predicted to contain GHL domains, we cannot rule out the possibility that effects of acarbose and glycogen on the PhK β and γ subunits are mediated by the binding of these ligands to the α subunit.
Other potential functions for the β subunit have been suggested from models of its C terminus from residues 671 to 1092. From comparing sequence alignments of the α and β subunits from diverse species, Carriere et al. (23) predicted for this region of β (as well as the homologous α) two structurally related α-helical domains as follows: C (residues 711–915) with no fold recognition, and D (residues 915–1092) with a calcineurin B-like (CBL) fold. The CBL fold, identified by hydrophobic cluster analysis (23), is typical of Ca2+-binding proteins containing two globular EF-hand motifs (two perpendicular α-helices tethered by a loop) that are connected by a flexible linker (70). Based on observed chemical cross-linking between the α subunit CBL domain and the catalytic γ subunit (6, 38), the latter of which also binds the EF-hand endogenous CaM (δ) subunit (5), it was proposed that the CBL domain targets known CaM-binding sites on γ and/or those predicted for the α and β subunits (23). Although we did not observe a CBL domain on β by threading, the predicted structural relatedness of domains C and D and their predominantly α-helical structure are consistent with our modeling of this region of the subunit as a single domain using the template PP2AA subunit. Crystal structure analysis of PP2AA revealed a molecule in which 15 tandem HEAT repeats (motifs comprising a pair of anti-parallel α-helices) formed a left-handed superhelical structure (65). HEAT-repeat domains have been shown to mediate protein-protein interactions in several proteins, including the PP2AA subunit (65). PP2AA tightly associates with the catalytic PP2A subunit to form a heterodimeric scaffold for binding diverse regulatory subunits that direct the functional phosphatase to specific subcellular sites (71). If present in the β subunit, the HRL domain may function similarly in the context of the PhK complex, forming a scaffold with the GHL domain that promotes interactions with either the catalytic γ subunit or other regulatory β subunits. Such interactions are consistent with the formation of ββ and βγ dimers by cross-linking PhK with DFDNB and GMBS (29, 30), with the observed structural coupling between β and γ in the intact PhK complex (16), and with two-hybrid analyses of β subunit truncation mutants, which demonstrated that the C-terminal half of the HRL domain is required for self-association of the β subunits (7). In addition to a potential correspondence in function, the predominantly helical HEAT-repeat structure agreed well with the 2° structural content estimated for the β subunit and complemented the shape of the bridge structure and interior lobe face after docking β into the PhK cryo-EM envelope.
The first direct evidence for a β4 subcomplex in the (αβγδ)4 PhK complex revealed herein by top-down MS and inter-subunit cross-linking of the β subunits by DFDNB provided the rationale for docking these subunits in the central connecting bridge region of the PhK cryo-EM structure. This arrangement of the β subunits is consistent with the observed head-to-head packing of αβγδ protomers in D2 symmetry (15), which requires any homotetrameric association of subunits to occur at known points of contact between the lobes, i.e. centrally in the complex (Fig. 5). Additionally, our direct results correspond to several reports indirectly linking the β subunits to a central location of the complex by partial proteolysis and EM immunolocalization (16, 17). Taken together, the results herein indicate the presence of a β4 core in PhK and suggest a structural role for these subunits as a scaffold upon which the α, γ, and δ subunits are arrayed. It is likely that the β subunits position the lobes, and thus the catalytic γ subunits, differently with respect to one another in nonactivated and activated conformers of PhK. For example, we have demonstrated that the conformation of the β subunits correlates with activation of PhK by several mechanisms (10, 26, 29), that the β and γ subunits are structurally coupled to one another during enzyme activation (16), and that changes in the dihedral angles between the lobes and altered bridge structures are readily observed between nonactivated and Ca2+-activated forms of PhK by EM and SAXS analyses (10, 19).
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK32953 (to G. M. C.) and Grants GM083107 and GM084222 from NIGMS (to Y. Z.). This work was also supported by National Science Foundation Grant DBI 1027394 and the University of Michigan-Shanghai Jiao Tong University Joint Institute Foundation (to Y. Z.) and the Royal Society and the Engineering and Physical Sciences Research Council (to C. V. M.).
This article was selected as a Paper of the Week.
