Abstract
Chronic kidney disease is a common disease with increasing prevalence in the western population. One common reason for chronic kidney failure is diabetic nephropathy. Diabetic nephropathy and hyperglycemia are characteristics of the mouse inbred strain KK/HlJ, which is predominantly used as a model for metabolic syndrome due to its inherited glucose intolerance and insulin resistance. We used KK/HlJ, an albuminuria-sensitive strain, and C57BL/6J, an albuminuria-resistant strain, to perform a quantitative trait locus (QTL) cross to identify the genetic basis for chronic kidney failure. Albumin-creatinine-ratio (ACR) was measured in 130 F2 male offspring. One significant QTL was identified on chromosome (Chr) X and four suggestive QTLs were found on Chrs 6, 7, 12, and 13. Narrowing of the QTL region was focused on the X-linked QTL and performed by incorporating genotype and expression analyses for genes located in the region. From the 485 genes identified in the X-linked QTL region, a few candidate genes were identified using a combination of bioinformatic evidence based on genomic comparison of the parental strains and known function in urine homeostasis. Finally, this study demonstrates the significance of the X chromosome in the genetic determination of albuminuria.
Keywords: chronic kidney failure, QTL, genetic cross, ACR, diabetic nephropathy
Introduction
Chronic kidney disease, a common disease with increasing prevalence in the western population (Zhang and Rothenbacher 2008), is caused by environmental (i.e., diet) and genetic risk factors (Iyengar et al. 2007). Understanding the genetic basis of chronic kidney disease will ultimately help to identify novel treatment targets and preventative strategies through personalized medicine. Human studies are difficult due to their length and costs. However, mouse models of human diseases are commonly used to investigate the genetic basis of complex traits due to their short life span, high reproductive rate, easy-to-control environment, and the availability of large-scale genomic and gene expression databases for many inbred and wild-derived strains (DiPetrillo et al. 2005; Peters et al. 2007; Shockley et al. 2009). In addition, complex trait QTL are often concordant between humans and mice and, therefore, mouse genetics can help to accelerate the identification of genes underlying human diseases (Leduc et al. 2011; Tsaih et al. 2010; Wang and Paigen 2005).
Elevated albumin-to-creatinine ratio (ACR), a condition also called albuminuria, is a phenotypic marker of chronic kidney disease in both humans and mice (Grindle et al. 2006; Iyengar et al. 2007). Albuminuria varies widely among mouse inbred strains (Tsaih et al. 2010). While most strains have low ACR, a few strains (e.g., A/J, KK/HlJ (KK) and PWD/PhJ) show consistently elevated ACR. Such phenotypic variations can be investigated in quantitative trait locus (QTL) studies to identify genomic regions that modulate the disease trait. Indeed, previous QTL studies have successfully identified genomic regions influencing ACR using albuminuria-susceptible and resistant strains, including 129S1/SvImJ, 129S6, A/J, C57BL/6J (B6), DBA/2J, FGS/Kist, MRL/MpJ, NZM/Aeg2410, NZW/LacJ, and SM/J (Doorenbos et al. 2008; Hageman et al. 2011; Kato et al. 2008; Kim et al. 2005; Morel et al. 1994; Salzler et al. 2007; Sheehan et al. 2007; Tsaih et al. 2010). However, to our knowledge, there is no report on a QTL study using the highly susceptible strain KK.
KK substrains are predominantly used for investigating the metabolic syndrome due to the inherited glucose intolerance and insulin resistance resulting in hyperglycemia (Herberg and Coleman 1977). In KK mice hyperglycemia is often accompanied by a strong tendency to develop type 2 diabetes (T2D) (Ikeda 1994) and diabetic nephropathy (characterized by increased kidney weight, albuminuria, and proteinuria) (Qi et al. 2005; Reddi et al. 1990). A detailed histopathological investigation of aged KK mice showed that the most common aberrations in the kidneys were membraneous glomerulonephritis and chronic interstitial nephritis (personal communications, JPS). KK mice are also susceptible to aging-related vascular mineralization in heart and kidney, which may contribute to the strain’s “pre-sensitized” state to develop albuminuria in response to chronic hyperglycemia (Leiter 2009). Although the phenotypic observations of elevated ACR and histopathological kidney aberrations have long been recognized for KK, their genetic bases have yet to be investigated.
The aim of this study was to investigate the underlying genetics that determine albuminuria in KK mice. An F2 intercross between KK and B6 — an albuminuria-resistant mouse strain — was generated and the offspring population was investigated for QTL that determined elevated ACR. A statistically significant X-linked QTL with molecular evidence for several candidate genes for albuminuria was identified based on QTL analysis, bioinformatics, and gene expression analyses.
Materials and methods
Mice
KK/HlJ (KK) and C57BL/6J (B6) mice were obtained from The Jackson Laboratory (Bar Harbor, ME). KK females were mated to B6 males to produce F1 mice ([KKxB6]F1), which were then intercrossed by sister–brother matings to produce 309 F2 mice ([(KKxB6)x(KKxB6)]F2). Reciprocal F1 mice were generated by crossing B6 females with KK males ([B6xKK]F1) to identify non-Mendelian type of inheritance. Differences between F1 and reciprocal F1 mice indicate that the trait is partially inherited through genes that are imprinted or that are located on the sex chromosomes or mitochondrial DNA. Only male offspring were used in the present study due to the low or undetectable ACR in female mice (n=130). At 10 days of age, tail tissue was collected for subsequent genotyping analysis and at 21 days of age mice were weaned. Mice were raised and maintained on a 6% fat-containing chow diet (LabDiet® 5K52, PMI Nutritional International, Bentwood, MO) and had ad libitum access to acidified water (pH 2.8–3.2). All mice were kept under a controlled climate (i.e., room temperature and 12:12h light-dark cycle). Regular monitoring for viruses, bacteria, parasites, and microsporidium showed that the colonies were free of infestation by any known mouse pathogen (http://jaxmice.jax.org/html/health/quality_control.shtml#Animalhealth). All protocols were reviewed and approved by The Jackson Laboratory Institutional Animal Care and Use Committee.
