Abstract
The goal of genome-wide association studies is to identify SNPs unique to disease. It usually involves a single sampling from subjects' lifetimes. While primary DNA sequence variation influences gene-expression levels, expression is also influenced by epigenetics, including the ‘somatic epitype’ (GSE), an epigenotype acquired postnatally. While genes are inherited, and novel polymorphisms do not routinely appear, GSE is fluid. Furthermore, GSE could respond to environmental factors (such as heavy metals) and to differences in exercise, maternal care and dietary supplements – all of which postnatally modify oxidation or methylation of DNA, leading to altered gene expression. Change in epigenetic status may be critical for the development of many diseases. We propose a ‘longitudinal epigenome-wide association study’, wherein GSE are measured at multiple time points along with subjects' histories. This Longitudinal epigenome-wide association study, based on the ‘dynamic’ somatic epitype over the ‘static’ genotype, merits further investigation.
Keywords: association study, environment, epigenetics, epigenomics, genomics, GWAS, LEWAS
The current widely used genome-wide association study (GWAS) aims at determining SNPs that are unique to a human disease and that usually requires a single sampling from subjects' lifetimes. Using the GWAS to work out genetic factors behind different sporadic diseases follows logically from the Human Genome Project. The goal of GWAS is to use genomic data to understand complex mechanisms, diagnosis, prevention and treatment of many disorders. In this context, efforts by different groups have been fruitful, but improvements in understanding the nature and etiology of human diseases have, nevertheless, been modest. The present article focuses on the limitations of the GWAS approach, and proposes a new framework to circumvent the problems involved in the study of complex human disorders.
The partly fulfilled promise of GWAS
One of the major reasons for this shortcoming has been the failure of GWAS to integrate contributions of environment and environmental interaction with genes in disease processes. In short, GWAS is essentialist. It presumes that an organism's state of health is solely and fundamentally determined by its ‘fixed’ genome. After many years and much effort, there is still controversy over how useful GWAS has actually turned out to be. Their utility has been questioned [1] and defended [2,3]. In its defense, GWAS has been able to elucidate genetic associations such as macular degeneration with complement H [4], and it has suggested molecular pathways to explore, even with genes that explain relatively low phenotypic variation in a population, such as PPAR- γ in Type 2 diabetes, which has become an effective drug target.
The GWAS model is an end point assay, comparing a specific genetic sequence with a difference in disease state at the time the GWAS is run. Under the simplest form of this model, if a person possesses a pathogenic allele, the person will develop the associated disorder. While prima facie elegant, this model often does not apply. One conventional solution to this short-coming would be to invoke cumulative effects of endophenotypes or other ‘subclinical’ genetic contributions to disease. Unfortunately, the concept can be of limited use, such as when applied to diabetes. As Goldstein has noted, the strongest SNP-related sibling relative risk for Type 2 diabetes (found on the TCF7L2 gene) amounts to only 1.02, and the contribution of further common risk alleles falls off dramatically from that point [1]. Thus, ‘lucky strikes’, such as PPAR-γ, cannot necessarily be relied upon and may actually only represent the low hanging fruit of gene–disorder interactions. An even more stark contrast can be drawn with a fundamental, readily visible, nonpathogenic trait, such as adult height. In 1886, a method to predict adult height based entirely upon parental height was proposed by Galton [5]. Nearly 150 years later, Galton's method still surpasses methods based on genomic inference [6].
In addition, even a well-known genetic risk factor may turn out to be strongly subject to nongenetic influence. For example, the APOE ε4 allele is a generally accepted genetic risk factor for Alzheimer's disease (AD). However, while it has been shown that AD risk can correlate with APOEε4 allele dose (non-ε4/non-ε4 <ε4/non-ε4 <ε4/ε4) [7], other studies have shown no association between APOEε4 allele status and AD in a different population [8]. In addition, levels of circulating ApoE protein correlate with accumulation of the AD-associated amyloid-β peptide (Aβ), irrespective of APOEε4 status [9].
Genetic variation does not explain pathogenesis of ‘sporadic’ disorders
Both critique and defense of GWAS may be missing a biological ‘elephant in the room’. They do not look beyond the theoretical frame-work of GWAS to question whether some fundamental feature aside from, or in addition to, genomic DNA sequence is behind a significant proportion of sporadic disorders. Primary DNA sequence variation is certainly behind some diseases, and this is most obvious in autosomally inherited disorders such as hemophilia or sickle cell. Likewise, early-onset forms of AD are autosomally inherited and traceable to specific DNA variations [10], but these are still a minority of AD cases. An alternative for those disorders that involve protein configuration changes, such as AD, would be to invoke prions. However, the prion hypothesis does not explain disease progression or take into account a wide variety of environmental influence, particularly not latent influences [11]. It is now generally acknowledged that variation in regulatory regions influences gene expression and may contribute to pathogenesis of some disorders, but expression is also influenced by other structural elements, such as presence or absence of DNA methylation at CpG dinucleotides or oxidation of guanine residues (particularly at GG dinucleotides) [12–14]. Further more, a direct relationship has been found between guanine oxidation and interference with adjacent CpG methylation [15]. Methyl-cytosine can undergo a further modification to hydroxymethylcytosine. Impairment of this process may play a role in cancer development [16]. In addition, acetyl and methyl modification of the histone structural elements of chromatin plays a significant role in gene expression [17]. Significant differences in methylation between control and disease states have been found in several disorders, including schizophrenia, bipolar disorder, suicide following abuse during childhood and AD, among others [18 –20]. Differences in guanine oxidation that correlate with symptoms have been found in Parkin-son's disease models [21]. Histone acetylation differences have been found in Waldenstrom's macroglobulinemia [22]. Influence of epigenetic status is also very well known in oncology and is beginning to be recognized in inflammatory and neurodegenerative disorders [23]. In other words, genotype plus epigenetic status is a closer model for determining phenotype, rather than genotype alone. We have referred to this combination as the ‘somatic epitype’ (GSE), an epigenotype acquired postnatally, after methylation ‘reset’ and imprintation. While our original definition of GSE was restricted to methylation status, we now consider other epigenetic DNA modifications, such as oxidation and hydroxy-methylation, to also fall in this category [201]. We would like to make it plain that the GSE is not a somatic mutation. It is a purely epigenetic phenomenon and does not involve alteration of the primary DNA sequence.
Such a model cannot exclude the possibility that some ‘hidden variable’ might still, some-how, be valid, and that GWAS has merely not been given enough time to produce results. However, there is no epistemologically conclusive way, short of screening every genomic and mitochondrial DNA base of every cell of every human presently or previously in existence (or at least having had lived since a specific disease was first formally characterized), to exclude such unknown DNA sequence variations. Given the power of current GWAS assays and associated data analysis, and given that these powerful assays have, upon diligent application by skilled researchers, come up short to fulfill their original goal, it is reasonable to propose that probability of such discoveries in the future is low enough to justify exploring alternative methods.
How would GSE-based screening improve upon GWAS? First, it would look at the direct substrate of transcription. Epigenetic changes that influence transcription are invisible to a GWAS. Direct measurement of GSE would overcome this shortfall. However, a screen for somatic epitypes that uses an end point model such as GWAS would still not be sufficient. A high-throughput methylome analysis has been proposed in detail elsewhere [24]. If combined with epigenome-wide histone modification, this gives rise to an ‘epigenome-wide association study’ (EgWAS) [17], although it ignores the ‘oxidome’ (oxidative modification of DNA), the ‘hydroxymethylome’ (hyodroxymethylated DNA) and other likely components of the epigenome as a whole. Even overlooking the lack of attention paid to the oxidome, the EgWAS still does not directly consider environmental input. A different protocol, the ‘environment-wide association study’ (EWAS), which, despite its broad name, actually measures circulating chemical traces of various potential exposures and other environmental effects, has recently been tested with some measure of success for Type 2 diabetes [25].
