Abstract
The replication timing of 9 genes commonly involved in cancer was investigated in the MCF10 cell lines for human breast cancer progression. Six of these nine genes are part of a constellation of tumor suppressor genes that play a major role in familial human breast cancer (TP53, ATM, PTEN, CHK2, BRCA1 and BRCA2). Three other genes are involved in a large number of human cancers including breast as either tumor suppressors (RB1 and RAD51) or as an oncogene (cMYC). Five of these nine genes (TP53, RAD51, ATM, PTEN and cMYC) show significant differences (p< 0.05) in replication timing between MCF10A normal human breast cells and the corresponding malignant MCF10CA1a cells. These differences are specific to the malignant state of the MCF10CA1a cells since there were no significant differences in the replication timing of these genes between normal MCF10A cells and the non-malignant cancer MCF10AT1 cells. Microarray analysis further demonstrated that three of these five genes (TP53, RAD51 and cMYC) showed significant changes in gene expression (≥ 2-fold) between normal and malignant cells. Our findings demonstrate an alteration in the replication timing of a small subset of cancer related genes in malignant breast cancer cells. These alterations partially correlate with the major transcriptional changes characteristic of the malignant state in these cells.
Keywords: REPLICATION TIMING, BREAST CANCER PROGRESSION, MCF10 CELL LINES, TP53, ATM, PTEN, CHK2, BRCA1, BRCA2, RB1, RAD51, PTEN, COL1A1, PPFIA2, ANO1, TCN1
INTRODUCTION
DNA replication and transcription have structural and temporal organization within the cell nucleus [Spector et al., 1993; Berezney 2002; Stein et al., 2003; Berezney et al., 2005; Cremer et al., 2006; Zaidi et al., 2007; Misteli, 2007; Lanctôt et al., 2007; Stein et al., 2008; Malyavantham et al., 2008a; Malyavantham et al.2008b; Malyvantham et al., 2010]. Regions within the nucleus are zoned for either transcription or replication during the S phase of the cell cycle [Wei et al., 1998; Berezney, 2002; Malyavantham et al., 2008b]. Moreover these regions within the genome are temporally organized so that genes which are highly active in transcription predominantly replicate earlier than those which are not [Schübeler et al., 2002; White et al., 2004; Woodfine et al., 2004]. For instance, housekeeping genes consistently replicate early in all cells [Zhang et al., 2002], while tissue specific genes replicate early only in their respective tissues [Groudine and Weintraub, 1981; Goldman et al., 1984; Hatton et al., 1988; Groudine et al., 1989; Kitsberg et al., 1993a; Hansen et al., 1996; Hansen et al., 2000; Hiratani et al., 2004] and the beta globin gene replicates earlier in correlation with transcriptional activity during erythropoiesis [Simon et al., 2001; Goren et al., 2008]. Such temporal organization occurs not only within single genes, but is correlated with the genome-wide expression profile maintained across evolution in Drosophila, mouse, and human cells [Schübeler et al., 2002; MacAlpine et al., 2004; White et al., 2004; Woodfine et al., 2004; Jeon et al., 2005; Karnani et al., 2007; Farkas h-Amar et al., 2008]. While no correlation of replication to transcriptional activity was identified in Saccharomyces cerevisiae [Raghuraman et al., 2001; Yabuki et al., 2002], the temporal organization of large gene clusters was coupled to the redox-state in synchronized budding yeast when expression levels were elevated [Klevecz et al., 2003].
An important role of higher order nuclear architecture in replication timing is indicated by numerous reports of the temporal arrangement of replication into mbp-sized replication zones [Selig et al., 1992; Kitsberg et al., 1993a; Simon and Cedar, 1996; White et al., 2004; Farkash-Amar, 2008; Göndör and Ohlsson, 2009]. Consistent with these findings, higher order chromatin domains of replication have been directly visualized using 3-D microscopy followed by computer imaging analysis [Wei et al., 1998; Wei et al.,1999; Berezney, 2002] and persist following the extraction of cells for nuclear matrix [Wei et al.,1999; Berezney, 2002]
Of fundamental importance is whether replication timing can be reorganized during cancer progression. Some studies have found that replication timing is asynchronous in multiple types of cancer [Amiel, 1998; Amiel, 1999; Korenstein-Ilan, 2002; Reish, 2003; Dotan et al., 2004]. Other studies have found late replication in diseases characterized by chromosome instability [Kuhn et al.,1987; Otto et al.,1981] and translocations [Breger et al., 2005].
In this study we used the MCF10 breast cancer progression model to investigate whether progression towards cancer involves changes in the replication timing of genes and the extent to which these changes are due to alterations in the transcriptional state of these genes and/or progression toward the malignant state. This model consists of three cell lines: (a) Normal ductal epithelium human breast cells termed MCF10A (10A) [Dawson et al., 1996]; (b) Premalignant transformed MCF10AT1 (10AT1) [Dawson et al., 1996]; and (c) Malignant MCF10CA1a (10CA1a) [Santner et al., 2001]. We investigated a total of thirteen genes. Six are included among the top ten genes for inherited breast cancer (p53, ATM, PTEN, CHK2, BRCA1 and BRCA2; [Walsh and King, 2007; Lee and Muller, 2010]). Three other selected genes are known to be involved in multiple cancers including breast cancer (RB1 [Jiang et al., 2011], RAD51 [Lose et al., 2005], and cMYC [Figueiredo et al., 2007; Worsham et al., 2006]. ANO1 and TCN1 were selected from our microarray analysis as being among those genes which showed large changes in transcription levels from 10A normal breast epithelial cells to the malignant 10CA1a cells [Marella et al, 2009]. Also included in the analysis as controls is the PPFAI2 gene, which was shown to replicate late in lymphoblastoid cells [Woodfine et al., 2004] and the COL1A1 collagen gene which is highly expressed in fibroblast cell lines but not in the breast epithelial cells used in this study. Our findings demonstrate replication timing alterations which are specific to the malignant state of the 10CA1a cells and partially correlate with transcriptional alterations.
RESULTS
SELECTION OF GENES
The replication timing of thirteen genes were studied across the MCF10 human breast cancer progression model cell lines. Of the 13 genes studied, 9 were chosen due to their relevance to cancer in human patients. Six of these nine genes are part of a constellation of tumor suppressor genes that play a major role in familial human breast cancer (TP53, ATM, PTEN, CHK2, BRCA1, BRCA2 [Walsh and King, 2007; Lee and Muller, 2010]. Three other genes are involved in a large number of human cancers including breast as either tumor suppressors (RB1 [Jiang et al., 2011] and RAD 51[Lose et al., 2006]) or as an oncogene (cMYC [Kishimoto et al., 2005]). In addition, PPFAI2 (a tyrosine phosphatase) was chosen as a gene that - based on previous studies [Woodfine et al., 2004] - replicated late in S phase. ANO1 and TCN1 were selected from our microarray analysis as being among those genes which showed large changes in transcription levels from 10A normal breast epithelial cells to the malignant 10CA1a cells [Marella et al., 2009]. Finally, COL1A1 is an alpha 1A1 collagen gene that is expressed at high levels in human fibroblast cells such as WI38, but is not expressed significantly in normal breast epithelial cells.
