Abstract
APOBEC3 proteins inhibit HIV-1 replication in experimental systems and induce hypermutation in infected patients; however, the relative contributions of several APOBEC3 proteins to restriction of HIV-1 replication in the absence of the viral Vif protein in human primary CD4+ T cells and macrophages are unknown. We observed significant inhibition of HIV-1Δvif produced in 293T cells in the presence of APOBEC3DE (A3DE), APOBEC3F (A3F), APOBEC3G (A3G), and APOBEC3H haplotype II (A3H HapII) but not APOBEC3B (A3B), APOBEC3C (A3C), or APOBEC3H haplotype I (A3H HapI). Our previous studies showed that Vif amino acids Y40RHHY44 are important for inducing proteasomal degradation of A3G, whereas amino acids 14DRMR17 are important for degradation of A3F and A3DE. Here, we introduced substitution mutations of 40YRHHY44 and 14DRMR17 in replication-competent HIV-1 to generate vif mutants NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 to compare the antiviral activity of A3G to the combined antiviral activity of A3F and A3DE in activated CD4+ T cells and macrophages. During the first 15 days (round 1), in which multiple cycles of viral replication occurred, both the NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 mutants replicated in activated CD4+ T cells and macrophages, and only the NL4-3 YRHHY>A5 mutant showed a 2- to 4-day delay in replication compared to the wild type. During the subsequent 27 days (round 2) of cultures initiated with peak virus obtained from round 1, the NL4-3 YRHHY>A5 mutant exhibited a longer, 8- to 10-day delay and the NL4-3 DRMR>A4 mutant exhibited a 2- to 6-day delay in replication compared to the wild type. The NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 mutant proviruses displayed G-to-A hypermutations primarily in GG and GA dinucleotides as expected of A3G- and A3F- or A3DE-mediated deamination, respectively. We conclude that A3G exerts a greater restriction effect on HIV-1 than A3F and A3DE.
INTRODUCTION
The APOBEC3 family of proteins are cytidine deaminases and form an intracellular host defense mechanism that protects the cell from viral infections, including that with human immunodeficiency virus type I (HIV-1) (1–5). APOBEC3B (A3B), APOBEC3C (A3C), APOBEC3DE (A3DE), APOBEC3F (A3F), APOBEC3G (A3G), and some APOBEC3H haplotypes have been reported to inhibit HIV-1 in the absence of the accessory protein viral infectivity factor (Vif) (1–4, 6–14). The antiviral activities of A3B (9, 15), A3C (6), and A3DE (8, 16) have been reported in a limited number of studies and in some cases have been controversial (16–20); overall, A3G and A3F have been consistently reported to have strong antiviral activity in transient-transfection assays by numerous investigators and are thought to be the primary restriction factors that inhibit HIV-1 replication (21, 22).
APOBEC3H haplotype I (A3H HapI), the major haplotype in Caucasians, has low steady-state protein expression levels due to instability and consequently does not exhibit significant antiviral activity (7, 10, 13, 23). However, A3H HapII, which is more prevalent in people of African descent, expresses a stable protein and has been reported to exhibit antiviral activity (24). Endogenous A3H HapII along with A3DE may play a role in inhibiting HIV-1 during in vitro infection of A3F-null CEM2n cells (25), which suggests a potential role for these proteins in inhibiting HIV-1ΔVif in primary cells; however, the ability of HIV-1 Vif to overcome the inhibitory effects of A3H HapII appears to be subtype and even isolate dependent (26). For example, LAI Vif can partially rescue HIV-1Δvif infectivity in the presence of A3H HapII, but NL4-3 Vif cannot (10, 26–28).
During reverse transcription, A3G and A3F induce cytidine deamination of minus-strand DNA, which generates G-to-A hypermutation in the plus-strand DNA and inactivates the viral genome (1, 5, 29–32). HIV-1 expresses a 23-kDa viral infectivity factor (Vif), which counteracts the antiviral activity of A3DE, A3F, A3G, and A3H HapII by forming a Vif-E3 ubiquitin ligase-Cullin5/ElonginBC complex and targeting the degradation of the A3 factors (3, 12, 33–35). In the absence of Vif, A3G, A3F, A3DE, and A3H HapII are packaged into newly formed virus particles, resulting in hypermutation of the proviral genome (8, 10–12, 34, 36, 37); A3G and A3F have also been shown to inhibit DNA synthesis, integration, and proviral DNA formation (38–41). Distinct regions of Vif are important for binding to A3G and A3F/A3DE. The 40YRHHY44 region in Vif is important for binding to A3G, while the 14DRMR17 region in Vif is important for binding to A3F and A3DE (42–46). A 40YRHHY44 substitution mutant of Vif fails to block A3G activity but retains the ability to block A3F activity in a single-cycle assay; similarly, a 14DRMR17 substitution mutant of Vif fails to block A3F activity but not A3G activity in a single-cycle assay (44). The DRMR mutant of Vif also fails to degrade A3DE in transient-transfection assays (46).
CD4+ T cells and macrophages are the primary target cells for productive HIV-1 infection (47, 48). Vif is essential for HIV-1 replication in these cells (49–52). Different expression levels of A3G and A3F in monocytes and CD4+ T cells might result in different levels of HIV-1 restriction in these cells (53, 54). A3G mRNA is 3- to 10-fold more abundant in primary cells than A3F mRNA, suggesting that A3G protein levels and antiviral activity may be higher in these cells (53, 54). Alpha interferon (IFN-α) increases the mRNA expression levels of A3G, A3F, A3DE, and A3H, suggesting a greater potential for inhibition of virus replication in primary cells after IFN-α treatment in the absence of Vif-induced degradation (53–56).
Despite all of these studies, the relative contributions of different A3 proteins to HIV-1 restriction in primary activated CD4+ T cells and macrophages are unknown. This was emphasized by recent studies which concluded that A3F has minimal or no antiviral activity in primary cells or when expressed at levels similar to those present in primary cells (57, 58). The extent to which an A3 protein exerts restriction of HIV-1 depends on its level of expression, efficiency of virion incorporation, nucleic acid affinity, cytidine deaminase activity, and perhaps other factors. To date, it has not been possible to compare the steady-state levels of A3 proteins, since they are detected using different antibodies. While steady-state levels of A3 mRNAs have been determined, it is not known whether the mRNA levels correlate with the protein levels. Although it has been reported that increases in A3G and A3F mRNA levels are associated with increases in protein levels (54), it is not clear if increases in mRNA levels for the other A3 family members also correlate with increased protein levels. Finally, the intrinsic antiviral activities of the proteins in transient-transfection assays may not reflect their activities in primary cells, since the activities may be modulated by covalent modifications such as phosphorylation.
Here, we compared the antiviral activities of A3G and A3F/A3DE in CEM cells, activated primary CD4+ T cells, and macrophages by using vif mutants of replication-competent HIV-1 which do not rescue infection in the presence of A3G (NL4-3 YRHHY>A5) or A3F and A3DE (NL4-3 DRMR>A4) and compared the replication capacities and APOBEC3-induced hypermutation of these mutants over several weeks during which multiple cycles of viral replication occurred.
Over a 2-week period in which multiple cycles of viral replication occurred (round 1), only the A3G degradation-deficient mutant, NL4-3 YRHHY>A5, showed a delay in replication kinetics compared to wild-type NL4-3 (NL4-3 WT) in CEM cells, CD4+ T cells and macrophages. Over a subsequent 27-day period in cultures initiated with viruses from the first 2-week period (round 2), both mutants exhibited a delay in replication kinetics compared to NL4-3 WT. As expected, the dinucleotide context in which G-to-A hypermutation occurred in the NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 proviruses was consistent with cytidine deamination by A3G and A3F or A3DE, respectively. These results indicate that A3G exerts a stronger restriction of HIV-1 replication in CEM cells, primary CD4+ T cells, and macrophages than the combined antiviral activity of A3F and A3DE.
MATERIALS AND METHODS
Cell culture and plasmids.