D. Xu and Y. Zhang, manuscript in preparation.
- PhK
- phosphorylase kinase
- CBL
- calcineurin B-like
- γCRD
- C-terminal regulatory domain of the γ subunit
- cryo-EM
- cryoelectron EM
- DFDNB
- 1,5-difluoro-2,4-dinitrobenzene
- GH
- glycosyl hydrolase
- GHL
- GH-like
- GMBS
- N-[γ-maleimidobutyryloxy] succinimide ester
- HEAT
- Huntingtin elongation A subunit TOR
- HRL
- HEAT-repeat domain
- NB1
- β subunit N-terminal phosphorylatable domain
- PDB
- Protein Data Bank
- PhK
- the (αβγδ)4 phosphorylase kinase complex
- PP2A
- protein phosphatase 2A
- PP2AA
- PR65/A subunit of PP2A
- SAXS
- small angle x-ray scattering
- 2°
- secondary
- 3°
- tertiary
- 4°
- quaternary.
REFERENCES
- 1. Brushia R. J., Walsh D. A. (1999) Phosphorylase kinase. The complexity of its regulation is reflected in the complexity of its structure. Front. Biosci. 4, D618–641 [DOI] [PubMed] [Google Scholar]
- 2. Lowe E. D., Noble M. E., Skamnaki V. T., Oikonomakos N. G., Owen D. J., Johnson L. N. (1997) The crystal structure of a phosphorylase kinase peptide substrate complex. Kinase substrate recognition. EMBO J. 16, 6646–6658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Owen D. J., Noble M. E., Garman E. F., Papageorgiou A. C., Johnson L. N. (1995) Two structures of the catalytic domain of phosphorylase kinase. An active protein kinase complexed with substrate analogue and product. Structure 3, 467–482 [DOI] [PubMed] [Google Scholar]
- 4. Dasgupta M., Honeycutt T., Blumenthal D. K. (1989) The γ-subunit of skeletal muscle phosphorylase kinase contains two noncontiguous domains that act in concert to bind calmodulin. J. Biol. Chem. 264, 17156–17163 [PubMed] [Google Scholar]
- 5. Jeyasingham M. D., Artigues A., Nadeau O. W., Carlson G. M. (2008) Structural evidence for co-evolution of the regulation of contraction and energy production in skeletal muscle. J. Mol. Biol. 377, 623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rice N. A., Nadeau O. W., Yang Q., Carlson G. M. (2002) The calmodulin-binding domain of the catalytic γ subunit of phosphorylase kinase interacts with its inhibitory α subunit. Evidence for a Ca2+ sensitive network of quaternary interactions. J. Biol. Chem. 277, 14681–14687 [DOI] [PubMed] [Google Scholar]
- 7. Nadeau O. W., Anderson D. W., Yang Q., Artigues A., Paschall J. E., Wyckoff G. J., McClintock J. L., Carlson G. M. (2007) Evidence for the location of the allosteric activation switch in the multisubunit phosphorylase kinase complex from mass spectrometric identification of chemically cross-linked peptides. J. Mol. Biol. 365, 1429–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paudel H. K., Carlson G. M. (1987) Inhibition of the catalytic subunit of phosphorylase kinase by its α/β subunits. J. Biol. Chem. 262, 11912–11915 [PubMed] [Google Scholar]
- 9. Krebs E. G., Love D. S., Bratvold G. E., Trayser K. A., Meyer W. L., Fischer E. H. (1964) Purification and properties of rabbit skeletal muscle phosphorylase B kinase. Biochemistry 3, 1022–1033 [DOI] [PubMed] [Google Scholar]
- 10. Nadeau O. W., Carlson G. M., Gogol E. P. (2002) A Ca2+-dependent global conformational change in the three-dimensional structure of phosphorylase kinase obtained from electron microscopy. Structure 10, 23–32 [DOI] [PubMed] [Google Scholar]
- 11. Nadeau O. W., Gogol E. P., Carlson G. M. (2005) Cryoelectron microscopy reveals new features in the three-dimensional structure of phosphorylase kinase. Protein Sci. 14, 914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vénien-Bryan C., Lowe E. M., Boisset N., Traxler K. W., Johnson L. N., Carlson G. M. (2002) Three-dimensional structure of phosphorylase kinase at 22-Å resolution and its complex with glycogen phosphorylase b. Structure 10, 33–41 [DOI] [PubMed] [Google Scholar]
- 13. Norcum M. T., Wilkinson D. A., Carlson M. C., Hainfeld J. F., Carlson G. M. (1994) Structure of phosphorylase kinase. A three-dimensional model derived from stained and unstained electron micrographs. J. Mol. Biol. 241, 94–102 [DOI] [PubMed] [Google Scholar]
- 14. Traxler K. W., Norcum M. T., Hainfeld J. F., Carlson G. M. (2001) Direct visualization of the calmodulin subunit of phosphorylase kinase via electron microscopy following subunit exchange. J. Struct. Biol. 135, 231–238 [DOI] [PubMed] [Google Scholar]
