Abstract
Water permeability through single-file channels is affected by intrinsic factors such as their size and polarity and by external determinants like their lipid environment in the membrane. Previous computational studies revealed that the obstruction of the channel by lipid headgroups can be long-lived, in the range of nanoseconds, and that pore-length-matching membrane mimetics could speed up water permeability. To test the hypothesis of lipid-channel interactions modulating channel permeability, we designed different gramicidin A derivatives with attached acyl chains. By combining extensive molecular-dynamics simulations and single-channel water permeation measurements, we show that by tuning lipid-channel interactions, these modifications reduce the presence of lipid headgroups in the pore, which leads to a clear and selective increase in their water permeability.
Introduction
Increasing evidence shows that protein-lipid interactions are essential for the proper functioning of membrane proteins (1–4). For example, protein-lipid interactions are required for voltage-sensing in potassium channels (5,6), for the correct folding and in vivo functioning of transport proteins such as LacY (7) and multidrug transporter LmrP (8), or for the formation of stable signaling complexes (9) and microdomains in the membrane (10). In addition, covalently bound lipids in G-proteins ensure their correct incorporation in target membranes (11,12). Another example is the mitochondrial anion channel VDAC, which shows different gating characteristics in different types of phospholipid membranes (13). These studies strongly suggest that many integral membrane proteins, channels among them, do not function as isolated entities, but instead form a functional unit together with the membrane in which they are embedded.
In addition to their fundamental function in biology, channels also play an important role in nanotechnological applications due to their small size and potential tunable selectivity. For instance, aquaporin water channels have been proposed as highly potent water filters (14), whereas other channels could be used as detectors of protein-ligand interactions (15–17). Gramicidin A (gA) is a peptide antibiotic that forms a pore when inserted in a lipid membrane. It is well known as a cation channel, but water permeation studies indicated that it is also an excellent water channel (18–21). gA has been widely used as a template for multiple applications due to its limited complexity, e.g., to measure pH (22), construct voltage-gated (23) and light-gated channels (24), and as a sensor for chemical signals or changes in the mechanical properties of its surrounding membrane (25–27).
Single-channel conductance studies have shown that gA can be found in two different conformations that differ in their single-channel ion conductivities and water permeabilities (21,28). For the prevailing conformation, known as helical-dimer conformation (29), molecular dynamics (MD) simulations done by Chiu et al. (19), Kim et al. (30), and by us (de Groot et al. (20)) identified that in a 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine (DMPC) membrane, the lipid headgroups heavily interact with the pore entrance. These interactions have a direct impact on the access of water molecules to the pore lumen, transiently blocking the pore on the nanosecond timescale. The total thickness of the DMPC bilayer is somewhat larger than the length of the gA channel, such that lipid headgroups can easily interfere with water entry and exit to/from the channel. Indeed, by using a membrane mimetic that perfectly matches the length of a gramicidin-like peptidic pore, we computed water fluxes an order-of-magnitude larger than for gA in DMPC (31).
This observation has led to the hypothesis that it should be possible to modulate the water permeation rate by modifying the entrance of the channel, as it was previously demonstrated by the work of Pfeifer et al. (32) in terms of ion conductance. To test this hypothesis, instead of building a molecular funnel that assists the entrance of ions and water, our approach aims to replace the first shell of lipids around the peptide, effectively freeing the channel entrance of lipid headgroups. Inspired by the work of Vogt et al. (33), who coupled a fatty acid to the ethanolamine (ETA) group of gA, we designed four derivatives of gA that carry synthetic D-amino acids with acyl chains attached to the side chains (Fig. 1). These acyl-side chains are designed to align with the hydrophobic tails of the lipid membrane, thereby putatively displacing the most proximal lipids from the pore entrance and therefore preventing them from interfering with water permeation.
Figure 1.
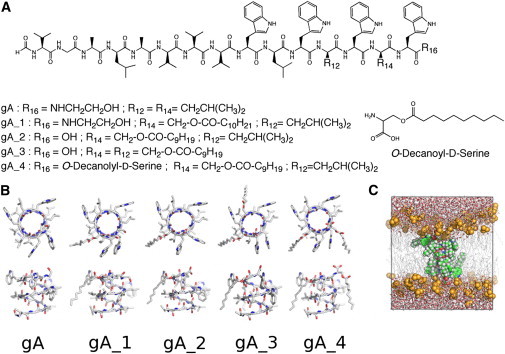
(A) Different gramicidin derivatives used in this study: Rn indicates residues and n the position in the sequence. (B) Top and side view of the corresponding molecular models extracted from our MD simulations. Only one monomer is displayed; hydrogen atoms were omitted for clarity. (C) Typical simulation box, showing a peptide (spheres) embedded in a hydrated DMPC bilayer (lipid tails as lines, lipid headgroups as spheres, water molecules as sticks).