The promising results of this study and similar work has led to a recent proposition that the older concept of the ‘exposome’ (the sum of all external exposures an organism undergoes) should be given greater consideration in ‘idiopathic’ disease etiology [26]. It has been pointed out that multiple samplings over time of the same subjects would be desirable [26], which we applaud. However, studies cited in advocating exposome research still presume an end point paradigm that contrasts subjects with a disorder against those without [25]. They do not test whether observed differences actually precede or follow upon a disorder. In addition, ‘pure’ exposome studies would have great difficulty discerning latent response to exposome perturbation, wherein any pathogenic material could have been cleared from the organism long before clinical symptoms might develop, as has been suggested by data linking early-life environmental exposures to AD and other neuropsychiatric disorders [18]. The possibility of ‘latent sequelae’ that appear long after exposure to a toxin such as lead was recognized decades ago [27]. Single time-point exposomics would be unable to detect such conditions. Furthermore, overall mortality is very likely linked to early-life inflammatory exposure and nutrition, and this link may be sufficiently strong to explain the general increase in lifespan observed in developed countries over the last century [28].
A diagnostically powerful study would need to fulfill several criteria. It would need to determine the ‘classic’ epigenome, which disregards the oxidome. It would need to explore the ‘envirome’, which combines chemistry-based exposome or toxicome with health history, sociocultural and behavioral factors. However, it would also need to measure somatic epitypes, that is, the epigenome including the oxidome, hydroxymethy-lome and histone modification. It would need to do this longitudinally and correlate changes in results from the repeated samplings with those disorders that emerge over a subject's lifetime (Figure 1A) – a kinetic approach. This research pathway is a viable alternative to the static model implied by GWAS and GWAS-like epigenomic and exposomic methods. We offer an alternative to the end point assay approach of GWAS, EgWAS and EWAS. Specifically, we suggest the use of a longitudinal combination of multiple time-point EgWAS (methylomic, hydroxymethylomic, oxidomic and histone based) and enviromics, in essence a ‘kinetic’ assay based on a dynamic, environment-responsive model of ‘sporadic’ disease etiology. Thus, ‘longitudinal epigenome/envirome-wide association study’ (LEWAS) would fulfill the unmet criteria.
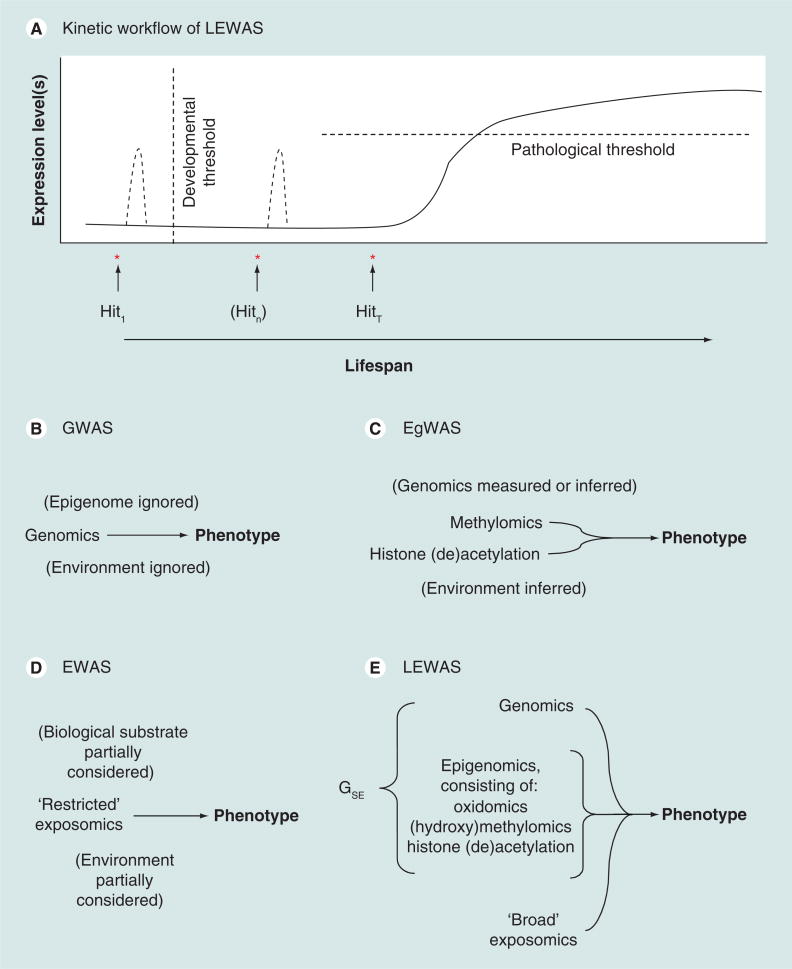
Figure 1. Comparison of the genome-wide association study (surveying genes) and other end point association assays versus the longitudinal epigenome/envirome-wide association study (measuring somatic epitype over time).
(A) Kinetic workflow of LEWAS. Expression of a gene or genes that are ‘pathogenic’ is schematically indicated by the line graph. This model presumes a ‘disease of excess’. However, a ‘disease of defciency’ could also be accounted for. Before an epigenetic event, GWAS would show alleles of genes while LEWAS would show somatic epitypes in a ‘fundamental’ state. After an H1 that could occur before a specific developmental threshold, such as formation of the blood–brain barrier in early life, GWAS would show genes in the same state as before, while LEWAS would indicate a somatic epitype or epitypes have been altered. After (possibly optional) Hn and a final HT, which leads to disease state, GWAS would still show genes in the same state as when there was no pathology, except for disorders associated with somatic mosaic mutations, while LEWAS would show changes in somatic epitypes corresponding to disease state. Detecting the changes over time would be critical to determining which somatic epitypes are important for a given disease. (B–E) Comparison of LEWAS versus other assays, highlighting specific elements of ‘organism’/biological substrate and ‘environment’ addressed by each. (B) GWAS. The genome is directly measured. Environmental input and epigenetic status are ignored: ‘genotype determines phenotype’. (C) EgWAS. Epigenomic status is directly measured. Genomic variation is measured or inferred. Environmental effects are inferred as contributing to epigenetic status. Single-time epigenotype determines phenotype. (D) EWAS. Chemical and biochemical traces of specific suspected exposures are directly measured. Environmental effects are partially inferred from circulating (bio)chemicals. Genomic and epigenomic effects might be partially inferred. Current (nonlatent) single-time evidence of past exposures correlates with phenotype. (E) LEWAS. Genomic analysis is combined with measuring epigenetic (oxidomic, methylomic, hydroxymethylomic and histone) status and ‘exposure’, which includes chemicals, previous infections and social influences. Sampling is performed at more than one time point to detect specific changes in an individual that correlate with disease state rather than indirectly infer such changes by comparing currently healthy with currently affected individuals. Changes in the somatic epitype lead to changes in phenotype, including idiopathic disorders. EgWAS: Epigenome-wide association study; EWAS: Environment-wide association study; GSE: Somatic epitype; GWAS: Genome-wide association study; Hit1: Initial hit; Hitn: Intermediate hit; HitT: Triggering hit; LEWAS: Longitudinal epigenome/envirome-wide association study.
We have recently published a short critique of the end point paradigm in disease research and suggested that a kinetic assay would overcome end point assay shortcomings [29]. Such a model has been foreshadowed in well-accepted theories such as Barker's fetal/neonatal basis of disease [30], and work that has shown behavioral effects associated with epigenetic reprogramming due to mammalian maternal behavior [31,32] and hypermethylation of the genes for ribosomal RNA in brains of subjects who have commited suicide [33]. These observations are part of a large body of evidence that has led to ‘life course epidemiology’ [34–37]. LEWAS would be an explicitly proposed assay based on these and similar epidemiological models.
Looking past disease
Interaction of biology and environment go beyond disease etiology. One example is the ‘Hispanic paradox’, in which immigrants to the USA from Latin American countries, such as Mexico, are more healthy than their socioeconomic status within the USA would predict [38]. Some explanation has been found for this from the finding that immigrants from Mexico who stay in the USA tend to be taller than Mexicans who do not immigrate and Mexicans who immigrate and return [39]. This suggests the explanation that healthier individuals are more likely to immigrate to the USA from Mexico. It has also been noted that a health disparity exists between immigrant and native-born residents of the USA. Immigrants tend to have longer life-spans and be less prone to specific disorders, and this trend has increased over time [40].
