Background: The interaction of the peptide VGVAPG with the elastin binding protein is critically involved in aneurysm progression.
Results: A molecular model of this interaction is proposed and explored using a site-directed mutagenesis strategy.
Conclusion: Three residues, Leu-103, Arg-107, and Glu-137, of elastin binding protein are critical players in this interaction.
Significance: Our data now allow the design of antagonists of VGVAPG.
Keywords: Computer Modeling, Elastin, Peptide Interactions, Peptides, Receptors
Abstract
The elastin binding protein (EBP), a spliced variant of lysosomal β-galactosidase, is the primary receptor of elastin peptides that have been linked to emphysema, aneurysm and cancer progression. The sequences recognized by EBP share the XGXXPG consensus pattern found in numerous matrix proteins, notably in elastin where the VGVAPG motif is repeated. To delineate the elastin binding site of human EBP, we built a homology model of this protein and docked VGVAPG on its surface. Analysis of this model suggested that Gln-97 and Asp-98 were required for interaction with VGVAPG because they contribute to the definition of a pocket thought to represent the elastin binding site of EBP. Additionally, we proposed that Leu-103, Arg-107, and Glu-137 were essential residues because they could interact with VGVAPG itself. Site-directed mutagenesis experiments at these key positions validated our model. This work therefore provides the first structural data concerning the interaction of the VGVAPG with its cognate receptor. The present structural data should now allow the development of EBP-specific antagonists.
Introduction
Elastin is the extracellular matrix protein responsible for the elasticity of vertebrate tissues and is more abundant in tissues where resilience is required, such as large vessels (1). It is deposited as elastic fibers during early prenatal and postnatal life. Elastin is constituted of tropoelastin molecules covalently bound to each other by covalent cross-links (2). Its hydrophobic and highly cross-linked nature makes it a very durable polymer, experiencing essentially no turnover in healthy tissues (3).
The elastin receptor complex (4–6) is derived from the lysosomal complex of acid β-galactosidase (β-gal, EC 3.2.1.23) (4). It is a heterotrimer comprising a peripheral elastin binding protein (EBP)5 and two membrane-associated proteins, neuraminidase-1 (EC 3.2.1.18) and the protective protein/cathepsin A (EC 3.4.16.1).
Although neuraminidase-1 and protective protein/cathepsin A are identical to the subunits found in the lysosome, EBP is an enzymatically inactive spliced variant of lysosomal β-gal (5, 7, 8). The alternative splicing of β-gal primary transcripts leading to EBP consists of two deletions. The first one deletes exons 3 and 4 and introduces a frameshift; the second one occurs in exon 6 and restores the initial reading frame. As a consequence, EBP possesses a unique 32-residue sequence, termed V32, which is thought to contain its elastin binding site: 83LPGSCGQVVGSPSAQDEASPLSEWRASYNSAG114. EBP has no enzymatic activity but retains the ability to bind galactosugars (6). When galactosugars bind EBP, the affinity of the elastin site for its ligand dramatically decreases, leading to their release and the dissociation of EBP from the complex (5).
The elastin receptor complex is a critical actor of elastogenesis (9). In this highly regulated and coordinated phenomenon, it plays a dual role. First, it protects the newly synthesized tropoelastin molecules from prematurely aggregating in the cell and behaves as a chaperone (10). Second, it accompanies and delivers the tropoelastin molecules to the cell surface where elastin assembly occurs (11).
Additionally, this receptor also serves as a functional sensor for its degradation. Elastin peptides are the result of elastin wear and tear due to mechanical fatigue or degradation by elastases during specific pathophysiological processes (12–14). Elastin peptide concentration in the blood flow is usually low, but it can reach high values in aged subjects, notably in those developing pathologies where elastin is massively degraded such as aneurysms (15, 16). Unlike insoluble fibrous elastin, elastin peptides promote cell proliferation, cell chemotaxis, angiogenesis, and proteases release in a wide variety of normal cells (13, 17–19). The effects promoted by VGVAPG, a sequence repeated six times in human tropoelastin, are the most documented. As a consequence, this peptide is regarded as the archetypal elastin peptide.
It is now established that the catalytic activity of neuraminidase-1 is essential for proper functioning of the receptor. It is required for both elastin assembly (9) and elastin peptide signaling (20). Importantly, this activity requires the presence of EBP. As a consequence, EBP seems to behave as a molecular switch allowing neuraminidase-1 activity. Elastin peptides promote this activity and subsequent signaling, whereas galactosugars block it.
The molecular functioning of the elastin receptor complex remains unclear. This is notably due to the lack of data concerning the structure of its individual subunits and notably that of EBP.
In this work, we used homology modeling to propose the first molecular model of EBP. According to this model, the EBP-specific V32 sequence defined a surface pocket stabilized by interactions between the QDEA and SYNS strings of V32. Additionally, the docking of VGVAPG on this model suggested that the Glu-137, Leu-103, and Arg-107 residues of EBP were critical elements for VGVAPG recognition. These hypotheses were confirmed by site directed mutagenesis of these individual residues as COS-7 expressing these mutant forms of EBP had lost their ability to induce human collagenase-1 (MMP-1) promoter activity following VGVAPG treatment. In conclusion, our data provide the first detailed structural data on the interaction of VGVAPG with EBP and open promising perspectives in the design of EBP antagonists.
EXPERIMENTAL PROCEDURES
Homology Modeling
A homology model of EBP was built using MODELLER software (21) and the crystallographic data available for the unbound Penicillium sp. β-galactosidase (Protein Data Bank code 1TG7) (22). A multiple sequences alignment was first generated using ClustalX software (23), which included all of the β-galactosidase sequences available in the SwissProt database and sharing a maximum E-value of 10 with the target. The global identity rate between 1TG7 and the target sequence (EBP formerly termed BGAM_HUMAN, accession no. P16279) was 30% (55% similar) with an E-value of 3.8 × 10−15. The main difference between the target and the reference structure resided in the inserted V32 peptide in the target model, which was built de novo. The resulting 100 models produced by the standard protocol of MODELLER were then ranked according to their objective function values, and the best one was conserved for the docking experiments.
Docking
Docking of the VGVAPG peptide against the built EBP structure was performed using the Autodock software (version 4.0) (24, 25). Lamarckian Genetic Algorithm was used, and 250 dockings were performed with a grid resolution of 0.25 Å in the region encompassing the whole V32 peptide. Docking runs were performed with the default parameters of Autodock except for population size (200), number of energy evaluations (106), and the maximum number of generations (500, 000). Molecular models were graphed with the PyMOL software available online.
Materials
Reagents for cell culture and transfection reagent Lipofectamine 2000 were from Invitrogen. The synthetic peptide VGVAPG was purchased from GeneCust (Luxembourg, Luxembourg). Rabbit polyclonal phospho-specific antibodies against active forms of ERK1/2 (phosphorylated on Thr-202 and Tyr-204) and anti-ERK1/2 were from Cell Signaling Technology, Inc. (Beverly, MA, distributed by Ozyme, Saint Quentin en Yvelines, France). Others reagents were from Sigma (Saint-Quentin Fallavier, France).
Expression Plasmids
The −512ColA-luc plasmid encoding luciferase under the control of the MMP-1 promoter was generated by cloning the 5′ upstream sequence of MMP-1 into the pGL3Basic reporter plasmid (Promega, Lyon, France), as described previously (26). The cDNA of human EBP cloned in a eukaryotic expression vector (pSC/rSV-EBP) was supplied by Dr. Aleksander Hinek (Hospital for Sick Children, Toronto, Canada).