In this work, we present a combined MD simulation and experimental study in which we report the water permeability for different acylated gramicidin derivatives. To quantify the water permeability, we employ the osmotic permeability coefficient pf (34) that can be accessed directly both from experiments and simulations (20,35,36). It is shown that, by modification of the channel-lipid interactions, the water permeability can be enhanced by up to a factor of three.
Methods
We have designed four different modifications of gA that contained residues with long acyl chains, schematically shown in Fig. 1. The length of the alkyl chains was designed to match the length of half a gA dimer in helical-dimer conformation. We have prepared the derivatives with increasing number of substitutions, different coupling positions, and charge states to elucidate which are the modifications that are most effective at increasing water permeability. We expect that distributing the acyl derivatives homogeneously around the peptide would be more effective at displacing the surrounding lipids from the pore entrance. We have not modified the positions occupied by tryptophan groups, which favor membrane anchoring. The groups gA_1 and gA_2 have one single modification, with gA_1 having one carbon atom more in the alkyl chain than gA_2. The derivatives gA_3 and gA_4 have two residue modifications in different positions, which bear chains of same length. Only the gA_1 modification contains ETA at the C-terminal part, and gA_2 to gA_4 carry a net charge at the C-terminal end.
Synthesis and characterization
All peptide syntheses were performed via solid-phase peptide synthesis using a peptide synthesizer (ABI 433A, Applied Biosystems) and based on the peptide sequence of gramicidin A. In the derivatives, nonnatural amino-acid residues substituted the amino acids D-Leu14 and/or D-Leu12 (Fig. 1). These new building blocks were synthesized as n-Fmoc-D-Ser(decanoyl)-OH and n-Fmoc-D-Ser(undecanoyl)-OH by coupling n-Fmoc-D-Ser-OH with decanoyl chloride and undecanoyl chloride, respectively. The synthesis for compounds gA_2 – gA_4 was performed on Fmoc-L-Trp(Boc)-wang resin. For the peptide gA_1, the synthesis was carried out on glycinol 2-chlorotrityl-resin. Formylation of the gA derivatives was performed on resin by using pentafluorophenyl formate in dichloromethane (37). Subsequently, the gA-derivatives were cleaved from the resin, the crude product was precipitated with cold diethyl ether (5 mL), purified by reverse phase HPLC, and characterized by high-resolution mass spectrometry. The synthesized acylgramicidin derivatives were incorporated into large unilamellar DMPC vesicles and its secondary structure was examined by circular dichroism spectroscopy (CD). All the details on the synthesis, purification, and characterization of the compounds can be found in the Supporting Material.
Water measurement studies
The single-channel osmotic permeability coefficients (pf) for the different derivatives in a solvent-free 1,2-diphytanoyl-sn-glycerol-3-phosphocholine (DPhPC) membrane were determined by establishing an osmotic gradient across the membrane, i.e., by adding urea into only one of the two aqueous compartments. Urea served as the osmolyte because it neither permeates gramicidin (38) nor alters the buffer viscosity. The resulting transmembrane water flow increases the solute concentration on the hypotonic side of the membrane (the trans side) and depletes it on the other side (the cis side) (39). The osmotic water flow decreased the concentration ci(x) of an impermeant ion, Mg+2 according to (40)
| (1) |
The values cs, x, a, Vw, cosm, and Di are, respectively, the solute concentration at the interface, the distance to the membrane, the stirring parameter, the molecular volume of water, the near-membrane concentration of the solute used to establish the transmembrane osmotic pressure difference, and the bulk solute diffusion coefficient. Fitting the experimental concentration profiles, obtained by scanning electrochemical microscopy, to Eq. 1 allowed calculation of Pf, the total osmotic membrane permeability coefficient. We obtained cosm by correcting the bulk urea concentration for dilution in the immediate membrane vicinity (41). Simultaneous measurement of membrane conductance G allowed determination of pf,
| (2) |
where g is the single ion channel conductance, measured using voltage-clamp. The single ion conductance was corrected for the lower electrolyte concentration used in the voltage-clamp experiments according to Hladky et al. (42). Further details can be found in the Supporting Material.
Computational studies
The structures of the acyl gramicidin derivatives were built using PyMOL (43) based on the gramicidin NMR structure 1MAG. The gramicidin derivatives were inserted in a previously equilibrated DMPC membrane of 124 lipid molecules, solvated with ∼4000 water molecules, and we generated 10 independent MD simulations of 50 ns each. As control, we have performed three sets of 50-ns MD simulations of the derivatives in a DPhPC membrane of 124 lipid molecules. All MD simulations were carried out using the GROMACS-4 software (44). All the relevant details of the simulation parameters can be found in the Supporting Material.