Adult height is used as ‘a proxy for a range of preadult exposures’ [41], as are mortality indexes versus certain chronic disorders [42]. However, specific effects of different exposures and events are not always well characterized but have been ‘inferred’ [42], and it has been recognized that research is needed to explore direct measures of diet, stress, illness and other factors on adult health instead of a single end point indirect measurement [41]. Likewise, explanations based on adult height gathered from body measurements and medical history do not consider potential genetic contributions to issues such as population movement. A multivariate kinetic study such as LEWAS could directly explore what is generally considered a purely ‘social’ phenomenon. Since it considers personal history, it would not suffer (or at least not suffer as much) from accusations of excessive reductionism that have be leveled at ‘purely’ biological studies. Such adoption of a systems biology approach has been suggested in oncology and theoretical biology [43,44]. LEWAS is a novel application of the systems biology approach to basic investigation of specific disorders.
The need for a ‘kinetic’ approach versus an 'end point' assay
A gene, including its regulatory (promoter) sequence, is presumed to be relatively static. It is not thought to undergo changes within an individual lifetime, although possible significant contributions of somatic DNA mutations/mosaicism to disease etiology has recently been mentioned in the literature for disorders as diverse as neurofibromatosis 1, Creutzfeldt–Jakob, Duchenne muscular dystrophy and AD [45–49]. A GSE, on the other hand, is by its nature dynamic, and any study based on the GSE would have to be a kinetic study. A dual study conducted on Ice-landic and US populations indicated significant amounts of DNA methylation drift over the entire genome [50]. Likewise, DNA methylation changes in response to stress persist late into life [51], although this study did not investigate possible effects of underlying deficiencies in DNA methylase and associated genes. DNA oxidation changes with age [52,53], but these changes can be environmentally modified [52]. The GSE is a product of environmental factors acting upon genes and presumes fluidity within a single organism's lifetime. Such factors could include differences in maternal grooming [32], exercise [54], direct oxidative stress [55], dietary levels of nutrients such as folate [56] or caloric restriction [52], all of which postnatally modify oxidation or methylation of specific genes (it must be noted that the above studies do not always report whether or not these differences might also correlate to sequence variations in genes that regulate DNA methylation or oxidation). Furthermore, childhood poverty and stress specifically alter adult working memory in humans [57]. Changes in behavior, such as decision-making, due to stress refect basic neuroanatomical alterations [58]. Such modification leads to changes in gene-expression levels. Somatic epitypes can explain the incomplete penetrance found for the vast majority of pathology-associated genes. A given gene's expression levels ordinarily would not be pathogenic. However, perturbation of the GSE would alter expression levels and induce pathology [59].
This is not to say that primary DNA sequence has no function in disease etiology nor that GWAS has outlived its usefulness. Two recent very large GWAS examining AD discovered a significant association with the CLU gene [60,61]. Epigenetic and epigenomic features rely upon the underlying genome to be able to exist. Long stretches of regulatory DNA with low levels of CpG (methylation) or GG (oxidation) dimers would be highly unlikely to have significant somatic epitypic variation. However, the GSE can determine affinity for a wide range of (hydroxy) methyl and oxidation sensitive transcription factors, including MeCP or inhibition of SP1 binding. In addition, DNA methylation has significant influence upon histone acetylation or methylation, which is a fundamental feature of chromatin rearrangement. GWAS ignore these fundamental features of gene expression, and it is the gene as expressed that contributes to an organism's health or illness.
From the point of view of disease etiology, simply suffering from a disease is, ultimately, not very interesting. What matters is how the organism changed or was changed in order to enter a disease state. For example, in some in stances of AD, such as Swedish familial AD (FAD), which is due to the ‘Swedish’ mutation of the Alzheimer's disease associated amyloid-β precursor (APP) gene (APPSWE), the disease mechanism is not subject to environmental influence [62]. However, there is no fundamental mechanistic change in individual members of the APPSWE population that produces AD. The APPSWE- AD relationship is fixed. Thus, while there may be a difference between APPSWE/+AD adults and non-APPSWE/non-AD adults, there is no realistically inducible change that could occur to the non-APPSWE/non-AD adults to convert them into APPSWE/+AD adults. Nevertheless, a great deal of AD research was expended on FAD under the presumption that differences seen in FAD from the general population are necessarily consistent with changes that convert a member of the general population into someone with sporadic AD. This presumption has met with, at very best, limited success [63].
How should research approach non-FAD/+AD versus non-FAD/non-AD subjects? For nearly all sporadic cases, the consensus in the feld seems to be that some change occurs that can ‘convert’ a non-FAD/non-AD into a non-FAD/+AD individual. For example, a pair of monozygotic twins had been raised together but were discordant for AD. They had differential DNA methylation in temporal neocortical neurons [64,65]. What produced AD in one but not the other? It could be proposed that it was some kind of environmental influence resulting in an epigenetic effect. To strip the question to fundamental theory of disease, something induced a change in one twin and not in the other. Whether this change was pathogenic or pathopreventative cannot be determined from one sample taken at one time point, but at least in theory, the twins began ‘the same’. At some point, a vital difference was induced, and this difference either led to or prevented AD. This is, of course, a simplification. A chain of inductions could have occurred in each twin, ‘nudging’ each toward or away from AD, until one twin either was pushed over a pathogenic threshold or was held back from that threshold, which would have otherwise been crossed.
The strength of older models to answer questions can instill a desire to apply those models beyond the limits of supporting evidence. For example, a measurable difference between diseased and nondiseased populations has been successfully used as a placeholder for changes that convert a specific nondiseased individual into a diseased individual. For simple infectious diseases, this theoretical substitution can work well. However, we can see from the literature that the ‘end point difference = necessary change’ presumption may not apply to epigenetic states. Some studies unite differences in DNA methylation with a disorder [66–68], others lead to the opposite conclusion [69–71]. This is where LEWAS could make a difference. To return to the example of the AD discordant twins, the study indicated an association between lower levels of methylation and AD [65], but that does not answer fundamental etiological questions: would DNA demethylation in the brain lead to AD, making the methylated brain healthy? Would DNA methylation in the brain prevent AD, meaning that, barring methylation, both of the twins would have developed AD? On the other hand, could AD lead to DNA demethylation in the brain, making a lower DNA methylation state a symptom and not a contributing factor? Similar questions can be asked about observed differences in DNA oxidation linked to disease states. A kinetic-paradigm study, such as LEWAS, would go far towards answering these questions.
The ‘demethylase issue’
One of the fundamental principles behind LEWAS is changes in epigenetic status that would include DNA methylation and demethylation. DNA hypomethylation is well attested in cancers [72], and we hypothesize that it would be an issue in other sporadic disorders. Unfortunately, while human DNA methylase is known, no specific mammalian DNA demethylase has been discovered to date. This has led some workers to insist that an etiological model based on changes in DNA methylation status cannot be viable until and unless a specific DNA demethylase is discovered. If there were no evidence of demethylation, this objection would be valid. However, overall loss of DNA methylation over time has been demonstrated in humans [50], and such loss varied from individual to individual in that study, suggesting specificity of demethylation activity operating on the individual level. In addition, active demethylation activity has been observed for specific genes in specific tissues [73], including induced rapid demethylation of the memory-critical reelin gene that followed contextual fear conditioning [74]. Thus, while the identity of a specific demethylase molecule is currently unknown, this can not be used to deny the existence of target- and stimulus-specific demethylation activity, any more than lack of conclusive demonstration of nucleic acids as the physical ‘genetic material’ could be used to dismiss the entirety of genetic science that was undertaken before the Avery– MacLeod–McCarty experiment in 1944 [75]. By more recent analogy, the cleavage of Aβ from its APP precursor was determined to occur through two activities. Specifically, β-secretase activity cleaves the APP protein to produce the soluble APPβ (sAPPβ) and C99 fragments. C99 is then cleaved by γ-secretase activity to produce the Aβ and p3 peptides [76]. The β-secretase activity was described and accepted as real nearly a decade before the actual β-secretase protein was discovered [76,77]. Likewise, some time elapsed between the identification of γ-secretase activity and identification of the presenilins in that activity, and the full list of components of the γ-secretase complex is still unknown [78]. Thus, while we freely admit that no specific molecule or complex has been found that drives mammalian DNA demethylation, we note that lack of such specific knowledge regarding other molecular actors has not previously invalidated all potential research on a particular biological activity. In any case, recent work has suggested that hydroxymethylation via methylcytosine dioxygenase TET1 plays an important role in maintaining hypomethylation of CpG islands in mammalian promoters [79]. Hydroxymethylcytosine would then be demethylated by the AID/APOBEC family of cytosine deaminases [80].