ACR measurements
At 8 weeks of age, urine was collected in the parental strains, F1, and F2 mice over a 3-day period. Albumin and creatinine levels were measured on a Beckman Synchron CX5 chemistry analyzer (GMI, Inc., Ramsey, MN) and individual albumin-creatinine-ratios (ACRs) were calculated. Albumin concentrations were adjusted by linear regression to a standard curve generated with standard mouse albumin (Grindle et al. 2006). ACR allows the measurement of kidney efficiency as a quantitative trait without the trait being affected by the concentration of the urine. This quantitative trait has been validated in multiple studies (Doorenbos et al. 2008; Grindle et al. 2006; Hageman et al. 2011).
Genotyping
DNA was extracted from tail tissues using a phenol-chloroform-based method. DNA samples were sent to the Partners Genotyping Facility (Massachusetts General Hospital, Boston, MA) for genotyping using the Illumina platform, and 385 unique polymorphic markers were analyzed.
Liver collection
At 8 weeks of age 3 male KK and 3 male B6 mice were housed individually for 3 consecutive days prior to the tissue collection. On the third day mice were fasted for 4 hours prior to sacrifice. Mice were perfused with 60 mL phosphor-buffered saline via the right ventricle of the heart. The large lobe of the liver was collected and preserved in RNAlater (Ambion, Applied Biosystems, Foster City, CA) overnight and stored at −80°C prior to the gene expression analyses. Because we also examined lipid level with this cross, liver was collected. Liver was used as a surrogate for kidney tissue gene expression in this study.
QTL analysis
QTL analysis was performed using R/qtl (v1.09-43) (www.rqtl.org) (Broman et al. 2003) using the EM algorithm. Because ACR was not normally distributed, QTL analysis was performed on log10(ACR+1) transformed values as described previously (Hageman et al. 2011) and by using ACR as a binary trait (ACR = 0 or ACR > 0). QTL×QTL interaction was investigated using the scantwo function within R/qtl. Pairwise scans were performed using 2 cM spacing. If the interactive LOD score between two QTL was above 4, both QTL were recognized as interactive as suggested by Broman and Sen (2009). Significant (P < 0.05) and suggestive (P < 0.63) thresholds were based on 1,000 permutations for the autosomes and 16,404 permutations for the X chromosome (Broman et al. 2006). All single QTL and interactive QTL were added in a multi regression model and the proportion of variance explained by the QTL was determined. X chromosome QTL cannot be added to the regression model at this time as the corrections for chromosome number and dosage compensation have not been implemented in the function within R/qtl.
Microarray expression analysis
Microrarray experiments were performed using liver RNA from KK and B6 male mice (n = 3 per strain). Array hybridization was performed by Gene Expression Services at The Jackson Laboratory. RNA was hybridized to the Mouse Gene 1.0 ST microarray (1M) (Affymetrix, Santa Clara, CA). Average signal intensities for each probe set within arrays were calculated by the RMA function provided within the affy package from Bioconductor for R using a custom Entrez Gene file (Dai et al. 2005). The RMA method incorporates convolution background correction, quantile normalization, and summarization based on a multi-array model fit robustly using the median polish algorithm, and calculates intensities in a base 2 logarithm scale. Pairwise comparisons were used to statistically resolve gene expression differences between KK and B6 mice using the R/maanova analysis package (Wu et al. 2003). Specifically, differentially expressed genes were detected by using Fs, a modified F-statistic incorporating shrinkage estimates of variance components from within the R/maanova package (Cui et al. 2005; Wu et al. 2002). Statistical significance levels of the pairwise comparisons were calculated by permutation analysis (1000 permutations) and adjusted for multiple testing using the false discovery rate (FDR), Q-value, method (Storey 2002). Differentially expressed genes are declared at an FDR Q-value threshold of 0.05. All microarray data have been deposited in the Gene Expression Omnibus (GEO accession number: GSE37429). Pathway analysis was performed using Ingenuity Pathway Analysis (IPA).
Bioinformatics approach
First, the high-density single nucleotide polymorphism (SNP) panel at the Mouse Phenome Database (MPD; http://phenome.jax.org) was searched for non-synonymous coding polymorphisms segregating between KK and B6. The functionality of the non-synonymous SNP was estimated using the SIFT algorithm provided by the Craig J. Venter Institute (Ng and Henikoff 2001). Second, microarrays were analyzed to identify differentially expressed genes between the parental strains. Finally, the Genomic Institute of the Novartis Foundation (GNF) BioGPS website was searched for genes highly expressed in kidneys compared to other tissues. A gene was considered highly expressed in kidney if its expression was greater than twice the median calculated over the 75 tissues available on the BioGPS website (http://biogps.org).
Statistical analysis
All statistical analyses were performed using R (http://www.r-project.org/)
Results
ACR in parental strains, F1, reciprocal F1, and F2 male mice
At 8 weeks of age, ACR was detectable in only one out of eight (12.5%) parental B6 males, while ACR was detectable in all parental KK males. This observation confirms the susceptibility of KK males to develop albuminuria as opposed to B6 males. ACR was observed in both F1 (KKxB6) and reciprocal F1 (B6xKK) males and in about half of the F2 male population (54.6%) (Table 1).