From the MD simulations we computed the osmotic permeability coefficient using: the methodology previously described in de Groot et al. (20) and Portella et al. (31), briefly explained in the Supporting Material, and the lipid, water, peptide densities, and occupancy statistics using in-house software. We computed the order parameters SCD for the acyl chains of DMPC and for the new residues incorporated in gA as using the C-H bond as previously described in Vermeer et al. (45). Hydrogen-bonding energies were extracted from MD trajectories using the empirical formula of Espinosa et al. (46).
Results
Conformation of the derivatives embedded in a lipidic membrane
The CD spectra of gA and its acylated analogs gA_1 to gA_4 are characteristic for a single-stranded, right-handed β6.3-helical gA structure known as the thermodynamically preferred conformation for gA (47) and the conducting state. The CD spectra of the native gramicidin and the acylgramicidins gA_1 to gA_3 are almost superimposable, indicating no structural change of the modified gA compared to the native peptide (see Fig. S1 in the Supporting Material). Only gA_4 shows an insignificant deviation at the second maxima, which is positioned to longer wavelengths, and a minimum showing negative values. However, gA_4 also indicates similar conformational behavior and forms a helical β6.3-dimer. As it was previously shown for gA (23), the conformation of the acylated analogs in the membrane varies with the peptide/lipid ratio, most likely as a result of intermolecular gramicidin-gramicidin interactions at a higher peptide/lipid ratio favoring the β6.3-helical conformation.
In agreement with the results of CD spectroscopy, MD simulations indicate that the four acylated gA derivates are stable in their β-helix fold. On average, the gA backbone shows a root-mean-square deviation of 0.11 nm from the NMR model, and the backbone of the derivatives shows a root-mean-square deviation smaller than 0.14 nm from the same reference structure (see Fig. S2). The main deviations take place in positions occupied by the introduced residues with attached lipophilic tails, as indicated by an increase of ∼0.16 nm in the residue-averaged root-mean-square fluctuation (RMSF) with respect to gA (see Fig. S3 a). These fluctuations mainly take place in the side chains, as indicated by very similar backbone RMSF profiles for all channels (see Fig. S3 b). However, it is clear from both residue-averaged and backbone residue-averaged RMSF that gA_2 and gA_3 are slightly more flexible than the rest of the derivatives. The difference between gA_2 and gA_1 is related to the presence of the net charge at the C-termini in gA_2, which tends to move toward the solvent seeking better solvation, decreasing also the rigidity of the hydrogen-bond partners one β-helix turn below. The reason for the difference between gA_3 and gA_4 is due to the anchoring effect of the acyl chain in position 16, which counteracts the destabilizing effect of the net charge.
The simulations also show that all derivatives tend to adopt similar tilt angles with respect to the membrane plane, with tilt angles between 9 and 11°. On average, the presence of the channel on the membrane does not affect the ordering of lipid tails, as measured by the lipid order parameter SCD (see Fig. S4). Remarkably, the acyl chains attached to the peptide behave qualitatively similar to the lipid tails. Except when positioned on the very last amino acid, the first atoms of the acyl chains tend to be perpendicular to the membrane normal. As we move toward the end of the acyl chains there is an increase and then a decrease of the ordering of the chain with respect to the membrane normal. This behavior is in agreement with the work of Vogt et al. (49) with palmitic acid incorporated at the C-terminal part of gA, where it was noticed that the carbon atoms at the end of the acyl chain tend to behave similarly to the lipid tails.
The presence of the channels in the lipid bilayer imposes short-range order on the radial positioning of the lipids around the pore (30), as shown in Fig. 2. Oscillations in the lipid density become more pronounced as we approach the channels, with a clear decay starting at ∼0.9 nm from the pore main axis, due to the volume occupied by the peptide side chains (see, for example, Fig. S5).
Figure 2.
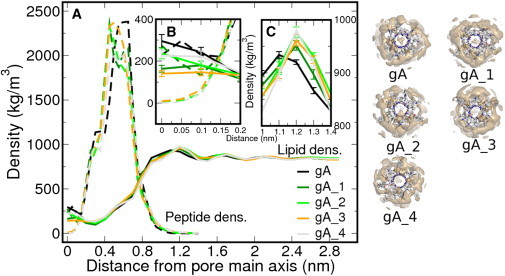
(A) Radial density profile from the pore main axis for DMPC lipids (straight) and the peptidic channels (dash), color-coded according to the derivative (left panel). All the atoms contribute to the density estimate. (Insets) Region corresponding to the pore lumen (B), where the occlusion takes place, and the shift in the first density maxima for the acyl-derivatives with respect to gA (C). In panels B and C, we show the standard error of the lipid density, omitted from panel A for clarity. On the right-hand side of the panel, the images show the peptidic channels surrounded by their lipid density isosurface of 0.5 atoms/nm3.