REPLICATION TIMING ACROSS THE MCF10 HUMAN BREAST CANCER MODEL
To estimate replication timing, we used the doublet/singlet method [Selig et al., 1992]. In this approach, genes which are not replicated in S-phase cells (BrdU positive cells) appear as one signal (singlets) (Figs. 1C, 1F, 1I) following FISH labeling with a probe complimentary to the gene of interest, while those that have replicated can be visualized as two closely aligned signals (doublets) (Figs. 1A, 1D,1G). Cells in which replication has occurred at one allele but not in the other appear as one singlet and one doublet (Figs. 1B, 1E, 1H)). This is termed asynchronous replication and typically is found in 10–15% of the population [Selig et al.,1992].
Fig.1. Singlet/Doublet Images.
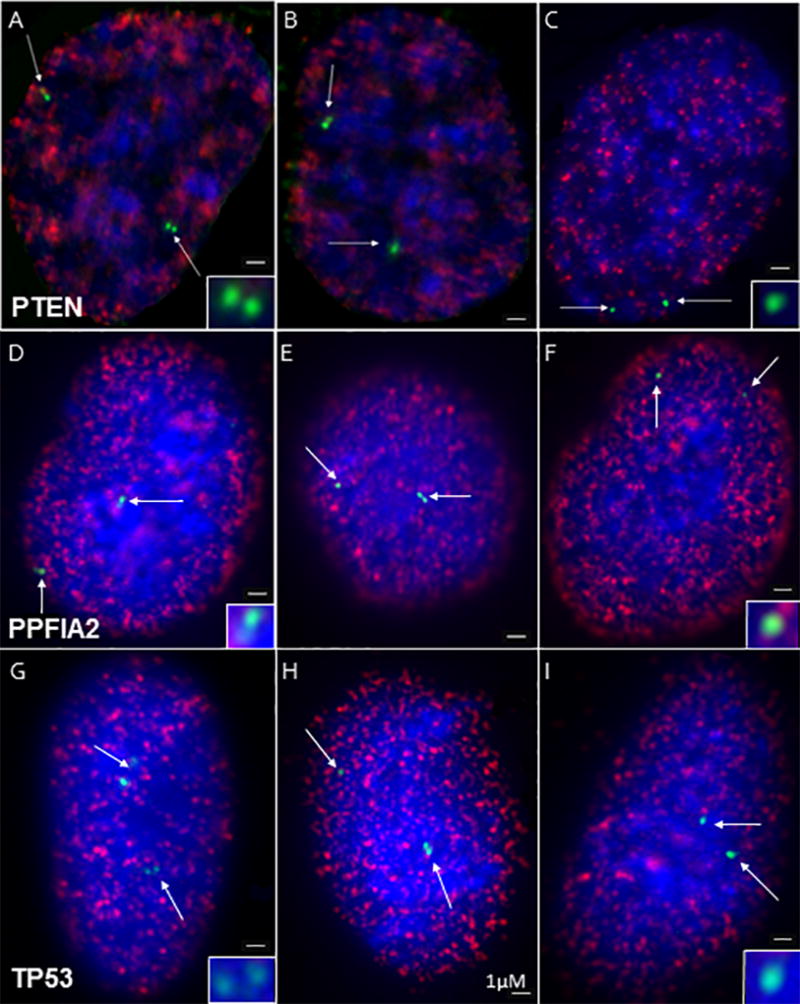
Representative FISH images of doublets (left row), singlet/ doublets (center row), and singlets (right row) are shown in BrdU positive cells (Blue = DAPI, red = BrdU, green = FISH signal) for the PTEN (A–C), PPF1A2 (D–F) and p53 (G–I) genes. Insets show FISH signals at higher magnification. Bars denotes 1 µM.
The time period that a gene replicates in S-phase (i.e., replication timing) is inversely related to the percent of doublets plus singlet/doublets in the population. Assuming that the S-phase period is 10 hours, a gene replicating in early S, e.g. hour 2, would show ~80% of doublets plus singlet/doublets. Conversely, a gene replicated in hour 7 of S would only show ~30% doublets plus singlet/doublets. The results of this analysis are summarized in Figure 2 for all 13 genes. 140 – 300 BrdU positive cells were counted for each gene in 3 separate experiments. Of the nine cancer related genes, five showed significant differences in replication timing (p< 0.05) between normal 10A and the malignant 10CA1a cell lines. These included three of the five inherited breast cancer related genes p53, ATM and PTEN as well as RAD51 and cMYC. The corresponding control studies for the ANO1 and TCN1 genes showed large and highly significant changes in replication timing (Fig. 2) that correlate with their changes in transcriptional levels in the normal versus malignant cell lines (Table I). As a second control, the PPFA12 gene, known to replicate in late S-phase in lymphoblastoid cells [Woodfine et al., 2004], also replicate in late S in all the human breast cell lines. Moreover, the COL1A1 collagen gene replicated very early in S phase in the WI38 human fibroblast cell line but later in mid S phase in the human breast cell lines (Fig. 2).
Fig. 2. Singlet doublet percentages for breast cancer related genes.

The distribution of cells in S phase are shown for each gene that have both alleles replicated (doublets, white), only one allele replicated (singlet/doublet, dark gray) and both alleles not replicated (singlets, light gray). Nine of these genes (TP53, RAD51, PTEN, cMYC, ATM, RB1, CHK2, BRCA1, and BRCA2) are known to be involved in breast cancer progression (A–I). Two of these genes (ANO1 and TCN1) were chosen from a microarray from among those genes which changed the most in expression from 10A to CA1a (J–K). PPFIA2 is a known late replicating gene (L) and COL1A1 is a highly expressed gene in fibroblasts that is compared between WI38 fibroblasts and the MCF10 lines (M). The number of determinants for each gene ranged from 140 to 300. Error bars represent SEM.
TABLE I.