Table 1.
Percentage of mice with ACR > 0 in the B6 and KK parental strains, F1, reciprocal F1, and F2 mice
| Mice | N | N (%) of mice with ACR>0 |
|---|---|---|
| B6 | 8 | 1 (12.5) |
| KK | 8 | 8 (100) |
| (B6xKK) F1 | 14 | 2 (14.3) |
| (KKxB6) F1 | 15 | 1 (6.7) |
| (KKxB6) F2 | 13 | 71 (54.6) |
| 0 |
N, number of mice; ACR, albumin-creatinine ratio
ACR QTL analysis
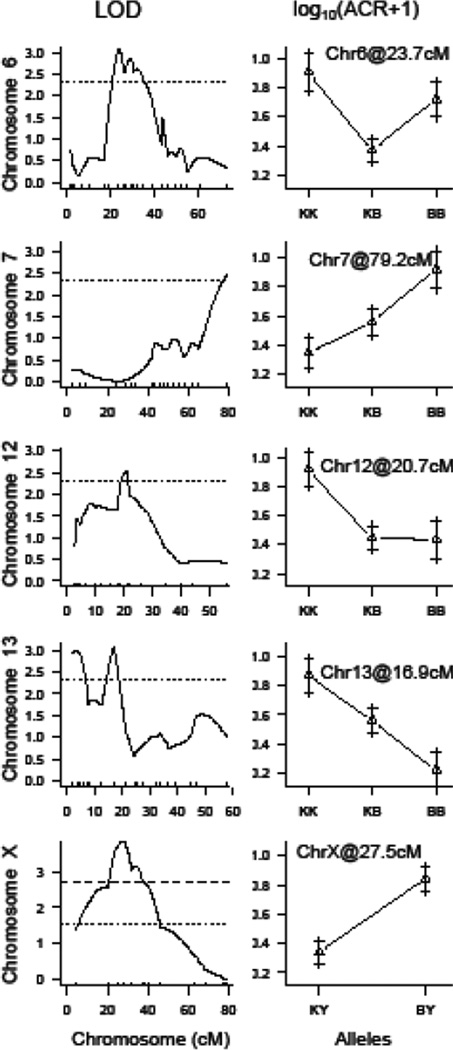
The results of the QTL analyses are shown in Fig. 1 and Table 2. We identified one significant QTL on chromosome (Chr) X at 27.5 cM, and four suggestive QTL on Chr 6 at 23.7 cM, Chr 7 at 79.2 cM, Chr 12 at 20.7 cM, and Chr 13 at 16.9 cM (Figure 1). Due to the shape of the LOD curve on Chr 13, we suspected the presence of two QTL, but they were not statistically distinguishable. On Chr 6, homozygous B6 (i.e., BB) and KK (i.e., KK) mice had higher ACR compared to heterozygous mice (i.e., BK; Fig. 2). On Chr 7, homozygous B6 mice had higher ACR compared to heterozygous and homozygous KK mice and showed an additive mode of inheritance. On Chrs 12 and 13, homozygous KK mice had higher ACR compared to heterozygous and homozygous B6 mice in a recessive and additive manner, respectively (Figure 2). Finally, on Chr X, B6 mice carried the high ACR allele and KK mice carried the low ACR allele (Figure 2). The scantwo function of R/qtl was performed and we identified a QTL interaction between Chr 6@23.7cM and Chr 8@27.4cM. The Chr 8 locus by itself does not exert an effect strong enough to be identified by a single QTL scan but the QTL is significant with regard to an epistatic effect with Chr 6: homozygous KK mice had higher ACR compared to homozygous B6 mice if they were also heterozygous at the Chr 6 QTL (Supplementary Fig. 1). To validate the log-transformed phenotype, QTL analysis was also performed using ACR as a dichotomized trait (ACR = 0 or ACR > 0), which confirmed the QTL on Chrs 6, 13, and X (Supplementary Fig. 2). Regression analysis was applied and each QTL explained between 7.7 and 24.7% of the ACR variation (Table 3).
Fig. 1.
Genome scan for log(ACR+1) in 130 F2 male progeny from an intercross between the strains KK and B6. Analysis was performed in males only. The thresholds of significance were calculated based on 1,000 permutation tests for the autosomes and 16,404 permutation tests for Chr X. The dashed line represents the threshold for significant QTLs (P = 0.05, LOD = 3.82 for the autosomes, LOD = 2.74 for Chr X.) The dotted line represents the threshold for suggestive QTLs (P = 0.63, LOD = 2.33 for the autosomes, LOD = 1.54 for Chr X)
Table 2.
Genome-wide QTL scan for ACR in KKxB6 F2 mice
| Chr | Peak (cM) (95% CI)a |
Peak (Mb) (95% CI)a |
LOD score |
Closest marker |
High strain; mode of inheritance |
|---|---|---|---|---|---|
| 6 | 23.7 (19.7–37.7) | 58.9 (51.2–93.5) | 3.0 | rs30220123 | KK & B6 |
| 7 | 79.2 (43.0–79.2) | 142.3 (87.5–142.3) | 2.5 | rs31680975 | B6, add |
| 8b | 27.4 | 74.8 | ns | rs3722665 | |
| 12 | 20.7 (0–30.7) | 68.5 (0–81.5) | 2.5 | rs6383133 | KK, rec |
| 13 | 16.9 (0–27.3) | 48.0 (0–68.3) | 3.1 | rs29533855 | KK, dom |
| X | 27.5 (16.5–40.5) | 48.2 (20.5–89.0) | 3.8 | rs13483751 | B6, na |
Chr, chromosome; CI, confidence interval; rec, recessive; dom, dominant; ns, non-significant; na, not available
95% CI. Genome-wide significance levels were determined by permuting the observed data 1,000 times. CI was calculated with the Bayesian method
Interactive QTL with the Chr 6 QTL
Fig. 2.