Site-directed Mutagenesis
The EBP mutants were obtained from the wild type plasmid using site-directed mutagenesis with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The sequences of primers used to introduce single point mutations within pSC/rSV-EBP are presented in Table 1. The reaction was performed in a final volume of 50 μl. The sample reaction contained 1 μl of Pfu Turbo DNA polymerase, 5 μl of 10× buffer (20 mm MgSO4), 50 ng of pSC/rSV-EBP preparation, 125 ng of each oligonucleotide, 10 mm dNTP, and H2O. The reaction was performed using a Mastercycler gradient thermocycler (Eppendorf, Le Pecq, France) and subjected to the following program: denaturation of 30 s at 95 °C, 16 cycles of 30 s at 95 °C, 1 min at 55 °C, and 8 min at 68 °C. The sample was digested by 1 μl of DpnI (10 units/μl) for 3 h at 37 °C and chemically transformed into the XL1 Blue strain. Cells were applied on a LB agar plate containing 50 μg/ml ampicillin and incubated overnight at 37 °C. The obtained clones were sequenced (Institut de Biologie Moléculaire des Plantes, Strasbourg). The different plasmids were then concentrated and purified by High Speed Maxi Prep kit (Qiagen, Courtaboeuf, France).
TABLE 1.
Primers used to introduce single point mutations within EBP
The underlined nucleotides correspond to the introduced mutation.
| Q97A | 5′-AGT CCT TCT GCC GCA GAT GAA GCC TCT CC-3′ |
| D98A | 5′-GGG AGT CCT TCT GCC CAA GCT GAA GCC TCT CCT CTA-3′ |
| A100L | 5′-GTC CTT CTG CCC AAG ATG AAC TCT CTC CTC TAT CAG AAT GGA GGG-3′ |
| L103A | 5′-GAT GAA GCC TCT CCT GCA TCA GAA TGG AGG GCC-3′ |
| R107A | 5′-TCT CCT CTA TCA GAA TGG GCG GCC AGT TAT AAC AGT GC-3′ |
| R107K | 5′-CC TCT CCT CTA TCA GAA TGG AAG GCC AGT TAT AAC AGT GCA G-3′ |
| E137A | 5′-GGA CCC TTG ATC AAT TCT GCA TTC TAT ACT GGC TGG CTA G-3′ |
| E137D | 5′-GGA GCA CTG ATC AAT TCT GAC TTC TAT ACT GGC TGG CTA G-3′ |
| E137Q | 5′-GGA GCA CTG ATC AAT TCT CAA TTC TAT ACT GGC TGG CTA G-3′ |
| Y200A | 5′-GCA CAG CCC ACC AGC TAC GCC GAC TAT GAT GCC CC-3′ |
Western Blotting
To validate the use of cultured COS-7 cells for further transfection experiments, we first evaluated the ability of transfected cells to transduce intracellular signals in the presence of elastin peptides. This was achieved by assessing the level of ERK1/2 activation after elastin peptide treatment. Being derived from mammalian cells, COS-7 cells harbor a simian elastin receptor complex. The ability of these cells to respond to the presence of elastin peptides was checked when they were transfected with a plasmid bearing the human wild type EBP construct.
COS-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g/liter of glucose, glutamax I, and pyruvate, supplemented with 10% fetal calf serum and in the presence of 5% CO2. Cells were then incubated for 24 h in 200 μl of serum-free DMEM with a mixture of Lipofectamine 2000 and DNA plasmids for 4 h. Cells were further cultured in a serum-free medium for 24 h before a 30 min VGVAPG stimulation (200 μg/ml) at 37 °C.
106 COS-7 cells were washed twice in ice-cold PBS containing 50 μm Na3VO4, scrapped, and sonicated in lysis buffer (PBS, pH 7.4, 0.5% Triton X-100, 80 mm glycerophosphate, 50 mm EGTA, 15 mm MgCl2, 1 mm Na3VO4, and protease inhibitor mixture). Insoluble material was removed by centrifugation (20,000 × g, 20 min, 4 °C). Protein concentrations were determined by bicinchoninic acid protein assay (Pierce, Rockford, IL; distributed by Interchim, Montluçon, France). Equal amounts of proteins were heated for 5 min at 100 °C in Laemmli sample buffer, resolved by SDS-PAGE under reducing conditions and transferred to nitrocellulose membranes. The membranes were placed in blocking buffer (5% (w/v) nonfat dry milk in Tris-buffered saline/Tween 20 (50 mm Tris, pH 7.5, 150 mm NaCl, and 0.1% (v/v) Tween 20)) for 1 h at room temperature and incubated overnight at 4 °C with anti-phospho-ERK1/2 (1:1000) and anti-ERK1/2 (1:500) antibodies. After five washings with Tris-buffered saline/Tween 20, the membranes were incubated for 1 h at room temperature in the presence of horseradish peroxidase-coupled anti-rabbit antibodies (1:4000 in blocking buffer). Immunocomplexes were detected by chemiluminescence. Blots were quantitated by densitometry using PhosphorAnalyst software (Bio-Rad).
EBP Mutant Addressing
Human EBP-FLAG mutant constructs were obtained by PCR-based approach using the Phusion High-Fidelity DNA polymerase (Finnzymes, Vantaa, Finland). Restriction sites for HindIII and KpnI were introduced by PCR immediately adjacent to the initiation and termination codons of EBP mutants, respectively. After digestion, the resulting insert was ligated in a p3xFLAG CMV14 vector encoding a FLAG tag. After sequencing of the constructs, COS-7 cells were grown in six-well plates and were transiently transfected by 2 μg of EBP-FLAG mutant cDNA. Two wells were pooled for each mutant. 24 h post transfection, cells were biotinylated using 500 μg/ml of biotin. Three washes were then performed with cold PBS containing Na3VO4 (50 μm), and cells were lysed in cold lysis buffer (50 mm Na2HPO4 (pH 8), 300 mm NaCl, 1% Triton X-100 containing 1 mm Na3VO4 and a proteinase inhibitor mixture). After sonication, the human EBP-FLAG mutants were solubilized for 1 h at 4 °C. Lysates were centrifuged at 20,000 × g for 30 min at 4 °C. The protein content of the resulting surpernatant was assayed, and the biotinylated proteins were precipitated for 45 min at 4 °C with 20 μl of streptavidin beads. Precipitated proteins were eluted for 5 min at 100 °C with 100 μl Tris/HCL 0.5 m (pH 6.8), 0.5% bromphenol blue, 10% SDS, 20% saccharose, and 25% β-mercaptoethanol. Proteins resolved by SDS-PAGE were transferred onto nitrocellulose membrane (Dutscher, Brumath, France). After incubation in TBS supplemented with 5% skimmed milk and 1% Tween 20 for 1 h at room temperature, membranes were incubated overnight at 4 °C with a monoclonal anti-FLAG M2 primary antibody (Sigma) solution (1:1000) in TBS supplemented with 5% skimmed milk and 1% Tween 20. After washes in TBS supplemented with 1% Tween 20, the biotinylated proteins were revealed using a HRP-conjugated secondary antibody (1:3000) and the ECL chemiluminescent reagent (Millipore, Molsheim, France).
Determination of EBP Functionality
COS-7 cells were grown in DMEM containing 4.5 g/liter of glucose, GlutaMAX I, and pyruvate, supplemented with 10% fetal calf serum and in the presence of 5% CO2. For transfection experiments, COS-7 cells were grown on 96-well plates to 80% confluence (16,000 cells) in medium containing 10% serum. They were further incubated for 24 h in 200 μl of serum-free DMEM with a mixture of Lipofectamine 2000 and DNA plasmids for 4 h. The ratio used was 0.2 μl of Lipofectamine 2000 for 0.2 μg of total plasmidic DNA divided in 50% of mutant plasmid and 50% of plasmid containing a MMP-1 luciferase promoter. After 3 h, cells were incubated in DMEM for 24 h and stimulated with or without VGVAPG peptide (150 μg/ml). When lactose (EBP antagonist) was used, cells were preincubated with 1 mm lactose for 3 h before stimulation with VGVAPG. After 16 h, 100 μl of SteadyLite reporter lysis buffer were added (PerkinElmer Life Sciences), and luciferase activities were measured with a PerkinElmer TopCount microplate counter.
Statistical Analysis
Experiments were performed in triplicate. Results are expressed as mean ± S.E. Comparisons were made using two-tailed (stimulated versus control) or one-tailed (stimulated versus own control) Student's t test. The results were considered significantly different at p < 0.05.