293T and HeLa-derived HIV-1 reporter TZM-bl cell lines (59, 60) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (FCS), penicillin (50 units/ml), and streptomycin (50 μg/ml). CEM cells were maintained in RPMI 1640 medium with 10% FCS and the antibiotics penicillin (50 units/ml) and streptomycin (50 μg/ml). Peripheral blood mononuclear cells (PBMCs) were isolated from healthy donors using Ficoll-Hypaque (Sigma). CD4+ T cells were harvested from human donor peripheral blood cells using a CD4+ positive isolation kit (Dynal, Invitrogen) according to the manufacturer's instructions. For cell activation, CD4+ T cells were treated with 5 μg/ml phytohemagglutinin (PHA) and 200 U/ml interleukin-2 (IL-2) for 3 days. Activated cells were maintained in RPMI 1640 medium containing only IL-2. The purity and the stimulation efficiency of the cells were analyzed by fluorescence-activated cell sorter (FACS) analysis staining for CD4-allophycocyanin (APC) (ranging from 81.2% to 97%) and the activation marker CD25 (∼90%), respectively. Macrophages were generated from CD14+ monocytes obtained from healthy donors by culturing with granulocyte-macrophage colony-stimulating factor and 5% human serum. Nonattached cells were removed after 6 days. The replication analysis was performed with CD4+ T cells and macrophages from three different donors as described in Results. For CD4+ T cells, the cells were infected 3 days after stimulation and the medium was changed every second day.
HIV-1 mutants YRHHY>A5 and DRMR>A4 were previously described (44). For generation of the replication-competent NL4-3 YRHHY>A5 mutant, amino acids 40 to 44 encoded by the vif gene were replaced with five alanines (61). Amino acids 14 to 17 encoded by the vif gene were replaced with AKTK to generate the replication-competent NL4-3 DRMR>A4 mutant. The first 19 amino acids of Vif overlap the C-terminal end of integrase, and replacement of DRMR residues with AKTK resulted in an R284S substitution in the integrase protein, which did not cause a replication kinetic defect during preliminary experiments in CEM-SS or CEM cells (data not shown). To generate mutant viruses containing the envelope of the R5-tropic AD8 virus, NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 were digested with EcoRI and AgeI and cloned into the EcoRI- and AgeI-digested plasmid pNL4-3 (AD8) (62), resulting in a plasmid containing the NL4-3 backbone and an AD8 envelope. The structures of the resulting plasmids were confirmed by restriction enzyme mapping and DNA sequencing.
pFLAG-A3G was generated by modification of pcDNA-APO3G (kindly provided by Klaus Strebel, NIAID, National Institutes of Health) (44). pFLAG-A3F was constructed by modification of pcDNA3.1-APOBEC3F (1, 44). pFLAG-A3C and pFLAG-A3DE (46) were constructed using pcDNA3.1-APOBEC3C-V5-6×His and pcDNA3.1-APOBEC3DE-V5-6×His, respectively, which were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from B. Matija Peterlin and Yong-Hui Zheng (5, 8). pApobec3B-HA was also obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Bryan R. Cullen (9). The A3H HapI cDNA in pINCY (Open Biosystems) was PCR amplified and cloned to generate FLAG-A3H HapI, a plasmid that expresses A3H HapI with an N-terminal FLAG epitope. FLAG-A3H HapII was generated by introducing substitutions G105R, K121D, and E178D into FLAG-A3H HapI by site-directed mutagenesis with the QuikChange II site-directed mutagenesis kit (Stratagene).
Virus production and infection.
For virus production, 293T cells were transfected in 100-mm-diameter dishes by the modified CaCl2-phosphate method (63) or the polyethylenimine method (64) with modifications as previously described (65). Viruses were harvested at 48 h after transfection, filtered through a 0.45-μm filter (Millipore), and stored at −80°C. To determine the amount of virus released from transfected 293T cells, the p24 capsid (CA) content in harvested supernatants was analyzed using a p24 CA enzyme-linked immunosorbent assay (ELISA) kit (Perkin-Elmer or XpressBio). TZM-bl cells (4 × 103) were infected using culture supernatants containing 5 ng of p24 CA protein in a 96-well plate for 3 h. Luciferase activities were measured after 24 h using a luciferase kit (Perkin-Elmer) and LUMIstar Galaxy luminometer.
Virus replication analysis.
CEM cells (1 × 106) and CD4+ T cells (1 × 107) were placed in 75-cm flasks, and monocytes (4 × 106 cells) were differentiated into macrophages in 12-well plates; these cells were infected with NL4-3 viruses containing 10 ng p24 CA. For infection of macrophages, viruses expressing the AD8 envelope were used. Virus supernatants were harvested every 2 days and analyzed for p24 CA content. For round 2 infections, viruses from the peak time points from round 1 infections (100 ng of p24 CA) were used to infect fresh CEM cells, CD4+ T cells, and macrophages. The CD4+ T cells and macrophages from the same donors were used for both round 1 and round 2 infections.
DNA extraction and sequence analysis.
DNA from 1 × 106 infected cells was extracted with a DNA isolation kit (DNeasy blood and tissue kit; Qiagen) according to the manufacturer's instructions. Extracted DNA was used to amplify a region containing the vif gene using forward primer VifF (5′CAGGGAGATTCTAAAAG3′) and reverse primer VifR (5′GGATAAACAGCAGTTGTTGC3′) and cloned into a pGEM vector (Promega). Cloned PCR products were analyzed by sequencing using the primer NL4-3-seq-4921F (5′GAGATCCAGTTTGGAAAGGAC3′), spanning a 730-bp region containing vif and a portion of the vpr gene, as previously described (61).
Western blot analysis of A3G and A3F expression in T cell lines and primary cells.
Western blotting was performed to detect endogenous A3G and A3F in CD4+ T cells, CEM cells, and macrophages. CD4+ T cells were resuspended in lysis buffer (50 mM Tris-HCl [pH 7.4] with 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and protease inhibitor cocktail [Roche]) 3 days after activation with PHA and IL-2. Macrophages were differentiated with granulocyte-macrophage colony-stimulating factor and human serum for 6 days before they were harvested. Anti-A3G Apo-C17 antibody (1:5,000 dilution) and rabbit anti-human A3F (C-18; 1:500 dilution) (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) (anti-ApoC17 from Klaus Strebel and anti-human APOBEC3F from Michael Malim) were used to detect endogenous A3G and A3F, respectively, in T cell lines and primary human cells.
RESULTS
Sensitivity of NL4-3 WT, NL4-3 YRHHY>A5, and NL4-3 DRMR>A4 to restriction by human A3 proteins in transient-transfection assays.
Previously, we reported that the Vif protein in which alanine amino acid substitutions were introduced in the 40YRHHY44 and 14DRMR17 regions lost their activity against A3G and A3F, respectively, in single-cycle assays (44, 61) and that the DRMR>4A mutant lost its ability to degrade A3DE (46). To compare the relative contributions of A3 proteins to inhibition of HIV-1 replication, we generated replication-competent NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 mutants and analyzed their replication kinetics in primary CD4+ T cells and macrophages, which are the natural target cells for HIV-1 infection.