Chromosome scans and allele effect plots for main effect QTLs. The chromosome-specific scan of each main effect QTL is depicted followed by the effect plot at the peak location. The thresholds for significant and suggestive QTL are represented by the dashed and dotted lines, respectively, as described in Fig. 1. In the allele effect plots, KK indicates mice homozygous for the KK allele, BK indicates mice heterozygous with both a KK and B6 allele, and BB indicates mice homozygous for the B6 allele. ACR is depicted as the mean of log10(ACR+1) ± SE per genotype at each locus
Table 3.
Regression analysis of variance for ACR in KKxB6 F2 mice
| Chr (cM) | d.f. | % variance | F-value | P-valuea |
|---|---|---|---|---|
| Chr6@23.7cM | 6 | 24.7 | 7.5 | 7.8 × 10−7 |
| Chr12@20.7M | 2 | 7.7 | 7.0 | 1.3 × 10−3 |
| Chr8@27.6cM | 6 | 16.6 | 5.0 | 1.2 × 10−4 |
| Chr6@23.7cM × Chr8@27.6cM | 4 | 15.7 | 7.1 | 3.5 × 10−5 |
P < 0.05 was used as a threshold for the QTL inclusion in the model. Each P-value is estimated taking one QTL at a time
X-linked QTL: Narrowing of the QTL interval and candidate gene identification
The X-linked QTL was the only significant QTL identified in this cross. The QTL extended from 20.5 to 89 Mb (million base pairs) and contained 458 genes. To identify candidate genes within the region, a variety of bioinformatics tools were applied and integrated, including a search for non-synonymous coding region polymorphisms with functional evidence, gene expression differences, and tissue expression. The corroboration of multiple evidential factors increases the confidence regarding the functionality of a particular gene in regulating ACR.
Genes with non-synonymous coding polymorphisms
To identify non-synonymous coding region polymorphisms segregating between KK and B6, the high density SNP database available on the MPD was searched. Eighteen non-synonymous coding polymorphisms were located in 14 genes (Table 4). According to SIFT, two of these polymorphisms are likely to change the function of their gene products: M22L in Gm6880 and Q454R in 4930595M18Rik.
Table 4.
Bioinformatic support for the candidate gene on the X chromosome
| Gene name | Position (Mb) |
Non-synonymous coding polymorphisma |
Expression Fold difference (KK vs. B6) (P- value)b |
Q- value b |
Kidney expressionc |
|---|---|---|---|---|---|
| Pgrmc1 | 34.1 | - | +1.13 (0.034) |
ns | >2X |
| Rhox4g | 35.1 | - | −2.00 (0.002) |
0.031 | na |
| Cul4b | 35.9 | - | −1.30 (0.026) |
ns | >1X |
| Stag2 | 39.5 | - | −1.43 (0.01) |
ns | ne |
| Ocrl | 45.3 | - | +1.21 (0.025) |
ns | >1X |
| Xpnpep2 | 45.5 | I13V | ns | ns | >1X |
| Utp14a | 45.6 | - | −1.31 (0.026) |
ns | ne |
| Elf4 | 45.8 | N344S | na | na | >1X |
| Rab33a | 45.9 | - | +1.09 (0.017) |
ns | ne |
| Gpc4 | 49.4 | - | −1.24 (0.004) |
0.048 | >2X |
| Hprt1 | 50.3 | - | +1.25 (0.007) |
ns | ne |
| Plac1 | 50.4 | R4L | ns | ns | ne |
| Mospd1 | 50.7 | - | −1.52 (0.001) |
0.024 | >1X |
| 3830403N18Rik | 53.4 | T62R | ns | ns | ne |
| Htatsf1 | 54.3 | - | −1.20 (0.024) |
ns | ne |
| Rbmx | 54.6 | - | −1.51 (0.005) |
ns | ne |
| Aff2 | 67.0 | T229A | ns | ns | ne |
| P432S | |||||
| N730S | |||||
| BC023829 | 67.7 | - | −1.40 (0.001) |
0.025 | ne |
| Mamld1 | 68.3 | - | +1.09 (0.041) |
ns | na |
| Mtm1 | 68.5 | - | −1.35 (0.009) |
ns | >1X |
| Mtmr1 | 68.6 | - | −1.33 (0.009) |
ns | >2X |
| Magea4 | 69.4 | L145F | ns | ns | ne |
| Magea10 | 69.6 | S277F | ns | ns | na |
| Gabra3 | 69.7 | - | −1.22 (0.032) |
ns | >2X |
| Zfp185 | 70.2 | R105L | ns | ns | >2X |
| I212V | |||||
| Idh3g | 71.0 | - | −1.18 (0.002) |
0.036 | >2X |
| Arhgap4 | 71.1 | - | −1.47 (0.01) |
ns | ne |
| Avpr2 | 71.1 | - | +1.17 (0.032) |
ns | >2X |
| Irak1 | 71.3 | - | +1.20 (0.006) |
ns | >2X |
| Tktl1 | 70.4 | K37R | ns | ns | >1X |
| Flna | 70.4 | A2245V | ns | ns | ne |
| Taz | 71.5 | - | −1.14 (0.036) |