RESULTS
Homology Modeling of EBP
To model EBP, we first generated a multiple alignment of the sequences of β-gal family members, among which figured Penicillium sp. β-galactosidase, which structure had been solved by x-ray crystallography (22). The sequence of EBP was further aligned, and the corresponding alignment (supplemental Fig. 1) was used to construct EBP models.
The most optimized model, derived from homology modeling, is presented in Fig. 1. It encompasses residues 28 to 238 of EBP (accession no. P16279). Its atomic coordinates are given as supplemental data. Due to the lack of significant homology between EBP and Penicillium sp. β-galactosidase beyond EBP residue 239, our model does not account for the whole structure of EBP but correctly modeled the supposed region of elastin binding. This is why our model was further refined and optimized by classical molecular mechanics approaches.
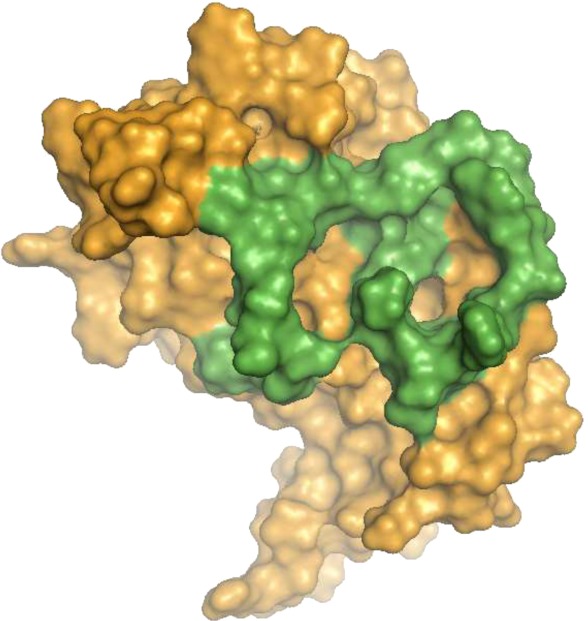
FIGURE 1.
Homology model of EBP. This model was built using the MODELLER package as described under “Experimental Procedures.” The surface of EBP is colored orange. The surface corresponding to V32 is colored green.
As shown (Fig. 1), the EBP-specific V32 sequence (green) was located at the surface of the molecule and delimited a horseshoe-like pocket. Consequently, we hypothesized that this pocket could represent the elastin binding site of the molecule, and we examined it closely.
Previous works (5, 6, 27) had emphasized that the VVGSPSAQDEASPL sequence of EBP was important for elastin peptide binding. In particular, the hydrophilic QDEA string (residues 97–100) was identified as a potential interaction site (28). We thus hypothesized that these residues could participate to elastin peptide recognition.
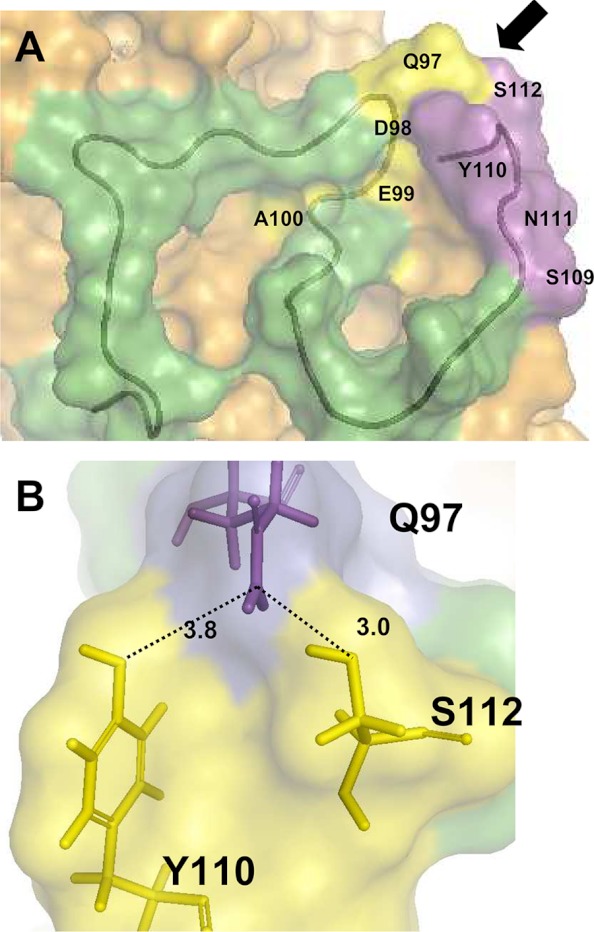
The QDEA motif (Fig. 2A, yellow) was located in the upper part of the potential elastin binding site and contributed to the definition of its external limits. Interestingly, the SYNS segment of V32 (Fig. 2A, residues 109–112, magenta) was very close. In the model, these sequences defined the right-hand external limit of the potential elastin binding site. When observed from another position (Fig. 2B), it appeared that the amide group of Gln-97 and the hydroxyls of Tyr-110 and Ser-112 were positioned in a geometry compatible with H-bonding.
FIGURE 2.
Analysis of the elastin binding site. A, conformation of the V32 sequence in EBP (green tube). The surface defined by the QDEA (yellow) and SYNS (magenta) strings are labeled. The arrow indicates the orientation permitting to observe B. B, stick representation of Gln-97 (magenta) and the SYNS string (yellow) local organization. The thick sticks correspond to residues (Gln-97 (Q97), Tyr-110 (Y110), Ser-112 (S112)) orienting groups in a local geometry favorable to H-bonding. The distance between these groups are reported in angstroms.
Comparison of EBP and β-gal Models
The crystal structure of human β-gal has been resolved very recently (29). The comparison between our predictive EBP model and the experimental one of β-gal is provided in Fig. 3.
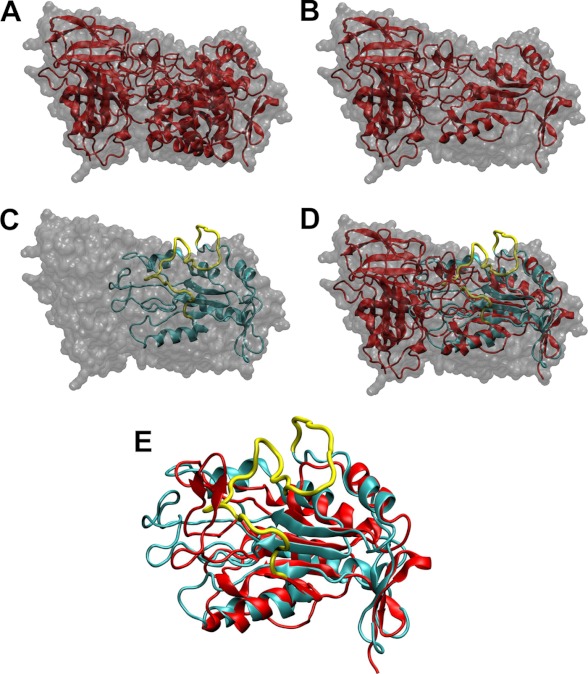
FIGURE 3.
Comparison of the homology model of EBP with the crystal structure of human β-galactosidase. A, crystal structure of human β-gal (Protein Data Bank code 3THC). The surface of the molecule is colored gray. The secondary structure of the enzyme is represented as a red schematic. The catalytic TIM barrel is on the right side. B, same representation than A in which the sequences spliced in EBP have been removed. C, molecular model of EBP overlaid on the surface envelope of human β-gal. The molecule is represented as a cyan schematic. The yellow line corresponds to the backbone trace of V32. D, superposition of B and C. E, overlay of β-gal TIM barrel after splicing (from B) and our EBP model.