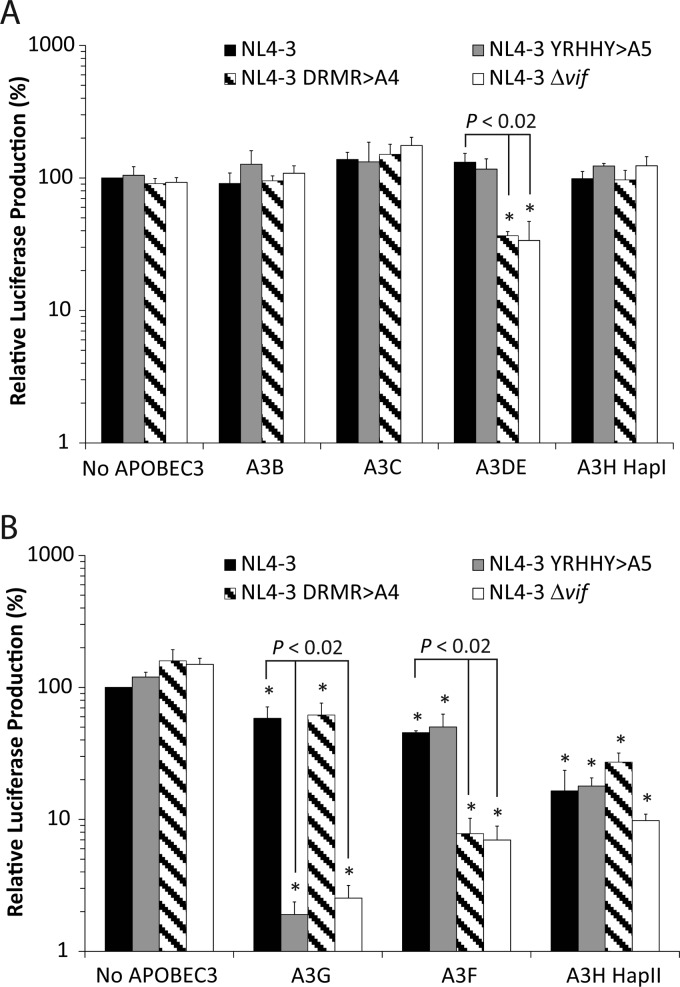
First, to determine which A3 proteins can inhibit wild-type or mutant NL4-3 viruses, we analyzed the ability of several A3 proteins to inhibit NL4-3 WT and vif mutants pNL4-3 YRHHY>A5, pNL4-3 DRMR>A4, or NL4-3Δvif in a transient-transfection assay (Fig. 1). The A3 protein expression plasmids and the wild-type or mutant HIV-1 expression plasmids were cotransfected into 293T cells, and viruses were harvested at 48 h posttransfection. Equal amounts of viruses, as determined by their p24 CA content, were then used to infect TZM-bl indicator cells; subsequently, luciferase activity was measured at 24 h postinfection. In the absence of any A3 proteins, the wild-type and vif mutants of HIV-1 produced similar amounts of luciferase in the TZM-bl cells, indicating that none of the mutations in NL4-3 vif had an effect on the viral infectivity. The infectivity of NL4-3 WT in the presence of all of the A3 proteins except A3H HapII was not significantly different from its infectivity in the absence of A3 proteins. A3B, A3C, and A3H HapI did not reduce the infectivity of the NL4-3 YRHHY>A5, NL4-3 DRMR>A4, or NL4-3Δvif viruses compared to the NL4-3 WT. A3DE and A3F significantly reduced the infectivity of the NL4-3 DRMR>A4 and HIV-Δvif viruses but not the infectivity of the NL4-3 YRHHY>A5 virus. A3G reduced the infectivity of the NL4-3 YRHHY>A5 and NL4-3Δvif viruses but not the infectivity of the NL4-3 DRMR>A4 virus. A3H HapII significantly reduced the infectivity of the NL4-3Δvif virus to <10% of that of NL4-3 WT in the absence of any A3 proteins (P ≪ 0.02). Interestingly, there was no significant difference between NL4-3 WT and any of the NL4-3 vif mutants produced in the presence of A3H HapII, which agrees with previous studies indicating that the NL4-3 Vif does not significantly rescue viral infectivity in the presence of A3H HapII (66, 67). These results indicate that the infectivity of the mutant virus NL4-3 YRHHY>A5 was impaired in the presence of A3G and A3H HapII but not other A3 proteins; in contrast, infectivity of the NL4-3 DRMR>A4 mutant virus was diminished in the presence of A3F, A3DE, and A3H HapII but not A3G or other A3 proteins (Fig. 1). The infectivity of NL4-3Δvif viruses, as assessed by luciferase production in TZM-bl cells, was almost completely abrogated in the presence of A3G, A3F, or A3H HapII and was reduced to about 34% in the presence of A3DE compared to that of NL4-3 WT. The lack of inhibition of all virus types produced in the presence of A3B, A3C, and A3H HapI suggests that endogenous forms of these A3 proteins are unlikely to contribute to differences in NL4-3 replication, regardless of which Vif variant is used. Likewise, A3H HapII inhibits all NL4-3 virus variants independent of Vif, indicating that it is not a major contributor to replication differences between the NL4-3 YRHHY>A5, and NL4-3 DRMR>A4 variants.
Fig 1.
Single-cycle assays to determine sensitivities of NL4-3 mutants to A3 proteins. Plasmids encoding NL4-3 WT, NL4-3 YRHHY>A5, NL4-3 DRMR>A4, or NL4-3 Δvif were transfected into 293T cells in the presence of the A3 expression plasmids. The ratio of NL4-3 to A3 plasmid DNAs was either 3:1 (A) or 12:1 (B). After 48 h, culture supernatants were harvested, the amounts of p24 CA in the supernatants were determined, and viruses containing equivalent amounts of p24 CA were used to infect TZM-bl indicator cells. Infectivity was determined by measuring the luciferase activity produced in the infected cells at 48 h postinfection. Infectivity of NL4-3 WT in the absence of any A3 protein (no APOBEC3) was set to 100%. The average from three to five independent experiments is shown. Error bars represent the standard error of the mean. Asterisks indicate statistically significant decreases in infectivity compared to the NL4-3 WT/no APOBEC3 control (P < 0.02 by Student's t test).
Replication of NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 vif mutants is delayed in CD4+ T cells and macrophages.
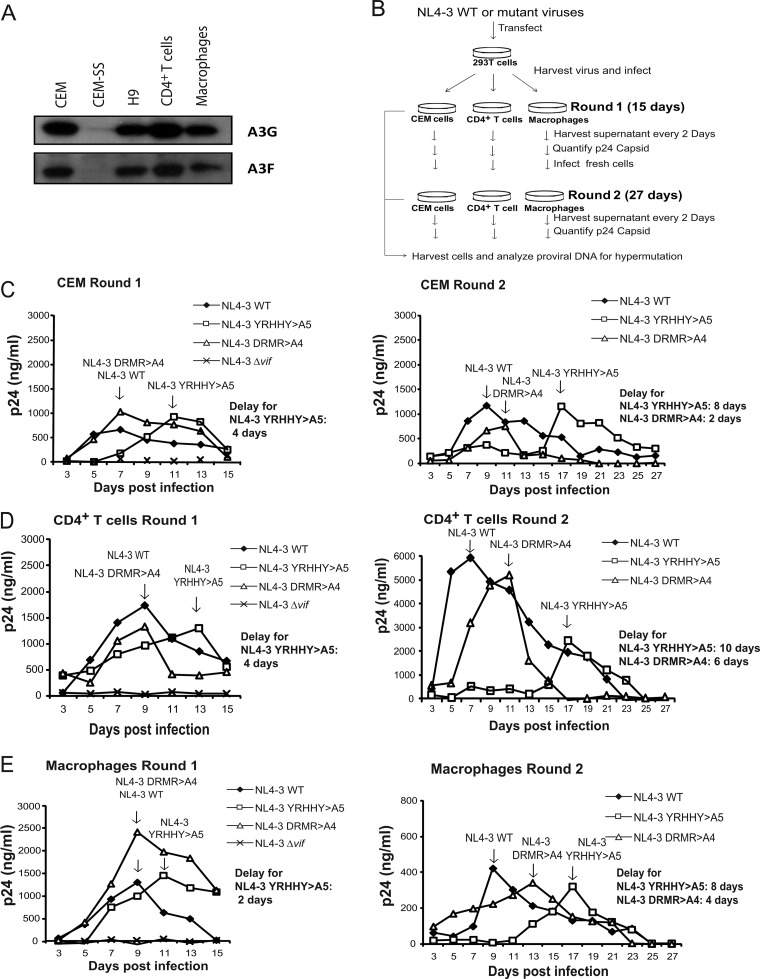
To analyze the antiviral activities of A3 proteins in primary CD4+ T cells, we first analyzed activated primary CD4+ T cells, monocyte-derived macrophages, and T cell lines by Western blotting. Primary CD4+ T cells were stimulated with PHA and IL-2 for 3 days, whereas macrophages were differentiated for 6 days before cell lysates were prepared. The results showed that CEM, H9, and CD4+ T cells and macrophages expressed detectable amounts of A3G and A3F, whereas little or no A3G and A3F could be detected in CEM-SS cells (Fig. 2A).