Replication timing (RT) of genes is shown. Changes in RT and % differences between 10A and 10AT1 or 10CA1a demonstrate larger differences between 10A and 10CA1a compared to 10AT1. Differences in expression show an inverse relationship to RT. The first 9 genes are known to be involved in breast cancer progression, the next two are genes among the top changing in expression in MCF10, and the last two are controls for late replication (PPFIA2) and expression relating to replication timing in MCF10 versus WI38 (COL1A1).
| Average RT (h) | Change in RT (h) | % difference | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||
| Gene | 10A | 10AT1 | 10CA1a | 10A vs. 10AT1 |
10A vs. 10CA1a |
10A vs. 10AT1 |
10A vs. 10CA1a |
Expression fold change |
|
|
|
|
|
|
|
|
|
|
| TP53 | 2.6 | 2.7 | 4.5 | 0.1 | 1.9*** | 3.85 | 73.1 | −2.00 |
| RAD51 | 4.6 | 3.6 | 1.0 | −1.0 | −3.6*** | −21.7 | −78.3 | 2.00 |
| ATM | 2.1 | 1.4 | 1.1 | −0.7 | −1.0** | −33.3 | −47.6 | 1.03 |
| PTEN | 2.0 | 1.2 | 1.0 | −0.8 | −1.0** | −40.0 | −50.0 | 1.10 |
| cMYC | 1.9 | 1.0 | 1.0 | −0.9 | −0.9** | −47.4 | −47.4 | 3.00 |
| RB1 | 1.4 | 1.6 | 1.0 | 0.2 | −0.4 | 14.3 | −28.6 | −2.24 |
| CHK2 | 1.0 | 1.0 | 0.8 | 0.0 | −0.2 | 0.00 | −20.0 | 1.47 |
| BRCA1 | 2.3 | 3.0 | 2.6 | 0.7 | 0.3 | 30.4 | 13.0 | 1.10 |
| BRCA2 | 1.6 | 2.5 | 2.3 | 0.9* | 0.7 | 56.3 | 43.8 | 3.10 |
|
|
|
|
|
|
|
|
|
|
| ANO1 | 4.8 | 4.7 | 1.4 | −0.1 | −3.4*** | −2.08 | −70.8 | 81.0 |
| TCN1 | 2.6 | 2.7 | 4.8 | 0.1 | 2.2*** | 3.85 | 84.6 | −104 |
|
|
|
|
|
|
|
|
|
|
| PPFIA2 | 6.0 | 5.7 | 5.3 | −0.3 | −0.7 | −5.00 | −11.7 | 1.30 |
| COL1A1 | 4.0 | 4.5 | 4.0 | 0.5 | 0.0 | 12.5 | 0.00 | 1.02 |
chi squared test determines
p<.05,
p<.01,
p<.001
To express the average replication timing as the hours into S-phase, we determined the doubling time of each cell line as well as the percent of BrdU positive cells (S-phase cells). As shown in Figure 3A, the average percentages of cells in S phase in unsynchronized populations were: 27% (10A), 39% (10AT1), and 42% 10CA1a. From the growth curves of these cell lines (Fig. 3B), we estimated the doubling times as: 39, 24 and 23.4 h for 10A, 10AT1 and 10CA1, respectively. By multiplying the percent of BrdU positive cells by the doubling times for each cell line, we estimated the lengths of S phase as 10.5 h ± 0.7 SEM for 10A, 9.4 h ± 0.6 for 10AT1, and 9.8 h ± 0.7 for 10CA1a. Since the average S-period for each of these three cell lines were similar and averaged 9.9 hours, we estimated replication timing in hours based on a 10 hour S phase for all three cell lines. For this calculation, the percent of doublet plus singlet/doublets for each gene is multiplied by the 10 hour S phase period. This value is then subtracted from 10 h to give the average time for replication of doublets plus doublet/singlets (Fig. 4 and Table I).
Fig. 3. Cell cycle characterization of MCF10A lines.
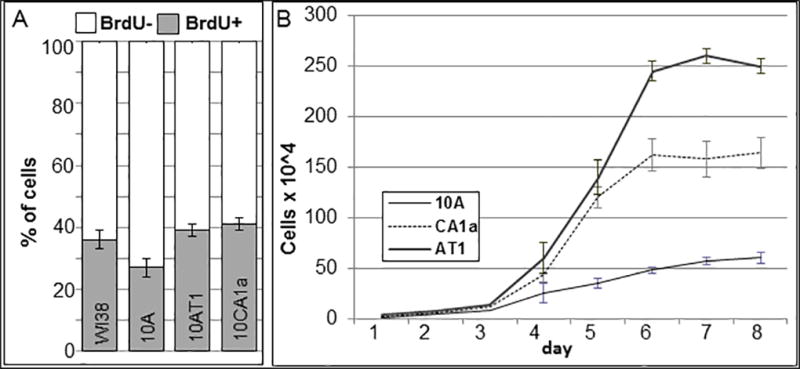
(A) Percentages of BrdU+/ BrdU− based on 300 cells ± SEM for each cell line. (B) Growth curves for each cell line. Average of 3 determinations ± SEM
Fig. 4. Average Replication Timing of Breast Cancer Related Genes.
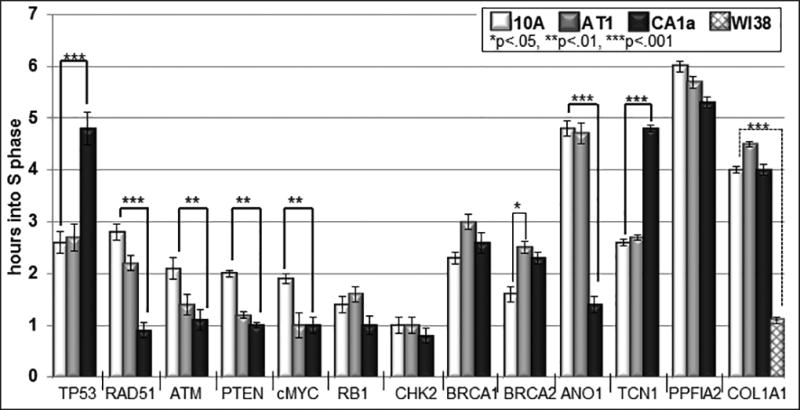
The average replication timing in hours was calculated based on a 10 hour S phase. The percentage of doublets + singlets/doublets was calculated for each gene shown in Figure 2, multipled by 10 and subtracted from 10 hours to estimate the average replication timing in hours. Based on the Chi squared test, 7 of 13 genes showed significant differences between 10A and 10CA1a (thick brackets), while only BRCA2 (thin bracket) was significantly different between 10A and 10AT1, and COL1A1 (dotted bracket) is statistically significant comparing MCF10 to WI38; *p<.05, **p<.01, ***p<.001.