The crystal structure of human β-gal (Protein Data Bank code 3THC) is presented in Fig. 3A. Fig. 3B provides the same view in which the sequences spliced in EBP have been removed. This splicing only impacts the catalytic TIM barrel of the enzyme (right side of the view). Our EBP model was compared with the molecular surface of the unspliced enzyme (Fig. 3C) and further overlaid on the structure of β-gal where the spliced domains had been removed. Strikingly, the agreement between both models is very good as evidenced in Fig. 3E. Notably, the sheet structure of the barrel constituting the core of this fold seems to be barely affected by the splicing. As a consequence the external helices found in both structures align very well. This observation greatly supported the validity of our molecular model of EBP and allowed further docking experiments.
Docking
The VGVAPG peptide is well characterized at the biological and molecular level (30). This string is repeated six times in human tropoelastin sequence, and it presents the consensus GXXPG sequence, which is thought to stabilize a type VIII β-turn required for bioactivity of the peptide (30, 31). As a consequence, we docked this peptide on our molecular model of EBP. The docking procedure yielded several models in which the VGVAPG sequence was bound onto EBP. Their validities were assessed by examining their overall potential energy, the lowest energy meaning the highest affinity.
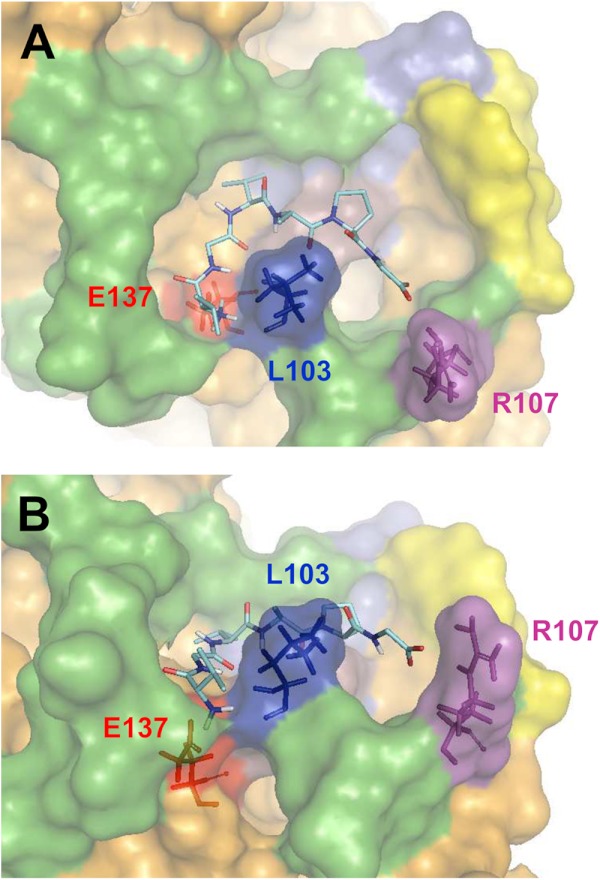
Most high affinity models (in the μm range) placed VGVAPG near or inside of the pocket defined by the V32 sequence. The best of these models is presented in Fig. 4, A and B, and corresponded to an affinity of 4.85 μm for the EBP/VGVAPG interaction. This solution placed the VGVAPG peptide inside of the pocket defined by the V32 sequence of EBP, thus supporting our former hypothesis that this horseshoe-like depression could constitute the elastin binding site.
FIGURE 4.
Model of VGVAPG docked on EBP. A, the VGVAPG peptide, which is located in the pocket defined by the V32 sequence (green), is represented as sticks. The three residues stabilizing the interaction are colored in red (Glu-137), dark blue (Leu-103), and violet (Arg-107). The yellow surface corresponds to QDEA, the magenta surface corresponds to SYNS. B, other view of the docked peptide permitting to better appreciate the position of Glu-137, Leu-103, and Arg-107.
In our docking model, the GVAP string of the bound VGVAPG adopted a conformation close to a type VIII β-turn. Additionally, the Glu-137, Leu-103, and Arg-107 residues of EBP (Fig. 3, A and B) seemed to play an important role in the interaction with VGVAPG. For instance, the hexapeptide coiled around the hydrophobic Leu-103 residue, whereas Glu-137 and Arg-107 were orientated to allow ionic interactions with the charged NH3+ and COO− extremities of the peptide, respectively.
The theoretical stability of this EBP/VGVAPG interaction model was checked by molecular dynamics in explicit solvent conditions. The results (data not shown) maintained that the interaction of VGVAPG with EBP at the supposed elastin binding site was stable. Indeed, the hexapeptide remained located inside the binding pocket throughout the three 50-ns simulations performed.
Validation of the Biological System
Following elastin peptide stimulation, the MEK/ERK pathway is activated in COS-7 cells, and this activation could be monitored using a luciferase assay of MMP-1 promoter activity (20).
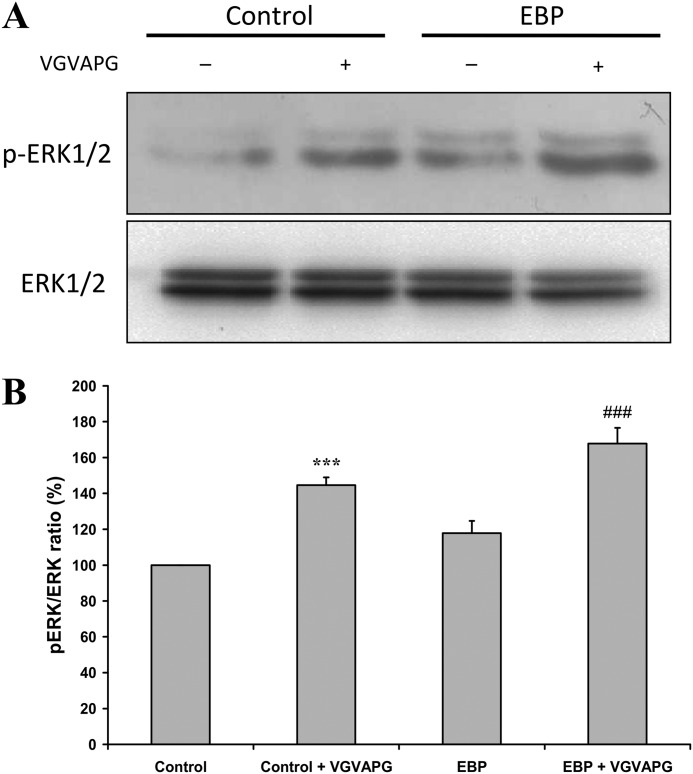
As we intended to assay MMP-1 promoter activity in conditions necessitating another transfection (i.e. EBP mutant constructs), we first checked the ability of COS-7 to activate their MEK/ERK signaling cascade when human wild-type EBP was also expressed. Indeed, in these conditions, the human EBP competes with the simian EBP from COS-7, and we had to check that this “hybrid” elastin receptor complex, containing the transfected EBP associated with endogenous neuraminidase-1 and protective protein/cathepsin A, could be correctly assembled so as to efficiently promote MEK/ERK activation after interaction with VGVAPG. The data presented in Fig. 5 showed that when COS-7 cells were transfected with an empty vector and treated with VGVAPG, ERK phosphorylation was significantly increased as compared with their basal level in the absence of stimulation (100 to 145%). This indicated that these cells possessed a functional elastin receptor complex. The transfection of the WT-EBP construct in these cells in the absence of stimulation increased slightly ERK activation status (100 to 116%). We therefore concluded that WT-EBP transfection did not substantially modify intrinsic COS-7 ERK activation when these cells were not stimulated by VGVAPG. However, increased phosphorylation of ERK was observed in these transfected cells following VGVAPG treatment (116 to 163%). This finding strongly suggested that a functional elastin receptor complex could be assembled at the surface of these transfected cells.
FIGURE 5.
VGVAPG activates the ERK pathway in transfected COS-7 cells. A, Western blots of pERK and ERK levels in COS-7 transfected with an empty vector or with WT-EBP, after or without VGVAPG treatment. Cells were incubated for 24 h in 200 μl of serum-free DMEM with a mixture of Lipofectamine 2000 and DNA plasmids for 4 h. Cells were allowed to rest for 24 h in a serum-free medium before a 30-min VGVAPG stimulation (150 μg/ml) at 37 °C. B, corresponding densitometric analysis. The pERK/ERK ratio is compared with the corresponding basal situation in the absence of VGVAPG treatment. The excitability of the system was evaluated using a two-tailed Student's t test. Control COS-7 cells versus VGVAPG-treated COS-7 cells (***, p < 0.001); EBP-transfected cells versus VGVAPG-stimulated EBP-transfected cells (###, p < 0.001).