ns | ne |
| Atp6ap1 | 71.5 | - | −1.18 (0.016) |
ns | >1X |
| Gdi1 | 71.6 | I58M | ns | ns | ne |
| Plxna3 | 71.6 | - | −1.20 (0.009) |
ns | ne |
| Gm6880 | 71.7 | M22L | na | na | na |
| Dkc1 | 72.3 | - | −1.24 (0.048) |
ns | na |
| 4930428E23Rik | 72.4 | - | −1.10 (0.039) |
ns | na |
| Fundc2 | 72.6 | - | −1.50 (0.008) |
ns | >1X |
| Mtcp1 | 72.7 | - | −1.25 (0.028) |
ns | >1X |
| Pls3 | 73.0 | - | −1.54 (0.001) |
0.018 | ne |
| 4930468A15Rik | 73.8 | T17A | na | na | ne |
| 4930480E11Rik | 75.6 | - | 1.12 (0.009) |
ns | ne |
| 4930595M18Rik | 78.6 | N514D | ns | ns | na |
| Q454R |
Mb, million base pairs; ns, not significant; na, not available
The high density SNP database on MPD was used to identify non-synonymous coding polymorphisms segregating between KK and B6. Bold indicates that the amino-acid change is damaging as detected by SIFT
Gene expression differences between KK and B6 were identified in liver tissue and are depicted with fold-change, P-value, and Q-value. A P and Q = 0.05 were used as the threshold for significance
Genes highly expressed in kidney were identified by searching the GNF database. A gene was considered highly expressed in kidney if the expression of the gene was twice as high as the median value calculated across 75 tissues (2X), but genes for which the expression reached the median are also reported (1X). Genes for which the expression was lower than the median among 75 tissues are considered not expressed (ne)
Differential gene expression between KK and B6
Microarray analysis was performed in liver tissue to search for differentially expressed genes between 8-week-old KK and B6 males. In the region of interest on the X chromosome, 30 differentially expressed genes were identified (P < 0.05) (Table 4), six of which were significant at the genome-wide level (Q < 0.05): Rhox4g (reproductive homeobox 4G), Gpc4 (glypican 4), Mospd1 (motile sperm domain containing 1), BC023829, Idh3g (isocitrate dehydrogenase 3 (NAD+), gamma), and Pls3 (plastin 3, T-isoform).
Differential gene expression across multiple tissues
Our experimental design limited microarray analyses to liver tissue only. Because genes that may affect ACR are more likely to be highly expressed in kidneys, GNF’s BioGPS website, which contains genome-wide expression levels of over 75 tissues in mice, was examined for genes with high expression patterns in kidneys. A gene was considered highly expressed if its expression was twice the median of the expression among all tissues (Burgess-Herbert et al. 2008). We identified 31 genes highly expressed in kidney tissue (Supplementary Table 1). Eight of those were also differentially expressed between KK and B6, two of them at the genome-wide level: Gpc4 (−1.24 fold change KK versus B6, P = 0.004, Q = 0.048) and Idh3g (−1.18 fold change KK versus B6, P = 0.002, Q = 0.036). In addition, Avpr2, arginine vasopressin receptor 2, was highly expressed in kidney and also differentially expressed between KK and B6 (+1.17 fold change KK versus B6, P = 0.032, Q > 0.05).
Pathway analysis
We used Ingenuity Pathway Analysis (IPA) to identify genes involved in kidney function at the genome-wide level. A total of 1182 genes were differentially expressed at the genome wide level, 1112 genes were recognized by IPA, and we identified 47 genes related to nephrotoxicity (Table 5). We surveyed interactions (in vitro and in vivo) between these 47 genes and all candidate genes located within the X-linked QTL. We identified 14 genes showing a relationship with a gene located within the X-linked QTL. Among them, four genes belong to the list of candidate genes for which we identified bioinformatic evidence: Cul4b and Irak1 with differential expression between B6 and KK, and Fln1 and Xpnpep with a non-synonymous polymorphism segregating between B6 and KK (Table 4).
Table 5.