Precursors to LEWAS: studies that have partially implemented principles
The experimental concepts that comprise the LEWAS are not completely untested. Several experiments and surveys have used some element or another of this proposed method. These fall into three categories, specifically:
Epigenetic induction of phenotype experiments in animals;
Targeted epigenetic studies of humans;
Survey studies in humans.
For example, bisphenol A was used to induce hypomethylation of the DNA sequence upstream of the mouse Agouti gene, resulting in a shift of coat color distribution toward yellow. Both the epigenetic and epiphenotypic results were prevented by maternal nutritional supplementation [81]. Placing mice pups in an enriched ‘communal nest’ that provides a highly stimulating early social environment altered histone acetylation at the BDNF gene, and this was accompanied by increased BDNF expression after environmental challenge compared with nonenriched mice [82]. Mice were fed a high-fat diet from weaning to 18 weeks. These mice had reduced preference for sucrose and decreased µ-opioid mRNA levels specific to the ventral tegmental area, nucleus accumbens and prefrontal cortex, but not the hypothalamus. Such change in expression was accompanied by an increase in specific receptor gene promoter methylation in these brain regions, among several other specific epigenetic differences induced by diet [83].
These, and many other studies, have features that support implementing the LEWAS, such as:
Induction of specific epigenetic changes owing to environmental stress or enhancement;
In some cases epiphenotypic differences requiring additional environmental stimulus;
Complex (behavioral) phenotype responses that accompanied the induced epigenetic changes.
Of course, such studies differ from the LEWAS in that such animal studies:
Use defined, inbred subjects with well-controlled ‘life paths’;
Target specific genes and epigenetic target regions;
Track epiphenotypes after specific, intentional treatment.
This would be in contrast to the LEWAS, which, for humans, would be concerned with ‘wild’ subjects and conditions not under laboratory control. However, the controlled rodent studies give ample evidence to the critical underlying principle: environment induces epigenetic changes that have important consequences for the organism as a whole.
Many targeted epigenetic studies have been performed in humans, including several already mentioned herein. To reiterate, significant differences in DNA methylation have been found in schizophrenia, bipolar disorder, suicide following childhood abuse and AD [18–20], and histone acetylation aberrations exist in Waldenstrom's macroglobulinemia [22]. These differ from the LEWAS in that they either measure nonspecific changes in epigenetic markers or predetermined epigenetic markers. Furthermore, they are non-longitudinal and do not measure changes in the epigenome, only compare different epigenes and epigenomes. Nevertheless, they establish epigenomic links to disease in humans. Finally, epigenome and exposome-wide studies have been performed on human subjects, such as the demonstration that DNA methylation changes over time in the human genome [50] and recent ‘exposome’ studies of Type 2 diabetes [25]. Such documented changes in the human epigenome over time and the exposome link to Type 2 diabetes each illustrate two fundamental principles of LEWAS: longitudinal epigenome alterations and environmental induction of sporadic disease in humans. In short, although the LEWAS has not yet been implemented, its individual elements have already been well documented. LEWAS unites these threads into a cohesive approach.
Novelty of LEWAS: applying epigenomics longitudinally
Large-scale methylomic epigenetic programs are under way, such as the Alliance for the Human Epigenome and Disease (AHEAD) [84] and the Human Epigenome Project [85]. Both of these are devoted to discovering the ‘baseline’ human methylome down to specific tissue and cell types. These could be used as baselines for comparison with methylomic surveys of diseased individuals and to compare with nondisease controls in an individual EWAS. These studies would still, unfortunately, fall short of measuring the impact of changes in various GSE within a person's life-time. They would still presume an end point model, treating a difference in methylomic state as a static marker for disease. The critical element that may not have been considered is that these markers change over time, and that these changes are clinically meaningful. In addition, the studies do not take changes in the oxidome or histone modifcation into account, nor do they consider environmental interaction.
We propose that it would be more fruitful to explore the kinetic aspect of epigenetic changes with the LEWAS, which measures GSE of multiple genes and applies (hydroxy)methylomics, oxidomics and histone studies, at the very least, at multiple time points in subjects' lives and correlates them with subjects' histories of environmental conditions such as social status, known exposures, nutrition, rearing conditions (such as presence or absence of abuse) and others. For neuropsychiatric disorders, such a study would be particularly strong were it to begin in early childhood, before events such as final formation of the blood–brain barrier. This is suggested by studies that led to the ‘latent early-life associated regulation’ model [18]. However, early-life stress could also be important in epigenetic control of cancers [86,87] and cardiovascular disease [88], although the emphasis in cardiovascular disease has still been mostly on prenatal factors. LEWAS could differentiate between early natal, early childhood and later effects on disease development. This is not to say that LEWAS could only be conducted on populations sampled in early childhood. What would be important is finding an initial cohort sampling time that would sufficiently predate the development of a condition of interest. We admit that this may require some calibration, as evidence has amassed that even ‘senescent’ disorders, such as AD, may actually see their start very early in the human lifespan [89,90]. Of further note is the level of interaction that exists between the genome and epigenome. Differences in specific DNA sequences influence histone methylation [91]. DNA methylation and oxidation depend upon specific underlying primary sequence. The contribution of DNA oxidation and methylation to mutations in the DNA primary sequence has been long known [92,93]. In addition, histone acetylation plays an important role in DNA strand repair [94], and its disruption may also contribute to DNA sequence mutation. A LEWAS, if genomic information was collected at each stage, could track such two-way interaction and correlate it to changes in disease state.
We admit that until and unless a new molecular target or a new way of detecting a molecular target is discovered, there will be no such thing as a fully novel assay. Instead, there would be novel applications of currently available assays. Methylome-wide survey assay protocols already exist, as do histone acetylation and methylation assays. Broad toxicological screens are well known. Statistical techniques to handle very large (e.g., GWAS) data sets, which use a multivariate approach necessary for analyzing LEWAS, already exist [95,96]. Patient histories are already used in medical research. In some cases, patient histories already include family disease histories, environmental exposures and conditions, and other ‘broad background’ data points, but this is not a universal practice and depends on the focus of the study. In some cases, patient histories used in research follow a person through a useful portion of life stages. Thus, the largest technical innovation would be development and optimization of an oxidome-wide survey assay, for which the necessary technology already exists. In addition to measurement of overall levels of 8-oxo-dG in circulating lymphocytes [97] and excreted in urine [98], immune assays based on monoclonal antibody to 8-oxo-dG have been used to visualize concentrations of oxidized deoxyguanine on chromosomes [99], and combined activity of the enzymes MutM, which excises cytosine paired with 8-oxo-dG, MutT, which hydrolyzes 8-oxo-dG, and MutY, which excises adenine mispaired with 8-oxo-dG [100], could be used to provide ‘oxidome cleavage product’ probes for judiciously designed DNA microarrays. Major differences among LEWAS, GWAS, EgWAS and EWAS are summarized in Table 1 & Figure 1B–E.
Table 1.
Comparison of association studies.
| Assay | Biochemical substrate | Time points | Additional data | Correlation | Ref. |
|---|---|---|---|---|---|
| GWAS | DNA | One | Disease state | Disease state versus genotype | [4] |
| EgWAS | DNA methylation, histone acetylation | One | Disease state | Disease state versus end point methylation/acetylation states | [13] |
| EWAS | NA | One | Disease state, general medical history, circulating chemicals | Disease state versus end point circulating environmental chemicals | [22] |
| LEWAS | DNA, and its(hydroxy)methylation andoxidation, histone acetylation | Multiple | Disease state, general medical history, social and other environmental factors | Change in disease state versus change in GSE, and environmental factors, referenced to known exposures or stressors and to previous changes in epigenotype | Present study |
EgWAS: Epigenome-wide association study; EWAS: Environment-wide association study; GSE: Somatic epitype; GWAS: Genome-wide association study; LEWAS: Longitudinal epigenome/exposome-wide association study; NA: Not applicable.