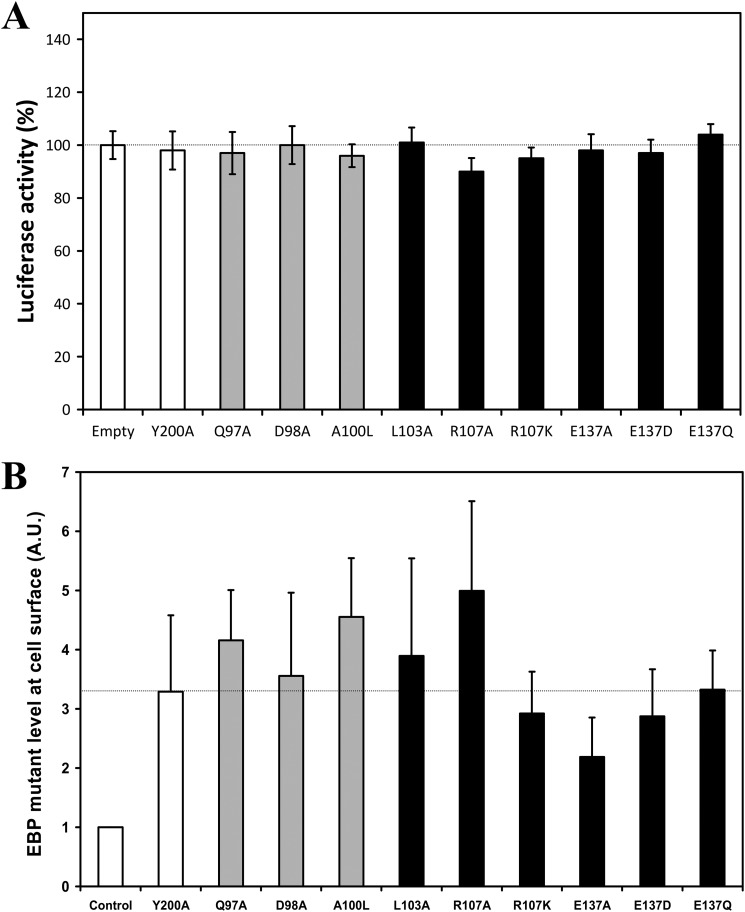
As our further experiments required that the luciferase activity could be compared between transfected cells, we first analyzed the ability of these cells to express our various EBP constructs. COS-7 cells were transfected with the −512ColA-luc plasmid, and their basal luciferase activity were determined and compared (Fig. 6A). We observed that the luciferase basal level was consistent independently of the transfected plasmid. We thus concluded that the transfection yield was equivalent whatever the plasmid considered, suggesting that our further data could be compared quantitatively.
FIGURE 6.
Effect of EBP mutant transfection on basal luciferase activity and EBP mutant levels at cell surface in transfected COS-7 cells. A, COS-7 cells were co-transfected with constructs encoding luciferase under the control of MMP-1 promoter, the indicated mutant EBP constructs, or with an empty vector. Cells were then stimulated with the VGVAPG peptide (150 μg/ml). Basal luciferase activity is expressed as a percentage of the control condition (empty vector). B, COS-7 cells were transiently transfected by 2 μg of EBP-FLAG mutant cDNA for 24 h. Cells were further biotinylated with 500 μg/ml of biotin, washed three times, and sonicated. Human EBP-FLAG mutants were then solubilized for 1 h at 4 °C and centrifuged at 20,000 × g for 30 min at 4 °C. The biotinylated proteins were precipitated for 45 min at 4 °C with 20 μl of streptavidin beads. After elution, retained proteins were analyzed by Western blots using an anti-FLAG antibody. The presented results correspond to the densitometric analysis of three experiments after normalization. Control, COS-7 cells transfected with the Y200A EBP-FLAG construct without biotinylation. Gray bars, mutations in the QDEA string; black bars, mutations of Leu-103, Arg-107, and Glu-137. No statistical difference was observed between the estimated EBP-FLAG levels in transfected cells as compared with the Y200A level (two-tailed Student's t test). A.U., arbitrary units.
Finally, we checked the impact of EBP mutations on its plasma membrane targeting. To this end, FLAG-tagged EBP mutants were constructed and transfected in COS-7. After biotinylation of the cell surface, membrane proteins were isolated and the level of EBP-FLAG mutants found at the cell surface was evaluated by Western blot. As shown in Fig. 6B, the transfection of EBP constructs in COS-7 cells permitted us to detect EBP-FLAG proteins at their surface. Importantly, the overall level of EBP protein detected was consistent as no significant differences were observed between the mutants as compared with Y200A. This point suggested that our EBP mutants were addressed correctly, allowing us to further analyze their functionality.
Functionality of EBP Mutants
As we had established that transfection of human EBP cDNA did not impede the ability of COS-7 cells to trigger their ERK pathway and that the different EBP mutants localized well to the plasma membrane, we further assessed the functionality of EBP mutants using a luciferase assay of MMP-1 activity in the presence of VGVAPG. To this end, COS-7 cells were transfected with both the −512ColA-luc plasmid and a mutated EBP construct, and their ability to develop a luciferase signal following VGVAPG treatment was analyzed.
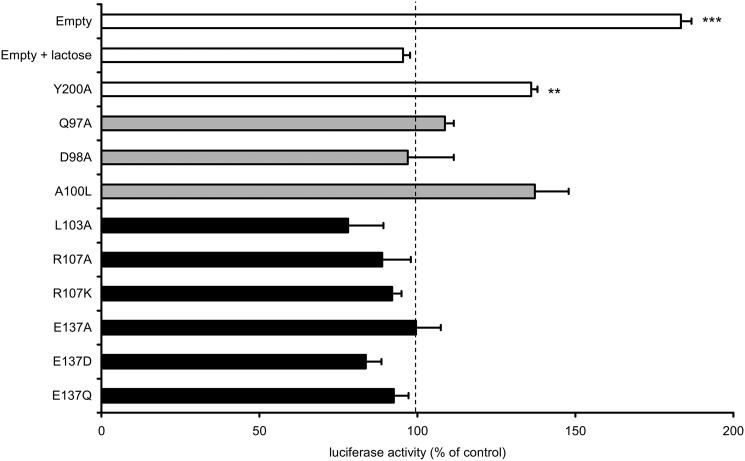
As shown in Fig. 7, COS-7 cells transfected with a mock plasmid (empty vector) were responsive to the presence of VGVAPG, leading to an 80% increase of MMP-1 promoter activity as compared with their basal rate. This effect could be blocked by preincubating cells in the presence of lactose indicating the involvement of the elastin receptor complex. This experiment also showed that doubly transfected COS-7 cells retained their ability to activate their intrinsic ERK pathway following VGVAPG treatment (Fig. 5) explaining the onset of the luciferase signal.
FIGURE 7.
Site-directed mutagenesis exploration of VGVAPG binding site. COS-7 cells were co-transfected with constructs encoding luciferase under the control of MMP-1 promoter, the indicated mutant EBP constructs, or with an empty vector. Cells were then stimulated with the VGVAPG peptide (150 μg/ml) in the absence or presence of 1 mm lactose (EBP antagonist). The results correspond to the luciferase activity measured after VGVAPG treatment. They are expressed as a percentage of luciferase activity observed for the same situation without treatment. This control value was set to 100% for all experiments (dashed line). White bars, control experiments; gray bars, mutations in the QDEA string; black bars, mutations of Leu-103, Arg-107, and Glu-137. The reported significance compares the treated versus untreated cells (one-tailed Student's t test). **, p < 0.01; ***, p < 0.001.