Only one of the five cancer related genes that showed statistically significant differences (p ≤ 0.05) in replication timing, shifted to a later replication time in the S-phase of the malignant 10CA1a compared to the normal breast 10A cells (Fig. 4 and Table I). This was the tumor suppressor gene p53 which shifted from 2.6 h (early S) to 4.5 h (mid S). All the other genes shifted to earlier replication times including: RAD51 (4.6 to 1.0 h), ATM (2.1 to 1.0 h), PTEN (2.0 to 1.0 h), and cMYC (1.9 to 1.0 h). The RB1, CHK2, BRCA1 and BRCA2 genes also showed differences in replication timing (Fig. 4, Table I), but these differences were not statistically significant. ANO1 and TCN1 showed very significant changes in replication timing from 4.8 h (mid S) and 2.6 h (early S) to 1.4 h (early S) and 4.8 h (mid S), respectively. The collagen gene specific for expression in fibroblasts, replicated early in WI38 fibroblasts (1.3 h) but much later with no significant differences in the MCF10 cell lines (4–4.5 h in S-phase; Fig. 4 and Table I).
In contrast to these major differences in replication timing between normal breast epithelial 10A cells and their malignant counterpart 10CA1a, no significant differences were detected between normal cells and the corresponding non-malignant transformed 10AT1 cells in this subset of five cancer related genes. The only significant difference (p= 0.04) was in the BRCA2 gene where the replication timing increased from 1.6 to 2.5 hours in the 10AT1 cells. Moreover, the 10CA1a cells differed from their non-malignant transformed counterpart 10AT1 in the replication timing of two of the five cancer genes that are altered in the malignant versus normal breast cells. These were the tumor suppressor gene p53 and the gene for the DNA repair protein RAD51.
Correlating Gene Expression With Replication Timing Across Breast Cancer Progression
Numerous studies have demonstrated that chromatin replicated in early S is enriched in actively transcribed genes, while late S replicating chromatin contains much lower levels of transcribed genes [Simon et al., 2001; Schübeler et al., 2002; Zhang et al., 2002; White et al., 2004; Woodfine et al., 2004; Goren et al., 2008]. With this as a basis, we determined whether the genes which changed their replication timing in the malignant state also displayed changes in expression. Gene expression profiles between the 10A and 10CA1a cells were previously performed using a dye swap based microarray approach [Marella et al., 2009].
As shown in Table I, the two genes selected for their large changes in gene expression in the malignant 10CA1a cells, ANO1 (81 fold up-regulated) and TCN1 (104 fold down-regulated), showed correspondingly large changes in replication timing with the up-regulated ANO1 shifting to a lower replication timing from mid S (4.8 h) to early S (1.4 h), and the down-regulated TCN1 increasing from early S (2.6 h) to mid S (4.8 h). The collagen gene COL1A1 showed no change in gene expression in the malignant 10CA1a compared to the normal 10A cells and correspondingly no change in expression levels, while PPFA12 showed a decrease in replication timing albeit not statistically significant from 6.0 to 5.3 h and a slight increase in transcription level (1.3 fold).
Among the 5 cancer related genes that show significant differences in replication timing between normal and malignant breast cells, three showed corresponding changes in gene expression that correlate with the replication timing changes (2-fold decrease in p53, 3-fold increase in RAD51 and a 3-fold increase in cMYC). In the other two genes that showed significant shifts in replication timing (PTEN and ATM), there were no significant changes in gene expression levels (Table I).
DISCUSSION
REPLICATION TIMING, TRANSCRIPTION AND CHROMATIN ORGANIZATION
Earlier investigations demonstrated that housekeeping, cell and tissue-specific, and developmentally regulated genes replicate earlier upon active expression [Goldman et al., 1984; Hatton et al., 1988; Hansen et al., 1996; Hansen et al., 2000; Groudine et al., 1989; Groudine and Weintraub, 1981; Kitsberg et al., 1993a; Hiratani et al., 2004]. In contrast, silenced genes, such as those within the inactive X chromosome, yeast silenced mating type genes [Fangman and Brewer, 1992; Raghuraman et al., 1997] and genes transfected into late S phase cells [Zhang et al., 2002] typically replicate later. Few studies, however, have identified a change in replication timing prior to changes in transcription [Cimbora et al., 2000; Zhou et al., 2002]. Hiratani et al. (2004), demonstrated that replication timing can change during the differentiation of neurons, and that this change in replication timing was confined to GC poor regions of the genome. Within the genes which were GC low and that also changed transcription, more than half demonstrated changed in replication timing [Hiratani et al., 2004].
While a relationship between replication timing and gene expression was found using high-throughput studies in higher metazoans such as Drosophila melanogaster [Schübeler et al., 2002; MacAlpine et al., 2004] and in human cells [Woodfine et al., 2004; White et al., 2004] it was not in yeast [Raghuraman et al., 2001; Yabuki et al., 2002]. This coordination of replication timing has been suggested to be due to the need for replication to occur faithfully once and only once during S phase. Replication is therefore a synchronous process in that both copies of a given allele will replicate at approximately the same time during S phase. This is, however, not the case for genes which display monoallelic expression due to X inactivation [Ensminger and Chess 2004] or imprinting [Kitsberg et al., 1993b] which replicate asynchronously.
Other genomic features have also been correlated with replication timing. At the sequence level, replication timing has been shown to be influenced by the cis-elements within the DNA (e.g., GC%, LINEs, telomeric DNA, [Hiratani et al., 2004]). Regions which are either high in GC content, gene density, and/or low in LINE density generally replicate early [Woodfine et al., 2004]. Still other factors at the DNA level can influence replication timing. Proximity to telomeres influences genes to replicate late [Stevenson and Gottschling, 1999; Ofir et al., 1999]. Furthermore, the length of the DNA of interest was determined to be important since replication timing varies from the 5’ side to 3’ side of extremely large genes [Watanabe et al., 2008]. The tendency for highly active genes to replicate early may be independent of any effect that these cis- elements have on replication timing. In contrast, other studies indicate that a change in replication timing does not occur in regions that are below 41% GC (Hiratani et al., 2008).
At a higher level of organization, replication timing has been related to location with respect to the nuclear periphery [Zhou et al., 2002] which is greater in heterochromatin content than the nuclear interior [Towbin et al., 2012]. This is coincident with the finding that early replicating genes are more internal and late replicating genes have a preference for the nuclear periphery [Grasser et al., 2008]. Moreover, these patterns were maintained across evolution [Grasser et al., 2008; Ryba et al., 2010] and in cancer cells [Grasser et al., 2008]. It has also been demonstrated that the nuclear localization of some genes change during the progression of human breast cancer [Meaburn et al., 2009]. While comparing normal breast cells to cancerous cells, a subset of genes were found more peripheral upon carcinogenesis [Meaburn et al., 2009]. In particular HES1 was found more peripheral in all breast cancer samples.
