Background: Cause of the complete loss of CDC73 in a subset of parathyroid tumors remains to be elucidated in the absence of a second mutation and promoter methylation.
Results: Oncogenic miR-155 causes down-regulation of tumor suppressor CDC73 in OSCC.
Conclusion: miR-155 up-regulation adds yet another mechanism for CDC73 down-regulation in tumors.
Significance: Targeting miR-155 adds novelty to OSCC therapeutics.
Keywords: Apoptosis, Cancer Therapy, HNSCC, MicroRNA, Tumor Suppressor Gene
Abstract
The CDC73 gene is mutationally inactivated in hereditary and sporadic parathyroid tumors. It negatively regulates β-catenin, cyclin D1, and c-MYC. Down-regulation of CDC73 has been reported in breast, renal, and gastric carcinomas. However, the reports regarding the role of CDC73 in oral squamous cell carcinoma (OSCC) are lacking. In this study we show that CDC73 is down-regulated in a majority of OSCC samples. We further show that oncogenic microRNA-155 (miR-155) negatively regulates CDC73 expression. Our experiments show that the dramatic up-regulation of miR-155 is an exclusive mechanism for down-regulation of CDC73 in a panel of human cell lines and a subset of OSCC patient samples in the absence of loss of heterozygosity, mutations, and promoter methylation. Ectopic expression of miR-155 in HEK293 cells dramatically reduced CDC73 levels, enhanced cell viability, and decreased apoptosis. Conversely, the delivery of a miR-155 antagonist (antagomir-155) to KB cells overexpressing miR-155 resulted in increased CDC73 levels, decreased cell viability, increased apoptosis, and marked regression of xenografts in nude mice. Cotransfection of miR-155 with CDC73 in HEK293 cells abrogated its pro-oncogenic effect. Reduced cell proliferation and increased apoptosis of KB cells were dependent on the presence or absence of the 3′-UTR in CDC73. In summary, knockdown of CDC73 expression due to overexpression of miR-155 not only adds a novelty to the list of mechanisms responsible for its down-regulation in different tumors, but the restoration of CDC73 levels by the use of antagomir-155 may also have an important role in therapeutic intervention of cancers, including OSCC.
Introduction
The CDC73 (cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (Saccharomyces cerevisiae)), also known as HRPT2 (hyperparathyroidism 2), is a tumor suppressor gene whose inactivation and mutation cause the hyperparathyroidism-jaw tumor syndrome. This syndrome is an autosomal dominant disorder characterized by the occurrence of parathyroid adenoma or carcinoma, fibro-osseous jaw tumors of the mandible or maxilla, and renal neoplastic and non-neoplastic abnormalities such as Wilm's tumor, hamartoma, and cystic renal disease (1–3). The 531-amino acid parafibromin encoded by CDC73 forms polymerase-associated factor1 (Paf1) complex (4–6). As a part of Paf1, it remains associated with ribonucleic acid (RNA) polymerase II, regulates global gene expression, and is involved in coupling of transcriptional and posttranscriptional events (7–10).
CDC73 overexpression is documented to inhibit colony formation and cellular proliferation and induces cell cycle arrest in the G1 phase, indicating that it has a critical role in cell growth and proliferation (11). Furthermore, BTK (Bruton's tyrosine kinase) has been found to increase the abundance of CDC73 in the absence of WNT3A stimulation, and in turn CDC73 acts as a repressor of β-catenin-mediated transcription in human colorectal cancer cells and B cells (12). These findings suggest the potential role of CDC73 in pathogenesis and progression of malignancies.
Besides mutations (predominantly), the loss of heterozygosity and promoter methylation of CDC73 in tumors has been reported as having different mechanisms for its down-regulation (13, 14). But there are also reports where methylation was not identified in any specimens despite a complete loss of parafibromin expression in parathyroid carcinomas with a single detectable CDC73 mutation and retention of the wild-type CDC73 allele (15). Furthermore, no mutations of possibly pathogenic nature were identified in the 5′-UTR of CDC73 (15). These data strongly suggest that some other mechanisms such as mutations in CDC73 intronic regions, alternate epigenetic regulation (e.g. histone modifications), or other regulatory inactivation mechanisms (e.g. targeting by microRNAs) may play a role in the loss of CDC73 expression. Regulation of CDC73 expression has been studied at transcriptional and posttranscriptional levels. Although BTK has been found to increase CDC73 level, it has been shown by quantitative polymerase chain reaction analysis of patient-derived B cells that CDC73 transcripts were unexpectedly more abundant in cells lacking BTK, but its protein level was low (12). This suggests that the availability of its protein level is determined by some posttranscriptional mechanisms (12). Therefore, a clear understanding of the post-transcriptional regulation of CDC73 will be of tremendous scientific and clinical significance.
Squamous cell carcinoma accounts for more than 95% of the carcinomas of the oral cavity (16). Oral squamous cell carcinoma (OSCC)2 in developing countries is the sixth most common cancer in males and tenth in females (16). In India it is the leading cancer in males and third most common malignancy in females (17). The five-year survival rate of OSCC lacks any improvements, and the efforts to develop an appropriate therapeutic strategy still lies in catch 22, demanding the identification of molecular markers and signaling pathways for better prognosis and therapeutic intervention of OSCC (17).
Being a multigenic disease, the single gene-based therapy of OSCC may fall short in therapy (18). Recent studies have shown that a growing class of noncoding RNAs called microRNAs (miRNAs) are involved in posttranscriptional regulation of genes (19). There is a growing body of literature supporting the potential role of miRNAs in regulation of cancer (20). The importance of CDC73 in the orchestration of several cellular functions and the prediction for its posttranscriptional regulation makes it an attractive candidate for miRNA-mediated regulation of cell growth and proliferation.
Here we report the identification of an oncogenic microRNA-155 (miR-155) that regulates the expression of tumor suppressor gene CDC73. Furthermore, restoration of CDC73 levels by the use of antagomir-155 warrants a prolific strategy to treat cancers, including OSCC.
MATERIALS AND METHODS
Sample Collection
A total of 22 matched OSCC samples and normal oral tissues from the patients were ascertained at the Bangalore Institute of Oncology, Bangalore, India. All tumor samples were mostly from the tongue and cheek areas of the mouth. This study was performed with informed consent from the patients and approval from the ethics committee of the Bangalore Institute of Oncology. The clinicopathological data for 22 patients are given in supplemental Table S1. Tumors were classified according to tumor, node, and metastasis criteria (21).
In Silico Identification of miRNA Binding Sites
We used a consensus approach by employing a total of six established target prediction programs (Table 1) to perform target prediction (22). We selected only miRNA, which is detected by all the six programs, to bind to the same site in the target 3′-UTR. Because inaccessibility to the region of binding may be due to strong local secondary structure, we therefore carried out the ΔG analysis for 70 bases on each side flanking to the miRNA binding site using the mFold software (23).
TABLE 1.
A list of predicted miRNAs having potential binding sites in the 3-UTR of CDC73 and binding free energy around the potential binding site of hsa-miR-155
hsa, Homo sapiens.