Fig 2.
Replication of NL4-3 WT, NL4-3 YRHHY>A5, NL4-3 DRMR>A4, and NL4-3Δvif in CEM T cells, primary CD4+ T cells, and macrophages. (A) Expression of A3G and A3F in CEM, CEM-SS, H9, and CD4+ T cells and macrophages. Endogenous A3G and A3F were detected by using the anti-A3G Apo-C17 antibody and the anti-A3F C18 antibody, respectively. (B) Protocol for evaluation of viral replication. (C to E) Replication of NL4-3 WT, NL4-3 YRHHY>A5, NL4-3 DRMR>A4, and NL4-3Δvif mutants in CEM cells (C), CD4+ T cells (D), and macrophages (E). Cells were infected with 10 ng of p24 CA-normalized viruses, supernatants were harvested every 2 days, and the p24 CA amounts were determined by ELISA. Multiple cycles of viral replication occurred over a 15-day period during round 1. The viruses from round 1 (left panels) peak time point samples were then p24 CA normalized and used to infected fresh cells (right panels) for round 2 infections, during which multiple cycles of viral replication occurred over an additional 27-day period. Two or three independent experiments were performed to determine the average delay in replication kinetics; results from one representative experiment are shown.
Multiple rounds of HIV-1 replication were examined as outlined in Fig. 2B. Macrophages, CD4+ T cells, and CEM cells were infected with culture supernatants containing 10 ng of p24 CA, and supernatants were harvested from the infected cells every 2 days beginning on day 3 (round 1). Replication of the viruses was monitored for 15 days and was quantified as the amount of p24 CA released in the supernatant for each time point. Viruses from the peak time points were then normalized for p24 CA amounts and used to infect fresh CEM cells, CD4+ T cells, and macrophages (round 2), and culture supernatants were harvested from the infected cells every 2 days. The results of these studies may be dependent on the individual donor genotype and the state of cell activation. To minimize the effects of donor variation, we performed independent experiments with cells obtained from two or three different donors.
In round 1 (Fig. 2C to E, left panels), which includes multiple rounds of virus replication, HIV-1 WT virus peaked at day 7 in CEM cells and at day 9 in CD4+ T cells and macrophages, whereas replication of the NL4-3 YRHHY>A5 mutant displayed a 2- to 4-day delay in all cell types tested; the delays were 4 days for CEM cells and CD4+ T cells and 2 days for macrophages. These delays in replication kinetics were consistent in independent experiments; the average delay for the NL4-3 YRHHY>A5 mutant in CEM cells was 4 days (4 days each for two experiments), the average delay in CD4+ T cells was 3 days (2, 4, and 4 days for three experiments), and the average delay in macrophages was 3 days (2, 2, and 4 days for three experiments). In contrast, the NL4-3 DRMR>A4 mutant did not display any delay in viral growth compared to wild-type virus in CEM cells, CD4+ T cells, and macrophages, indicating that there is little to no antiviral activity by A3 proteins other than A3G, causing no significant impact on viral replication. If these cells had low antiviral activity from NL4-3 DRMR>A4-nonsusceptible A3 family members (A3F and A3DE), the NL4-3 DRMR>A4 mutant might exhibit delayed growth kinetics after prolonged culturing and additional cycles of viral replication. To test this hypothesis, peak viruses from the round 1 infection were normalized for p24 CA content, and fresh CEM cells, CD4+ T cells, or macrophages were infected with equal amounts of virus (round 2); subsequently, culture supernatants were harvested from the infected cells every 2 days for 27 days, during which many additional cycles of viral replication occurred (Fig. 2C to E, right panels). In the round 2 infections, WT virus peaked at day 9 in CEM cells, day 7 in CD4+ T cells, and day 9 in macrophages. The delay for the NL4-3 YRHHY>A5 mutant was greater in the round 2 infections than that for NL4-3 WT (8-day delay for CEM cells, 10-day delay for CD4+ T cells, and 8-day delay for macrophages). The average delay for NL4-3 YRHHY>A5 was 8 days in CEM cells (8 days each in two experiments), 10 days in CD4+ T cells (10 days each in two experiments), and 9 days in macrophages (10 and 8 days in two experiments). In the round 2 infections, the NL4-3 DRMR>A4 mutant also displayed a 2- to 6-day delay compared to NL4-3 WT virus, which was consistent in all cell types tested; the average delay in two independent experiments was 4 days for CEM cells (6 and 2 days), 5 days for CD4+ T cells (4 and 6 days), and 5 days for macrophages (4 and 6 days) compared to NL4-3 WT virus, confirming the presence of low A3F and/or A3DE antiviral activity in these cells. Taken together, these results showed that the restriction capacity of A3G is greater than the combined antiviral activity of A3F and A3DE.
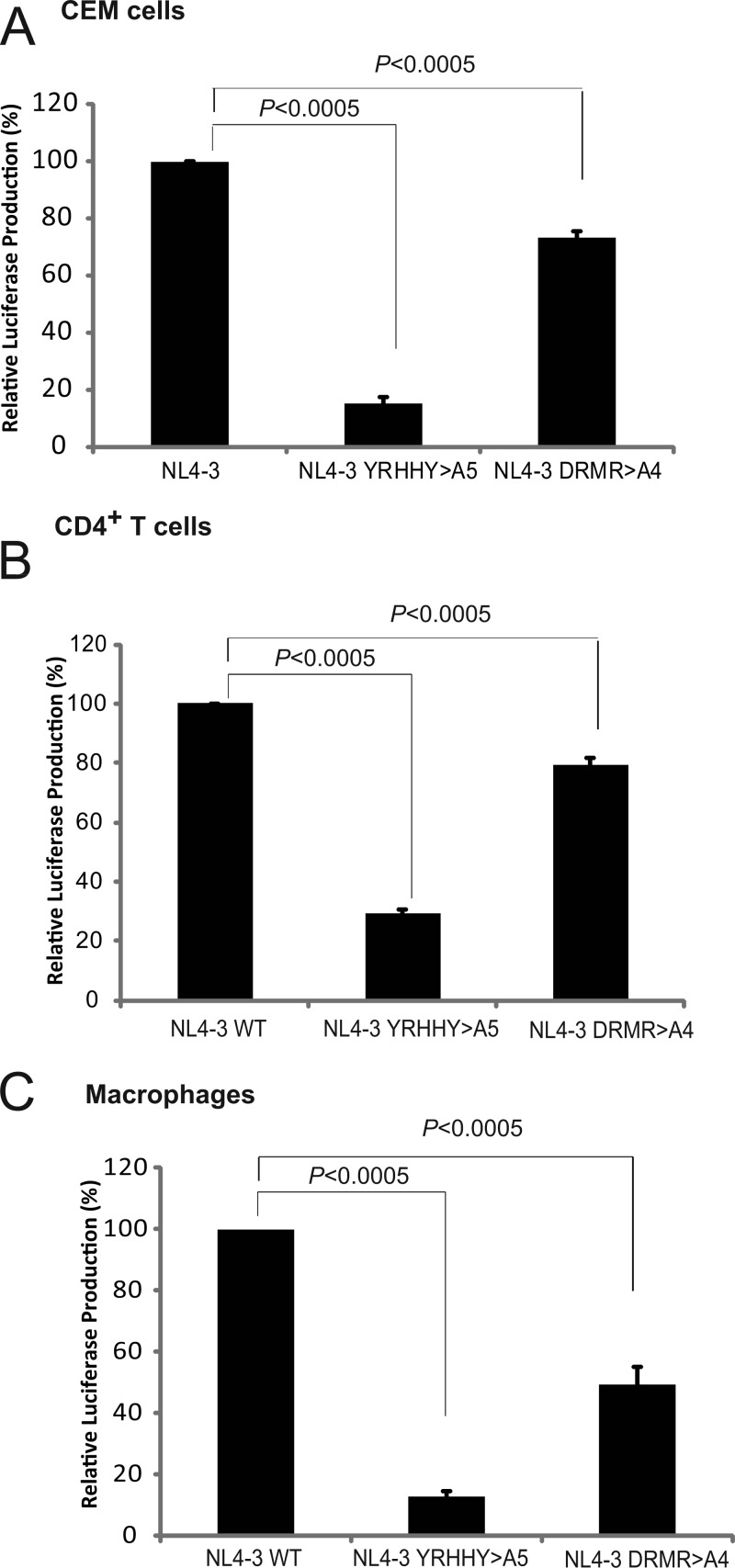
The infectivities of the viruses taken from round 1 peak time points in CEM cells, CD4+ T cells, and macrophages were determined by first quantifying the p24 CA amounts in the virus stocks. Equal amounts of p24 CA were then used to infect TZM-bl cells. Infectivity of the NL4-3 YRHHY>A5 virus was reduced to ∼10 to 25% of that of the NL4-3 control in CEM, CD4+ T cells, and macrophages (Fig. 3A, B, and C). Interestingly, even though no significant delay in replication was observed for the NL4-3 A4 mutant in round 1, the infectivity of virus was reduced to ∼50 to 80% of that of the NL4-3 WT control in CEM, CD4+ T cells, or macrophages. This observation suggested that A3F and/or A3DE antiviral activity did have an impact on the infectivity of the NL4-3 DRMR>A4 mutant in round 1. However, this reduction in infectivity was less than 2-fold and was not sufficient to produce a noticeable delay in replication kinetics in round 1. As expected, the NL4-3 YRHHY>A5 mutant virus from the peak time point exhibited a significant reduction in infectivity (4- to 10-fold), indicating that the antiviral activity of A3G had a substantial impact on viral infectivity, which resulted in a replication delay.