Genome-wide differentially expressed genes related to nephrotoxicity and their relationship with X linked QTL genes
| Gene name a |
Chr | Position (Mb) |
Expression Fold difference (KK vs. B6) (P-value)b |
Q- value b |
X linked QTL genes related to the primary nephrotoxic gene c |
|---|---|---|---|---|---|
| Acaa1b | 9 | 119.1 | 1.3 (0.003) | 0.042 | |
| Adora1 | 1 | 136.1 | 1.67 (0) | 0.013 | |
| Aga | 8 | 54.6 | −1.37 (0.002) | 0.028 | |
| Agt | 8 | 127.1 | −1.24 (0) | 0.011 | Agtr2 |
| Aqp11 | 7 | 104.9 | 1.31 (0.004) | 0.047 | |
| Ar | X | 95.3 | 3.24 (0) | 0.002 | Cul4b, Flna, F9, Gdl1, Pls3, Nr0b1, Pbsn, Usp26 |
| Bcl6 | 16 | 24 | −3.44 (0) | 0.002 | Nkrf |
| Hc | 2 | 34.8 | −2.25 (0) | 0.003 | |
| Ccbl1 | 2 | 30 | −1.37 (0.004) | 0.048 | |
| Cd44 | 2 | 102.7 | −1.38 (0.002) | 0.028 | Prkx |
| Cd59a | 2 | 103.9 | −1.39 (0.001) | 0.023 | |
| Crym | 7 | 127.3 | 1.72 (0.001) | 0.017 | |
| Eif2ak2 | 17 | 79.3 | −1.38 (0.002) | 0.034 | |
| Erbb4 | 1 | 68.1 | −1.64 (0) | 0.010 | |
| F7 | 8 | 13 | −1.21 (0.004) | 0.047 | F8a1, F8, F9, Ikbkg |
| Fcer1g | 1 | 173.2 | −1.64 (0.002) | 0.033 | |
| Fgfr4 | 13 | 55.3 | 1.33 (0.001) | 0.021 | |
| Gm6614 | 6 | 141.9 | 1.79 (0.002) | 0.030 | |
| Gstm2 | 3 | 107.8 | 1.5 (0.001) | 0.016 | |
| Gstp1 | 19 | 4 | −1.58 (0) | 0.009 | Mecp2 |
| Hexb | 13 | 97.9 | −1.61 (0.002) | 0.029 | |
| Igfbp3 | 11 | 7.1 | 1.54 (0.004) | 0.047 | Mecp2 |
| Igfbp5 | 1 | 72.9 | 1.3 (0.001) | 0.024 | Fmr1 |
| Irf1 | 11 | 53.6 | −1.61 (0.002) | 0.026 | Hcfc1 |
| Itga1 | 13 | 115.8 | −1.34 (0.002) | 0.035 | |
| Klb | 5 | 65.7 | −1.66 (0) | 0.007 | |
| Kng1 | 16 | 23.1 | −1.11 (0.003) | 0.036 | Xpnpep2 |
| Mafb | 2 | 160.2 | 1.72 (0) | 0.011 | |
| Npr2 | 4 | 43.6 | −2.17 (0) | 0.004 | |
| Ntrk2 | 13 | 58.9 | 1.62 (0) | 0.011 | |
| Pde4c | 8 | 73.2 | 1.29 (0) | 0.009 | |
| Pde4d | 13 | 109.7 | 1.48 (0) | 0.005 | |
| Pde5a | 3 | 122.4 | 1.22 (0.002) | 0.030 | |
| Pfkm | 15 | 97.9 | −1.22 (0) | 0.006 | |
| Ppp3r1 | 11 | 17.1 | −1.33 (0.002) | 0.034 | |
| Prdx3 | 19 | 60.9 | −1.38 (0.001) | 0.018 | Arhgef6, Magea11 |
| Rassf4 | 6 | 116.6 | 8.79 (0) | 0.002 | |
| Rgs4 | 1 | 171.7 | 1.45 (0.002) | 0.035 | |
| Sdhc | 1 | 173.1 | 1.28 (0) | 0.007 | |
| Slc27a5 | 7 | 13.6 | −1.53 (0) | 0.003 | |
| Spp1 | 5 | 104.9 | −1.48 (0.002) | 0.036 | Irak1, Elk1 |
| Srxn1 | 2 | 151.9 | 1.37 (0) | 0.004 | |
| Sult1a1 | 7 | 133.8 | −1.71 (0) | 0.007 | |
| Tfrc | 16 | 32.6 | 1.56 (0.001) | 0.018 | L1cam |
| Trp53inp1 | 4 | 11.1 | −1.53 (0) | 0.006 | |
| Txnip | 3 | 96.4 | −2.06 (0) | 0.005 | Fhl1 |
| Uxt | 18 | 37.8 | −1.29 (0.003) | 0.043 |
Mb, million base pairs; ns, not significant; na, not available
Genes differentially expressed at Q ≤ 0.05 and annotated as “nephrotoxic” using Ingenuity
Gene expression differences between KK and B6 were identified in liver tissue and are depicted with fold-change, P-value, and Q-value. A P and Q = 0.05 were used as the threshold for significance
Genes within the X linked QTL confidence interval showing a relationship (in vitro or in vivo) with the primary gene annotated as nephrotoxic. Genes in bold also show bioinformatic evidence (Table 4)
Discussion
This study indicates that genomic regions on Chrs 6, 7, 12, 13, and X are linked to phenotypic variations in ACR among male progeny of the intercross between mouse strains KK and B6. ACR is an important indicator for chronic kidney disease and has been investigated in multiple genetic QTL crosses in the mouse (Doorenbos et al. 2008; Hageman et al. 2011; Kato et al. 2008; Kim et al. 2005; Morel et al. 1994; Salzler et al. 2007; Sheehan et al. 2007; Tsaih et al. 2010). However, none of the QTL identified here has been reported previously. This identification of exclusively novel QTL regions was most likely due to the use of the albuminuria-susceptible, phenotypically unique inbred strain KK. For example, histopathological investigation of KK mice found that this strain commonly develops lesions in the kidneys such as membranous glomerulonephritis and chronic interstitial nephritis (personal communications, JPS). In addition, KK is most commonly used as a model for T2D due to its intrinsic hyperglycemia (Leiter 2009); Tsaih et al. 2010), which eventually can lead to kidney failure. Finally, mineralization of the kidney tissue and kidney-associated blood vessels have been reported (Leiter 2009). Altogether, these kidney aberrations render KK mice especially susceptible to decreased functional capacity with the likely result of elevated ACR levels. Therefore, these outstanding phenotypic characteristics of KK compared to other previously investigated strains in ACR QTL studies may explain the lack of overlap between QTL regions.
Due to the lack of common QTL regions between our current and other genetic studies on ACR in mice, it is difficult to narrow the large-sized QTL regions to single genes or genetic variations. A previous study in rats identified a QTL on the X chromosome for ACR, but the orthologous confidence interval does not overlap with the X-linked QTL presented here (Schulz et al. 2003). Nevertheless, integration of bioinformatics tools can help to accelerate QTL narrowing and gene identification (Peters et al. 2007). Here, the QTL on the X chromosome was narrowed from 458 potential genes to four likely candidate genes by using a combination of bioinformatic tools. The candidate genes were identified based on the presence of non-synonymous coding region polymorphisms, gene expression differences between the parental strains, and high expression levels in kidneys compared to other tissues. Other molecular mechanisms, such as microRNA regulation and imprinting events, could also have explained the QTL. We could not explore these possibilities, however, due to the lack of both knowledge and high throughput databases, which is a limitation of our approach.