Plasmid Constructs
To generate the pcDNA3.1(+)-bic construct expressing the miR-155, BIC (B-cell integration center) exon 3 sequence was amplified using primers as described by Eis et al. (24) with certain modification that included incorporation of different restriction enzyme sites in the primers to facilitate directional cloning and human genomic DNA as a template. Similarly, the pcDNA3.1(+)-dme-mir-4 construct harboring the Drosophila melanogaster mir-4 was generated using D. melanogaster genomic DNA as a template in amplification reactions. To generate constructs with CDC73 3′-UTR at the 3′ end of the luciferase ORF in the pMIR-REPORT vector (Invitrogen), a 1214-base pair-long CDC73 3′-UTR from position +1595 to +2809 was amplified from human genomic DNA and cloned both in sense and antisense manner using a standard laboratory method. The sense, mutant, and antisense constructs were designated as pMIR-REPORT-3′-UTR-sense, pMIR-REPORT-3′-UTR-mutant, and pMIR-REPORT-3′-UTR-antisense, respectively. The mutant 3′-UTR was generated by site-directed mutagenesis by deleting bases 2–7 from the seed region of miR-155 binding site using pMIR-REPORT-3′-UTR-sense as a template. The pcDNA3-HA-Pf (pcDNA3-HA-CDC73) construct harboring a full-length CDC73 ORF was a kind gift from Dr. F. Kowalke from ETH, Zurich. The pcDNA3-HA-CDC73–3′-UTR-sense construct harboring the intact 3′-UTR was generated by amplifying and cloning of the 3′-UTR sequence downstream of the CDC73 ORF at XhoI and ApaI sites in pcDNA3-HA-CDC73. The pcDNA3-HA-CDC73-3′-UTR-mutant construct was generated by site-directed mutagenesis using pcDNA-HA-CDC73–3′-UTR-sense as a template and specific primers. All the constructs were sequenced on an ABIprism A310-automated sequencer (Invitrogen) to ascertain directionality and error free sequence of the inserts. Details of the primers used for cloning are available on request from the authors.
Cell Culture, Transient Transfection, and Reporter Assays
Human cell lines HEK293 (embryonic kidney), HEK293T (embryonic transformed kidney), A549 (lung adenocarcinoma), HeLa (cervical carcinoma), KB (oral squamous cell carcinoma), SCC084 (oral squamous cell carcinoma), and SCC131 (oral squamous cell carcinoma) were maintained in DMEM supplemented with 10% fetal bovine serum and 1x antibiotic/antimycotic solution (Sigma) in a humidified chamber with 5% CO2 at 37 °C. HEK293, HEK293T, A549, HeLa, and KB cells were procured from the National Centre for Cell Science, Pune, India. SCC084 and SCC131 cells were kind gifts from Prof. Susanne Gollin, University of Pittsburgh, Pittsburgh, PA.
For overexpression studies, 2 × 106 cells/well in a 6-well plate were transfected with an appropriate construct or a combination of constructs using LipofectamineTM 2000 (Invitrogen) according to the manufacturer's instructions. Luciferase reporter gene assays were performed post 24 or 48 h of transfection using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI) according to the manufacturer's protocol. Cells were also co-transfected with the pRL-TK control vector (Promega), encoding Renilla luciferase for normalizing transfection efficiency. Different stable transfectants in KB cells were generated by the pooling of clones acquired after KB cells were continuously grown in G418 containing selection medium post 48 h of transfection.
Total RNA Extraction and RT-PCR
Total RNA was isolated from different cell lines using the TRI-ReagentTM (Sigma) according to the manufacturer's instructions. RNA was stored at −70 °C until further use. cDNA was made using a first-strand cDNA synthesis kit (Fermentas, Ontario, Canada). RT-PCR was performed using primers specific to CDC73 and GAPDH following a standard protocol. Details of the primers used for RT-PCR are available on request from the authors.
miR-155 Northern Hybridization
Total RNA (50 μg) samples were run on a 15% acrylamide gel under denaturing conditions, transferred onto a N+Biodyne nylon membrane (Pall, Port Washington, NY), and hybridized with a γ-32P-labeled probe (5′-ACCCCTATCACGATTAGCATTAA-3′) specific to the antisense strand of human miR-155. Hybridization was performed overnight at 42 °C in a hybridization buffer containing 5× SSC, 20 mm NaH2PO4, pH 7.5, 7% SDS, 2× Denhardt's solution, and 40 ng/ml sheared salmon sperm DNA. Subsequently, the membrane was washed in 3× SSC, 25 mm NaH2PO4, pH 7.5, 5% SDS, and 10× Denhardt's solution twice for 5 min each followed by washing in 1× SSC and 1% SDS once for 5 min. The membrane was dried and exposed to a phosphorimaging screen (Fujifilm, Japan) for 48 h. As a loading control, a γ-32P-labeled probe (5′-GCTAATCTTCTCTGTATCGTTCCA) specific to the U6 RNA was used.
Western Hybridization
Whole protein lysates were prepared using the CelLyticTM M Cell Lysis Reagent (Sigma). They were resolved on a SDS-PAGE and transferred to a BioTrace PVDF nylon membrane (Pall, Port Washington, NY) using a semi-dry transfer apparatus. The membrane was blocked using 5% fat-free milk powder in 1× PBS-Tween, and the signal was visualized using a 1:750 dilution of an anti-CDC73 antibody either from Santa Cruz Biotechnology (catalog #2H1) or from Sigma (product #C3620). After incubation with a horseradish peroxidase linked secondary antibody, the signal was detected using the ImmobilonTM Western Chemiluminescent HRP substrate (Millipore, Billerica, MA) and x-ray films. The mouse β-actin and HA epitope antibodies were purchased from Sigma.
miR-Q RT-PCR
The miR-155 expression analysis was carried out by miR-Q, a method developed by Sharbati-Tehrani et al. (25) for the expression analysis of miRNA by RT-PCR. The expression of miR-155 in OSCC samples was compared with normal tissues using the 2−ΔΔCT method and 5 S rRNA as a normalizing control (26).
Mutation Analysis
The entire coding region of CDC73 including its exon/intron junctions was amplified using gene-specific primers and sequenced on an ABIprism A310-automated sequencer. Details of the primers used for mutation analysis are available on request from the authors.
LOH Analysis at the CDC73 Locus
For LOH studies, total genomic DNA from blood, and tissue samples was isolated using the Wizard® Genomic DNA purification kit (Promega) according to the manufacturers' instructions. Matched normal and tumor DNA samples were genotyped using four highly polymorphic microsatellite markers: three extragenic markers (D1S384, D1S422, and D1S408) and one intragenic marker (para14M) present in intron 14 of CDC73. Details of the first three markers were retrieved from the UCSC Genome Bioinformatics site. Details of the internal marker were adapted from a previous report (1).
Combined Bisulfite Restriction Analysis
Methylation status of the CDC73 promoter described by Hewitt et al. (14) was examined by combined bisulfite restriction analysis (COBRA) using AciI and BstUI enzymes following a standard procedure in our laboratory (27).
Cell Proliferation Assay
The cell proliferation was measured by the MTT assay as described previously (28). In brief, 48 h of post transfection of HEK293 or KB cells in 6-well plates with different constructs, antagomir-155 (catalog #AM17000; Invitrogen) or scrambled oligo (catalog #AM17010, Invitrogen), transfection medium in each well was replaced by 100 μl of fresh serum-free medium with 0.5 g/liter MTT. After incubation at 37 °C for 4 h, the MTT medium was aspirated out, and 50 μl of DMSO was added to each well. After incubation at 37 °C for another 10 min, the absorbance value of each well was measured using an ELISA plate reader (Bio-Rad) at 595 nm. The relative cell proliferation rate was calculated according to Jiang et al. (28).
Apoptosis Assay
The rate of apoptosis was measured using the CaspGLOWTM Fluorescein Active Caspase-3 Staining kit (BioVision, Milpitas, CA) using the manufacturer's protocol. The kit provides a sensitive detection of activated caspase-3 (CASP3) in living cells. It utilizes the CASP3 inhibitor, DEVD-fluoromethyl ketone, conjugated to FITC (FITC-DEVD-FMK) as a marker. Briefly, 1 × 106 cells were grown in 6-well plates to about 60% confluence and transiently transfected with pcDNA3.1(+)-dme-mir-4, pcDNA3.1(+)-bic, antagomir-155, or scrambled oligo. Post 48 h of transfection, cells were resuspended in a complete DMEM and incubated with FITC-DEVD-fluoromethyl ketone conjugate for 1.5 h in dark. Post incubation, cells were washed and finally suspended in 300 μl of wash buffer. Cells were then analyzed with the FACSCalibur flow cytometer (BD Biosciences). The relative cell apoptosis rate was calculated according to Jiang et al. (28). The same protocol was used to assess the rate of apoptosis of different stable KB transfectants.