Recent studies of replication timing across the genome has led to the emerging concept that replication timing is an epigenetic property of the genome which is more closely related to the transcriptional poised state of chromatin rather than transcription per se [Barton and Crowe, 2001; Goren et al., 2008; Schwaiger et al., 2009]. It is proposed that replication of genes in early S phase promotes an “open state” of the chromatin and facilitates transcription while replication in late S phase leads to a “closed state” of chromatin and gene silencing [Aran et al., 2011]. In this view, DNA replication may function as a global modifier of chromatin structure and transcriptional potential and provide the basis for chromatin to alter its transcriptional state [Barton and Crowe, 2001; Goren et al., 2008]. It is further proposed that replication timing is both a cause and consequence of chromatin structure by providing a means to inherit chromatin states that in turn regulate replication timing in subsequent cell generations [McNairn and Gilbert, 2003; Lucas and Feng, 2003; Esteller et al., 2008; Hanson et al., 2010; Ryba et al., 2011].
The findings that the temporal patterns of replication timing at the genome level are arranged in a series of mbp–sized replication zones [Selig et al., 1992; Kitsberg et al., 1993a; Simon and Cedar, 1996; White et al., 2004; Farkash-Amar, 2008; Göndör and Ohlsson, 2009] provides a basis to study this regulation at the level of higher order nuclear architecture. For example, these replication zones have been directly visualized and persist following extraction of cells for nuclear matrix architecture [Wei et al., 1999; Ma et al.,1999; Berezney, 2002].
REPLICATION TIMING, CHROMOSOMAL ABERRATIONS AND CANCER
It is fundamentally important to determine whether replication timing can be altered in cancer cells and the possible role of multiple chromosomal aberrations that are characteristic of cancer cells in mediating these alterations. Previous results indicated that replication becomes asynchronous under conditions where there are extra autosomes [Amiel et al., 1999]. The replication timing of RB1, TP53, cMYC, and Her2 are asynchronous in tissues of patients suffering from autosomal aneuploidy such as Down’s syndrome [Amiel et al., 1997; Amiel et al., 1998] or in patients suffering from lymphoma [Amiel et al., 1997]. Changes in replication timing were attributed either to monoallelic expression or loss of replication control. It was further found that genes that were involved in X chromosome aneuploidy had two alleles which replicate synchronously while a third allele replicated later [Ensminger and Chess, 2004]. Identical results were found for aneuploid autosomes. In our experiments, however, we did not observe this as the two genes that were investigated on aneuploid chromosomes (cMYC on chromosome 8 and PTEN on chromosome10) replicated synchronously (Fig. 2C–D) in the various MCF10 cell lines.
Cohen et al. (2007) found that genes involved in the apoptosis pathway, which is important in cancer progression, replicate early. Consistent with these findings many of the genes we studied are implicated in apoptosis and replicated early. Recent results suggest that replication timing of several genes including those of some cell cycle and apoptosis genes are altered in colon carcinoma cells that are p53−/− compared with wild type [Watanabe et al., 2007]. These authors attributed this change to the effects of p53 on replication. P53, however, could alter the expression of many genes and thus alter their replication timing. Other studies have determined that cis-elements have an effect on replication timing in cancer. Watanabe et al. (2009) showed that several genes on chromosomes 11q and 21q that are involved in cancer progression, were located near regions that switch in sequence from high to low GC content. These regions have high genomic instability and vary widely in replication timing (the high GC content regions replicated early whereas the low GC content regions replicated later [Watanabe et al., 2009]). Furthermore, the timing of replication is related to heterochromatin in diseased cells. For example, the Epstein-Barr virus genome replicates late after it has incorporated into the host genome’s heterochromatin [Zhou et al., 2009].
REPLICATION TIMING ALTERATIONS AND GENE EXPRESSION IN THE MCF10 HUMAN BREAST CANCER CELL LINES
It is well known that transcriptional profiles are altered as cells progress towards malignancy including breast cancer [Marella et al., 2009; Jones et al., 2012; Kuo et al., 2012; Need et al., 2012]. A fundamental question is whether progression towards a cancerous state results in replication timing changes in genes and if this can be related to the transcriptional status. Our previous microarray expression analysis revealed significant changes (≥ 2-fold) in the transcriptional levels of ~17% of the genes in the CA1a cells compared to the normal 10A human breast cells [Marella et al., 2009]. This prompted us to examine the replication timing of cancer related genes in the MCF10 cell culture breast cancer model.
Our investigation demonstrated alterations in the replication timing of a subset of genes commonly involved in cancer. Of the nine cancer related genes analyzed, five showed significant changes (p ≤ 0.05) in replication timing in the malignant 10CA1a cells compared to their counterpart 10A normal human breast cells. These alterations are specific to the malignant 10CA1a cells since the corresponding non-malignant 10AT1 breast cancer cells, which proliferate at a nearly identical rate as the CA1a cells, had no significant changes in replication timing for this subset of 5 genes. The only significant difference in replication timing between the non-malignant AT and normal 10A cells was in the BRCA2 gene. While the malignant 10CA1a cells also showed an alteration in replication timing of the BRCA2 gene (Table I), this change was not statistically significant (p= 0.24).
Of the five cancer related genes that showed changes in replication timing in the CA1a cells, three showed corresponding changes in transcriptional levels with p53 decreasing in transcription with increasing replication timing (2.7 to 4.5 hours, Table I) and the RAD51 and cMYC genes decreasing in replication timing (4.6 to 1.0 hours and 1.9 to 1.0 hours, respectively, Table 1) with increasing transcriptional levels. Despite significant decreases in the replication timing of the ATM and PTEN, however, there was no corresponding increase in the transcriptional levels of these genes (Table I). Moreover, BRCA2 and RB1 changed in expression levels, but did not change significantly in their replication timing. One explanation of these findings is that these genes may be located in higher order replication zones where the transcriptional level, chromatin state or DNA sequences may dominant in controlling the replication timing [Selig et al., 1992; Kitsberg et al., 1993a; Simon and Cedar, 1996; White et al., 2004; Farkash-Amar, 2008; Göndör and Ohlsson, 2009].
In conclusion, our findings demonstrate changes in the replication timing of several cancer related genes in the MCF10 human breast cancer model cell lines. While changes in transcriptional levels of genes in cells progressing into malignancy may be one factor mediating these alterations in replication timing, other factors are likely involved. We propose that progression towards the malignant state of the genome may involve alterations in global epigenetic properties of chromatin such as the transcriptionally poised state and/or higher order replication zoning.
MATERIALS AND METHODS
CELL CULTURE
MCF10A, MCF10AT1 and MCF10CA1a cells were obtained from the Barbara Ann Karamanos Cancer Institute, Detroit, Michigan. MCF10A and MCF10AT1 were grown in DMEM/F-10 media supplemented with 5% horse serum, 2% insulin, EGF, hydrocortisone, cholera enterotoxin, and 1% penicillin/streptomycin. MCF10CA1a was cultured in DEME/F-10 media with 5% horse serum and 1% penicillin/streptomycin. All cell lines were grown at 37C in a 5% CO2 incubator.