Fig 3.
Relative infectivities of NL4-3 WT, NL4-3 YRHHY>A5, and NL4-3 DRMR>A4 mutant viruses in CEM cells (A), CD4+ T cells (B), and macrophages (C). The infectivities of the peak viruses from round 1 were normalized for p24 CA, and equal amounts of p24 CA were used to infect TZM-bl cells. After 72 h, the infectivity, as determined by the luciferase activity, was measured. The average from 3 independent experiments is shown. Error bars represent the standard error of the mean. Statistical significance was determined by Student's t test (P < 0.0005).
vif mutant proviral DNA in infected CEM cells, CD4+ T cells, and macrophages exhibits G-to-A hypermutation.
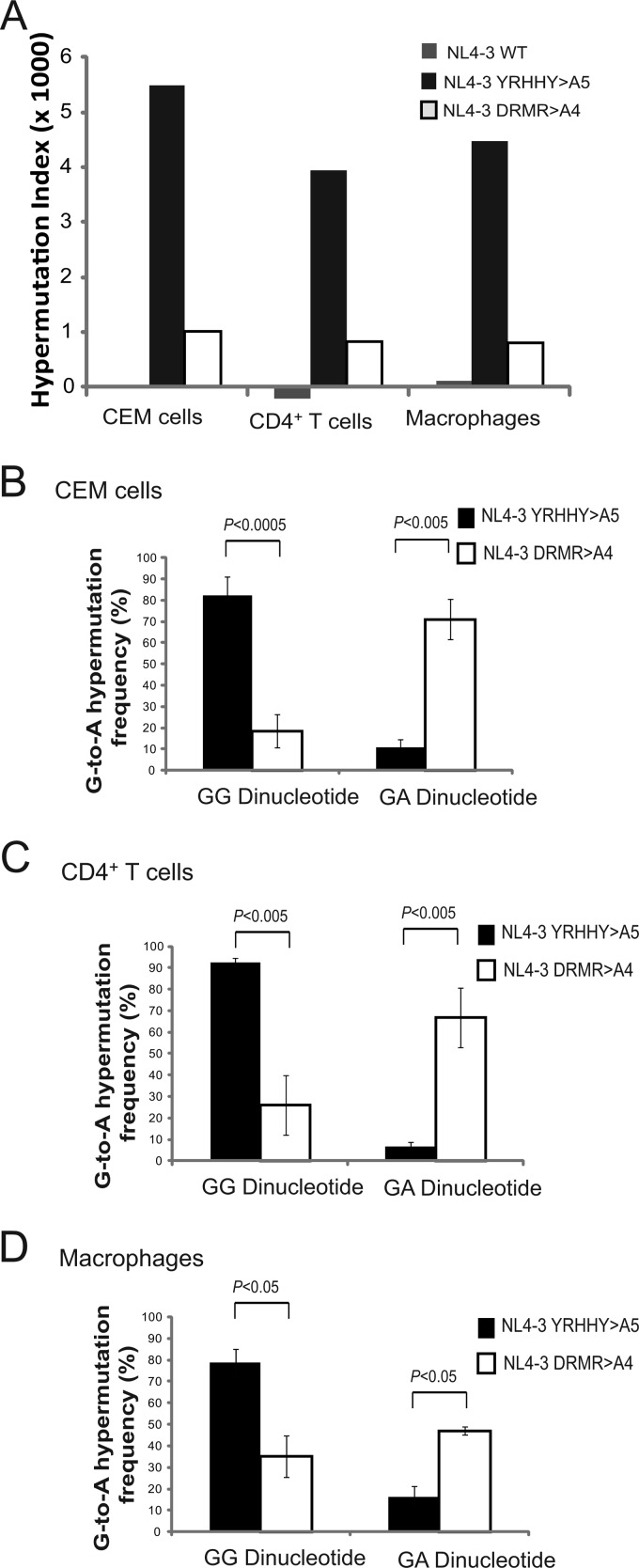
To determine whether inhibition of HIV-1 replication in primary CD4+ T cells and macrophages was associated with G-to-A hypermutation, we isolated DNA from cells infected with NL4-3 WT, NL4-3 YRHHY>A5, and NL4-3 DRMR>A4 from round 1 CEM cells and CD4+ T cells and round 2 CEM cells, CD4+ T cells, and macrophages. We then PCR amplified a portion of the viral genome and sequenced a 730-bp region containing the whole vif region and a portion of vpr and analyzed the sequences for evidence of G-to-A hypermutation as previously described (61). The hypermutation index (HI) was calculated as previously described (68) by subtracting the A-to-G mutations from the G-to-A mutations and dividing by the sequence length.
As summarized in Table 1, in round 1 infection, which included multiple rounds of viral replication in 15 days, the NL4-3 YRHHY>A5 mutant displayed a high HI (1.07 in CEM cells and 13.85 in CD4+ T cells) compared to that of NL4-3 WT (−0.2 in CEM cells and −0.7 in CD4+ T cells). The NL4-3 DRMR>A4 mutant also displayed an increased HI (0.27 in CEM cells and 1.51 in CD4+ T cells) compared to that of NL4-3 WT. Similarly, in round 2, the NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 mutants displayed significant increases in HI, while NL4-3 WT displayed no significant increase (Fig. 4A and Table 1). The very low HI for NL4-3 WT produced during multiple rounds of CEM or CD4+ T cell infection indicates that no A3 proteins that can overcome NL4-3 Vif to induce significant hypermutation are present. Our single-cycle infectivity experiments (Fig. 1) indicate that only A3H Hap II significantly inhibits replication of NL4-3 WT and therefore could potentially induce hypermutation. Interestingly, CEM2n cells were recently reported to be homozygous for A3H HapII (25), suggesting that our CEM cells may express this A3 protein. The absence of hypermutation in NL4-3 WT indicates that the A3H HapII levels present in CEM cells are not sufficient to induce significant hypermutation, which is essential for A3H Hap II to inhibit HIV-1 (10). Furthermore, the low HI for NL4-3 WT in CD4+ T cells and macrophages indicates that no A3 proteins are present at sufficient levels to overcome NL4-3 WT Vif to induce cytidine deamination and hypermutation.