Soft Agar Assay
Tumor cells have the propensity to grow in an anchorage-independent manner and hence can form colonies in a semi-solid medium such as soft agar. The anchorage-independent growth of the stable CDC73 and vector clones was analyzed by the soft agar assay in 35-mm tissue culture dishes by following a standardized protocol.
Propidium Iodide Analysis
The proliferation of stable vector and CDC73 clones was analyzed by the FACSCalibur flow cytometer after staining the cells with propidium iodide (Sigma) following a standard protocol.
In Vivo Assay for Tumor Growth
To see the effect of restoration of CDC73 expression by inhibition of miR-155 on tumor growth, 2 × 106 KB cells were transfected separately with 200 nm antagomir-155 or mock (scrambled oligos). Post 24 h of transfection, 3 × 106 cells were suspended in 150 μl of Dulbecco's modified phosphate buffer and then subcutaneously injected into the right posterior flank of each female BALB/c athymic 6-week-old nude mouse. Tumors of measurable size appeared post 1 week of injection. Tumor growth was monitored, and its volume was measured using a digital caliper for every 3 days till 23 days. Tumor volume (V) was calculated as V = L × W2 × 0.5, where L and W represent length and width, respectively. Tumor weight was taken at the end of the study. All nude mice experiments were approved by the institutional animal ethics committee.
Statistical Analysis
An independent two-tailed Student's t test was performed to determine the significance of difference between two experiments.
RESULTS
Identification of miR-155 as a Regulator of CDC73
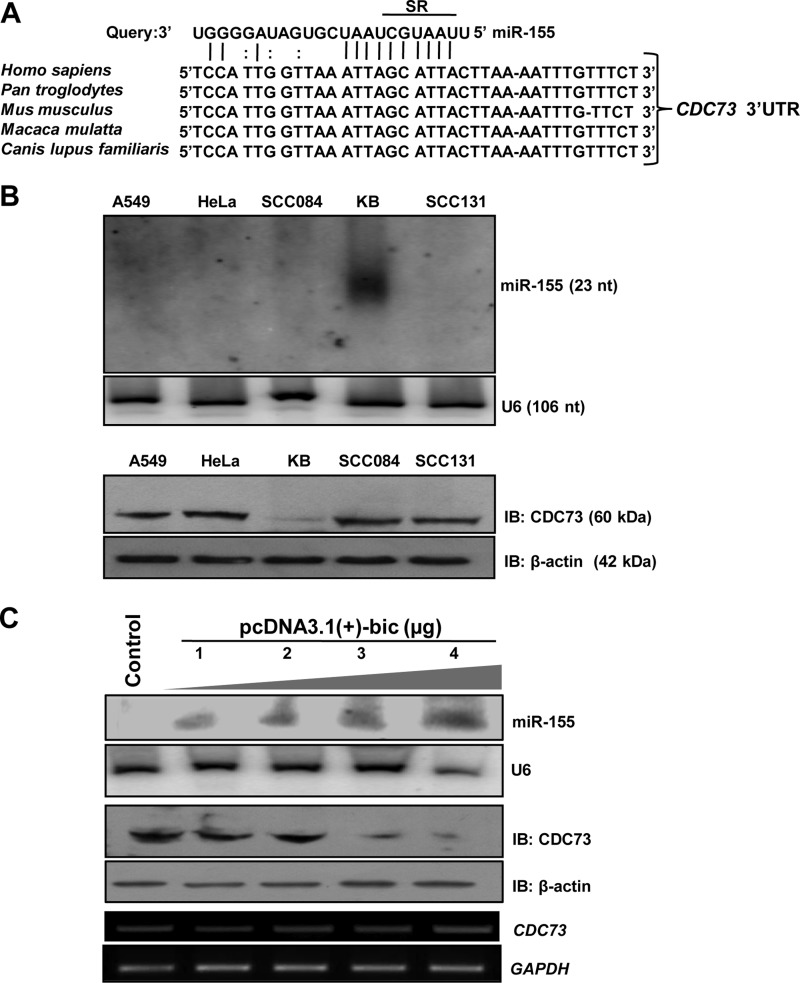
To predict the miRNAs that may regulate CDC73, we used a consensus approach by employing six different miRNA target prediction algorithms such as DIANA-microT, PicTar, Miranda, TargetScan, MirTarget2, and Microcosm. We found that several miRNAs have potential binding sites in the 3′-UTR of CDC73 (Table 1). These miRNA binding sites are evenly scattered all over the 3′-UTR. We preferentially picked up miR-155 for its validation because all target prediction algorithms predicted it targeting the CDC73 (Table 1). Our bioinformatics analysis predicted the binding of miR-155 to the 3′-UTR of CDC73 in a region encompassing +1831 to +1854 bases (Fig. 1A). The ΔG analysis showed that the miR-155 binding region in the 3′-UTR has a very weak secondary structure (Table 1). The ClustalW alignment of miRNA seed region binding site in the 3′-UTR of CDC73 showed that it is conserved in different species (Fig. 1A), underscoring that miR-155 may have a basal role to play in regulating CDC73 expression.
FIGURE 1.
Identification of miR-155 as a regulator of CDC73 expression. A, shown is a putative miR-155 binding site within the CDC73 3′-UTR (GenBankTM accession no. NM_024529.0). Perfect matches and G:U (or G:T) pairs are indicated by vertical lines and colons, respectively. The ClustalW alignment shows that the putative miR-155 binding site is conserved in different species. SR (seed region) denotes the seed region. B, shown is expression analysis of miR-155 and CDC73 in A549, HeLa, SCC084, KB, and SCC131 cells. Note the expression of miR-155 only in KB cells (upper panel). Also note the reduced expression of CDC73 in KB cells as compared with other cell lines (lower panel), suggesting that miR-155 negatively regulates CDC73. The expression of miR-155 and CDC73 was analyzed by Northern and Western hybridizations, respectively. C, shown is the effect of miR-155 overexpression on CDC73 expression in HEK293T cells. For overexpression of miR-155, 1–4 μg of pcDNA3.1(+)-bic was transfected in HEK293T cells. The Control lane represents transfection of HEK293T cells with the D. melanogaster miRNA dme-mir-4-expressing construct pcDNA3.1(+)-dme-mir-4 that does not have any mammalian target as shown by prediction programs used in this study. Note the overexpression of miR-155 led to reduced CDC73 expression in a dose-dependent manner. U6 RNA, GAPDH, and β-actin were used as loading controls for Northern, RT-PCR, and Western hybridization, respectively. IB, immunoblot; nt, nucleotides.
Effect of miR-155 Overexpression on CDC73 Expression
To validate if miR-155 regulates CDC73 expression, a panel of human cell lines (viz., A549, HeLa, KB, SCC084, and SCC131) was examined for miR-155 expression by Northern blot analysis. The expression of miR-155 was easily detectable in KB cells, whereas it was not detectable in other cell lines even after loading 50 μg of total RNA (Fig. 1B, upper panel). To see whether this differential expression of miR-155 in KB cells is reflected with a low expression of its predicted target CDC73, total protein lysates were prepared from these cell lines and examined for CDC73 expression by Western blotting. CDC73 was barely detectable in KB cells as compared with other cell lines (Fig. 1B). Also, KB cells did not show any CDC73 promoter methylation and mutation upon analysis, suggesting the regulation of CDC73 by miR-155.
The pcDNA3.1(+)-bic construct coding for miR-155 was transiently transfected into HEK293T cells followed by Northern and Western blot analyses to check for the expression of miR-155 and CDC73, respectively. The results showed that, as expected, miR-155 down-regulated CDC73 in a dose-dependent manner, whereas the control pcDNA3.1(+)-dme-mir-4 construct coding for an irrelevant D. melanogaster mir-4 had no effect (Fig. 1C). It is interesting to note that the level of CDC73 transcript remained constant (Fig. 1C). Thus, it is most likely that miR-155 decreases CDC73 expression by inhibiting its translation.