Among those candidate genes, Gpc4 — a heparan sulfate proteoglycan — was identified. The gene product of Gpc4 is present on the cell surface where it interacts with Col18a1. Interestingly, mutations in Col18a1 have been found in patients with Knobloch Syndrome, some of which develop kidney defects (Williams et al. 2008). Other likely candidate genes include Avpr2, which is involved in urine homeostasis, as well as four genes that likely interact with functionally relevant genes differentially expressed within the genome. KK mice are used as a model of metabolic disease (Qi et al. 2005; Reddi et al. 1990) and mutations in Avpr2 in human populations lead to X-linked nephrogenic diabetes insipidus (Birnbaumer 2000). Future investigations should be directed towards the dissection of the potential functionality of those genes for ACR variability.
Finally, this study highlights the importance of analyzing X-linked associations between phenotype and genotype. As reported in the MPD and apparent in our investigations, ACR is changed predominantly in male mice. Potentially, this evidence of X-linked variations may be an important factor. In the past, linkage analysis often omitted reporting the results on the X chromosome either due to a lack of genotyping of X-linked markers, a lack of accurate statistical methods to handle dosage compensation, or a lack of confidence that genes located on the X chromosome are involved in complex traits. For instance, in human studies, very few genome-wide association studies for complex traits reported results for the X chromosome. Formerly, when markers were genotyped individually, the X chromosome was often not prioritized in QTL crosses because most QTL were found on autosomes. More recent genotyping platforms and arrays now include markers for the X chromosome, and our study shows the importance of analyzing the X chromosome in relation to complex traits such as ACR.
In summary, we identified novel genomic loci for kidney disease susceptibility in mice. Among them, we identified a novel and strong X-linked QTL for ACR. Further in vivo investigation will yield insight into the confirmation of this X-linked QTL and its potential role in sexual dimorphism of kidney disease in mice and humans.
Supplementary Material
Acknowledgments
The authors would like to thank Joanne Currer for editing the manuscript.
Grants: This study was funded by U.S. National Institutes of Health grants (PO083069, HL077796, and HL081162 to BP) and the National Cancer Institute Core grant (CA034196 to The Jackson Laboratory). Dr. Berndt is recipient of mentorships by the Parker B. Francis foundation and the North American Hair Research Society.
Abbreviations
- AB
Annerose Berndt
- BP
Beverly Paigen
- MSL
Magalie S. Leduc
- JPS
John P. Sundberg
REFERENCES
- Birnbaumer M. Vasopressin receptors. Trends Endocrinol Metab. 2000;11:406–410. doi: 10.1016/s1043-2760(00)00304-0. [DOI] [PubMed] [Google Scholar]
- Broman KW, Sen S. A guide to QTL mapping with R/qtl. Springer; 2009. [Google Scholar]
- Broman KW, Sen S, Owens SE, Manichaikul A, Southard-Smith EM, Churchill GA. The X chromosome in quantitative trait locus mapping. Genetics. 2006;174:2151–2158. doi: 10.1534/genetics.106.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Burgess-Herbert SL, Cox A, Tsaih SW, Paigen B. Practical applications of the bioinformatics toolbox for narrowing quantitative trait loci. Genetics. 2008;180:2227–2235. doi: 10.1534/genetics.108.090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPetrillo K, Wang X, Stylianou IM, Paigen B. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 2005;21:683–692. doi: 10.1016/j.tig.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Doorenbos C, Tsaih SW, Sheehan S, Ishimori N, Navis G, Churchill G, Dipetrillo K, Korstanje R. Quantitative trait loci for urinary albumin in crosses between C57BL/6J and A/J inbred mice in the presence and absence of Apoe. Genetics. 2008;179:693–699. doi: 10.1534/genetics.107.085142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindle S, Garganta C, Sheehan S, Gile J, Lapierre A, Whitmore H, Paigen B, DiPetrillo K. Validation of high-throughput methods for measuring blood urea nitrogen and urinary albumin concentrations in mice. Comp Med. 2006;56:482–486. [PubMed] [Google Scholar]
- Hageman RS, Leduc MS, Caputo CR, Tsaih SW, Churchill GA, Korstanje R. Uncovering genes and regulatory pathways related to urinary albumin excretion. J Am Soc Nephrol. 2011;22:73–81. doi: 10.1681/ASN.2010050561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberg L, Coleman DL. Laboratory animals exhibiting obesity and diabetes syndromes. Metabolism. 1977;26:59–99. doi: 10.1016/0026-0495(77)90128-7. [DOI] [PubMed] [Google Scholar]
- Ikeda H. KK mouse. Diabetes Res Clin Pract. 1994;24(Suppl):S313–S316. doi: 10.1016/0168-8227(94)90268-2. [DOI] [PubMed] [Google Scholar]