The acyl chains on the derivatives have a significant impact on the lipid density around and on top of the pore channel, although the designed peptides still retain a significant fraction of lipids disturbing the pore entrance. The first density maxima from the pore are only shifted 0.1 nm with respect to gA (Fig. 2 c), but nevertheless there is clear reduction in lipid density on top of the channel entrance for the new derivatives. Indeed, whereas gA has an average of one lipid headgroup disturbing either of its channel entrances, this value is reduced to 0.8 when we include one acyl chain, and drops below 0.5 for the doubly acylated derivatives (Fig. 3 c). In line with this result, there is a clear increase of the mean open state lifetime (Fig. 3 b; and see Table S1 in the Supporting Material) and a reduction of the residence time of the lipids: the quickly exchanging lipid headgroups drop their residence time over the channel entrance from ∼70 ps to ∼30 ps as we increase the number of substitutions (e.g., in Fig. 3 d, and see Table S1). For gA we identified rare lipid occlusion with long residence times, between 5 and 20 ns, which are reduced in the new derivatives, e.g., <3 ns for the doubly acylated channels. In addition to the lipid headgroups positioned on top of the membrane, the channels with the ETA capping group (gA and gA_1) display peptide density along the channel axis (Fig. 2 c), which has a nonnegligible contribution to the pore blockage (Fig. 3 a).
Figure 3.
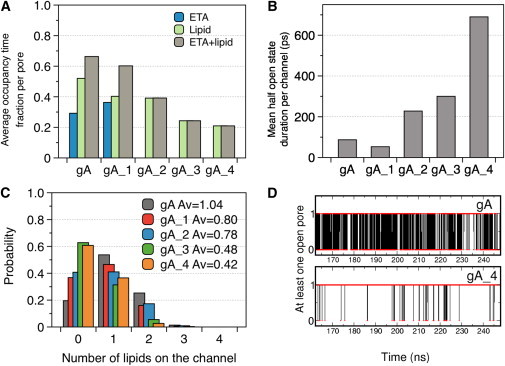
(A) Averaged fraction of time that the channels are blocked by ETA and lipid headgroups. (B) Mean duration time of the channels with at least one open pore entrance. (C) Normalized probability distribution of the number of lipids on the channel (both pore entrances). (D) Example of the time-dependent opening probability of the gA and gA_4 channels, showing a reduced number of transitions to the occluded state for the modified derivative. The data is extracted from a concatenated trajectory of 500 ns in total.
Water permeation
We added gramicidin A and derivatives from an ethanol stock solution to both sides of the black lipid membrane. Addition of 1 M urea to the cis side induced the transmembrane volume flux, which diluted the magnesium ions in the cis unstirred layer (cis-USL) and increased their concentration in the trans-USL. The effect was augmented while raising the peptide concentration due to the increase in water flow (see Fig. S6). The total membrane water permeability Pf is found to be a linear function of the total membrane conductance G (see Fig. S7). The slope is proportional to the single-channel water permeability pf of the peptidic channels, which can be extracted by considering the measured single file channel conductance g (results in Fig. 4 b) and Eq. 2. Acetylation of gA has a minor effect on the single channel potassium conductance. The derivatives gA_2 and gA_3 show a slightly higher conductance, respectively, 14 and 13.5 pS, whereas the derivatives gA_4 and gA_1 are characterized by a conductance of 9.2 and 10 pS, respectively (Fig. 4, and see Fig. S8 and Fig. S9).
Figure 4.

(A) Osmotic permeability coefficients pf for gA and its acylated derivatives experimentally determined by scanning electrochemical microscopy carried out in combination with voltage-clamp experiments and extracted from MD simulations. (B and C) Single-channel potassium conductance and the channel lifetime of the derivatives.
In Fig. 4 a, we plot both the experimentally determined osmotic permeation coefficient and the values obtained from our simulations for gA and its four derivatives. The pf value for gA is in agreement with previous estimates (21). The single modifications, with and without the ETA capping group, show similar experimental pf values, whereas the double modifications clearly allow an increase in water permeability. Double modifications on the 12 and 14 positions (gA_3) show a twofold increase in water permeation, whereas modifications on the 14 and 16 positions (gA_4) lead to a threefold increase.
In our MD simulations of the different derivatives in DMPC we found that gA_1 and gA_2 exhibit very similar pf values as gA, in quantitative agreement with the measured values. Interestingly, as in the experiments, gA_4 shows a significantly higher water permeability also in the simulations. Only for gA_3, that shows a twofold enhanced water permeability experimentally, the simulations report an unaltered pf. Overall, therefore, the data indicate that indeed the introduced membrane anchors can enhance water permeability. In analyzing the underlying mechanism, it should be noted that lipid-coverage of the channel entrance is not the only determinant of water permeability. Because the local conformation of the pore can modulate water binding affinity, we extracted the hydrogen-bond interaction energy for a water molecule along the pore axis (Fig. 5).