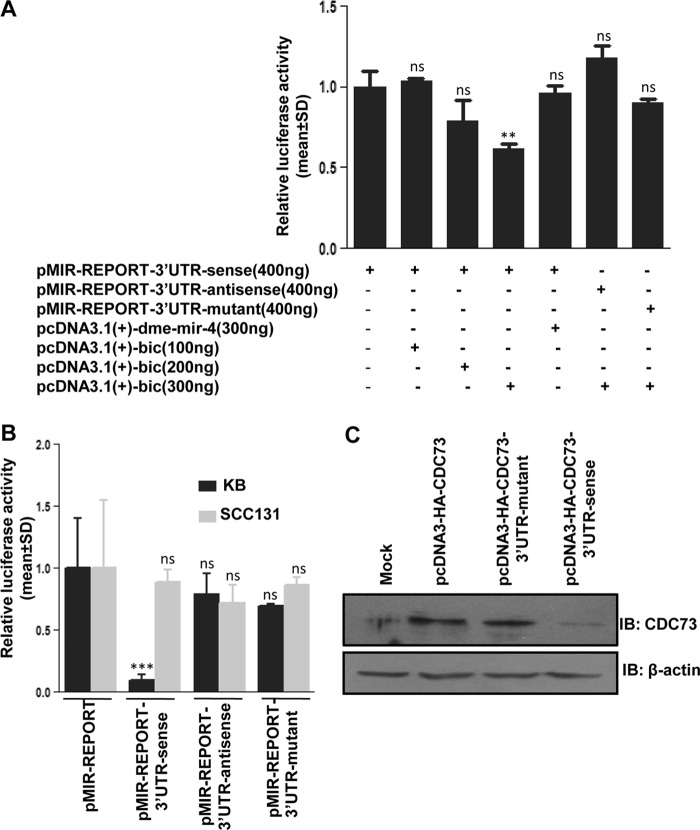
Confirmation of Target Site for miR-155 in CDC73 3′-UTR
When HEK293T cells were cotransfected with pcDNA3.1(+)-bic coding for miR-155 and pMIR-REPORT-3′-UTR-sense containing the intact CDC73 3′-UTR in a sense orientation, we observed a significantly reduced luciferase reporter expression compared with cells transfected with pMIR-REPORT-3′-UTR-sense (Fig. 2A). However, when HEK293T cells were cotransfected with pMIR-REPORT-3′-UTR-sense and pcDNA3.1(+)-dme-mir-4 constructs, there was no significant effect on luciferase expression compared with cells transfected with pMIR-REPORT-3′-UTR-sense (Fig. 2A). Most importantly, when pcDNA3.1(+)-bic was cotransfected with pMIR-REPORT-3′-UTR-mutant containing the mutated binding site or pMIR-REPORT-3′-UTR-antisense containing the intact CDC73 3′-UTR in an antisense orientation and lacking the miR-155 binding site in HEK293T cells, there was no significant reduction in the luciferase expression as compared with cells transfected with pMIR-REPORT-3′-UTR-sense (Fig. 2A). Also, when the pMIR-REPORT-3′-UTR-sense construct was transfected in KB cells with a high level of endogenous miR-155, the luciferase expression was significantly reduced as compared with KB cells with the control pMIR-REPORT vector (Fig. 2B). This down-regulation was not seen in KB cells transfected with either pMIR-REPORT-3′-UTR-antisense or pMIR-REPORT-3′-UTR-mutant (Fig. 2B). No change in the luciferase expression was observed in SCC131 cells with an undetectable expression of miR-155 after transfection of any of these constructs in comparison to SCC131 cells transfected with pMIR-REPORT (Fig. 2B). Furthermore, as expected, when KB cells having a high endogenous level of miR-155 were stably transfected with the CDC73 construct pcDNA3-HA-CDC73–3′-UTR-sense, the expression of CDC73 was reduced compared with cells transfected with either pCDNA3-HA-CDC73 lacking the 3′-UTR or pcDNA3-HA-CDC73–3′-UTR-mutant (Fig. 2C). These experiments clearly indicated that the mature miR-155 regulates CDC73 expression by interacting with its 3′-UTR in a site-specific manner.
FIGURE 2.
Confirmation of the miR-155 target site in the CDC73 3′-UTR. A, miR-155 reduces luciferase expression in HEK293T cells by binding to a target site in the CDC73 3′-UTR. Note that an increasing concentration of pcDNA3.1(+)-bic significantly reduces the luciferase expression in cells co-transfected with pMIR-REPORT-3′-UTR-sense construct compared with cells transfected with pMIR-REPORT-3′-UTR-sense alone, whereas the luciferase expression does not change in cells co-transfected with pcDNA3.1(+)-bic and pMIR-REPORT-3′-UTR-antisense or pMIR-REPORT-3′-UTR-mutant constructs compared with cells transfected with p-MIR-REPORT-3′-UTR-sense construct alone. Also note that the luciferase expression does not change in cells co-transfected with pMIR-REPORT-3′-UTR-sense and pcDNA3.1(+)-dme-mir-4 constructs compared with cells transfected with p-MIR-REPORT-3′-UTR-sense construct alone, suggesting that miR-155 reduces the luciferase expression by binding to a specific site in the CDC73 3′-UTR. The total DNA for each transfection was adjusted to 800 ng with the pcDNA3.1(+) vector. B, the effect of endogenous miR-155 on luciferase reporter expression is shown. The luciferase reporter vector pMIR-REPORT (control) and luciferase reporter constructs with CDC73 3′-UTR, pMIR-REPORT-3′-UTR-sense, pMIR-REPORT-3′-UTR-mutant, and pMIR-REPORT-3′-UTR-antisense were transfected individually in KB and SCC131 cells, and the luciferase expression was measured post-24 h of transfection. Note a reduced expression of luciferase reporter in KB cells with pMIR-REPORT-3′-UTR-sense construct as compared with cells transfected with the pMIR-REPORT vector, pMIR-REPORT-3′-UTR-mutant, or pMIR-REPORT-3′-UTR-antisense constructs. This is due to the fact that a high endogenous level of miR-155 is able to bind the CDC73 3′-UTR in a sense orientation and inhibit translation of the luciferase reporter. Also note that the luciferase expression does not change in SCC131 cells with an undetectable level of miR-155 when these cells were transfected with the pMIR-REPORT vector, pMIR-REPORT-3′-UTR-sense, pMIR-REPORT-3′-UTR-mutant, or pMIR-REPORT-3′-UTR-antisense constructs. C, shown is the effect of an endogenous level of miR-155 on the expression of CDC73 with or without its 3′-UTR in KB cells. Note a reduced expression of CDC73 in cells transfected with the construct pcDNA3-HA-CDC73–3′-UTR-sense having its 3′-UTR in a sense orientation as compared with cells transfected with the construct pcDNA3-HA-CDC73 without its 3′-UTR or the construct pcDNA3-HA-CDC73–3′-UTR-mutant with its mutated miR-155 binding site, underscoring that miR-155 targets CDC73 by binding to its 3′-UTR. Mock represents cells transfected with the pcDNA3-HA vector. n = 3; ns, data are statistically non-significant; **, p < 0.01; ***, p < 0.001. IB, immunoblot.