LEWAS is particularly novel in that it would combine these elements in a rational fashion to compare changes in epigenetic markers and in environment, upon the background of genetic variation, with changes in disease state. LEWAS depends upon its precursors. The LEWAS would include GWAS along with repeated EgWAS, EWAS and oxidome-wide association study from the same subjects at different times.
Potential limits of LEWAS are surmountable
While we propose that LEWAS can be a powerful tool, it must not be forgotten that the specific molecular target of LEWAS is the epigenome. Therefore, progress with the LEWAS would be greatly aided by profiles of ‘normal’ epigenomic states for multiple tissues, organ regions and cell types. Fortunately, much of this groundwork is currently being laid for the methylome by groups such as AHEAD [84] and the Human Epigenome Project [85]. Unfortunately, such large-scale projects are not currently up and running for the oxidome, histone modification or the envirome. Competent selection of controls will be able to compensate for a great deal of this lack of background information, much as significant strides have been made in methylomics before AHEAD and the Human Epigenome Project got off the ground. Ultimately, as the ‘kinetic organism within the environment’ paradigm of LEWAS becomes more popular, sufficient support should exist to fund large-scale methylome, histone modification and other such projects.
LEWAS would be more expensive than the same number of GWAS, EgWAS or EWAS on independent samples, owing to costs of tracking individual subjects for follow-up tests, loss of subjects over time and other challenges inherent to longitudinal studies. However, previous health cohort studies have been performed that include banking of biological tissues. These could be used as a sample source for LEWAS, depending on the completeness of environmental information that accompanied the samples. Pilot-scale LEWAS, at the very least, could mine these collections to circumvent these costs. However, comprehensively addressing funding sources for a large-scale LEWAS, beyond pointing to its utility and novel concept is beyond the scope of this article. Costs specific to an oxidome-wide study would be speculative until a large-scale assay has been optimized. However, avidin/streptavidin detection of 8-oxo-deoxy-guanosine has already been well demonstrated [101]. This could be combined with DNA sequence-specific immobilization in plate wells or on chips, analogous to current genome chip technology. In addition, the greater amounts of information needed from subjects for LEWAS would increase time and cost of collection and data analysis. LEWAS would ideally begin sampling its subjects before the appearance of disorder symptoms. This would pose a problem on a number of subjects basis. For example, the widest prevalence of autism spectrum disorders estimated has been no more than 2.64% of the general population [102]. Epidemiological studies have already developed acceptable ways of beginning with a broad sample population and reducing it to a ‘hypothesis relevant’ subsample [103]. Iceland, with its unique genomic and population information bank, may be particularly useful for some LEWAS, as it has already proved for demonstrating methylomic changes over time [50]. However, if there is sufficient will, even Iceland's' special situation would not be necessary. This has been shown for enormous, worldwide studies such as the second stage of the ADNI2. This study uses facilities and coprincipal investigators at 55 different facilities, with hundreds of subjects who will be imaged at multiple time points [202]. Given the broad range of sampling and observation a LEWAS will require, even sample reduction methods may still leave requirements for large study populations and the attending expenses and administrative diffculties. However, a LEWAS, which samples multiple time points, would be worth such inherent administrative costs, as it would measure changes in epigenetic status and environment over time, changes we consider critical for the development of many disease states that have not so far been elucidated through GWAS.
A fundamental trait of epigenetic variation is that it is very often tissue-type specific [104]. This opens the question of appropriate material to study diseases in organs that cannot be safely sampled repeatedly, or at all, for example, brain tissue. This could be partially addressed by finding appropriate proxy tissues. Indeed, for any disorders that involve internal organs, an accessible proxy tissue would be highly preferable. Initially, LEWAS could use circulating lymphocytes, fibroblasts or olfactory neuroepithelium, as these are the most convenient sources of ‘resampleable’ epigenetic material. The olfactory cells have already shown promise in cell adhesion research on schizophrenia [105]. Direct assay of tissues from live people for diseases such as AD, for example, such as the hippocampus and parietal lobes, is not possible, likely to become possible, or even appropriate to contemplate. We do not consider this difficulty to be sufficient grounds to summarily reject LEWAS. For example, while DNA for GWAS is normally taken from blood, this approach would appear to be rigorously valid only if one ignores the effect of local DNA mutation/mosaicism in a gene-based disorder, such as has been suggested to exist for Creutzfeldt–Jakob, Duchenne muscular dystrophy and AD [45–49]. Such a shortcoming has not eliminated the usefulness of GWAS in the study of any of these diseases, since conventional genetic contributions to disease at one time were unknown.
Discovery and validation of proxy tissues would be a necessary component of optimizing the LEWAS procedure, overall. In addition, rational study design would have to be carefully adhered to, with strong reference to what would already be known of the underlying biology for a disorder of interest. That is to say, a given LEWAS would be best performed based on results from more conventional studies. What would distinguish LEWAS from more conventional single-cause studies is that potential agents would be studied in a kinetic fashion, and the possibility of multivariate association among agent actions would be directly explored. Otherwise, the scope of a particular LEWAS would become an impossible to perform ‘everything and the kitchen sink’ affair. Not only would such a nondesigned study be a logistical nightmare, but useful information would be more likely to be lost in a large mass of data. The more comprehensive a study, the more it will benefit from rational study design based on known biological principles. While LEWAS might be used for fishing expeditions, its sheer scope would greatly limit its use in this fashion.
Conclusion & future perspective
We propose LEWAS as a solution to many of the shortcomings of currently used association studies. Its inclusion of multiple complementary measurements and kinetic approach will directly measure what has hitherto primarily been inferred. We consider it a strong potential addition to the pathology arsenal. We admit that LEWAS will not track down all complex genome-related disease etiology. For example, copy number variation (CNV) has been implicated in the etiology and progression of several disorders, including autism [106], schizophrenia [107] and lung cancers [108]. These variations alter gene dose without altering underlying gene sequence or epigenetic markers. However, the possibility exists that each copy of a gene in a specific organism's genome could have a distinct GSE, meaning that disease state could result in interaction between CNV and GSE. Accounting for epigenetic variation could inform and refine results of less than conclusive CNV results. Somatic mosaicism may contribute to several disorders [45–49]. Sampling the mosaic cells would be a matter of blind luck. However, given known connections between epigenomic modification and DNA misrepair, a LEWAS could suggest promising genome locations prone to giving rise to mosaicism.
Genotyping and GWAS will continue to have utility. For example, a recent study determined that significant neurotoxicity in mice from exposure to inhaled anesthetic was due to transgenic PSEN1 dE9 status [109]. In theory, such differences in reaction to environment could be seen as purely genetic and thus ‘visible’ to GWAS. EgWAS will certainly open up many avenues of research, and the EWAS holds distinct promise, especially given that toxins known to have decades long half lives, such as lead, still occur in the developed world, in food products targeted at children [203], and the long-term effects of demethylation caused by biphenyl-A in infant plastic products remains to be seen (although some remediation may be possible) [81]. Our proposed LEWAS takes into account both the contribution of environment, such as imbalance of dietary factors, rearing and metals, and it tracks changes in the GSE longitudinally at different life points of an individual. It should not only enhance the power of current association studies but also overcome the problems inherent to an end point assay.
Executive summary.
Background: the partly fulfilled promise of the genome-wide association study
Genome-wide association studies are end point assays that compare genetic sequences between diseased and nondiseased individuals.
In several diseases, genetic relative risk is remarkably low.
Nongenetic influence plays an important role in many disorders, such as Alzheimer's disease (AD).
Genetic variation does not explain pathogenesis of sporadic disorders
In disorders with known genetic components, such as AD, cases that can be explained solely by genetic mutation are a small minority.