Table 1.
Hypermutation index and dinucleotide context of G-to-A mutations
| Round | Cells | NL4-3 virus | No. of: |
Hypermutation index (%) × 1,000b | Dinucleotide context of G-to-A mutations (%c) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotides (sequences)a | A-to-G mutations | G-to-A mutations | GG | GA | GC | GT | ||||
| 1d | CEM | WT | 6,570 (9) | 2 | 1 | −0.2 | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| YRHHY>A5 | 6,570 (9) | 3 | 10 | 1.07 | 10 (100) | 0 (0) | 0 (0) | 0 (0) | ||
| DRMR>A4 | 7,300 (10) | 3 | 5 | 0.27 | 3 (60) | 2 (40) | 0 (0) | 0 (0) | ||
| CD4+ T | WT | 7,300 (10) | 6 | 1 | −0.7 | 1 (100) | 0 (0) | 0 (0) | 0 (0) | |
| YRHHY>A5 | 6,570 (9) | 7 | 98 | 13.85 | 77 (79) | 21 (21) | 0 (0) | 0 (0) | ||
| DRMR>A4 | 7,300 (10) | 6 | 17 | 1.51 | 2 (12) | 11 (65) | 3 (18) | 1 (6) | ||
| 2e | CEM | WT | 19,710 (27) | 2 | 0 | −0.1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| YRHHY>A5 | 10,950 (15) | 13 | 73 | 5.4 | 60 (82) | 8 (11) | 3 (4) | 1 (1) | ||
| DRMR>A4 | 22,630 (31) | 15 | 38 | 1.0 | 7 (18) | 27(71) | 1 (3) | 3(8) | ||
| CD4+ T | WT | 27,740 (38) | 9 | 3 | −0.2 | 1 (33) | 1 (33) | 1 (33) | 0 (0) | |
| YRHHY>A5 | 30,660 (42) | 25 | 146 | 3.9 | 135 (93) | 20 (7) | 0 (0) | 0 (0) | ||
| DRMR>A4 | 20,440 (28) | 10 | 27 | 0.83 | 7 (26) | 18 (67) | 2 (4) | 1 (4) | ||
| Monocyte-derived macrophages | WT | 17,520 (24) | 3 | 5 | 0.114 | 1 (20) | 2 (40) | 2 (40) | 0 (0) | |
| YRHHY>A5 | 25,550 (35) | 22 | 136 | 4.46 | 107 (79) | 22 (16) | 5 (4) | 2 (1) | ||
| DRMR>A4 | 12,410 (17) | 7 | 17 | 0.8 | 6 (35) | 8 (47) | 0 (0) | 2 (12) | ||
Total number of nucleotides sequenced from the total number of cloned PCR products sequenced.
Calculated as (G-to-A mutations) − (A-to-G mutations)/sequence length (bp) (68).
Percentage of mutated G nucleotides.
Round 1, replication period of 15 days.
Round 2, replication period of 27 days.
Fig 4.
Hypermutation in CEM cells, CD4+ T cells, and macrophages. Proviruses from round 1 and round 2 infections were isolated, and a 730-bp sequence containing vif and a portion of vpr were sequenced and analyzed for G-to-A hypermutation. (A) Hypermutation indices for round 2 infections of CEM cells, CD4+ T cells, and macrophages. The hypermutation index was calculated as G-to-A substitutions (bp) − A-to-G substitutions (bp)/sequence length (bp) in CEM cells, CD4+ T cells, and macrophages. (B to D) Dinucleotide contexts of G-to-A hypermutation in CEM cells (B), CD4+ T cells (C), and macrophages (D) for round 2 infections. The error bars represent the standard deviation. Statistical significance was determined by Student's t test (P < 0.0005 to 0.05).
The HIs for NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 apparently increased in CEM cells, and decreased in CD4+ T cells, when comparing round 1 to round 2 HIs. It is not clear whether these differences are biologically significant; we and others have noted that individual hypermutated proviral sequences can exhibit very large differences in the number of G-to-A changes, which can contribute to large differences in the HI. For example, 2 of the 9 NL4-3 YRHHY>A5 proviruses obtained from round 1 infections contained 53 of the 98 G-to-A substitutions, which greatly increased the HI for round 1 (data not shown). Finally, no hypermutation was observed in CEM-SS cells infected with either the NL4-3 YRHHY>A5 or the NL4-3 DRMR>A4 mutant virus (data not shown).
In the NL4-3 YRHHY>A5 proviruses, a high percentage of the G-to-A mutations occurred in the GG dinucleotide context (Table 1). In round 2, the percentages of G-to-A mutations that occurred in the GG dinucleotide context were 82% for CEM cells (Fig. 4B; Table 1), 93% for the CD4+ T cells (Fig. 4C; Table 1), and 79% for macrophages (Fig. 4D; Table 1). In contrast, in the NL4-3 DRMR>A4 proviruses (round 2) (Table 1), the majorities of the G-to-A mutations that occurred in the GA dinucleotides context were 71% in CEM cells (Fig. 4B; Table 1), 67% in CD4+ T cells (Fig. 4C; Table 1), and 47% in macrophages (Fig. 4D; Table 1). These results are consistent with A3G-mediated hypermutation of the NL4-3 YRHHY>A5 proviruses and A3F- and/or A3DE-mediated hypermutation of the NL4-3 DRMR>A4 proviruses.
It should be noted that in CD4+ T cells, 8/28 sequences for NL4-3 DRMR>A4 and 6/42 sequences for NL4-3 YRHHY>A5 mutants had no G-to-A hypermutation or other mutations. Very similar results were obtained for macrophages; 8/35 sequences for NL4-3 YRHHY>A5 and 7/17 sequences for NL4-3 DRMR>A4 displayed no mutations. The absence of hypermutation in some clones suggests that some viruses can escape from A3G, A3F, or A3DE activity, allowing replication of these mutant viruses. However, most clones that exhibit hypermutation are expected to have defects in expression of Vif and other viral proteins, which would result in a delay in virus growth in CD4+ T cells and macrophages. Overall, these results show the presence of A3G antiviral activity and, to a lesser extent, A3F and/or A3DE antiviral activity in CD4+ T cells and macrophages.
DISCUSSION
Among the human A3 proteins, A3B, A3C, A3DE, A3F, A3G, and A3H HapII have been shown to inhibit HIV-1 infectivity in single-cycle assays or in T cell lines (1–11, 13, 14, 32). However, it has not been established whether, and the extent to which, these A3 proteins restrict HIV-1 replication in its natural target cells of infection, specifically, activated CD4+ T cells and macrophages. Comparing the antiviral activities of A3 proteins in primary cells is challenging, since protein expression levels cannot be directly compared because of the use of different antibodies which bind with unknown affinities to different epitopes. A comparison of the mRNA levels of various A3 proteins in primary cells is informative, and it has been reported that human PBMCs express high levels of A3C and A3G but lower levels of A3F and A3DE (53, 54). However, it should be noted that the mRNA levels may not reflect the steady-state protein expression levels; furthermore, the antiviral activity of each A3 protein, which is a combination of its efficiency of virion incorporation, cytidine deaminase activity, and deaminase-independent antiviral activity, cannot be predicted on the basis of its expression levels. Therefore, we used NL4-3 vif mutants which specifically do not bind to A3G (NL4-3 YRHHY>A5) or A3F and A3DE (NL4-3 DRMR>A4) as tools to analyze the relative antiviral activities of these A3 proteins in primary HIV-1 target cells. In addition, our use of NL4-3 minimizes A3H as a contributing factor to observable differences between Vif mutants in our assays, as there are no distinct effects of A3H on infectivity with WT or Vif-deficient NL4-3 viruses. Thus, our results distinguish the effects on HIV replication of A3G and the combined antiviral activity of A3F and A3DE; these three A3 proteins are common in all human populations.