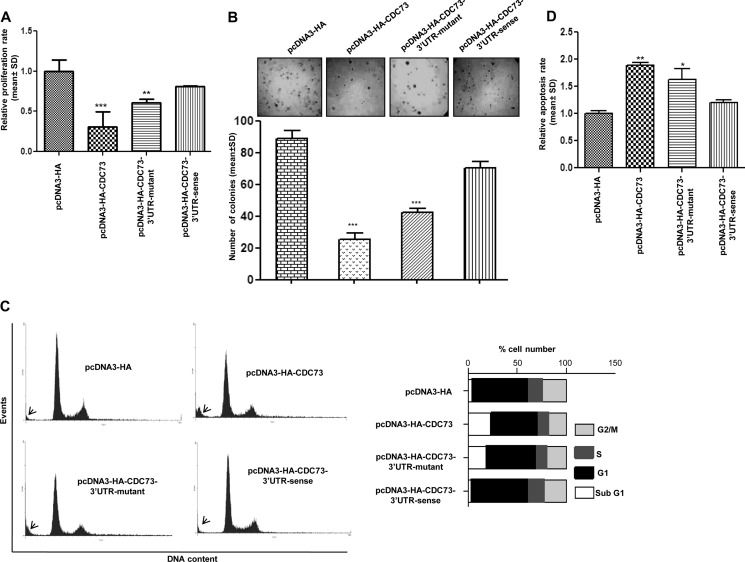
CDC73 Overexpression Is Dependent on the Presence or Absence of Its 3′-UTR
To assess the role of CDC73 3′-UTR in the regulation of its expression and hence its function, CDC73 constructs with and without 3′-UTR were stably transfected into KB cells. We chose this cell line because it has a high expression of miR-155 and a low level of CDC73 (Fig. 1B). We then assessed the rate of proliferation and apoptosis of KB cells stably transfected with different CDC73 constructs by FACS analysis of propidium iodide-stained cells, MTT, soft agar, and CASP3 assays. Compared with KB cells harboring the pcDNA3-HA vector or the pcDNA3-HA-CDC73–3′-UTR-sense construct with an intact 3′-UTR, proliferation and anchorage-independent growth of cells stably transfected with pcDNA3-HA-CDC73 (lacking 3′-UTR) and pcDNA3-HA-CDC73–3′-UTR-mutant were significantly decreased (Fig. 3, A and B), whereas the rate of apoptosis was significantly increased (Fig. 3, C and D). These experiments not only underscore the fact that CDC73 has a definite role in regulating cell growth and proliferation but also give an insight that miR-155 is exercising its oncogenic role by targeting CDC73.
FIGURE 3.
Anti-tumorigenic function of CDC73 in KB cells. A, shown is a quantitative analysis of cell proliferation by the MTT assay. Note a significantly reduced proliferation of KB cells transfected with either pcDNA3-HA-CDC73 or pcDNA3-HA-CDC73–3′-UTR-mutant constructs compared with cells transfected with either pcDNA3-HA (vector control) or pcDNA3-HA-CDC73–3′-UTR-sense construct. B, shown is an assessment of tumorigenic potential of KB cells by the soft agar assay. Note a reduction in the number of colony formation by KB cells transfected with either pcDNA3-HA-CDC73 or pcDNA3-HA-CDC73–3′-UTR-mutant constructs compared with cells transfected with either pcDNA3-HA or pcDNA3-HA-3′-UTR-CDC73-sense construct. The lower panel is a graphical representation of the soft agar data. C, shown is a FACS analysis of propidium iodide-stained KB cells stably transfected with the pcDNA3-HA vector and different CDC73 constructs. The sub-G1 population (cells undergoing death) is marked by arrows. Note an increase in sub-G1 population of cells transfected with either pcDNA3-HA-CDC73 or pcDNA3-HA-CDC73–3′-UTR-mutant constructs compared with cells transfected with either pcDNA3-HA or pcDNA3-HA-CDC73–3′-UTR-sense. A graphical representation of the data is shown on the right. D, shown is the assessment of the rate of apoptosis by the CASP3 assay. Note a significantly increased rate of apoptosis in KB cells transfected with either pcDNA3-HA-CDC73 or pcDNA3-HA-CDC73–3′-UTR-mutant constructs compared with cells transfected with either pcDNA3-HA or pcDNA3-HA-CDC73–3′-UTR-sense. n = 3; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
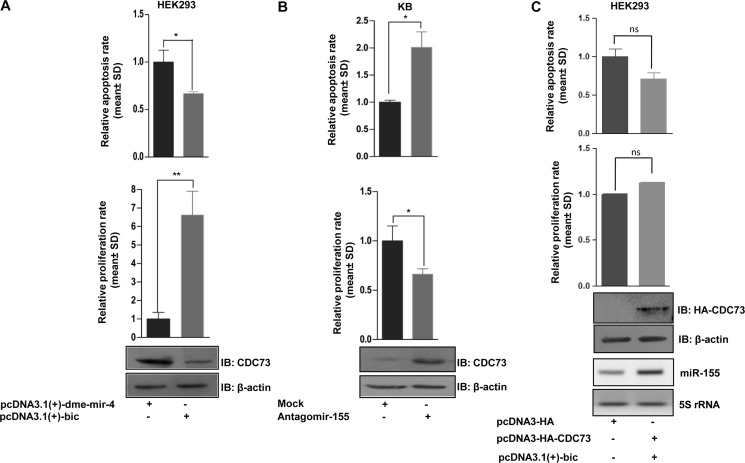
Regulation of Cell Growth and Proliferation by CDC73 as a Target of miR-155
To confirm further that the miR-155-mediated reduction of CDC73 has a functional relevance in cell growth and proliferation, we knocked down CDC73 expression in HEK293 cells using miR-155 expressing construct pcDNA3.1(+)-bic and quantitated cell proliferation and cell death (apoptosis) by MTT and CASP3 assays, respectively. As expected, the results demonstrated that the reduced CDC73 level is associated with an increased cell proliferation and decreased apoptosis (Fig. 4A). Because KB cells have a high endogenous level of miR-155 (Fig. 1B), we knocked it down using antagomir-155. The results showed that the CDC73 levels went up, resulting in decreased cell proliferation and enhanced apoptosis (Fig. 4B). Because a given miRNA can target several genes, we assessed the tumorigenic potential of miR-155 by targeting CDC73. The CDC73 construct (pcDNA3-HA-CDC73) was cotransfected with pcDNA3.1(+)-bic into HEK293 cells, and their proliferation and apoptosis were measured. Interestingly, the CDC73 abrogated the pro-oncogenic effect of miR-155 in HEK293 cells (Fig. 4C), although not completely. These results suggested that miR-155 regulates cell growth and proliferation largely by targeting CDC73.
FIGURE 4.
Regulation of cell proliferation and apoptosis by CDC73 as a target of miR-155. A, overexpression of miR-155 (pcDNA3.1(+)-bic) leads to a significantly reduced CDC73 expression, resulting in increased cell growth and proliferation and decreased apoptosis in HEK293 cells compared with cells transfected with a non-relevant Drosophila miRNA pcDNA3.1(+)-dme-mir-4 construct. IB, immunoblot. B, knockdown of miR-155 by antagomir-155 led to an increased CDC73 expression, resulting in reduced cell growth and proliferation and increased apoptosis in KB cells compared with cells transfected with a non-relevant scrambled oligo (mock). C, CDC73 overexpression abrogated the effect of miR-155 overexpression on proliferation and apoptosis in HEK293 cells. n = 3; ns, data are statistically non-significant; *, p < 0.05; **, p < 0.01.
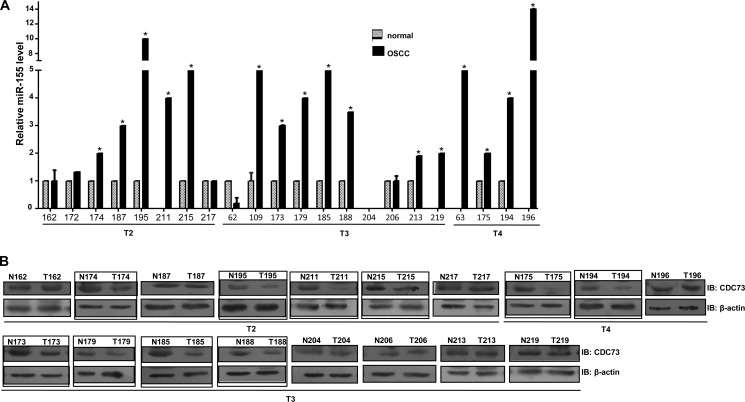
CDC73 Is Down-regulated in OSCC Samples with Up-regulation of miR-155
After validating CDC73 as a target of miR-155 by bioinformatics and in vitro assays, we analyzed the expression of miR-155 by qRT-PCR in OSCC samples. The results showed a significant up-regulation of miR-155 in 16/22 OSCC samples as compared with their matched normal oral tissues (Fig. 5A). Furthermore, we also analyzed the expression of CDC73 in 18 of these 22 samples. As expected, a majority of OSCC samples (10/18) having a high expression of miR-155 showed a low level of CDC73 (Fig. 5, A and B). To exclude the possibility of alternate mechanisms as the cause of CDC73 down-regulation in OSCC, all the 10 OSCC samples were analyzed for LOH at the CDC73 locus, somatic mutations, and promoter methylation. The results showed that these 10 OSCC samples did not show any mutation, promoter methylation, or LOH (supplemental Table S1), again validating the fact that CDC73 is a biological target of oncogenic miR-155 and possibly could be the major mechanism for CDC73 down-regulation in OSCC.