Epigenetic markers include cytosine methylation, guanine oxidation and histone acetylation/methylation. Changes in epigenetic markers can correspond to several disease states, including AD, schizophrenia, Parkinson's disease and other conditions.
We define changes in epigenetic markers acquired after imprinting as a ‘somatic epitype’ (GSE), thus, these disease-related changes are changes in the GSE. The GSE can change in response to environmental conditions.
Several assays currently exist that address parts of the GSE question, but nothing looks at the whole picture, particularly not changes over time.
We propose a longitudinal epigenome/exposome-wide association study (LEWAS), which unites genetic sequence, epigenomic markers, tracking environmental exposures and patient personal history taken at multiple time-points versus development of disorders.
Looking past disease
The concept of the GSE is not restricted to disease.
LEWAS could also investigate social phenomena, such as the Hispanic paradox.
The need for a ‘kinetic’ approach versus an ‘end point’ assay
Assays such as GWAS or even current single-sample point epigenomic studies compare healthy individuals with unhealthy individuals and presume that differences are causal to a disorder. However, in the case of single-point epigenomic studies, it is not trivial to infer whether or not a condition was caused by or contributed to a change in an epigenomic maker.
LEWAS, since it is longitudinal, can directly note changes that occur before symptoms appear and correlate them, over time, with later appearance of disease.
The ‘demethylase issue’
LEWAS presumes demethylation activity, even though no specific mammalian demethylase protein has been discovered.
This is not an actual setback, since demethylase activity has been confirmed in mammalian cells.
For other conditions, such as AD, β-secretase activity was accepted in the field long before the actual β-secretase protein was discovered.
Precursors to LEWAS: studies that have partially implemented principles
Several studies have partially implemented the LEWAS concept.
Mouse studies have measured changes in specific gene-associated epigenetic changes to environment and concomitant changes in expression for those specific genes.
Targeted epigenomic surveys have been carried out on human populations, comparing ‘end point’ differences in epigenomic markers and disease.
No study method currently unites multiple measurements over time with human populations and a genome-/epigenome-/environment-wide approach.
Novelty of LEWAS: applying the oxidome & methylome longitudinally
Large-scale epigenomic surveys are currently underway, but these are end point, not tracking changes in the epigenome.
The novel element of LEWAS is its longitudinal approach, attempting to measure, rather than infer, disease-critical epigenomic changes and relate those to previous environmental influences.
Potential limits of LEWAS are surmountable
LEWAS poses challenges in terms of scope and cost, particularly the need for large sample sizes in the face of unknown later development of disease and multiple sampling times.
Longitudinal studies have been successfully completed and are ongoing. Modern communication and travel methods have made these studies far more feasible than in the past.
Methods exist to begin studies with very large sample sizes and reduce them to a ‘hypothesis relevant’ subsample.
For disorders of the CNS or other vital organs, direct sampling will not be possible. However, proxy tissues may be developed. For example, olfactory neuroepithelial cells are easily accessed and can be repeatedly sampled with safety. These have been adequate proxies for other neurological studies.
Acknowledgments
The authors sincerely appreciate the grant support from the Alzheimer's Association (Zenith award and IIRG) and the National Institute on Aging/NIH (AG18379 and AG18884) to DK Lahiri.
Footnotes
Financial & competing interests disclosure: The authors have no other relevant affliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360(17):1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 2.Kraft P, Hunter DJ. Genetic risk prediction – are we there yet? N Engl J Med. 2009;360(17):1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 3.Hirschhorn JN. Genomewide association studies – illuminating biologic pathways. N Engl J Med. 2009;360(17):1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 4.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galton F. Regression towards mediocrity in hereditary stature. J Anthropol Inst. 1886;15:246–263. [Google Scholar]
- 6.Aulchenko YS, Struchalin MV, Belonogova NM, et al. Predicting human height by victorian and genomic methods. Eur J Hum Genet. 2009;17(8):1070–1075. doi: 10.1038/ejhg.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 8▪.Hall K, Murrell J, Ogunniyi A, et al. Cholesterol, APOE genotype, and Alzheimer disease: an epidemiologic study of Nigerian Yoruba. Neurology. 2006;66(2):223–227. doi: 10.1212/01.wnl.0000194507.39504.17. Examines a well-known risk factor for Alzheimer's disease, the ApoEε4 variant, and indicates a strong environmental influence on a trait that is usually presumed to be genetic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maloney B, Ge YW, Petersen RC, et al. Functional characterization of three single-nucleotide polymorphisms present in the human APOE promoter sequence: Differential effects in neuronal cells and on DNA-protein interactions. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(1):185–201. doi: 10.1002/ajmg.b.30973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selkoe DJ, Podlisny MB. Deciphering the genetic basis of Alzheimer's disease. Annu Rev Genomics Hum Genet. 2002;3:67–99. doi: 10.1146/annurev.genom.3.022502.103022. [DOI] [PubMed] [Google Scholar]
- 11.Lahiri DK. Prions: a piece of the puzzle? Science. 2012;337(6099):1172. doi: 10.1126/science.337.6099.1172-a. [DOI] [PubMed] [Google Scholar]
- 12.Zelko IN, Mueller MR, Folz RJ. CpG methylation attenuates Sp1 and Sp3 binding to the human extracellular superoxide dismutase promoter and regulates its cell-specific expression. Free Radic Biol Med. 2010;48(7):895–904. doi: 10.1016/j.freeradbiomed.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32(14):4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh R, Mitchell DL. Effect of oxidative DNA damage in promoter elements on transcription factor binding. Nucleic Acids Res. 1999;27(15):3213–3218. doi: 10.1093/nar/27.15.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zawia NH, Lahiri DK, Cardozo-Pelaez F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic Biol Med. 2009;46(9):1241–1249. doi: 10.1016/j.freeradbiomed.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maunakea AK, Chepelev I, Zhao K, Bruneau B. Epigenome mapping in normal and disease States. Circ Res. 2010;107(3):327–339. doi: 10.1161/CIRCRESAHA.110.222463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18▪▪.Lahiri DK, Maloney B, Zawia NH. The LEARn model: an epigenetic explanation for idiopathic neurobiological diseases. Mol Psychiatry. 2009;14(11):992–1003. doi: 10.1038/mp.2009.82. Explicitly lays out a testable model for the etiology of idiopathic disorders. Longitudinal epigenome/envirome-wide association study (LEWAS) would test this model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8(4):253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heindel JJ, McAllister KA, Worth L, Jr, Tyson FL. Environmental epigenomics, imprinting and disease susceptibility. Epigenetics. 2006;1(1):1–6. doi: 10.4161/epi.1.1.2642. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi Y, Yasuhara T, Agari T, et al. Urinary 8-OHdG elevations in a partial lesion rat model of Parkinson's disease correlate with behavioral symptoms and nigrostriatal dopaminergic depletion. J Cell Physiol. 2010;226:1390–1398. doi: 10.1002/jcp.22467. [DOI] [PubMed] [Google Scholar]
- 22.Sacco A, Issa GC, Zhang Y, et al. Epigenetic modifcations as key regulators of Waldenstrom's macroglobulinemia biology. J Hematol Oncol. 2010;3(1):38. doi: 10.1186/1756-8722-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23▪.Levenson VV. DNA methylation as a universal biomarker. Expert Rev Mol Diagn. 2010;10(4):481–488. doi: 10.1586/erm.10.17. The original definition of the somatic epitype concept upon which the LEWAS is based. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butcher LM, Beck S. Future impact of integrated high-throughput methylome analyses on human health and disease. J Genet Genomics. 2008;35(7):391–401. doi: 10.1016/S1673-8527(08)60057-0. [DOI] [PubMed] [Google Scholar]