- 15. Wilkinson D. A., Marion T. N., Tillman D. M., Norcum M. T., Hainfeld J. F., Seyer J. M., Carlson G. M. (1994) An epitope proximal to the carboxyl terminus of the α-subunit is located near the lobe tips of the phosphorylase kinase hexadecamer. J. Mol. Biol. 235, 974–982 [DOI] [PubMed] [Google Scholar]
- 16. Wilkinson D. A., Norcum M. T., Fizgerald T. J., Marion T. N., Tillman D. M., Carlson G. M. (1997) Proximal regions of the catalytic γ and regulatory β subunits on the interior lobe face of phosphorylase kinase are structurally coupled to each other and with enzyme activation. J. Mol. Biol. 265, 319–329 [DOI] [PubMed] [Google Scholar]
- 17. Trempe M. R., Carlson G. M., Hainfeld J. F., Furcinitti P. S., Wall J. S. (1986) Analyses of phosphorylase kinase by transmission and scanning transmission electron microscopy. J. Biol. Chem. 261, 2882–2889 [PubMed] [Google Scholar]
- 18. Vénien-Bryan C., Jonic S., Skamnaki V., Brown N., Bischler N., Oikonomakos N. G., Boisset N., Johnson L. N. (2009) The structure of phosphorylase kinase holoenzyme at 9.9 angstroms resolution and location of the catalytic subunit and the substrate glycogen phosphorylase. Structure 17, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Priddy T. S., MacDonald B. A., Heller W. T., Nadeau O. W., Trewhella J., Carlson G. M. (2005) Ca2+-induced structural changes in phosphorylase kinase detected by small angle x-ray scattering. Protein Sci. 14, 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kilimann M. W., Zander N. F., Kuhn C. C., Crabb J. W., Meyer H. E., Heilmeyer L. M., Jr. (1988) The α and β subunits of phosphorylase kinase are homologous. cDNA cloning and primary structure of the β subunit. Proc. Natl. Acad. Sci. U.S.A. 85, 9381–9385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zander N. F., Meyer H. E., Hoffmann-Posorske E., Crabb J. W., Heilmeyer L. M., Jr., Kilimann M. W. (1988) cDNA cloning and complete primary structure of skeletal muscle phosphorylase kinase (α subunit). Proc. Natl. Acad. Sci. U.S.A. 85, 2929–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carrière C., Jonic S., Mornon J. P., Callebaut I. (2008) Three-dimensional mapping of glycogenosis-causing mutations in the large regulatory α subunit of phosphorylase kinase. Biochim. Biophys. Acta 1782, 664–670 [DOI] [PubMed] [Google Scholar]
- 23. Carrière C., Mornon J. P., Venien-Bryan C., Boisset N., Callebaut I. (2008) Calcineurin B-like domains in the large regulatory α/β subunits of phosphorylase kinase. Proteins 71, 1597–1606 [DOI] [PubMed] [Google Scholar]
- 24. Pallen M. J. (2003) Glucoamylase-like domains in the α- and β-subunits of phosphorylase kinase. Protein Sci. 12, 1804–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chattopadhyaya R., Meador W. E., Means A. R., Quiocho F. A. (1992) Calmodulin structure refined at 1.7 Å resolution. J. Mol. Biol. 228, 1177–1192 [DOI] [PubMed] [Google Scholar]
- 26. Trempe M. R., Carlson G. M. (1987) Phosphorylase kinase conformers. Detection by proteases. J. Biol. Chem. 262, 4333–4340 [PubMed] [Google Scholar]
- 27. Cheng A., Fitzgerald T. J., Carlson G. M. (1985) Adenosine 5′-diphosphate as an allosteric effector of phosphorylase kinase from rabbit skeletal muscle. J. Biol. Chem. 260, 2535–2542 [PubMed] [Google Scholar]
- 28. Cohen P., Watson D. C., Dixon G. H. (1975) The hormonal control of activity of skeletal muscle phosphorylase kinase. Amino acid sequences at the two sites of action of adenosine-3′:5′-monophosphate-dependent protein kinase. Eur. J. Biochem. 51, 79–92 [DOI] [PubMed] [Google Scholar]
- 29. Fitzgerald T. J., Carlson G. M. (1984) Activated states of phosphorylase kinase as detected by the chemical cross-linker 1,5-difluoro-2,4-dinitrobenzene. J. Biol. Chem. 259, 3266–3274 [PubMed] [Google Scholar]
- 30. Nadeau O. W., Wyckoff G. J., Paschall J. E., Artigues A., Sage J., Villar M. T., Carlson G. M. (2008) CrossSearch, a user-friendly search engine for detecting chemically cross-linked peptides in conjugated proteins. Mol. Cell. Proteomics 7, 739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. King M. M., Carlson G. M. (1981) Synergistic activation by Ca2+ and Mg2+ as the primary cause for hysteresis in the phosphorylase kinase reactions. J. Biol. Chem. 256, 11058–11064 [PubMed] [Google Scholar]