FIGURE 5.
Up-regulation of miR-155 in OSCC samples with reduced expression of CDC73. A, mir-Q RT-PCR was performed to assess the level of miR-155 in matched OSCC and normal oral tissues (n = 22). Note a significant up-regulation of miR-155 levels in 16/22 OSCC samples compared with their matching normal samples (p < 0.05). No miR-155 expression was detected in normal samples from patients #211, 219, 63, and 196. The relative expression of miR-155 in tumor samples from these patients was calculated relative to the average expression of miR-155 in the rest of the normal samples. Both normal and tumor tissues of patient #204 did not show miR-155 expression upon analysis by mir-Q RT-PCR. B, shown is Western blot (IB) analysis to assess the expression of CDC73 in 18 matched OSSC and normal oral tissue samples. Note the down-regulation of CDC73 in 10 samples (marked by a rectangle) in comparison to their matching normal counterparts; this down-regulation occurs even in the early stages of OSCC development (see tumor samples from patient #174, 195, 211, and 215). Also note that all the 10 OSCC samples showing reduced CDC73 expression have a significant up-regulation of miR-155. The numbers represent different patients. T2, T3, and T4 denote stages of the tumors.
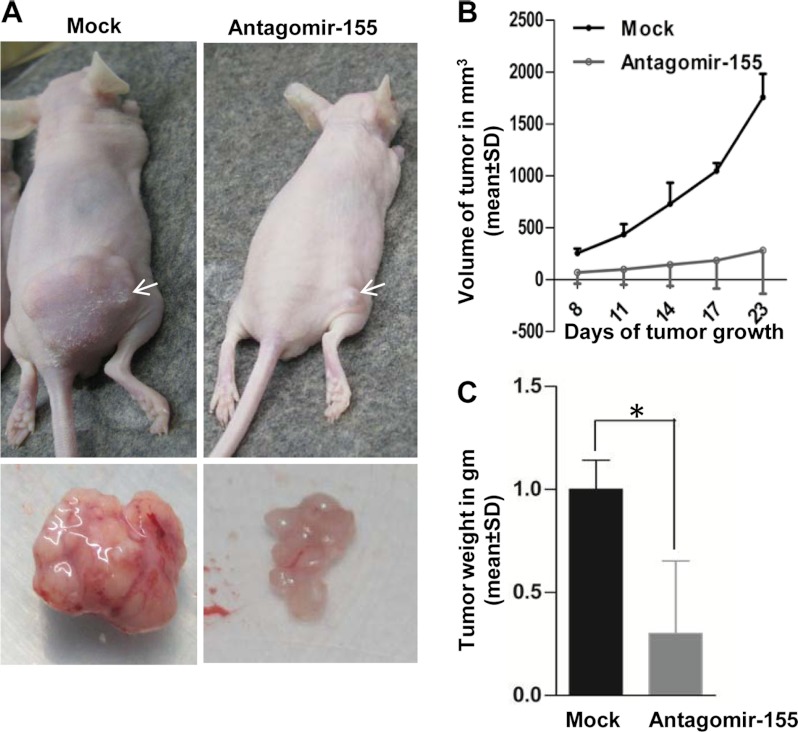
Restoration of CDC73 Expression by miR-155 Inhibition Suppresses Tumor Growth in Vivo
Based on the results of in vitro studies, we hypothesized that the abolition of miR-155 and the restoration of tumor suppressor CDC73 expression in KB cells might have an anti-tumor effect in vivo. To address this critical question, we used an in vivo pretreated xenograft nude mouse model. Briefly, we transfected the antagomir-155 and scrambled oligos (mock) separately in KB cells. Post 24 h of transfection, these cells were injected separately into nude mice. The mice were observed for the growth of tumors till 23 days. As expected, pretransfection of antagomir-155 into KB cells led to a significant reduction in both tumor volume and weight as compared with mock transfectant (Fig. 6).
FIGURE 6.
miR-155 inhibition suppresses tumorigenicity in vivo in BALB/c nude mice. A, shown is the effect ofantagomir-155 pretreatment to KB cells on tumor formation in a nude mouse xenograft model. Scrambled oligo-transfected (mock, left panel) and antagomir-155-transfected KB cells (right panel) were subcutaneously injected into posterior right flanks of nude mice (n = 6). Photographs illustrate representative features of tumor growth on day 23 after injection. Arrows mark the xenografts. Excised xenografts are shown below. B, shown is the effect of the antagomir-155 on tumor volume as compared with mock-treated KB cells during a time course of 23 days. C, shown is the effect of the antagomir-155 on tumor weight as compared with mock-treated KB cells. The graph illustrates the weight difference on day 23 after injection. Note that the pretreatment of KB cells with antagomir-155 significantly reduces the tumor volume and weight in nude mice xenografts. *, p < 0.05.
DISCUSSION
The 3′-UTR of the human CDC73 gene is almost 4 kb long, whereas the mean size of 3′-UTRs of human genes is around 800 nucleotides (29). It also contains Alu sequences that have been recently reported as the donors of potential miRNA target sites (30). As mentioned before, a complete loss of CDC73 expression has been reported in a subset of parathyroid carcinomas in the absence of promoter methylation with a single detectable somatic mutation and retention of the wild-type allele (15). These observations suggested that CDC73 expression could be regulated through its 3′-UTR. In this study we found that miR-155 has a key role in regulating CDC73 expression. We provide a combination of in silico, in vitro, and in vivo evidence including a series of miRNA transfection experiments in support of this conclusion.
Our study focused on miR-155, a microRNA that has been shown to be highly overexpressed in lymphomas of activated-B-cell origin, including Hodgkin's lymphoma (31, 32) and diffuse large B-cell lymphoma (33). Not an exclusive bane of lymphoid cells, miR-155 has also been detected at elevated levels in the bone marrow of some patients suffering with acute myeloid leukemia (34). miR-155 overexpression has additionally been observed in solid tumors of diverse origin (e.g. breast, lung, stomach, prostrate, colon, thyroid, pancreatic, and head and neck squamous cell carcinoma) (35–39). All these lines of evidence are consistent with the notion that miR-155 functions as an oncogenic miRNA in human cancers.
As mentioned above, miR-155 is overexpressed in many human cancers. But the mechanisms by which it functions as a putative oncogenic miRNA are largely unknown. Oncogenic miRNAs, which are overexpressed in tumors, function in tumor development mainly via repressing the expression of genes having anti-tumoral activity (40, 41). Of note, miR-155 promotes pancreatic tumor development and mammary gland epithelial cell migration and invasion via targeting the tumor suppressors TP53INP1 and RhoA, respectively (42, 43). In addition, miR-155 induces B-cell malignancies by targeting Ship and C/EBPβ in a transgenic mouse model (44). Given the scenario that miRNAs usually regulate a large set of targets, there might be more miR-155 targets having anti-tumoral activity. CDC73, having anti-tumoral activity, could be one of them.