- 25▪.Patel CJ, Bhattacharya J, Butte AJ. An environment-wide association study (EWAS) on Type 2 diabetes mellitus. PLoS One. 2010;5(5):e10746. doi: 10.1371/journal.pone.0010746. Describes the environment-wide association study, which would be adopted as part of LEWAS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rappaport SM, Smith MT. Environment and disease risks. Science. 2010;330:460–461. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De la Burde B, Choate MS., Jr Does asymptomatic lead exposure in children have latent sequelae? J Pediatr. 1972;81(6):1088–1091. doi: 10.1016/s0022-3476(72)80236-1. [DOI] [PubMed] [Google Scholar]
- 28.Finch CE, Crimmins EM. Infammatory exposure and historical changes in human life-spans. Science. 2004;305(5691):1736–1739. doi: 10.1126/science.1092556. [DOI] [PubMed] [Google Scholar]
- 29.Lahiri DK. An integrated approach to genome studies. Science. 2011;331(6014):147. doi: 10.1126/science.331.6014.147-a. [DOI] [PubMed] [Google Scholar]
- 30.Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ. Weight in infancy and death from ischaemic heart disease. Lancet. 1989;2(8663):577–580. doi: 10.1016/s0140-6736(89)90710-1. [DOI] [PubMed] [Google Scholar]
- 31.Diorio J, Meaney MJ. Maternal programming of defensive responses through sustained effects on gene expression. J Psychiatry Neurosci. 2007;32(4):275–284. [PMC free article] [PubMed] [Google Scholar]
- 32.Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 33▪.McGowan PO, Sasaki A, Huang TC, et al. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS One. 2008;3(5):e2085. doi: 10.1371/journal.pone.0002085. Demonstrates a molecular, epigenetic feature of a perplexing negative psychiatric outcome that transcends specifc disorders. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rusby JS, Tasker F. Long-term effects of the British evacuation of children during World War 2 on their adult mental health. Aging Ment Health. 2009;13(3):391–404. doi: 10.1080/13607860902867750. [DOI] [PubMed] [Google Scholar]
- 35.Gluckman PD, Hanson MA, Beedle AS. Early life events and their consequences for later disease: a life history and evolutionary perspective. Am J Hum Biol. 2007;19(1):1–19. doi: 10.1002/ajhb.20590. [DOI] [PubMed] [Google Scholar]
- 36.Ben-Shlomo Y, Kuh D. A life course approach to chronic disease epidemiology: conceptual models, empirical challenges and interdisciplinary perspectives. Int J Epidemiol. 2002;31(2):285–293. [PubMed] [Google Scholar]
- 37.Lynch J, Smith GD. A life course approach to chronic disease epidemiology. Annu Rev Public Health. 2005;26:1–35. doi: 10.1146/annurev.publhealth.26.021304.144505. [DOI] [PubMed] [Google Scholar]
- 38.Singh GK, Miller BA. Health, life expectancy, and mortality patterns among immigrant populations in the United States. Can J Public Health. 2004;95(3):I14–I21. doi: 10.1007/BF03403660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crimmins EM, Soldo BJ, Kim JK, Alley DE. Using anthropometric indicators for Mexicans in the United States and Mexico to understand the selection of migrants and the “Hispanic paradox”. Soc Biol. 2005;52(3–4):164–177. doi: 10.1080/19485565.2005.9989107. [DOI] [PubMed] [Google Scholar]
- 40.Singh GK, Hiatt RA. Trends and disparities in socioeconomic and behavioural characteristics, life expectancy, and cause-specific mortality of native-born and foreign-born populations in the United States, 1979–2003. Int J Epidemiol. 2006;35(4):903–919. doi: 10.1093/ije/dyl089. [DOI] [PubMed] [Google Scholar]
- 41.Batty GD, Shipley MJ, Gunnell D, et al. Height, wealth, and health: an overview with new data from three longitudinal studies. Econ Hum Biol. 2009;7(2):137–152. doi: 10.1016/j.ehb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 42.Vijh AK. Inverse trend between estimated worldwide frequency of major cancers and inferred infectious burdens of populations: possible role of adaptive immune system. Med Hypotheses. 2004;62(6):880–888. doi: 10.1016/j.mehy.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 43.Majumder D, Mukherjee A. A passage through systems biology to systems medicine: adoption of middle-out rational approaches towards the understanding of therapeutic outcomes in cancer. Analyst. 2010;136:663–678. doi: 10.1039/c0an00746c. [DOI] [PubMed] [Google Scholar]
- 44.Sternberg RV. DNA codes and information: formal structures and relational causes. Acta Biotheor. 2008;56(3):205–232. doi: 10.1007/s10441-008-9049-6. [DOI] [PubMed] [Google Scholar]
- 45.Erickson RP. Somatic gene mutation and human disease other than cancer. Mutat Res. 2003;543(2):125–136. doi: 10.1016/s1383-5742(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 46.Erickson RP. Somatic gene mutation and human disease other than cancer: an update. Mutat Res. 2010;705(2):96–106. doi: 10.1016/j.mrrev.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 47.Alzualde A, Moreno F, Martinez-Lage P, et al. Somatic mosaicism in a case of apparently sporadic Creutzfeldt–Jakob disease carrying a de novo D178N mutation in the PRNP gene. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(7):1283–1291. doi: 10.1002/ajmg.b.31099. [DOI] [PubMed] [Google Scholar]
- 48.Beck JA, Poulter M, Campbell TA, et al. Somatic and germline mosaicism in sporadic early-onset Alzheimer's disease. Hum Mol Genet. 2004;13(12):1219–1224. doi: 10.1093/hmg/ddh134. [DOI] [PubMed] [Google Scholar]
- 49.Sgaramella V. Variability of our somatic (Epi) genomes. Science. 2010;239:32–33. doi: 10.1126/science.329.5987.32-c. [DOI] [PubMed] [Google Scholar]
- 50▪▪.Bjornsson HT, Sigurdsson MI, Fallin MD, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299(24):2877–2883. doi: 10.1001/jama.299.24.2877. Explicitly lays out that changes in DNA methylation occur well after imprinting has ended and further suggests interesting genetic/epigenetic interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51▪.Murgatroyd C, Patchev AV, Wu Y, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12(12):1559–1566. doi: 10.1038/nn.2436. Describes explicit biochemical correlations among postnatal, early-life environmental stress to specific DNA hypomethylation, gene-expression differences, protein modifcation and later-life behavior. [DOI] [PubMed] [Google Scholar]
- 52.Hamilton ML, Van Remmen H, Drake JA, et al. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci USA. 2001;98(18):10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu T, Pan Y, Kao SY, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429(6994):883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 54.Buehlmeyer K, Doering F, Daniel H, Kindermann B, Schulz T, Michna H. Alteration of gene expression in rat colon mucosa after exercise. Ann Anat. 2008;190(1):71–80. doi: 10.1016/j.aanat.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 55.Campos AC, Molognoni F, Melo FH, et al. Oxidative stress modulates DNA methylation during melanocyte anchorage blockade associated with malignant transformation. Neoplasia. 2007;9(12):1111–1121. doi: 10.1593/neo.07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Vliet J, Oates NA, Whitelaw E. Epigenetic mechanisms in the context of complex diseases. Cell Mol Life Sci. 2007;64(12):1531–1538. doi: 10.1007/s00018-007-6526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evans GW, Schamberg MA. Childhood poverty, chronic stress, and adult working memory. Proc Natl Acad Sci USA. 2009;106(16):6545–6549. doi: 10.1073/pnas.0811910106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dias-Ferreira E, Sousa JC, Melo I, et al. Chronic stress causes frontostriatal reorganization and affects decision-making. Science. 2009;325(5940):621–625. doi: 10.1126/science.1171203. [DOI] [PubMed] [Google Scholar]
- 59.Lahiri DK, Maloney B. The “LEARn” (latent early-life associated regulation) model integrates environmental risk factors and the developmental basis of Alzheimer's disease, and proposes remedial steps. Exp Gerontol. 2010;45(4):291–296. doi: 10.1016/j.exger.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lambert JC, Heath S, Even G, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 62.Almqvist E, Lake S, Axelman K, Johansson K, Winblad B. Screening of amyloid precursor protein gene mutation (APP 717 Val-->Ile) in Swedish families with Alzheimer's disease. J Neural Transm Park Dis Dement Sect. 1993;6(2):151–156. doi: 10.1007/BF02261009. [DOI] [PubMed] [Google Scholar]