In round 1 infections, the NL4-3 YRHHY>A5 mutant exhibited a delay in replication kinetics, while the NL4-3 DRMR>A4 mutant replicated with the same kinetics as NL4-3 WT. These observations indicated that A3G restricted HIV-1 replication during a 2-week period in which multiple cycle of viral replication occurred, while the combined antiviral activity of A3F and A3DE did not have a measurable effect on viral replication kinetics during the same 2-week period. Despite the absence of any effect on replication kinetics, the virus produced at the peak time points of infection with NL4-3 DRMR>A4 mutants exhibited reduced infectivity in a single-cycle assay to about 50 to 80% of that of NL4-3 WT. Round 2 infections that were initiated with virus obtained from the peak time points did exhibit a delay in replication kinetics for both mutants, indicating that the combined A3F and A3DE antiviral activity had a cumulative effect that resulted in a delay in replication kinetics over a prolonged period. Since the NL4-3 YRHHY>A5 mutant exhibited a much greater delay in kinetics, we conclude that the antiviral activity of A3G is greater than the combined antiviral activity of A3F and A3DE.
Our studies clearly show that the cumulative antiviral activity of A3F and A3DE results in a delay in replication kinetics over a prolonged period of time, and they provide new insights into the relative contributions of A3G, A3F, and A3DE in the natural target cells of HIV-1 infection. Although our results agree with the reports of Mulder et al. (58) and Miyagi et al. (57), our conclusions differ because we analyzed viral replication over a much longer period of time than in these studies, leading us to clearly observe the combined antiviral activity of A3F and A3DE.
Our findings are consistent with the report from Mulder et al. (58) indicating that single-amino-acid substitution mutants of Vif that are defective in inducing degradation of A3F exhibit no delay in replication kinetics in primary PBMCs compared to the WT during a 2-week period (equivalent to round 1 in these studies). Our results also agree with the findings of Miyagi et al. (57), who observed that when A3F was expressed in HeLa cells at a level similar to that in H9 cells, no significant antiviral activity could be detected in a single round of replication. In contrast, our studies were performed with primary CD4+ T cells and macrophages over a prolonged period of time and more closely reflect natural HIV-1 infection. Furthermore, our studies clearly show that the cumulative antiviral activity of A3F and A3DE results in a delay in replication kinetics over a prolonged period of time.
Recent papers that quantitatively analyzed mRNA expression levels in several tissues have reported that A3G expression in lymphoid tissues is higher than A3F and A3DE expression (53, 54). In addition, Mulder et al. reported that protein expression levels in PBMCs for different donors suggest higher levels of A3G than of A3F expression and that there is also variation in A3G and A3F expression in different donors (58). The apparently higher A3G protein expression compared to A3F expression might be responsible for its apparently stronger antiviral activity in CD4+ T cells and macrophages, but the use of different antibodies precludes accurate quantification of protein expression. Furthermore, a comparison of the intrinsic antiviral activities of A3G and A3F tagged with the FLAG epitope and expressed at the same level suggested that A3G has a stronger antiviral activity than A3F (40). In addition, a lower antiviral effect of A3F has also been suggested in several publications (1, 45, 69). Thus, a combination of higher levels of expression as well as stronger intrinsic antiviral activity might be responsible for the greater restriction of HIV-1 in primary cells by A3G than by A3F and A3DE.
Our results showed that the NL4-3 YRHHY>A5 mutant proviruses displayed more G-to-A hypermutation than NL4-3 DRMR>A4 mutant proviruses, suggesting that A3G has a stronger cytidine deaminase activity than the combined activity of A3F and A3DE. While more G-to-A hypermutation in a GG dinucleotide context has been observed in a humanized mouse model (70), the dinucleotide context in which hypermutation occurs in HIV-1-infected human patients is controversial (68). While Kijak et al. (68) observed more hypermutations in a GG dinucleotide context, there are reports observing more hypermutations in a GA dinucleotide context (71, 72). Hypermutation in both GG and GA dinucleotide contexts in patients indicates that multiple A3 proteins contribute to HIV-1 restriction; however, the levels of hypermutation do not directly correlate with antiviral activity, since A3G induced more G-to-A hypermutations than A3F in a transient-transfection assay when comparable levels of inhibition were attained (40).
These studies do not directly address the extent to which A3B, A3C, A3H HapI, and A3H HapII restrict HIV-1 replication in primary CD4+ T cells and macrophages. However, the results of single-cycle assays in these studies as well as others imply that A3B, A3C, and A3H HapI possess little or no intrinsic antiviral activity against HIV-1. We observed statistically significant inhibition of all NL4-3 virus variants produced in the presence of A3H-HapII in single-cycle assays, which is in agreement with recent studies (10, 26). A3H HapII is more prevalent in African populations than in Caucasians, but it remains possible that it is expressed in the primary cells obtained from human donors used in these studies (24). Importantly for these studies, the sensitivity of the NL4-3 YRHHY>A5 and NL4-3 DRMR>A4 mutants to A3H HapII was not different from that of NL4-3 WT or NL4-3Δvif in single-cycle assays, and there was no significant hypermutation of NL4-3 WT in any of the cell types we tested. Therefore, even if A3H HapII was expressed in our human donor cells, it was unlikely to be expressed at levels that were sufficient to contribute to the delay in replication kinetics observed for these vif mutants.
A few publications have reported the antiviral activity of A3DE in a single-cycle experimental system and in T cell lines (8, 16), but no reports of activity in human primary cells have been published so far. Overall, our results suggest that A3G antiviral activity is much greater than the combined antiviral activity of A3F and A3DE, and they provide an experimental approach to deciphering the relative contributions of different A3 proteins to the restriction of HIV-1 in human primary target cells of infection.