Figure 5.
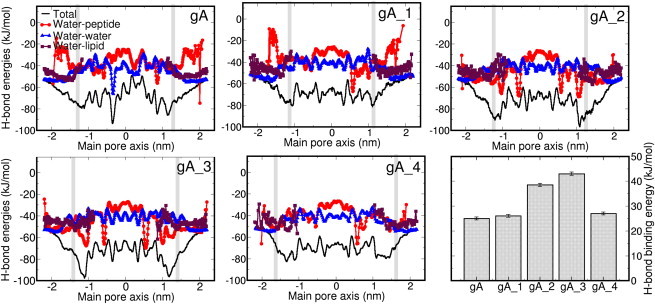
Hydrogen-bond energy per water molecule along the main pore axis (black curve) and its components: water-water (triangles), water-peptide (spheres), and water-lipid (squares). (Gray vertical lines) Pore entrance. (Bar graph) Hydrogen-bond binding energies at the entrance of the channel with respect to the bulk. (Error bars) Standard error of the mean value.
The clear minimum at the pore entrance for gA_2 and gA_3, compared to the rest of the derivatives, implies stronger binding energies, and therefore less mobile water molecules and lower pf values. Although gA_4 also has a net negative charge at the channel entrance, the anchoring effect of the acyl chain of residue 16 positions the carboxy group away from the channel entrance, thus weakening its interaction with water. Indeed, neutralizing the C-termini with an amino terminal group increases the pf for gA_3 (2.23 vs. 1.8 10−14 cm3/s) and does not affect the pf of gA_4 much (see Supporting Material and see Table S2). The mechanism emerging from these results, therefore, is that the channel permeation rate is the result of a subtle balance of channel-water affinity, channel conformation, and lipid-channel interactions. The designed acylation modifications of gA showed the predicted increase in water permeability of up to threefold, mostly, on the one hand, by preventing lipid block at the channel entry, and on the other hand, by providing sufficiently weak interactions to water. These interactions can be modulated as well by differences in channel flexibility (50,51), but the small differences observed in the backbone RMSF (see Fig. S3) seem to indicate that this is not the origin of the different water permeabilities in our systems.
Discussion
Molecular-dynamics simulations of unmodified gramicidin A in the nanosecond timescale reveal that water permeation through the channel can be transiently blocked by two different means: capping groups deviating from their reference positions (this effect is most pronounced for the ETA group), and lipid headgroups protruding at the top of the channel entrance. Typically, deviations of the capping groups from their positions are reestablished after a few picoseconds, but the effect of the lipids is more frequent and can be long-lived, above 10 ns. Here, we have shown that introducing lipid anchors in the form of acylated side chains to the gA channel can weaken the interference of lipid headgroups with water permeation through the channel, thereby increasing water permeability. These acyl chains behave similarly to the surrounding lipids, effectively pushing them away from the channel and reducing the amount and residence time of the occlusions.
For wild-type gA and three out of four derivatives, we obtained semiquantitative agreement between measured and computed pf values. Only for gA_3 we found a twofold increased water permeation rate experimentally, whereas an unaltered pf was found in MD simulations. The reason for this discrepancy could lie in invalid assumptions about the structure that was modeled as starting point for the simulations, in inaccuracies in the employed simulation parameters, or in the different membrane that was used in the simulation (DMPC) as compared to the experiment (DPhPC). As control, we performed three sets of 50-ns simulations of the derivatives in DPhPC. It should be noted that the applied DPhPC simulation parameters were validated on a rudimentary level only and were included for the sake of completeness rather than as a quantitative analysis.
The DPhPC results are largely consistent with the results of the DMPC simulations, with the exception of gA_1, which shows abnormally large permeability in DPhPC simulations (3.7 vs. 1.4 10−14 cm3/s) due to trapping of ETA by DPhPC headgroup in a fully extended conformation (see the Supporting Material and see Table S2). It should be noted that the comparison of rates is challenged by the fact that inaccuracies in the permeation barrier of just 2 kJ/mol already result in a rate error of a factor of two. Against this background, the agreement for the majority of the peptides, overall, suggests a valid simulation approach for the phenomena observed experimentally.
Covalent coupling of fatty acids to ETA increases channel lifetime substantially (33). The effect was missing in our experiments, most probably because we have used solvent-free planar bilayers, which due to their decreased thickness impose less mechanical tension on the open state of the channel (52). In addition, acyl chain anchoring to amino acids 12 and/or 14 may be less effective in increasing channel lifetime. Varying the coupling positions may also be responsible for the modest decrease in single channel conductance (30% for gA_1) which was not present when the fatty acids were attached to ETA (33).