To check that the site-directed mutagenesis process did not interfere with the cell response, we first co-transfected COS-7 with a human EBP mutant construct bearing the Y200A mutation. This point mutation was chosen because Tyr-200 is an extremely conserved residue within the β-gal family and because this residue is not located in the V32 sequence (see supplemental Fig. 1). COS-7 cells expressing this plasmid could be stimulated with VGVAPG (Fig. 7). The stimulation level attained was lower than that observed with the empty construct. This lowered stimulation level could be explained by fact that the assembly of a human EBP with endogenous simian protective protein/cathepsin A and neuraminidase-1 subunits could lead to a less efficient signaling by this hybrid complex. Nevertheless, we observed that COS-7 transfected with the human EBP construct could still be significantly stimulated by the addition of VGVAPG in the medium (45% increase of luciferase signal). This validated the method used, and we proceeded with other constructs. Two types of mutants were constructed.
First, we designed mutants bearing a mutation in the QDEA domain because this region is supposed to be critically involved in the interaction with elastin peptides (28). These EBP constructs bore the following mutations: Q97A, D98A, and A100L. When cells were transfected with the corresponding mutant constructs (Fig. 7, gray bars), the luciferase signal following VGVAPG treatment was severely attenuated except for the A100L construct. In this case, the stimulation level was consistent with that observed for the Y200A mutation control construct, but the observed increase was not significant. These findings suggested that Gln-97 and Asp-98 were important for VGVAPG binding.
Second, mutants bearing point mutations on residues possibly important for VGVAPG binding were devised on the ground of the analysis of our VGVAPG docking results. These EBP constructs bore the following mutations: L103A, R107A, R107K, E137A, E137D, and E137Q. Strikingly, cells transfected with constructs bearing one of these mutants (Fig. 7, black bars) invariably lost their ability to induce luciferase activity suggesting that these peculiar residues were of utmost importance for the recognition of VGVAPG by EBP. For instance, the charge-conservative R107K and E137D mutations did not allow MMP-1 promoter induction, suggesting that the local geometry at these sites was extremely important for proper VGVAPG binding. These observations greatly supported our docking model.
DISCUSSION
This work reports the first attempt at describing the structure and function of the EBP subunit of the elastin receptor complex. Using structural characteristics of β-galactosidases, a homology model of EBP is proposed. This model (Fig. 1) was built using a limited portion of EBP sequence (residues 28–238) that matched the sequence of the TIM barrel active site of Penicillium sp. β-galactosidase. As a consequence, the influence of the rest of the sequence on this peculiar region of the protein was overlooked.
The examination of human β-gal structure reveals that it possesses three distinct regions (29): a catalytic TIM barrel domain followed by two β-domains. In the crystal structure, these two regions are structurally independent from the TIM barrel to which they are connected by a large loop (29). As our EBP model is derived from the TIM barrel domain sequence, we feel that the incidence of the rest of the sequence on this region should be limited.
In our model (Fig. 1), the V32 sequence, which is specific of EBP, defines a pocket at the surface of the protein. The analysis of these data has allowed us to propose individual residues that could be involved in EBP function. A site-directed mutagenesis analysis of these amino acids was undertaken. Most mutants failed to trigger intracellular signals, suggesting that these residues were important for VGVAPG binding and also validated our model. As a consequence, we propose that the pocket defined by V32 at the surface of our model is likely the elastin binding site of EBP.
Our model presents several discrepancies as compared with the work of Moroy and co-workers (28). In their interaction model of VGVAPG with the VVGSPSAQDEASPLS peptide, the QDEA segment of the latter adopts a type I β-turn conformation thought to be very important because it exposes Gln so that it can form three H-bonds with the VGVAPG skeleton that coils around it. In our model, the QDEA string adopts a similar turn structure but, in this conformation, Gln is less exposed to the solvent. In fact, it interacts with the SYNS string of the V32 sequence. In their initial model, the authors could not see this interaction as their experiments were performed on a limited sequence of V32. In our interaction model, the VGVAPG does not coil around the Gln-97 residue of QDEA but around the hydrophobic Leu-103 of V32. This possibility could not be seen in the model of Moroy and co-workers (28) as the corresponding leucine was at the very C terminus of their peptide and, consequently, highly exposed to the solvent and thereby flexible. As a consequence, we suggest that our model would be more relevant that the former one as it brings additional information and presents a good agreement with both experimental site-directed mutagenesis data and the crystal structure of human β-galactosidase (29).
When residues located in V32 were mutated, a loss of signaling ability was observed in most cases. In the light of the docking of VGVAPG on EBP (Fig. 4), we suggest that these residues can contribute to EBP proper functioning in two ways. They can either participate to VGVAPG interaction (Arg-107 for example) or contribute to the structural stability of the elastin binding site. This later situation holds especially for the QDEA string that could interact with SYNS (Fig. 2B) and contribute to the overall stability of the site.
Among the tested mutants, the A100L substitution apparently leads to an EBP mutant that still exhibits elastin peptide affinity. In fact, cells transfected with this construct somehow retain their ability to be stimulated with VGVAPG (Fig. 7). This could be explained by the fact that, in our model, the side chain of this residue is turned toward the center of the protein. As a consequence, it would not participate in ligand binding, and mutations at these sites can be tolerated.
The importance of the QDEA string in VGVAPG interaction has already been suggested (5, 27). Our work supports this view but also points at the possible importance of the SYNS string of V32. As suggested by the geometry adopted by these peptides in our model (Fig. 2B), we propose that they interact together and thus doing, prevent the elastin binding site from opening up. That way, the site can accommodate a VGVAPG peptide.
The detailed analysis of the interaction of VGVAPG with EBP underlines that three residues are very important: Glu-137, Leu-103, and Arg-107. Indeed, our mutation experiments show that these three residues are essential for VGVAPG biological activity. This proposal is supported by the fact that even conservative mutations (E137D, R107K) resulted in a loss of EBP functionality. Indeed, the loss of a carbon along the acidic side chain in the E137D mutation shows that the local side chain positioning is extremely important for VGVAPG recognition. Likewise, changing the nature of the basic group at position 107 is not tolerated. These two observations strongly suggest that these residues are essential because they directly contribute to VGVAPG binding.
Glu-137 is in an ideal position to form an electrostatic bridge with the N terminus of the peptide. Its side chain points inside the pocket where it can establish either electrostatic or H-bond interactions with the backbone of the docked peptide. Similarly, Arg-107 seems to be able to cap its C terminus, whereas the hydrophobic Leu-103 forces the peptide to adopt a turn conformation. Thus doing, the two valyl residues of VGVAPG are orientated so that they can optimize hydrophobic contacts with Leu-103 for the first one and V32 VVG sequence for the second.
EBP is known to interact with various agonists (13, 31–35). In previous work, we have established that hexapeptides interacting with EBP share the XGXXPG consensus sequence that is thought to stabilize a type VIII β-turn (30, 31). Importantly, the conformation adopted by VGVAPG in our docking model is close to a type VIII β-turn. In fact, it corresponds to the semi-open form of the turn described formerly in long term molecular simulations of VGVAPG (30).
Besides the VGVAPG hexapeptide, EBP also binds the tropoelastin PGAIPG sequence (31), LGTIPG from laminin-1 (31, 35), and EGFEPG from fibrillin-1 (36). These sequences all share the XGXXPG consensus and are mostly hydrophobic. Thus, although the corresponding docking experiments have not been performed yet, what holds for VGVAPG could also explain the biological activity of these peptides and they could adopt the same position than VGVAPG. However, several differences could exist notably for the more hydrophilic EGFEPG fibrillin sequence. Further experiments are required.
Peptides of various lengths are known to bind to EBP, for example peptides bearing the XGXXPG consensus pattern can bind efficiently to EBP and can be removed by lactose (37). Interestingly, the N terminus of this pattern does not have to be free to allow bioactivity of these longer peptides. Our explanation is that, in this case, the peptide enters deeply in the pocket, probably guided by Leu-103 and Arg-107 interactions, and folds to adjust its conformation to the pocket constraints. In this case, the Glu-137 side chain, which points to the floor of the pocket, could make strong contributions by H-bonds and/or electrostatic terms between EBP and the backbone of the agonist.