- 32. Cohen P. (1973) The subunit structure of rabbit skeletal muscle phosphorylase kinase, and the molecular basis of its activation reactions. Eur. J. Biochem. 34, 1–14 [DOI] [PubMed] [Google Scholar]
- 33. King M. M., Fitzgerald T. J., Carlson G. M. (1983) Characterization of initial autophosphorylation events in rabbit skeletal muscle phosphorylase kinase. J. Biol. Chem. 258, 9925–9930 [PubMed] [Google Scholar]
- 34. Priddy T. S., Middaugh C. R., Carlson G. M. (2007) Electrostatic changes in phosphorylase kinase induced by its obligatory allosteric activator Ca2+. Protein Sci. 16, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lobley A., Whitmore L., Wallace B. A. (2002) DICHROWEB. An interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 18, 211–212 [DOI] [PubMed] [Google Scholar]
- 36. Sreerama N., Woody R. W. (1994) Protein secondary structure from circular dichroism spectroscopy. Combining variable selection principle and cluster analysis with neural network, ridge regression, and self-consistent methods. J. Mol. Biol. 242, 497–507 [DOI] [PubMed] [Google Scholar]
- 37. Sreerama N., Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra. Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 38. Nadeau O. W., Traxler K. W., Fee L. R., Baldwin B. A., Carlson G. M. (1999) Activators of phosphorylase kinase alter the cross-linking of its catalytic subunit to the C-terminal one-sixth of its regulatory α subunit. Biochemistry 38, 2551–2559 [DOI] [PubMed] [Google Scholar]
- 39. Schilling B., Row R. H., Gibson B. W., Guo X., Young M. M. (2003) MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically cross-linked peptides. J. Am. Soc. Mass Spectrom. 14, 834–850 [DOI] [PubMed] [Google Scholar]
- 40. Brill T. B., James K. J., Chawla R., Nicol G., Shukla A., Futrell J. H. (1999) Influence of the substituent on the major decomposition channels of the NO2 group in para-substituted nitrobenzenes. A tandem mass spectrometric study. J. Phys. Organic Chem. 12, 819–826 [Google Scholar]
- 41. Sigman M. E., Clark C. D. (2005) Two-dimensional correlation spectroscopy techniques applied to ion trap tandem mass spectrometric analysis. Nitroaromatics. Rapid Commun. Mass Spectrom. 19, 3731–3736 [DOI] [PubMed] [Google Scholar]
- 42. Chalkley R. J., Hansen K. C., Baldwin M. A. (2005) Bioinformatic methods to exploit mass spectrometric data for proteomic applications. Methods Enzymol. 402, 289–312 [DOI] [PubMed] [Google Scholar]
- 43. Sobott F., Hernández H., McCammon M. G., Tito M. A., Robinson C. V. (2002) A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal. Chem. 74, 1402–1407 [DOI] [PubMed] [Google Scholar]
- 44. Hernández H., Robinson C. V. (2007) Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protoc. 2, 715–726 [DOI] [PubMed] [Google Scholar]
- 45. Taverner T., Hernández H., Sharon M., Ruotolo B. T., Matak-Vinković D., Devos D., Russell R. B., Robinson C. V. (2008) Subunit architecture of intact protein complexes from mass spectrometry and homology modeling. Acc. Chem. Res. 41, 617–627 [DOI] [PubMed] [Google Scholar]
- 46. Zhang Y. (2008) I-TASSER server for protein three-dimensional structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu S., Skolnick J., Zhang Y. (2007) Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu S., Zhang Y. (2007) LOMETS. A local meta-threading server for protein structure prediction. Nucleic Acids Res. 35, 3375–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y., Skolnick J. (2004) SPICKER. A clustering approach to identify near-native protein folds. J. Comput. Chem. 25, 865–871 [DOI] [PubMed] [Google Scholar]
- 50. Li Y., Zhang Y. (2009) REMO. A new protocol to refine full atomic protein models from C-α traces by optimizing hydrogen-bonding networks. Proteins 76, 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 52. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 53. Kneller D. G., Cohen F. E., Langridge R. (1990) Improvements in protein secondary structure prediction by an enhanced neural network. J. Mol. Biol. 214, 171–182 [DOI] [PubMed] [Google Scholar]