It appears counterintuitive that the removal of the obstacle decreases single-channel ion conductivity. The removal of zwitterionic lipid headgroups from the channel entrance may impede partial ion dehydration, which is necessary for entering the single file of transported molecules, or affect the orientation of the tryptophan residues, known to influence the ion conductivity (53). The effect must be smaller when only one acyl chain is bound, which would explain why it was not observed in former studies. Because water dehydration is less costly (32,54), the effect is limited to ions.
Transient closure of the channel by a lipid lid may be expected to decrease both ion and water conductivities, especially if the obstruction is long-lived. Consequently, removal of that lid must affect g and pf in a similar fashion. In contrast, the experiment shows solely an increase in pf. A comparison of the mean obstruction time, in the range of hundreds of picoseconds, with the mean residence times of ions and water in the channel provides an explanation. The pf values indicate an approximate turnover number of water molecules between 6.6 × 108 s−1 and 2 × 109 s−1. The ion conductivity of ∼10 pS allows us to calculate an ion turnover number of only 6 × 106 s−1 at a transmembrane voltage of 100 mV. That is, the residence time of the ions in the channel exceeds the residence time of water molecules by two orders of magnitude. Consequently, ion movement is too slow to be impaired by short lipid blockages. In contrast, the much faster moving water molecules are arrested by their constant bumping into slowly moving lipid headgroups. Interestingly, this allows the specific tuning of the water permeability.
Conclusions
To increase the water transport through single-file channels we have designed, synthesized, and characterized four gramicidin-A derivatives with attached acyl chains. Single-channel ion and water transport, together with molecular-dynamics simulations, indicate that indeed the introduced membrane anchors can displace the lipids around the peptidic channels, enhancing their water permeability up to a factor of 3, while leaving ion-conductance mostly unaffected. This work highlights the importance of the lipid environment in modulating the activity of membrane peptide/proteins and contributes to its understanding. Modulation of lipid-channel interactions provides an attractive way to tune channel permeation characteristics, a prerequisite for the design of channels with a desired permeability and specificity.
Acknowledgments
We thank Erik Lindahl for the DMPC topology and Camilo Aponte-Santamaria for stimulating discussions.
The German research foundation Deutsche Forschungsgemeinschaft is gratefully acknowledged for funding via grant No. SFB803. This work was also supported by grant No. P23679 from the Austrian Science Fund (FWF) (to P.P.) and by a fellowship from the Sara Borrell Program (to G.P.).
Supporting Material
References
- 1.Phillips R., Ursell T., Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee A.G. How lipids and proteins interact in a membrane: a molecular approach. Mol. Biosyst. 2005;1:203–212. doi: 10.1039/b504527d. [DOI] [PubMed] [Google Scholar]
- 3.Nyholm T.K.M., Ozdirekcan S., Killian J.A. How protein transmembrane segments sense the lipid environment. Biochemistry. 2007;46:1457–1465. doi: 10.1021/bi061941c. [DOI] [PubMed] [Google Scholar]
- 4.Jensen M.O., Mouritsen O.G. Lipids do influence protein function-the hydrophobic matching hypothesis revisited. Biochim. Biophys. Acta. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Valiyaveetil F.I., Zhou Y., MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt D., Jiang Q.-X., MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- 7.Bogdanov M., Heacock P., Dowhan W. Plasticity of lipid-protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli. Proc. Natl. Acad. Sci. USA. 2010;107:15057–15062. doi: 10.1073/pnas.1006286107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hakizimana P., Masureel M., Govaerts C. Interactions between phosphatidylethanolamine headgroup and LmrP, a multidrug transporter: a conserved mechanism for proton gradient sensing? J. Biol. Chem. 2008;283:9369–9376. doi: 10.1074/jbc.M708427200. [DOI] [PubMed] [Google Scholar]
- 9.Jastrzebska B., Goc A., Palczewski K. Phospholipids are needed for the proper formation, stability, and function of the photoactivated rhodopsin-transducin complex. Biochemistry. 2009;48:5159–5170. doi: 10.1021/bi900284x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Bogaart G., Meyenberg K., Jahn R. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wedegaertner P.B., Wilson P.T., Bourne H.R. Lipid modifications of trimeric G proteins. J. Biol. Chem. 1995;270:503–506. doi: 10.1074/jbc.270.2.503. [DOI] [PubMed] [Google Scholar]
- 12.Moffett S., Brown D.A., Linder M.E. Lipid-dependent targeting of G proteins into rafts. J. Biol. Chem. 2000;275:2191–2198. doi: 10.1074/jbc.275.3.2191. [DOI] [PubMed] [Google Scholar]
- 13.Rostovtseva T.K., Kazemi N., Bezrukov S.M. Voltage gating of VDAC is regulated by nonlamellar lipids of mitochondrial membranes. J. Biol. Chem. 2006;281:37496–37506. doi: 10.1074/jbc.M602548200. [DOI] [PubMed] [Google Scholar]
- 14.Jensen, P. H., and D. Keller. 2006. Membrane for filtering of water. International Patent Application No. WO 2006/122566.