DNA FISH (FLUORESCENCE IN-SITU HYBRIDIZATION) AND IMMUNOFLUORESCENCE
Cells were grown on coverslips and pulsed with BrdU (20 uM) for 30 min. Hypotonic treatment to swell the cells was performed by incubating the cells in O.075M KCl for 10 min. Cells were fixed with (3:1) methanol:acetic acid at −20°C overnight. Coverslips were stored in 70% ethanol at 4°C for several days and transferred to 100% ethanol for >1 min prior to denaturation. Denaturation of cells was performed at 75° C for 30 to 90 s in 50% formamide/ 2XSSC. BAC probes that span the genes were obtained from Health Research Incorporated at Roswell Park Cancer Institute, labeled with biotin with a biotin nick translation kit (Invitrogen, Carlsbad, CA) and denatured for 10 minutes at 75 °C. The cells were then hybridized with the probe overnight followed by three post hybridization washes of 30 min each (wash I: 50% formamide in 2XSSC & 0.05% Tween; wash II: 2XSSC with 0.05% Tween; and wash III:1XSSC). Coverslips were then immunolabeled with anti-biotin rabbit (1:100) and anti-BrdU mouse (1:100) antibodies for 45 min followed by incubation with anti-rabbit-alexa 594 (1:100, Molecular Probes) and anti-mouse 488 (1:100, Molecular Probes) for 45 min. DAPI was used to visualize the nuclei. Cells were mounted in prolong gold or vectashield/ DAPI (1:2000, Vecta Laboratories) and visualized with fluorescence microscopy.
STATISTICS
SEMs were calculated using Microsoft excel’s STDEV function and dividing by the square root of n. P-values were calculated using chi squared analysis using Microsoft excel’s CHITEST function.
MICROSCOPY IMAGE ANALYSIS
Images were acquired with an Olympus BX51 upright microscope (100 X plan-apo, oil,1.4 NA) equipped with a Sensicam QE (Cooke Corporation, USA) digital charge-coupled device (CCD) camera, motorized z-axis controller (Prior) and Slidebook 4.0 software (Intelligent Imaging Innovations, Denver, CO). Optical sections were collected at 0.5 µm intervals through the z-axis. Nearest neighbor deconvolution was performed using Slidebook 4.0.
Acknowledgments
These studies were supported by National Institutes of Health grant GM 072131 awarded to R.B.
References
- Amiel A, Avivi L, Gaber E, Fejgin MD. Asynchronous replication of allelic loci in Down syndrome. Eur J Hum Genet. 1998;6(4):359–64. doi: 10.1038/sj.ejhg.5200199. [DOI] [PubMed] [Google Scholar]
- Amiel A, Korenstein A, Gaber E, Avivi L. Asynchronous replication of alleles in genomes carrying an extra autosome. Eur J Hum Genet. 1999;7(2):223–30. doi: 10.1038/sj.ejhg.5200267. [DOI] [PubMed] [Google Scholar]
- Amiel A, Litmanovich T, Gaber E, Lishner M, Avivi L, Fejgin MD. Asynchronous replication of p53 and 21q22 loci in chronic lymphocytic leukemia. Hum Genet. 1997;101(2):219–22. doi: 10.1007/s004390050619. [DOI] [PubMed] [Google Scholar]
- Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet. 2011;20(4):670–80. doi: 10.1093/hmg/ddq513. [DOI] [PubMed] [Google Scholar]
- Barton MC, Crowe AJ. Chromatin alteration, transcription and replication. What’s the opening line to the story? Oncogene. 2001;20(24):3094–9. doi: 10.1038/sj.onc.1204334. [DOI] [PubMed] [Google Scholar]
- Berezney R. Regulating the mammalian genome: the role of nuclear architecture. Adv Enzyme Regul. 2002;42:39–52. doi: 10.1016/s0065-2571(01)00041-3. [DOI] [PubMed] [Google Scholar]
- Berezney R, Malyavantham KS, Pliss A, Bhattacharya S, Acharya R. Spatio-temporal dynamics of genomic organization and function in the mammalian cell nucleus. Adv Enzyme Regul. 2005;45:17–26. doi: 10.1016/j.advenzreg.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Breger KS, Smith L, Thayer MJ. Engineering translocations with delayed replication: evidence for cis control of chromosome replication timing. Hum Mol Genet. 2005;14:2813–27. doi: 10.1093/hmg/ddi314. [DOI] [PubMed] [Google Scholar]
- Cimbora DM, Schübeler D, Reik A, Hamilton J, Francastel C, Epner EM, Groudine M. Long-distance control of origin choice and replication timing in the human beta-globin locus are independent of the locus control region. Mol Cell Biol. 2000;20(15):5581–91. doi: 10.1128/mcb.20.15.5581-5591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Cordeiro-Stone M, Kaufman DG. Early replication and the apoptotic pathway. J Cell Physiol. 2007;213(2):434–9. doi: 10.1002/jcp.21156. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S. Chromosome territories-a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol. 1996;148(1):313–9. [PMC free article] [PubMed] [Google Scholar]
- Dotan ZA, Dotan A, Ramon J, Avivi L. Altered mode of allelic replication accompanied by aneuploidy in peripheral blood lymphocytes of prostate cancer patients. Int J Cancer. 2004;111(1):60–6. doi: 10.1002/ijc.20237. [DOI] [PubMed] [Google Scholar]
- Ensminger AW, Chess A. Coordinated replication timing of monoallelically expressed genes along human autosomes. Hum Mol Genet. 2004;13(6):651–8. doi: 10.1093/hmg/ddh062. [DOI] [PubMed] [Google Scholar]
- Esteller M. Molecular Origins of Cancer, Epigenetics in Cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fangman WL, Brewer BJ. A question of time: replication origins of eukaryotic chromosomes. Cell. 1992;71(3):363–6. doi: 10.1016/0092-8674(92)90505-7. [DOI] [PubMed] [Google Scholar]
- Farkash-Amar S, Lipson D, Polten A, Goren A, Helmstetter C, Yakhini Z, Simon I. Global organization of replication time zones of the mouse genome. Genome Res. 2008;18(10):1562–70. doi: 10.1101/gr.079566.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo JC, Knight JA, Cho S, Savas S, Onay UV, Briollais L, Goodwin PJ, McLaughlin JR, Andrulis IL, Ozcelik H. Polymorphisms cMyc-N11S and p27-V109G and breast cancer risk and prognosis. BMC Cancer. 2007;7:99. doi: 10.1186/1471-2407-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MA, Holmquist GP, Gray MC, Caston LA, Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Göndör A, Ohlsson R. Replication timing and epigenetic reprogramming of gene expression: a two-way relationship? Nat Rev Genet. 2009;10(4):269–76. doi: 10.1038/nrg2555. [DOI] [PubMed] [Google Scholar]
- Goren A, Tabib A, Hecht M, Cedar H. DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes Dev. 2008;22(10):1319–24. doi: 10.1101/gad.468308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser F, Neusser M, Fiegler H, Thormeyer T, Cremer M, Carter NP, Cremer T, Müller S. Replication-timing-correlated spatial chromatin arrangements in cancer and in primate interphase nuclei. J Cell Sci. 2008;121(11):1876–86. doi: 10.1242/jcs.026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M, Weintraub H. Activation of globin genes during chicken development. Cell. 1981;24(2):393–401. doi: 10.1016/0092-8674(81)90329-9. [DOI] [PubMed] [Google Scholar]