Our finding of a differential level of miR-155 in various cell lines (Fig. 1B) and a subset of paired normal and OSCC samples (Fig. 5A) supports that the miR-155-mediated control of CDC73 is operational in cells that actively produce it. Furthermore, the expression analysis of miR-155 showed that it is highly enriched in a majority of OSCC samples compared with their normal counterparts and had an inverse effect on CDC73 expression, suggesting its key role in the pathogenesis of OSCC by targeting CDC73 (Fig. 5, A and B). Overexpression of miR-155 down-regulated CDC73 expression at the protein level in HEK293T cells, but there was no change at the RNA level (Fig. 1C). Pertinently, this change at the protein level, but not at the transcript level, has also been reported for other validated targets of miR-155 like AT1R (45). Furthermore, the effect of CDC73 differential overexpression was reflected on the proliferation, anchorage-independent growth, and apoptosis of KB cells stably transfected with different CDC73 constructs (Fig. 3). Overexpression of miR-155 not only led to the knockdown of CDC73 but also inversed the effect of CDC73 by promoting proliferation and suppression of apoptosis (Fig. 4A). Restoration of CDC73 by knockdown of miR-155 using antagomir-155 was accompanied by decreased cell proliferation, enhanced apoptosis, and reduction of tumor growth in xenograft pretreated model (Figs. 4B and 6). Furthermore, the combined overexpression of CDC73 and miR-155 into HEK293 cells abrogated the oncogenic potential of miR-155 (Fig. 4C). Thus, our findings highlight the key role that CDC73 plays in orchestration and functional maintenance of oral tissue.
High expression levels of miR-155 have been found to correlate with poor prognoses of lung cancer and pancreatic tumor (38, 46). Our results on therapeutic delivery of antagomir-155 in vivo provide a key to therapeutics of malignant OSCC. Being accessible from outside, the use of nucleic acid drugs such as miRNA inhibitors can be easily and directly injected at the tumor site. This kind of treatment could be more promising in the treatment of OSCC compared with its available therapy because a single-gene-based siRNA therapy aimed against a global gene regulator like miR-155 can be highly specific, more effective due to maximum availability of drug at the tumor site, and possibly free of any harmful side effects. Furthermore, to increase the efficiency of this kind of treatment, nucleic acid drugs can be modified to be more lipophilic that improves their intracellular penetration. Although modest compared with pretreatment model, the intratumoral injection of cholesterylated tumor suppressor miRNA mimics has been successful in regression of tumors in the post treatment xenograft model (18). Nonetheless, combined with the ease and simplicity of the drug delivery to a given tumor site in oral cavity, the use of antagomir-155 could have enormous potential to evolve for further drug development.
Taken together, our study validates tumor suppressor CDC73 as a target of oncogenic miRNA-155 (miR-155). Furthermore, we suggest that the reversal of pro-oncogenic properties of miR-155 due to its inhibitor puts a promise to the use of this inhibitor in cancer therapeutics.
This work was supported by the University Grants Commission (New Delhi).

This article contains supplemental Table S1.
- OSCC
- oral squamous cell carcinoma
- miRNA
- microRNA
- miR-155
- microRNA-155
- CASP3
- caspase-3
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- Alu
- Alu sequence.
REFERENCES
- 1. Shattuck T. M., Välimäki S., Obara T., Gaz R. D., Clark O.H., Shoback D., Wierman M. E., Tojo K., Robbins C. M., Carpten J. D., Farnebo L. O., Larsson C., Arnold A. (2003) Somatic and germ line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. N. Engl. J. Med. 349, 1722–1729 [DOI] [PubMed] [Google Scholar]
- 2. Aldred M. J., Talacko A. A., Savarirayan R., Murdolo V., Mills A. E., Radden B. G., Alimov A., Villablanca A., Larsson C. (2006) Dental findings in a family with hyperparathyroidism. Jaw tumour syndrome and a novel HRPT2 gene mutation. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 101, 212–218 [DOI] [PubMed] [Google Scholar]
- 3. Pimenta F. J., Gontijo Silveira L.F., Tavares G. C., Silva A. C., Perdigão P. F., Castro W. H., Gomez M. V., Teh B. T., De Marco L., Gomez R. S. (2006) HRPT2 gene alterations in ossifying fibroma of the jaws. Oral. Oncol. 42, 735–739 [DOI] [PubMed] [Google Scholar]
- 4. Carpten J. D., Robbins C. M., Villablanca A., Forsberg L., Presciuttini S., Bailey-Wilson J., Simonds W. F., Gillanders E. M., Kennedy A. M., Chen J. D., Agarwal S. K., Sood R., Jones M. P., Moses T. Y., Haven C., Petillo D., Leotlela P. D., Harding B., Cameron D., Pannett A. A., Höög A., Heath H., 3rd, James-Newton L. A., Robinson B., Zarbo R. J., Cavaco B. M., Wassif W., Perrier N. D., Rosen I. B., Kristoffersson U., Turnpenny P. D., Farnebo L. O., Besser G. M., Jackson C. E., Morreau H., Trent J. M., Thakker R. V., Marx S. J., Teh B. T., Larsson C., Hobbs M. R. (2002) HRPT2, encoding parafibromin, is mutated in hyperparathyroidism. Jaw tumour syndrome. Nat. Genet. 32, 676–680 [DOI] [PubMed] [Google Scholar]
- 5. Bradley K. J., Cavaco B. M., Bowl M. R., Harding B., Young A., Thakker R. V. (2005) Utilisation of a cryptic non-canonical donor splice site of the gene encoding PARAFIBROMIN is associated with familial isolated primary hyperparathyroidism. J. Med. Genet. 42, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang P. F., Tan M. H., Zhang C., Morreau H., Teh B. T. (2005) HRPT2, a tumour suppressor gene for hyperparathyroidism-jaw tumour syndrome. Horm. Metab. Res. 37, 380–383 [DOI] [PubMed] [Google Scholar]
- 7. Krogan N. J., Dover J., Wood A., Schneider J., Heidt J., Boateng M. A., Dean K., Ryan O. W., Golshani A., Johnston M., Greenblatt J. F., Shilatifard A. (2003) The Paf1 complex is required for histone H3methylation by COMPASS and Dot1p. Linking transcriptional elongation to histone methylation. Mol. Cell 11, 721–729 [DOI] [PubMed] [Google Scholar]
- 8. Wood A., Krogan N. J., Dover J., Schneider J., Heidt J., Boateng M. A., Dean K., Golshani A., Zhang Y., Greenblatt J. F., Johnston M., Shilatifard A. (2003) Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11, 267–274 [DOI] [PubMed] [Google Scholar]
- 9. Rozenblatt-Rosen O., Hughes C. M., Nannepaga S. J., Shanmugam K. S., Copeland T. D., Guszczynski T., Resau J. H., Meyerson M. (2005) The parafibromin tumour suppressor protein is part of a human Paf1 complex. Mol. Cell. Biol. 25, 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yart A., Gstaiger M., Wirbelauer C., Pecnik M., Anastasiou D., Hess D., Krek W. (2005) The HRPT2 tumour suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol. Cell. Biol. 25, 5052–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang C., Kong D., Tan M. H., Pappas D. L., Jr., Wang P. F., Chen J., Farber L., Zhang N., Koo H. M., Weinreich M., Williams B. O., Teh B. T. (2006) Parafibromin inhibits cancer cell growth and causes G1 phase arrest. Biochem. Biophys. Res. Commun. 350, 17–24 [DOI] [PubMed] [Google Scholar]
- 12. James R. G., Biechele T. L., Conrad W. H., Camp N. D., Fass D. M., Major M. B., Sommer K., Yi X., Roberts B. S., Cleary M. A., Arthur W. T., MacCoss M., Rawlings D. J., Haggarty S. J., Moon R. T. (2009) Bruton's tyrosine kinase revealed as a negative regulator of Wnt-β-catenin signaling. Sci. Signal. 2, ra25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cetani F., Pardi E., Borsari S., Viacava P., Dipollina G., Cianferotti L., Ambrogini E., Gazzerro E., Colussi G., Berti P., Miccoli P., Pinchera A., Marcocci C. (2004) Genetic analyses of the HRPT2 gene in primary hyperparathyroidism. Germline and somatic mutations in familial and sporadic parathyroid tumors. J. Clin. Endocrinol. Metab. 89, 5583–5591 [DOI] [PubMed] [Google Scholar]