EBP has also been reported to bind the shorter VGV and VAPG peptides (32, 38). The fact that these shorter segments exhibit biological activities related to those VGVAPG possesses suggest that they share a common binding site. We have noticed that most of the stabilizing interactions between VGVAPG and EBP are found on the left part of the pocket, i.e. where the VGV of VGVAPG occurs. This observation let us suppose that the tripeptide VGV could dock to this site as the local conformation appears to properly fit its physicochemical properties. Similarly, the tetrapeptide VAPG could possibly bind on the left part of the pocket with stabilizing interactions with Leu-103 and Glu-137. These hypotheses need further investigations.
Altogether, our results suggest that Glu-137 and Leu-103 could be essential for VGVAPG binding because they mostly contribute to the stabilization of the complex. Besides, Arg-107 appears to contribute to maintaining the peptide within the binding site by bridging the C terminus of VGVAPG. This possibility has been confirmed using molecular dynamics of the EBP-VGVAPG complex (data not shown).
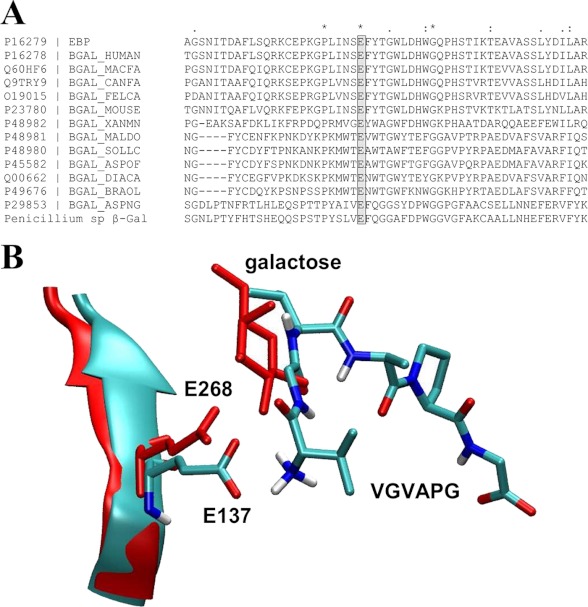
Finally, EBP is known for its ability to bind lactose, a compound blocking elastin peptide binding. Our data allow us to propose the following explanation. Of the residues important for VGVAPG biological activity, Glu-137, which provides the ionic interaction at the N terminus of VGVAPG, is the only one that does not belong to V32. This residue is strictly conserved in the multiple sequence alignment of the β-galactosidase family (Fig. 8A). In fact, according to the alignment and the splicing pattern of β-gal, that residue corresponds to Glu-268 of β-gal, a residue directly involved in the glycosidase activity of the enzyme. In fact, Glu-137 is the unique vestige of the catalytical site of β-gal (7) in EBP. It corresponds to the nucleophile (Glu-268) in this enzyme. Glu-188, the proton donor of β-gal, is absent from EBP as it is removed by the alternative splicing.
FIGURE 8.
Possible role of Glu-137 in the interplay between the elastin and galactolectin sites. A, sequence alignment showing the strict conservation of Glu-137 in β-galactosidases. Alignment ruler, *, identity' :, homology; ., similarity. B, after alignment of the protein backbone of their TIM barrels (human β-Gal and human EBP), the relative positions of Glu-268 interacting with galactose (in red, from Protein Data Bank code 3THC) and Glu-137 interacting with VGVAPG (from our model) suggest a possible competition between galactose and VGVAPG for Glu-137 binding. This competition at a common site could explain why EBP galactolectin occupancy blocks elastin peptide effects.
The crystallographic structure of human β-galactosidase bound to galactose is now available (Protein Data Bank code 3THC) (29). In β-gal, Glu-188 and Glu-268 are positioned to interact with galactosugars during the catalysis. Following splicing, one of these two residues, Glu-138 is removed, explaining why EBP lacks any enzymatic activity. However, the residue corresponding to Glu-268 of human β-gal (Glu-137 in EBP) remains. As we proposed that this residue is critical for binding VGVAPG and because crystallographic data show that it interacts with galactose, we aligned the TIM barrel backbone of the human β-gal interacting with galactose with that of EBP docked with VGVAPG. Furthermore, we extracted the atomic coordinates of Glu-268 and galactose in β-gal crystal (Protein Data Bank code 3THC) and the positions of Glu-137 and VGVAPG from our model.
The relative position of these compounds, reported in Fig. 8B, strongly suggests that the local presence of a galactose moiety could compete with VGVAPG for Glu-137 binding. This competition could explain why the elastin and galactolectin sites of EBP are mutually exclusive. Further numerical simulations and docking experiments of lactose are now underway to test this hypothesis.
This work presents the first functional model of the subunit EBP interacting with its VGVAPG ligand. As this interaction is important in several pathophysiological conditions, this knowledge now permits to envisage the rational design of elastin peptide antagonists that could either block the elastin binding site of EBP or selectively target Glu-137 as this residue could have a pivotal role in EBP functions. These molecules could be used to limit the progression of diseases where elastin degradation products are abundant and suspected of contributing to the disease progression.
Finally, our work suggests that Glu-137 of EBP could interact with both galactosugars and elastin peptides. As such, its involvement could possibly explain why galactosugars can remove elastin peptides from EBP. Further experimental and theoretical experiments are needed to fully understand this point.
Supplementary Material
Acknowledgment
We acknowledge the Molecular Modeling Platform and the ROMEO High Performance Computing Center for providing central processing unit time.
This work was supported by Fond National pour la Santé Grants ACI 2004 and ACI 2007 (Cancéropôle Grand-Est project), CNRS and Région Champagne Ardenne (CPER 2007 Reparation).
This article contains supplemental Fig. S1 and supplemental data.
- EBP
- elastin binding protein
- MMP-1
- collagenase-1.