- Groudine M, Forrester WC, Novak U, Epner E. Replication and activation of the human beta-globin gene domain. Prog Clin Biol Res. 1989;316A:15–35. [PubMed] [Google Scholar]
- Hatton KS, Dhar V, Brown EH, Iqbal MA, Stuart S, Didamo VT, Schildkraut CL. Replication program of active and inactive multigene families in mammalian cells. Mol Cell Biol. 1988;8:2149–2158. doi: 10.1128/mcb.8.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Canfield TK, Fjeld AD, Gartler SM. Role of late replication timing in the silencing of X-linked genes. Hum Mol Genet. 1996;5:1345–1353. doi: 10.1093/hmg/5.9.1345. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Stöger R, Wijmenga C, Stanek AM, Canfield TK, Luo P, Matarazzo MR, D'Esposito M, Feil R, Gimelli G, Weemaes CM, Laird CD, Gartler SM. Escape from gene silencing in ICF syndrome: Evidence for advanced replication time as a major determinant. Hum Mol Genet. 2000;9:2575–2587. doi: 10.1093/hmg/9.18.2575. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. PNAS. 2010;107(1):139–44. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Leskovar A, Gilbert DM. Differentiation-induced replication-timing changes are restricted to AT-rich/long interspersed nuclear element (LINE)-rich isochores. PNAS. 2004;101(48):16861–6. doi: 10.1073/pnas.0406687101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schübeler D, Gilbert DM. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Bekiranov S, Karnani N, Kapranov P, Ghosh S, MacAlpine D, Lee C, Hwang DS, Gingeras TR, Dutta A. Temporal profile of replication of human chromosomes. PNAS. 2005;102(18):6419–24. doi: 10.1073/pnas.0405088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Lechertier T, Mitter R, Herbert JM, Bicknell R, Jones JL, Li JL, Buffa F, Harris AL, Hodivala-Dilke K. Gene expression analysis in human breast cancer associated blood vessels. PLoS One. 2012;7(10):e44294. doi: 10.1371/journal.pone.0044294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Jones R, Liu JC, Deng T, Robinson T, Chung PE, Wang S, Herschkowitz JI, Egan SE, Perou CM, Zacksenhaus E. RB1 and p53 at the crossroad of EMT and triple-negative breast cancer. Cell Cycle. 2011;10(10):1563–70. doi: 10.4161/cc.10.10.15703. [DOI] [PubMed] [Google Scholar]
- Karnani N, Taylor C, Malhotra A, Dutta A. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res. 2007;17(6):865–76. doi: 10.1101/gr.5427007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto H, Wang Z, Bhat-Nakshatri P, Chang D, Clarke R, Nakshatri H. The p160 family coactivators regulate breast cancer cell proliferation and invasion through autocrine/paracrine activity of SDF-1alpha/CXCL12. Carcinogenesis. 2005;26(10):1706–15. doi: 10.1093/carcin/bgi137. [DOI] [PubMed] [Google Scholar]
- Klevecz RR, Bolen J, Forrest G, Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. PNAS. 2004;101(5):1200–5. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenstein-Ilan A, Amiel A, Lalezari S, Lishner M, Avivi L. Allele-specific replication associated with aneuploidy in blood cells of patients with hematologic malignancies. Cancer Genet Cytogenet. 2002;139(2):97–103. doi: 10.1016/s0165-4608(02)00610-6. [DOI] [PubMed] [Google Scholar]
- Kitsberg D, Selig S, Keshet I, Cedar H. Replication structure of the human beta-globin gene domain. Nature. 1993a;366(6455):588–90. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- Kitsberg D, Selig S, Brandeis M, Simon I, Keshet I, Driscoll DJ, Nicholls RD, Cedar H. Allele-specific replication timing of imprinted gene regions. Nature. 1993b;364(6436):459–63. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- Kuhn EM, Therman E, Buchler DA. Do individual allocyclic chromosomes in metaphase reflect their interphase domains? Hum Genet. 1987;77:210–3. doi: 10.1007/BF00284471. [DOI] [PubMed] [Google Scholar]
- Kuo WH, Chang YY, Lai LC, Tsai MH, Hsiao CK, Chang KJ, Chuang EY. Molecular characteristics and metastasis predictor genes of triple-negative breast cancer: a clinical study of triple-negative breast carcinomas. PLoS One. 2012;7(9):e45831. doi: 10.1371/journal.pone.0045831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Lee E, Muller W. Breast Cancer Oncogenes and Tumor Suppressor Genes. Cold Spring Harb Perspect Biol. 2010;2(10):a003236. doi: 10.1101/cshperspect.a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lose F, Lovelock P, Chenevix-Trench G, Mann GJ, Pupo GM, Spurdle AB the Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer Research. Variation in the RAD51 gene and familial breast cancer. Breast Cancer Res. 2006;8(3):R26. doi: 10.1186/bcr1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas I, Feng W. Spotlight on DNA replication, the essence of replication timing, determinants and significance. Cell Cycle. 2003;2(6):560–563. [PubMed] [Google Scholar]
- Ma H, Siegel AJ, Berezney R. Association of chromosome territories with the nuclear matrix. Disruption of human chromosome territories correlates with the release of a subset of nuclear matrix proteins. J Cell Biol. 1999;146(3):531–42. doi: 10.1083/jcb.146.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Rodríguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18(24):3094–105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyavantham KS, Bhattacharya S, Barbeitos M, Mukherjee L, Xu J, Fackelmayer FO, Berezney R. Identifying functional neighborhoods within the cell nucleus: proximity analysis of early S-phase replicating chromatin domains to sites of transcription, RNA polymerase II, HP1gamma, matrin 3 and SAF-A. J Cell Biochem. 2008a;105(2):391–403. doi: 10.1002/jcb.21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyavantham KS, Bhattacharya S, Alonso WD, Acharya R, Berezney R. Spatio-temporal dynamics of replication and transcription sites in the mammalian cell nucleus. Chromosoma. 2008b;117(6):553–67. doi: 10.1007/s00412-008-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyavantham KS, Bhattacharya S, Berezney R. The architecture of functional neighborhoods within the mammalian cell nucleus. Adv Enzyme Regul. 2010;50(1):126–34. doi: 10.1016/j.advenzreg.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella NV, Malyavantham KS, Wang J, Matsui S, Liang P, Berezney R. Cytogenetic and cDNA microarray expression analysis of MCF10 human breast cancer progression cell lines. Cancer Res. 2009;69(14):5946–53. doi: 10.1158/0008-5472.CAN-09-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Gudla PR, Khan S, Lockett SJ, Misteli T. Disease-specific gene repositioning in breast cancer. J Cell Biol. 2009;187(6):801–12. doi: 10.1083/jcb.200909127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- McNairn AJ, Gilbert DM. Epigenomic replication: linking epigenetics to DNA replication. Bioessays. 2003;25(7):647–56. doi: 10.1002/bies.10305. [DOI] [PubMed] [Google Scholar]