- 54. Nadeau O. W., Carlson G. M. (1994) Zero length conformation-dependent cross-linking of phosphorylase kinase subunits by transglutaminase. J. Biol. Chem. 269, 29670–29676 [PubMed] [Google Scholar]
- 55. Teo T. S., Wang T. H., Wang J. H. (1973) Purification and properties of the protein activator of bovine heart cyclic adenosine 3′,5′-monophosphate phosphodiesterase. J. Biol. Chem. 248, 588–595 [PubMed] [Google Scholar]
- 56. Kee S. M., Graves D. J. (1986) Isolation and properties of the active γ subunit of phosphorylase kinase. J. Biol. Chem. 261, 4732–4737 [PubMed] [Google Scholar]
- 57. Boulatnikov I. G., Peters J. L., Nadeau O. W., Sage J. M., Daniels P. J., Kumar P., Walsh D. A., Carlson G. M. (2009) Expressed phosphorylase b kinase and its αγδ subcomplex as regulatory models for the rabbit skeletal muscle holoenzyme. Biochemistry 48, 10183–10191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chan K. F., Graves D. J. (1982) Isolation and physicochemical properties of active complexes of rabbit muscle phosphorylase kinase. J. Biol. Chem. 257, 5939–5947 [PubMed] [Google Scholar]
- 59. Nadeau O. W., Carlson G. M. (2005) in Protein-Protein Interactions: A Molecular Cloning Manual (Golemis E., Adams P. D., eds) 2nd Ed., pp. 105–127, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 60. Xu D., Zhang J., Roy A., Zhang Y. (2011) Automated protein structure modeling in CASP9 by I-TASSER pipeline combined with QUARK-based ab initio folding and FG-MD-based structure refinement. Proteins 79, Suppl. 10, 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mizuno M., Tonozuka T., Suzuki S., Uotsu-Tomita R., Kamitori S., Nishikawa A., Sakano Y. (2004) Structural insights into substrate specificity and function of glucodextranase. J. Biol. Chem. 279, 10575–10583 [DOI] [PubMed] [Google Scholar]
- 62. Xu J., Zhang Y. (2010) How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 26, 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Svensson B., Sierks M. R. (1992) Roles of the aromatic side chains in the binding of substrates, inhibitors, and cyclomalto-oligosaccharides to the glucoamylase from Aspergillus niger probed by perturbation difference spectroscopy, chemical modification, and mutagenesis. Carbohydr. Res. 227, 29–44 [DOI] [PubMed] [Google Scholar]
- 64. Nadeau O. W., Liu W., Boulatnikov I. G., Sage J. M., Peters J. L., Carlson G. M. (2010) The glucoamylase inhibitor acarbose is a direct activator of phosphorylase kinase. Biochemistry 49, 6505–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Groves M. R., Hanlon N., Turowski P., Hemmings B. A., Barford D. (1999) The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96, 99–110 [DOI] [PubMed] [Google Scholar]
- 66. Benesch J. L., Robinson C. V. (2006) Mass spectrometry of macromolecular assemblies. Preservation and dissociation. Curr. Opin. Struct. Biol. 16, 245–251 [DOI] [PubMed] [Google Scholar]
- 67. Benesch J. L., Ruotolo B. T., Simmons D. A., Robinson C. V. (2007) Protein complexes in the gas phase. Technology for structural genomics and proteomics. Chem. Rev. 107, 3544–3567 [DOI] [PubMed] [Google Scholar]
- 68. Newsholme P., Angelos K. L., Walsh D. A. (1992) High and intermediate affinity calmodulin binding domains of the α and β subunits of phosphorylase kinase and their potential role in phosphorylation-dependent activation of the holoenzyme. J. Biol. Chem. 267, 810–818 [PubMed] [Google Scholar]
- 69. Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A., Finn R. D. (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nelson M. R., Chazin W. J. (1998) Structures of EF-hand Ca2+-binding proteins. Diversity in the organization, packing and response to Ca2+ binding. Biometals 11, 297–318 [DOI] [PubMed] [Google Scholar]
- 71. Shi Y. (2009) Serine/threonine phosphatases. Mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 72. Lane L. A., Nadeau O. W., Carlson G.M., Robinson C. V. (2012) Mass spectrometry reveals differences in stability and subunit interactions between activated and nonactivated conformers of the (αβγδ)4 phosphorylase kinase complex. Mol. Cell. Proteomics, in press [DOI] [PMC free article] [PubMed] [Google Scholar]