ACKNOWLEDGMENTS
We thank Tobias Paprotka, Narasimhan J. Venkatachari, and Taisuke Izumi for valuable discussions during manuscript preparation.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print 24 October 2012
REFERENCES
- 1. Liddament MT, Brown WL, Schumacher AJ, Harris RS. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385–1391 [DOI] [PubMed] [Google Scholar]
- 2. Rose KM, Marin M, Kozak SL, Kabat D. 2005. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS Res. Hum. Retroviruses 21:611–619 [DOI] [PubMed] [Google Scholar]
- 3. Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 [DOI] [PubMed] [Google Scholar]
- 4. Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bourara K, Liegler TJ, Grant RM. 2007. Target cell APOBEC3C can induce limited G-to-A mutation in HIV-1. PLoS Pathog. 3:1477–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dang Y, Siew LM, Wang X, Han Y, Lampen R, Zheng YH. 2008. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J. Biol. Chem. 283:11606–11614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dang Y, Wang X, Esselman WJ, Zheng YH. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 80:10522–10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doehle BP, Schafer A, Cullen BR. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339:281–288 [DOI] [PubMed] [Google Scholar]
- 10. Harari A, Ooms M, Mulder LC, Simon V. 2009. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 83:295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31 [DOI] [PubMed] [Google Scholar]
- 12. Marin M, Rose KM, Kozak SL, Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398–1403 [DOI] [PubMed] [Google Scholar]
- 13. OhAinle M, Kerns JA, Malik HS, Emerman M. 2006. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J. Virol. 80:3853–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tan L, Sarkis PT, Wang T, Tian C, Yu XF. 2009. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 23:279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bogerd HP, Wiegand HL, Doehle BP, Cullen BR. 2007. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology 364:486–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. 2011. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J. Virol. 85:11220–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duggal NK, Malik HS, Emerman M. 2011. The breadth of antiviral activity of Apobec3DE in chimpanzees has been driven by positive selection. J. Virol. 85:11361–11371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hache G, Shindo K, Albin JS, Harris RS. 2008. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr. Biol. 18:819–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Itaya S, Nakajima T, Kaur G, Terunuma H, Ohtani H, Mehra N, Kimura A. 2010. No evidence of an association between the APOBEC3B deletion polymorphism and susceptibility to HIV infection and AIDS in Japanese and Indian populations. J. Infect. Dis. 202:815–816 (Author reply, 202:816–817.) [DOI] [PubMed] [Google Scholar]
- 20. Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379–53386 [DOI] [PubMed] [Google Scholar]
- 21. Albin JS, Harris RS. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev. Mol. Med. 12:e4 doi:10.1017/S1462399409001343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith JL, Bu W, Burdick RC, Pathak VK. 2009. Multiple ways of targeting APOBEC3-virion infectivity factor interactions for anti-HIV-1 drug development. Trends Pharmacol. Sci. 30:638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gourraud PA, Karaouni A, Woo JM, Schmidt T, Oksenberg JR, Hecht FM, Liegler TJ, Barbour JD. 2011. APOBEC3H haplotypes and HIV-1 pro-viral vif DNA sequence diversity in early untreated human immunodeficiency virus-1 infection. Hum. Immunol. 72:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. 2008. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Refsland EW, Hultquist JF, Harris RS. 2012. Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog. 8:e1002800 doi:10.1371/journal.ppat.1002800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Binka M, Ooms M, Steward M, Simon V. 2012. The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H. J. Virol. 86:49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Larue RS, Lengyel J, Jonsson SR, Andresdottir V, Harris RS. 2010. Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J. Virol. 84:8193–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. LI MM, Wu LI, Emerman M. 2010. The range of human APOBEC3H sensitivity to lentiviral Vif proteins. J. Virol. 84:88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392–1396 [DOI] [PubMed] [Google Scholar]
- 30. Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809 [DOI] [PubMed] [Google Scholar]
- 31. Lecossier D, Bouchonnet F, Clavel F, Hance AJ. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 32. Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103 [DOI] [PubMed] [Google Scholar]
- 33. Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792–7798 [DOI] [PubMed] [Google Scholar]
- 34. Stopak K, de Noronha C, Yonemoto W, Greene WC. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591–601 [DOI] [PubMed] [Google Scholar]
- 35. Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060 [DOI] [PubMed] [Google Scholar]
- 36. Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404–1407 [DOI] [PubMed] [Google Scholar]
- 37. Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, Ono A, Freed EO, Hu WS, Pathak VK. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822–35828 [DOI] [PubMed] [Google Scholar]
- 38. Luo K, Wang T, Liu B, Tian C, Xiao Z, Kappes J, Yu XF. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 81:7238–7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mbisa JL, Barr R, Thomas JA, Vandegraaff N, Dorweiler IJ, Svarovskaia ES, Brown WL, Mansky LM, Gorelick RJ, Harris RS, Engelman A, Pathak VK. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mbisa JL, Bu W, Pathak VK. 2010. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J. Virol. 84:5250–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166–170 [DOI] [PubMed] [Google Scholar]
- 42. He Z, Zhang W, Chen G, Xu R, Yu XF. 2008. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 381:1000–1011 [DOI] [PubMed] [Google Scholar]
- 43. Mehle A, Wilson H, Zhang C, Brazier AJ, McPike M, Pery E, Gabuzda D. 2007. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding. J. Virol. 81:13235–13241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Russell RA, Pathak VK. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81:8201–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1:e6 doi:10.1371/journal.ppat.0010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith JL, Pathak VK. 2010. Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J. Virol. 84:12599–12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767 [DOI] [PubMed] [Google Scholar]
- 48. Orenstein JM, Fox C, Wahl SM. 1997. Macrophages as a source of HIV during opportunistic infections. Science 276:1857–1861 [DOI] [PubMed] [Google Scholar]
- 49. Fisher AG, Ensoli B, Ivanoff L, Chamberlain M, Petteway S, Ratner L, Gallo RC, Wong-Staal F. 1987. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science 237:888–893 [DOI] [PubMed] [Google Scholar]
- 50. Gabuzda DH, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine WA, Sodroski J. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66:6489–6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gabuzda DH, Li H, Lawrence K, Vasir BS, Crawford K, Langhoff E. 1994. Essential role of vif in establishing productive HIV-1 infection in peripheral blood T lymphocytes and monocyte/macrophages. J. Acquir. Immune Defic. Syndr. 7:908–915 [PubMed] [Google Scholar]
- 52. Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin MA. 1987. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature 328:728–730 [DOI] [PubMed] [Google Scholar]
- 53. Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. 2009. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 83:9474–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 38:4274–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang FX, Huang J, Zhang H, Ma X, Zhang H. 2008. APOBEC3G upregulation by alpha interferon restricts human immunodeficiency virus type 1 infection in human peripheral plasmacytoid dendritic cells. J. Gen. Virol. 89:722–730 [DOI] [PubMed] [Google Scholar]
- 56. Ying S, Zhang X, Sarkis PT, Xu R, Yu X. 2007. Cell-specific regulation of APOBEC3F by interferons. Acta Biochim. Biophys. Sin. (Shanghai) 39:297–304 [DOI] [PubMed] [Google Scholar]
- 57. Miyagi E, Brown CR, Opi S, Khan M, Goila-Gaur R, Kao S, Walker RC, Jr, Hirsch V, Strebel K. 2010. Stably expressed APOBEC3F has negligible antiviral activity. J. Virol. 84:11067–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mulder LC, Ooms M, Majdak S, Smedresman J, Linscheid C, Harari A, Kunz A, Simon V. 2010. Moderate influence of human APOBEC3F on HIV-1 replication in primary lymphocytes. J. Virol. 84:9613–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Russell RA, Moore MD, Hu WS, Pathak VK. 2009. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA. Retrovirology 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Freed EO, Englund G, Martin MA. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69:3949–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kingston RE, Chen CA, Okayama H. 2001. Calcium phosphate transfection. Curr. Protoc. Immunol. Chapter 10:Unit 10.13 [DOI] [PubMed] [Google Scholar]
- 64. Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U. S. A. 92:7297–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Russell RA, Smith J, Barr R, Bhattacharyya D, Pathak VK. 2009. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J. Virol. 83:1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ooms M, Majdak S, Seibert CW, Harari A, Simon V. 2010. The localization of APOBEC3H variants in HIV-1 virions determines their antiviral activity. J. Virol. 84:7961–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhen A, Wang T, Zhao K, Xiong Y, Yu XF. 2010. A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. J. Virol. 84:1902–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kijak GH, Janini LM, Tovanabutra S, Sanders-Buell E, Arroyo MA, Robb ML, Michael NL, Birx DL, McCutchan FE. 2008. Variable contexts and levels of hypermutation in HIV-1 proviral genomes recovered from primary peripheral blood mononuclear cells. Virology 376:101–111 [DOI] [PubMed] [Google Scholar]
- 69. Zennou V, Bieniasz PD. 2006. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology 349:31–40 [DOI] [PubMed] [Google Scholar]
- 70. Sato K, Izumi T, Misawa N, Kobayashi T, Yamashita Y, Ohmichi M, Ito M, Takaori-Kondo A, Koyanagi Y. 2010. Remarkable lethal G-to-A mutations in vif-proficient HIV-1 provirus by individual APOBEC3 proteins in humanized mice. J. Virol. 84:9546–9556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Armitage AE, Katzourakis A, de Oliveira T, Welch JJ, Belshaw R, Bishop KN, Kramer B, McMichael AJ, Rambaut A, Iversen AKN. 2008. Conserved footprints of APOBEC3G on hypermutated human immunodeficiency virus type 1 and human endogenous retrovirus HERV-K(HML2) sequences. J. Virol. 82:8743–8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Janini M, Rogers M, Birx DR, McCutchan FE. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J. Virol. 75:7973–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]