- 15.Cornell B.A., Braach-Maksvytis V.L., Pace R.J. A biosensor that uses ion-channel switches. Nature. 1997;387:580–583. doi: 10.1038/42432. [DOI] [PubMed] [Google Scholar]
- 16.Hirano A., Wakabayashi M., Sugawara M. A single-channel sensor based on gramicidin controlled by molecular recognition at bilayer lipid membranes containing receptor. Biosens. Bioelectron. 2003;18:973–983. doi: 10.1016/s0956-5663(02)00219-1. [DOI] [PubMed] [Google Scholar]
- 17.Futaki S., Zhang Y.J., Sugiura Y. Gramicidin-based channel systems for the detection of protein-ligand interaction. Bioorg. Med. Chem. 2004;12:1343–1350. doi: 10.1016/j.bmc.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Rosenberg P.A., Finkelstein A. Water permeability of gramicidin A-treated lipid bilayer membranes. J. Gen. Physiol. 1978;72:341–350. doi: 10.1085/jgp.72.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu S.W., Subramaniam S., Jakobsson E. Simulation study of a gramicidin/lipid bilayer system in excess water and lipid. II. Rates and mechanisms of water transport. Biophys. J. 1999;76:1939–1950. doi: 10.1016/S0006-3495(99)77353-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Groot B.L., Tieleman D.P., Grubmüller H. Water permeation through gramicidin A: desformylation and the double helix: a molecular dynamics study. Biophys. J. 2002;82:2934–2942. doi: 10.1016/S0006-3495(02)75634-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pohl P., Saparov S.M. Solvent drag across gramicidin channels demonstrated by microelectrodes. Biophys. J. 2000;78:2426–2434. doi: 10.1016/S0006-3495(00)76786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borisenko V., Zhang Z., Woolley G.A. Gramicidin derivatives as membrane-based pH sensors. Biochim. Biophys. Acta. 2002;1558:26–33. doi: 10.1016/s0005-2736(01)00415-1. [DOI] [PubMed] [Google Scholar]
- 23.Koeppe R.E., 2nd, Anderson O.S. Engineering the gramicidin channel. Annu. Rev. Biophys. Biomol. Struct. 1996;25:231–258. doi: 10.1146/annurev.bb.25.060196.001311. [DOI] [PubMed] [Google Scholar]
- 24.Banghart M.R., Volgraf M., Trauner D. Engineering light-gated ion channels. Biochemistry. 2006;45:15129–15141. doi: 10.1021/bi0618058. [DOI] [PubMed] [Google Scholar]
- 25.Capone R., Blake S., Mayer M. Designing nanosensors based on charged derivatives of gramicidin A. J. Am. Chem. Soc. 2007;129:9737–9745. doi: 10.1021/ja0711819. [DOI] [PubMed] [Google Scholar]
- 26.Andersen O.S., Bruno M.J., Koeppe R.E., 2nd Single-molecule methods for monitoring changes in bilayer elastic properties. Methods Mol. Biol. 2007;400:543–570. doi: 10.1007/978-1-59745-519-0_37. [DOI] [PubMed] [Google Scholar]
- 27.Lundbaek, J. A., R. E. Koeppe, and O. S.Andersen. Amphiphile regulation of ion channel function by changes in the bilayer spring constant. Proc. Natl. Acad. Sci. USA. 107:15427–15430. [DOI] [PMC free article] [PubMed]
- 28.Saparov S.M., Antonenko Y.N., Pohl P. Desformylgramicidin: a model channel with an extremely high water permeability. Biophys. J. 2000;79:2526–2534. doi: 10.1016/S0006-3495(00)76493-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersen O.S., Apell H.J., Woolley A. Gramicidin channel controversy—the structure in a lipid environment. Nat. Struct. Biol. 1999;6:609–612. doi: 10.1038/10648. [DOI] [PubMed] [Google Scholar]
- 30.Kim T., Lee K.I., Im W. Influence of hydrophobic mismatch on structures and dynamics of gramicidin a and lipid bilayers. Biophys. J. 2012;102:1551–1560. doi: 10.1016/j.bpj.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Portella G., Pohl P., de Groot B.L. Invariance of single-file water mobility in gramicidin-like peptidic pores as function of pore length. Biophys. J. 2007;92:3930–3937. doi: 10.1529/biophysj.106.102921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeifer J.R., Reiss P., Koert U. Crown ether-gramicidin hybrid ion channels: dehydration-assisted ion selectivity. Angew. Chem. Ind. Ed. 2006;45:501–504. doi: 10.1002/anie.200502570. [DOI] [PubMed] [Google Scholar]
- 33.Vogt T.C.B., Killian J.A., Andersen O.S. Influence of acylation on the channel characteristics of gramicidin A. Biochemistry. 1992;31:7320–7324. doi: 10.1021/bi00147a016. [DOI] [PubMed] [Google Scholar]