- 14. Hewitt K. M., Sharma P. K., Samowitz W., Hobbs M. (2007) Aberrant methylation of the HRPT2 gene in parathyroid carcinoma. Ann. Otol. Rhinol. Laryngol. 116, 928–933 [DOI] [PubMed] [Google Scholar]
- 15. Hahn M. A., Howell V. M., Gill A. J., Clarkson A., Weaire-Buchanan G., Robinson B., B., Delbridge L., Gimm O., Schmitt W. D., Teh B. T., Marsh D. J. (2010) CDC73/HRPT2 CpG island hypermethylation and mutation of 5′-untranslated sequence are uncommon mechanisms of silencing parafibromin in parathyroid tumors. Endocr. Relat. Cancer 17, 273–282 [DOI] [PubMed] [Google Scholar]
- 16. Mehrotra R., Yadav S. (2006) Oral squamous cell carcinoma. Etiology, pathogenesis, and prognostic value of genomic alterations. Indian J. Cancer 43, 60–66 [DOI] [PubMed] [Google Scholar]
- 17. Landis S. H., Murray T., Bolden S., Wingo P. A. (1999) Cancer statistics. CA Cancer J. Clin. 49, 8–31 [DOI] [PubMed] [Google Scholar]
- 18. Wu S., Lin Y., Xu D., Chen J., Shu M., Zhou Y., Zhu W., Su X., Zhou Y., Qiu P., Yan G. (2012) MiR-135a functions as a selective killer of malignant glioma. Oncogene 31, 3866–3874 [DOI] [PubMed] [Google Scholar]
- 19. Bartel D. P. (2004) MicroRNAs. Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 20. Zhang W., Dahlberg J. E., Tam W. (2007) MicroRNAs in tumorigenesis. Am. J. Pathol. 171, 728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sobin L. H., Fleming I. D. (1997) TNM classification of malignant tumors, fifth edition. Union Internationale Centre le Cancer and the American Joint Committee on Cancer. Cancer 80, 1803–1804 [DOI] [PubMed] [Google Scholar]
- 22. Vaz C., Ahmad H. M., Sharma P., Gupta R., Kumar L., Kulshreshtha R., Bhattacharya A. (2010) Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genomics 11, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuhn D. E., Martin M. M., Feldman D. S., Terry A. V., Jr., Nuovo G. J., Elton T. S. (2008) Experimental validation of miRNA targets. Methods 44, 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eis P. S., Tam W., Sun L., Chadburn A., Li Z., Gomez M. F., Lund E., Dahlberg J. E. (2005) Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. U.S.A. 102, 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharbati-Tehrani S., Kutz-Lohroff B., Bergbauer R., Scholven J., Einspanier R. (2008) miR-Q. A novel quantitative RT-PCR approach for the expression profiling of small RNA molecules such as miRNAs in a complex sample. BMC Mol. Biol. 9, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 27. Chakraborty S., Mohiyuddin S. M., Gopinath K. S., Kumar A. (2008) Involvement of TSC genes and differential expression of other members of the mTOR signaling pathway in oral squamous cell carcinoma. BMC Cancer 8, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang L., Dai Y., Liu X., Wang C., Wang A., Chen Z., Heidbreder C. E., Kolokythas A., Zhou X. (2011) Identification and experimental validation of G protein α inhibiting activity polypeptide 2 (GNAI2) as a microRNA-138 target in tongue squamous cell carcinoma. Hum. Genet. 129, 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mignone F., Gissi C., Liuni S., Pesole G. (2002) Untranslated regions of mRNAs. Genome Biology 3, S0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Daskalova E., Baev V., Rusinov V., Minkov I. (2006) 3′-UTR-located Alu elements. Donors of potential miRNA target sites and mediators of network miRNA-based regulatory interactions. Evol. Bioinform. Online 2, 103–120 [PMC free article] [PubMed] [Google Scholar]
- 31. van den Berg A., Kroesen B. J., Kooistra K., de Jong D., Briggs J., Blokzijl T., Jacobs S., Kluiver J., Diepstra A., Maggio E., Poppema S. (2003) High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer 37, 20–28 [DOI] [PubMed] [Google Scholar]
- 32. Kluiver J., Poppema S., de Jong D., Blokzijl T., Harms G., Jacobs S., Kroesen B. J., van den Berg A. (2005) BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 207, 243–249 [DOI] [PubMed] [Google Scholar]
- 33. Costinean S., Zanesi N., Pekarsky Y., Tili E., Volinia S., Heerema N., Croce C. M. (2006) Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E (mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 103, 7024–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Connell R. M., Rao D. S., Chaudhuri A. A., Boldin M. P., Taganov K. D., Nicoll J., Paquette R. L., Baltimore D. (2008) Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 205, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iorio M. V., Ferracin M., Liu C. G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., Ménard S., Palazzo J. P., Rosenberg A., Musiani P., Volinia S., Nenci I., Calin G. A., Querzoli P., Negrini M., Croce C. M. (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65, 7065–7070 [DOI] [PubMed] [Google Scholar]
- 36. Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nikiforova M. N., Tseng G. C., Steward D., Diorio D., Nikiforov Y. E. (2008) Micro-RNA expression profiling of thyroid tumors. Biological significance and diagnostic utility. J. Clin. Endocrinol. Metab. 93, 1600–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R. M., Okamoto A., Yokota J., Tanaka T., Calin G. A., Liu C. G., Croce C. M., Harris C. C. (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198 [DOI] [PubMed] [Google Scholar]
- 39. Xiqiang L., Zugen C., Jinsheng Y., James X., Xiaofeng Z. (2009) MicroRNA profiling and head and neck cancer. Comp. Funct. Genomics, 11 pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Esquela-Kerscher A., Slack F. J. (2006) Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 41. Kent O. A., Mendell J. T. (2006) A small piece in the cancer puzzle. Micro-RNAs as tumor suppressors and oncogenes. Oncogene 25, 6188–6196 [DOI] [PubMed] [Google Scholar]
- 42. Gironella M., Seux M., Xie M. J., Cano C., Tomasini R., Gommeaux J., Garcia S., Nowak J., Yeung M. L., Jeang K. T., Chaix A., Fazli L., Motoo Y., Wang Q., Rocchi P., Russo A., Gleave M., Dagorn J. C., Iovanna J. L., Carrier A., Pébusque M. J., Dusetti N. J. (2007) Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155 and its restoration inhibits pancreatic tumor development. Proc. Natl. Acad. Sci. U.S.A. 104, 16170–16175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kong W., Yang H., He L., Zhao J. J., Coppola D., Dalton W. S., Cheng J. Q. (2008) MicroRNA-155 is regulated by the transforming growth factor β/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 28, 6773–6784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Costinean S., Sandhu S. K., Pedersen I. M., Tili E., Trotta R., Perrotti D., Ciarlariello D., Neviani P., Harb J., Kauffman L. R., Shidham A., Croce C. M. (2009) Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein β are targeted by miR-155 in B cells of Eμ-MiR-155 transgenic mice. Blood 114, 1374–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng L., Xu C. C., Chen W. D., Shen W. L., Ruan C. C., Zhu L. M., Zhu D. L., Gao P. J. (2010) MicroRNA-155 regulates angiotensin II type 1 receptor expression and phenotypic differentiation in vascular adventitial fibroblasts. Biochem. Biophys. Res. Commun. 400, 483–488 [DOI] [PubMed] [Google Scholar]
- 46. Greither T., Grochola L. F., Udelnow A., Lautenschläger C., Würl P., Taubert H. (2010) Elevated expression of microRNAs 155, 203, 210, and 222 in pancreatic tumors associates with poorer survival. Int. J. Cancer 126, 73–80 [DOI] [PubMed] [Google Scholar]