REFERENCES
- 1. Kielty C. M. (2006) Elastic fibres in health and disease. Expert Rev. Mol. Med. 8, 1–23 [DOI] [PubMed] [Google Scholar]
- 2. Vrhovski B., Weiss A. S. (1998) Biochemistry of tropoelastin. Eur. J. Biochem. 258, 1–18 [DOI] [PubMed] [Google Scholar]
- 3. Shapiro S. D., Endicott S. K., Province M. A., Pierce J. A., Campbell E. J. (1991) Marked longevity of human lung parenchymal elastic fibers deduced from prevalence of d-aspartate and nuclear weapons-related radiocarbon. J. Clin. Invest. 87, 1828–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hinek A. (1994) Nature and the multiple functions of the 67-kD elastin-/laminin binding protein. Cell Adhes. Commun. 2, 185–193 [DOI] [PubMed] [Google Scholar]
- 5. Hinek A., Rabinovitch M., Keeley F., Okamura-Oho Y., Callahan J. (1993) The 67-kD elastin/laminin-binding protein is related to an enzymatically inactive, alternatively spliced form of β-galactosidase. J. Clin. Invest. 91, 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hinek A., Wrenn D. S., Mecham R. P., Barondes S. H. (1988) The elastin receptor: a galactoside-binding protein. Science 239, 1539–1541 [DOI] [PubMed] [Google Scholar]
- 7. Morreau H., Galjart N. J., Gillemans N., Willemsen R., van der Horst G. T., d'Azzo A. (1989) Alternative splicing of β-galactosidase mRNA generates the classic lysosomal enzyme and a β-galactosidase-related protein. J. Biol. Chem. 264, 20655–20663 [PubMed] [Google Scholar]
- 8. Privitera S., Prody C. A., Callahan J. W., Hinek A. (1998) The 67-kDa enzymatically inactive alternatively spliced variant of beta-galactosidase is identical to the elastin/laminin-binding protein. J. Biol. Chem. 273, 6319–6326 [DOI] [PubMed] [Google Scholar]
- 9. Hinek A., Pshezhetsky A. V., von Itzstein M., Starcher B. (2006) Lysosomal sialidase (neuraminidase-1) is targeted to the cell surface in a multiprotein complex that facilitates elastic fiber assembly. J. Biol. Chem. 281, 3698–3710 [DOI] [PubMed] [Google Scholar]
- 10. Hinek A., Rabinovitch M. (1994) 67-kD elastin-binding protein is a protective “companion” of extracellular insoluble elastin and intracellular tropoelastin. J. Cell Biol. 126, 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hinek A., Keeley F. W., Callahan J. (1995) Recycling of the 67-kDa elastin binding protein in arterial myocytes is imperative for secretion of tropoelastin. Exp. Cell Res. 220, 312–324 [DOI] [PubMed] [Google Scholar]
- 12. Lee S. H., Goswami S., Grudo A., Song L. Z., Bandi V., Goodnight-White S., Green L., Hacken-Bitar J., Huh J., Bakaeen F., Coxson H. O., Cogswell S., Storness-Bliss C., Corry D. B., Kheradmand F. (2007) Antielastin autoimmunity in tobacco smoking-induced emphysema. Nat. Med. 13, 567–569 [DOI] [PubMed] [Google Scholar]
- 13. Duca L., Floquet N., Alix A. J., Haye B., Debelle L. (2004) Elastin as a matrikine. Crit. Rev. Oncol. Hematol. 49, 235–244 [DOI] [PubMed] [Google Scholar]
- 14. Hornebeck W., Robinet A., Duca L., Antonicelli F., Wallach J., Bellon G. (2005) The elastin connection and melanoma progression. Anticancer Res. 25, 2617–2625 [PubMed] [Google Scholar]
- 15. Robert L. (1999) Aging of the vascular-wall and atherosclerosis. Exp. Gerontol. 34, 491–501 [DOI] [PubMed] [Google Scholar]
- 16. Kunecki M., Nawrocka A. (2001) Elastin-laminin receptor and abdominal aortic aneurysms. New subject to study? A review. Pathol. Biol. 49, 333–338 [DOI] [PubMed] [Google Scholar]
- 17. Jacob M. P., Fülöp T., Jr., Foris G., Robert L. (1987) Effect of elastin peptides on ion fluxes in mononuclear cells, fibroblasts, and smooth muscle cells. Proc. Natl. Acad. Sci. U.S.A. 84, 995–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Senior R. M., Griffin G. L., Mecham R. P. (1980) Chemotactic activity of elastin-derived peptides. J. Clin. Invest. 66, 859–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mochizuki S., Brassart B., Hinek A. (2002) Signaling pathways transduced through the elastin receptor facilitate proliferation of arterial smooth muscle cells. J. Biol. Chem. 277, 44854–44863 [DOI] [PubMed] [Google Scholar]
- 20. Duca L., Blanchevoye C., Cantarelli B., Ghoneim C., Dedieu S., Delacoux F., Hornebeck W., Hinek A., Martiny L., Debelle L. (2007) The elastin receptor complex transduces signals through the catalytic activity of its Neu-1 subunit. J. Biol. Chem. 282, 12484–12491 [DOI] [PubMed] [Google Scholar]
- 21. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 22. Rojas A. L., Nagem R. A., Neustroev K. N., Arand M., Adamska M., Eneyskaya E. V., Kulminskaya A. A., Garratt R. C., Golubev A. M., Polikarpov I. (2004) Crystal structures of β-galactosidase from Penicillium sp. and its complex with galactose. J. Mol. Biol. 343, 1281–1292 [DOI] [PubMed] [Google Scholar]
- 23. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huey R., Morris G. M., Olson A. J., Goodsell D. S. (2007) A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 28, 1145–1152 [DOI] [PubMed] [Google Scholar]
- 25. Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., Olson A. J. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19, 1639–1662 [Google Scholar]
- 26. Rutter J. L., Benbow U., Coon C. I., Brinckerhoff C. E. (1997) Cell-type specific regulation of human interstitial collagenase-1 gene expression by interleukin-1 β (IL-1β) in human fibroblasts and BC-8701 breast cancer cells. J. Cell Biochem. 66, 322–336 [PubMed] [Google Scholar]
- 27. Fahem A., Robinet A., Cauchard J. H., Duca L., Soula-Rothhut M., Rothhut B., Soria C., Guenounou M., Hornebeck W., Bellon G. (2008) Elastokine-mediated up-regulation of MT1-MMP is triggered by nitric oxide in endothelial cells. Int. J. Biochem. Cell Biol. 40, 1581–1596 [DOI] [PubMed] [Google Scholar]
- 28. Moroy G., Ostuni A., Pepe A., Tamburro A. M., Alix A. J., Héry-Huynh S. (2009) A proposed interaction mechanism between elastin-derived peptides and the elastin/laminin receptor-binding domain. Proteins 76, 461–476 [DOI] [PubMed] [Google Scholar]
- 29. Ohto U., Usui K., Ochi T., Yuki K., Satow Y., Shimizu T. (2012) Crystal structure of human β-galactosidase: structural basis of Gm1 gangliosidosis and morquio B diseases. J. Biol. Chem. 287, 1801–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Floquet N., Héry-Huynh S., Dauchez M., Derreumaux P., Tamburro A. M., Alix A. J. (2004) Structural characterization of VGVAPG, an elastin-derived peptide. Biopolymers 76, 266–280 [DOI] [PubMed] [Google Scholar]
- 31. Brassart B., Fuchs P., Huet E., Alix A. J., Wallach J., Tamburro A. M., Delacoux F., Haye B., Emonard H., Hornebeck W., Debelle L. (2001) Conformational dependence of collagenase (matrix metalloproteinase-1) up-regulation by elastin peptides in cultured fibroblasts. J. Biol. Chem. 276, 5222–5227 [DOI] [PubMed] [Google Scholar]
- 32. Faury G., Chabaud A., Ristori M. T., Robert L., Verdetti J. (1997) Effect of age on the vasodilatory action of elastin peptides. Mech. Ageing Dev. 95, 31–42 [DOI] [PubMed] [Google Scholar]
- 33. Hinek A., Boyle J., Rabinovitch M. (1992) Vascular smooth muscle cell detachment from elastin and migration through elastic laminae is promoted by chondroitin sulfate-induced “shedding” of the 67-kDa cell surface elastin binding protein. Exp. Cell Res. 203, 344–353 [DOI] [PubMed] [Google Scholar]
- 34. Mecham R. P., Hinek A., Griffin G. L., Senior R. M., Liotta L. A. (1989) The elastin receptor shows structural and functional similarities to the 67-kDa tumor cell laminin receptor. J. Biol. Chem. 264, 16652–16657 [PubMed] [Google Scholar]
- 35. Mecham R. P., Whitehouse L., Hay M., Hinek A., Sheetz M. P. (1991) Ligand affinity of the 67-kD elastin/laminin binding protein is modulated by the protein's lectin domain: visualization of elastin/laminin-receptor complexes with gold-tagged ligands. J. Cell Biol. 113, 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Booms P., Ney A., Barthel F., Moroy G., Counsell D., Gille C., Guo G., Pregla R., Mundlos S., Alix A. J., Robinson P. N. (2006) A fibrillin-1-fragment containing the elastin-binding-protein GxxPG consensus sequence upregulates matrix metalloproteinase-1: biochemical and computational analysis. J. Mol. Cell Cardiol. 40, 234–246 [DOI] [PubMed] [Google Scholar]
- 37. Heinz A., Jung M. C., Jahreis G., Rusciani A., Duca L., Debelle L., Weiss A. S., Neubert R. H., Schmelzer C. E. (2012) The action of neutrophil serine proteases on elastin and its precursor. Biochimie 94, 192–202 [DOI] [PubMed] [Google Scholar]
- 38. Pocza P., Süli-Vargha H., Darvas Z., Falus A. (2008) Locally generated VGVAPG and VAPG elastin-derived peptides amplify melanoma invasion via the galectin-3 receptor. Int. J. Cancer 122, 1972–1980 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.