- 34.Finkelstein A. Wiley & Sons; New York: 1987. Water Movement through Lipid Bilayers, Pores, and Plasma Membranes. [DOI] [PubMed] [Google Scholar]
- 35.de Groot B.L., Grubmüller H. The dynamics and energetics of water permeation and proton exclusion in aquaporins. Curr. Opin. Struct. Biol. 2005;15:176–183. doi: 10.1016/j.sbi.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 36.Zhu F., Tajkhorshid E., Schulten K. Theory and simulation of water permeation in aquaporin-1. Biophys. J. 2004;86:50–57. doi: 10.1016/S0006-3495(04)74082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kisfaludy L., Ötvös L., Jr. Rapid and Selective Formylation With Pentafluorophenyl Formate. Synthesis. 1987;3:510. [Google Scholar]
- 38.Finkelstein A. Water and nonelectrolyte permeability of lipid bilayer membranes. J. Gen. Physiol. 1976;68:127–135. doi: 10.1085/jgp.68.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fettiplace R., Haydon D.A. Water permeability of lipid membranes. Physiol. Rev. 1980;60:510–550. doi: 10.1152/physrev.1980.60.2.510. [DOI] [PubMed] [Google Scholar]
- 40.Pohl P., Saparov S.M., Antonenko Y.N. The effect of a transmembrane osmotic flux on the ion concentration distribution in the immediate membrane vicinity measured by microelectrodes. Biophys. J. 1997;72:1711–1718. doi: 10.1016/S0006-3495(97)78817-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pohl P., Saparov S.M., Antonenko Y.N. The size of the unstirred layer as a function of the solute diffusion coefficient. Biophys. J. 1998;75:1403–1409. doi: 10.1016/S0006-3495(98)74058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hladky S.B., Urban B.W., Haydon D.A. Raven Press; New York, NY: 1979. Ion Movements in Pores Formed by Gramicidin A. [Google Scholar]
- 43.DeLano W.L. DeLano Scientific; Palo Alto, CA: 2009. PyMOL Molecular Graphics System. [Google Scholar]
- 44.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 45.Vermeer L.S., de Groot B.L., Czaplicki J. Acyl chain order parameter profiles in phospholipid bilayers: computation from molecular dynamics simulations and comparison with 2H NMR experiments. Eur. Biophys. J. 2007;36:919–931. doi: 10.1007/s00249-007-0192-9. [DOI] [PubMed] [Google Scholar]
- 46.Espinosa E., Molins E., Lecomte C. Hydrogen bond strengths revealed by topological analyses of experimentally observed electron densities. Chem. Phys. Lett. 1998;285:170–173. [Google Scholar]
- 47.Killian J.A., Prasad K.U., Urry D.W. The membrane as an environment of minimal interconversion. A circular dichroism study on the solvent dependence of the conformational behavior of gramicidin in diacylphosphatidylcholine model membranes. Biochemistry. 1988;27:4848–4855. doi: 10.1021/bi00413a040. [DOI] [PubMed] [Google Scholar]
- 48.Reference deleted in proof.
- 49.Vogt T.C., Killian J.A., De Kruijff B. Structure and dynamics of the acyl chain of a transmembrane polypeptide. Biochemistry. 1994;33:2063–2070. doi: 10.1021/bi00174a012. [DOI] [PubMed] [Google Scholar]
- 50.Allen T.W., Andersen O.S., Roux B. On the importance of atomic fluctuations, protein flexibility, and solvent in ion permeation. J. Gen. Physiol. 2004;124:679–690. doi: 10.1085/jgp.200409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baştuğ T., Gray-Weale A., Kuyucak S. Role of protein flexibility in ion permeation: a case study in gramicidin A. Biophys. J. 2006;90:2285–2296. doi: 10.1529/biophysj.105.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lundbaek J.A., Andersen O.S. Spring constants for channel-induced lipid bilayer deformations. Estimates using gramicidin channels. Biophys. J. 1999;76:889–895. doi: 10.1016/S0006-3495(99)77252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu W., Cross T.A. Tryptophan hydrogen bonding and electric dipole moments: functional roles in the gramicidin channel and implications for membrane proteins. Biochemistry. 1995;34:14147–14155. doi: 10.1021/bi00043a020. [DOI] [PubMed] [Google Scholar]
- 54.Allen T.W., Andersen O.S., Roux B. Energetics of ion conduction through the gramicidin channel. Proc. Natl. Acad. Sci. USA. 2004;101:117–122. doi: 10.1073/pnas.2635314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