- Iyengar SK, Abboud HE, Goddard KA, Saad MF, Adler SG, Arar NH, Bowden DW, Duggirala R, Elston RC, Hanson RL, Ipp E, Kao WH, Kimmel PL, Klag MJ, Knowler WC, Meoni LA, Nelson RG, Nicholas SB, Pahl MV, Parekh RS, Quade SR, Rich SS, Rotter JI, Scavini M, Schelling JR, Sedor JR, Sehgal AR, Shah VO, Smith MW, Taylor KD, Winkler CA, Zager PG, Freedman BI. Family Investigation of Nephropathy and Diabetes Research Group. Genome-wide scans for diabetic nephropathy and albuminuria in multiethnic populations: the family investigation of nephropathy and diabetes (FIND) Diabetes. 2007;56:1577–1585. doi: 10.2337/db06-1154. [DOI] [PubMed] [Google Scholar]
- Kato N, Watanabe Y, Ohno Y, Inoue T, Kanno Y, Suzuki H, Okada H. Mapping quantitative trait loci for proteinuria-induced renal collagen deposition. Kidney Int. 2008;73:1017–1023. doi: 10.1038/KI.2008.7. [DOI] [PubMed] [Google Scholar]
- Kim EH, Lee CH, Hyun BH, Suh JG, Oh YS, Namikawa T, Ishikawa A. Quantitative trait loci for proteinuria in the focal glomerulosclerosis mouse model. Mamm Genome. 2005;16:242–250. doi: 10.1007/s00335-004-3023-7. [DOI] [PubMed] [Google Scholar]
- Leduc MS, Lyons M, Darvishi K, Walsh K, Sheehan S, Amend S, Cox A, Orho-Melander M, Kathiresan S, Paigen B, Korstanje R. The mouse QTL map helps interpret human genome-wide association studies for HDL cholesterol. J Lipid Res. 2011;52:1139–1149. doi: 10.1194/jlr.M009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiter EH. Selecting the "right" mouse model for metabolic syndrome and type 2 diabetes research. Methods Mol Biol. 2009;560:1–17. doi: 10.1007/978-1-59745-448-3_1. [DOI] [PubMed] [Google Scholar]
- Morel L, Rudofsky UH, Longmate JA, Schiffenbauer J, Wakeland EK. Polygenic control of susceptibility to murine systemic lupus erythematosus. Immunity. 1994;1:219–229. doi: 10.1016/1074-7613(94)90100-7. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LL, Robledo RF, Bult CJ, Churchill GA, Paigen BJ, Svenson KL. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat Rev Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- Qi Z, Fujita H, Jin J, Davis LS, Wang Y, Fogo AB, Breyer MD. Characterization of susceptibility of inbred mouse strains to diabetic nephropathy. Diabetes. 2005;54:2628–2637. doi: 10.2337/diabetes.54.9.2628. [DOI] [PubMed] [Google Scholar]
- Reddi AS, Velasco CA, Reddy PR, Khan MY, Camerini-Davalos RA. Diabetic microangiopathy in KK mice. VI. Effect of glycemic control on renal glycoprotein metabolism and established glomerulosclerosis. Exp Mol Pathol. 1990;53:140–151. doi: 10.1016/0014-4800(90)90038-f. [DOI] [PubMed] [Google Scholar]
- Salzler HR, Griffiths R, Ruiz P, Chi L, Frey C, Marchuk DA, Rockman HA, Le TH. Hypertension and albuminuria in chronic kidney disease mapped to a mouse chromosome 11 locus. Kidney Int. 2007;72:1226–1232. doi: 10.1038/sj.ki.5002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A, Standke D, Kovacevic L, Mostler M, Kossmehl P, Stoll M, Kreutz R. A major gene locus links early ons albuminuria with renal interstitial fibrosis in the MWF rat with polygenetic albuminuria. J Am Soc Nephrol. 2003;14:3081–3089. doi: 10.1097/01.asn.0000100126.62370.25. [DOI] [PubMed] [Google Scholar]
- Sheehan S, Tsaih SW, King BL, Stanton C, Churchill GA, Paigen B, DiPetrillo K. Genetic analysis of albuminuria in a cross between C57BL/6J and DBA/2J mice. Am J Physiol Renal Physiol. 2007;293:F1649–F1656. doi: 10.1152/ajprenal.00233.2007. [DOI] [PubMed] [Google Scholar]
- Shockley KR, Witmer D, Burgess-Herbert SL, Paigen B, Churchill GA. The Effects of Atherogenic Diet on Hepatic Gene Expression Across Mouse Strains. Physiol Genomics. 2009;39:172–182. doi: 10.1152/physiolgenomics.90350.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates under dependence. J R Stat Soc Ser. 2002;B:479–498. [Google Scholar]
- Tsaih SW, Pezzolesi MG, Yuan R, Warram JH, Krolewski AS, Korstanje R. Genetic analysis of albuminuria in aging mice and concordance with loci for human diabetic nephropathy found in a genomewide association scan. Kidney Int. 2010;77:201–210. doi: 10.1038/ki.2009.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Paigen B. Genetics of variation in HDL cholesterol in humans and mice. Circ Res. 2005;96:27–42. doi: 10.1161/01.RES.0000151332.39871.13. [DOI] [PubMed] [Google Scholar]
- Williams TA, Kirkby GR, Williams D, Ainsworth JR. A phenotypic variant of Knobloch syndrome. Ophthalmic Genet. 2008;29:85–86. doi: 10.1080/13816810701850041. [DOI] [PubMed] [Google Scholar]
- Wu H, Kerr MK, Cui X, Churchill GA. MAANOVA: A software package for the analysis of spotted cDNA microarray experiments. In: Parmigiani G, editor. The analysis of gene expression data. New York: Springer; 2003. pp. 313–341. [Google Scholar]
- Zhang QL, Rothenbacher D. Prevalence of chronic kidney disease in population-based studies: systematic review. BMC Public Health. 2008;11:8–117. doi: 10.1186/1471-2458-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.