- Need EF, Selth LA, Harris TJ, Birrell SN, Tilley WD, Buchanan G. Research Resource: Interplay between the Genomic and Transcriptional Networks of Androgen Receptor and Estrogen Receptor α in Luminal Breast Cancer Cells. Mol Endocrinol. 2012 doi: 10.1210/me.2011-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir R, Wong AC, McDermid HE, Skorecki KL, Selig S. Position effect of human telomeric repeats on replication timing. PNAS. 1999;96(20):11434–9. doi: 10.1073/pnas.96.20.11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto PG, Otto PA, Therman E. The behavior of allocyclic chromosomes in Bloom’s syndrome. Chromosoma. 1981;84:337–44. doi: 10.1007/BF00286023. [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294(5540):115–21. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Reish O, Orlovski A, Mashevitz M, Sher C, Libman V, Rosenblat M, Avivi L. Modified allelic replication in lymphocytes of patients with neurofibromatosis type 1. Cancer Genet Cytogenet. 2003;143(2):133–9. doi: 10.1016/s0165-4608(02)00858-0. [DOI] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20(6):761–70. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Sasaki T, Battaglia D, Kulik M, Zhang J, Dalton S, Gilbert DM. Replication timing: a fingerprint for cell identity and pluripotency. PLoS Comput Biol. 2011;(10):e1002225. doi: 10.1371/journal.pcbi.1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, Wolman SR, Heppner GH, Miller FR. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat. 2001;65(2):101–10. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schübeler D. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev. 2009;23:589–601. doi: 10.1101/gad.511809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet. 2002;32(3):438–42. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- Selig S, Okumura K, Ward DC, Cedar H. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 1992;11(3):1217–25. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I, Cedar H. Temporal Order of DNA Replication. Cold Spring Harbor Laboratory Press; 1996. pp. 387–408. [Google Scholar]
- Simon I, Tenzen T, Mostoslavsky R, Fibach E, Lande L, Milot E, Gribnau J, Grosveld F, Fraser P, Cedar H. Developmental regulation of DNA replication timing at the human beta globin locus. EMBO J. 2001;20(21):6150–7. doi: 10.1093/emboj/20.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Landon S, O'Keefe RT. Organization of RNA polymerase II transcription and pre-mRNA splicing within the mammalian cell nucleus. Biochem Soc Trans. 1993;21(4):918–20. doi: 10.1042/bst0210918. [DOI] [PubMed] [Google Scholar]
- Stein GS, Zaidi SK, Braastad CD, Montecino M, van Wijnen AJ, Choi JY, Stein JL, Lian JB, Javed A. Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol. 2003;13:584–592. doi: 10.1016/j.tcb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Stein GS, Zaidi SK, Stein JL, Lian JB, van Wijnen AJ, Montecino M, Young DW, Javed A, Pratap J, Choi JY, Ali SA, Pande S, Hassan MQ. Genetic and epigenetic regulation in nuclear microenvironments for biological control in cancer. J. Cell. Biochem. 2008;104:2016–2026. doi: 10.1002/jcb.21813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JB, Gottschling DE. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 1999;13(2):146–51. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiepolo L, Fraccaro M, Hultén M, Lindsten J, Mannini A, Ming PM. Timing of sex chromosome replication in somatic and germ-line cells of the mouse and the rat. Cytogenetics. 1967;6(1):51–66. doi: 10.1159/000129929. [DOI] [PubMed] [Google Scholar]
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-Wise Methylation of Histone H3K9 Positions Heterochromatin at the Nuclear Periphery. Cell. 2012;150(5):934–47. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Shibata K, Sugimura H, Maekawa M. p53-dependent change in replication timing of the human genome. Biochem Biophys Res Commun. 2007;364(2):289–93. doi: 10.1016/j.bbrc.2007.09.136. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Abe T, Ikemura T, Maekawa M. Relationships between replication timing and GC content of cancer-related genes on human chromosomes 11q and 21q. Gene. 2009;433(1–2):26–31. doi: 10.1016/j.gene.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Shibata K, Ikemura T, Maekawa M. Replication timing of extremely large genes on human chromosomes 11q and 21q. Gene. 2008;421(1–2):74–80. doi: 10.1016/j.gene.2008.06.016. [DOI] [PubMed] [Google Scholar]
- Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11(2):103–5. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Wei X, Samarabandu J, Devdhar RS, Siegel AJ, Acharya R, Berezney R. Segregation of transcription and replication sites into higher order domains. Science. 1998;281(5382):1502–6. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- Wei X, Somanathan S, Samarabandu J, Berezney R. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J Cell Biol. 1999;146(3):543–58. doi: 10.1083/jcb.146.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EJ, Emanuelsson O, Scalzo D, Royce T, Kosak S, Oakeley EJ, Weissman S, Gerstein M, Groudine M, Snyder M, Schübeler D. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. PNAS. 2004;101(51):17771–6. doi: 10.1073/pnas.0408170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP. Replication timing of the human genome. Hum Mol Genet. 2004;13(2):191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- Worsham MJ, Pals G, Schouten JP, Miller F, Tiwari N, van Spaendonk R, Wolman SR. High-resolution mapping of molecular events associated with immortalization, transformation, and progression to breast cancer in the MCF10 model. Breast Cancer Res Treat. 2006;96(2):177–86. doi: 10.1007/s10549-005-9077-8. [DOI] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7(8):781–9. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van Wijnen A, Lian JB, Stein JL, Stein GS. Nuclear microenvironments in biological control and cancer. Nat Rev Cancer. 2007;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H. Establishment of transcriptional competence in early and late S phase. Nature. 2002;420(6912):198–202. doi: 10.1038/nature01150. [DOI] [PubMed] [Google Scholar]
- Zhou J, Ermakova OV, Riblet R, Birshtein BK, Schildkraut CL. Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol Cell Biol. 2002;22(13):4876–89. doi: 10.1128/MCB.22.13.4876-4889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Snyder AR, Lieberman PM. Epstein-Barr virus episome stability is coupled to a delay in replication timing. J Virol. 2009;83(5):2154–62. doi: 10.1128/JVI.02115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]