- 63.Lahiri DK, Farlow MR, Sambamurti K, Greig NH, Giacobini E, Schneider LS. A critical analysis of new molecular targets and strategies for drug developments in Alzheimer's disease. Curr Drug Targets. 2003;4(2):97–112. doi: 10.2174/1389450033346957. [DOI] [PubMed] [Google Scholar]
- 64.Mastroeni D, Coleman PD, Grover A, Sue L, McKee A, Rogers J. Differential DNA methylation in neurons of identical twins discordant for Alzheimer's disease. Alzheimers Dement. 2009;5:P145–146. [Google Scholar]
- 65.Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer's disease. PLoS One. 2009;4(8):e6617. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grayson DR, Jia X, Chen Y, et al. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci USA. 2005;102(26):9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer's disease: decrements in DNA methylation. Neurobiol Aging. 2010;31(12):2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siegmund KD, Connor CM, Campan M, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2(9):e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tochigi M, Iwamoto K, Bundo M, et al. Methylation status of the reelin promoter region in the brain of schizophrenic patients. Biol Psychiatry. 2008;63(5):530–533. doi: 10.1016/j.biopsych.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 70.Barrachina M, Ferrer I. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J Neuropathol Exp Neurol. 2009;68(8):880–891. doi: 10.1097/NEN.0b013e3181af2e46. [DOI] [PubMed] [Google Scholar]
- 71.Schwob NG, Nalbantoglu J, Hastings KE, Mikkelsen T, Cashman NR. DNA cytosine methylation in brain of patients with Alzheimer's disease. Ann Neurol. 1990;28(1):91–94. doi: 10.1002/ana.410280117. [DOI] [PubMed] [Google Scholar]
- 72.Shen X, He Z, Li H, et al. Distinct functional patterns of gene promoter hypomethylation and hypermethylation in cancer genomes. PLoS One. 2012;7(9):e44822. doi: 10.1371/journal.pone.0044822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11(9):607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller CA, Sweatt JD. Covalent modifcation of DNA regulates memory formation. Neuron. 2007;53(6):857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 75.Avery OT, MacLeod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type Iii. J Exp Med. 1944;79(2):137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vassar R, Bennett BD, Babu-Khan S, et al. β-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286(5440):735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 77.Esch FS, Keim PS, Beattie EC, et al. Cleavage of amyloid β peptide during constitutive processing of its precursor. Science. 1990;248(4959):1122–1124. doi: 10.1126/science.2111583. [DOI] [PubMed] [Google Scholar]
- 78.Small DH, Klaver DW, Foa L. Presenilins and the γ-secretase: still a complex problem. Mol Brain. 2010;3(1):7. doi: 10.1186/1756-6606-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williams K, Christensen J, Pedersen MT, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA. 2007;104(32):13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Branchi I, Karpova NN, D'Andrea I, Castren E, Alleva E. Epigenetic modifications induced by early enrichment are associated with changes in timing of induction of BDNF expression. Neurosci Lett. 2011;495(3):168–172. doi: 10.1016/j.neulet.2011.03.038. [DOI] [PubMed] [Google Scholar]
- 83.Vucetic Z, Kimmel J, Reyes TM. Chronic high-fat diet drives postnatal epigenetic regulation of mu-opioid receptor in the brain. Neuropsychopharmacology. 2011;36(6):1199–1206. doi: 10.1038/npp.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.American Association for Cancer Research Human Epigenome Task Force; Network of Excellence, Scientific Advisory Board. European Union, Moving AHEAD with an international human epigenome project. Nature. 2008;454(7205):711–715. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brena RM, Huang TH, Plass C. Toward a human epigenome. Nat Genet. 2006;38(12):1359–1360. doi: 10.1038/ng1206-1359. [DOI] [PubMed] [Google Scholar]
- 86.Hughes LA, van den Brandt PA, de Bruine AP, et al. Early life exposure to famine and colorectal cancer risk: a role for epigenetic mechanisms. PLoS One. 2009;4(11):e7951. doi: 10.1371/journal.pone.0007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Waterland RA. Is epigenetics an important link between early life events and adult disease? Horm Res. 2009;71(Suppl. 1):13–16. doi: 10.1159/000178030. [DOI] [PubMed] [Google Scholar]
- 88.Symonds ME, Sebert SP, Hyatt MA, Budge H. Nutritional programming of the metabolic syndrome. Nat Rev Endocrinol. 2009;5(11):604–610. doi: 10.1038/nrendo.2009.195. [DOI] [PubMed] [Google Scholar]
- 89▪.Basha MR, Wei W, Bakheet SA, et al. The fetal basis of amyloidogenesis: exposure to lead and latent overexpression of amyloid precursor protein and β-amyloid in the aging brain. J Neurosci. 2005;25(4):823–829. doi: 10.1523/JNEUROSCI.4335-04.2005. Demonstrated a latent effect of early-life postnatal stress (lead exposure) on levels of amyloid β and activity of the SP1 transcription factor. This effect occurred despite clearance of the heavy metal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90▪.Wu J, Basha MR, Brock B, et al. Alzheimer's disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb), evidence for a developmental origin and environmental link for AD. J Neurosci. 2008;28(1):3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. Extended evidence of latent lead exposure effects from rodents to primates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Z, Willard HF. Evidence for sequence biases associated with patterns of histone methylation. BMC Genomics. 2012;13(1):367. doi: 10.1186/1471-2164-13-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Holliday R, Grigg GW. DNA methylation and mutation. Mutat Res. 1993;285(1):61–67. doi: 10.1016/0027-5107(93)90052-h. [DOI] [PubMed] [Google Scholar]
- 93.Jena NR. DNA damage by reactive species: mechanisms, mutation and repair. J Biosci. 2012;37(3):503–517. doi: 10.1007/s12038-012-9218-2. [DOI] [PubMed] [Google Scholar]
- 94.Kusch T, Florens L, Macdonald WH, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306(5704):2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 95.Guzzetta G, Jurman G, Furlanello C. A machine learning pipeline for quantitative phenotype prediction from genotype data. BMC Bioinformat. 2010;11(Suppl. 8):S3. doi: 10.1186/1471-2105-11-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96▪.Yang Q, Wu H, Guo CY, Fox CS. Analyze multivariate phenotypes in genetic association studies by combining univariate association tests. Genet Epidemiol. 2010;34(5):444–454. doi: 10.1002/gepi.20497. Describes a method for analyzing genome-wide association study data that can be translated to LEWAS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tanimura Y, Shimizu K, Tanabe K, Kono I, Ajisaka R. Effects of three consecutive days exercise on lymphocyte DNA damage in young men. Eur J Appl Physiol. 2010;110(2):307–314. doi: 10.1007/s00421-010-1499-2. [DOI] [PubMed] [Google Scholar]
- 98.Helbock HJ, Beckman KB, Shigenaga MK, et al. DNA oxidation matters: the HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proc Natl Acad Sci USA. 1998;95(1):288–293. doi: 10.1073/pnas.95.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ohno M, Miura T, Furuichi M, et al. A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome. Genome Res. 2006;16(5):567–575. doi: 10.1101/gr.4769606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fowler RG, White SJ, Koyama C, Moore SC, Dunn RL, Schaaper RM. Interactions among the Escherichia coli mutT, mutM, and mutY damage prevention pathways. DNA Repair (Amst) 2003;2(2):159–173. doi: 10.1016/s1568-7864(02)00193-3. [DOI] [PubMed] [Google Scholar]
- 101.Struthers L, Patel R, Clark J, Thomas S. Direct detection of 8-oxodeoxyguanosine and 8-oxoguanine by avidin and its analogues. Anal Biochem. 1998;255(1):20–31. doi: 10.1006/abio.1997.2354. [DOI] [PubMed] [Google Scholar]
- 102.Kim YS, Leventhal BL, Koh YJ, et al. Prevalence of autism spectrum disorders in a total population sample. Am J Psychiatry. 2011;168(9):904–912. doi: 10.1176/appi.ajp.2011.10101532. [DOI] [PubMed] [Google Scholar]
- 103.Ferguson CJ. Is psychological research really as good as medical research? Effect size comparisons between psychology and medicine. Rev Gen Psych. 2009;13(2):130–136. [Google Scholar]
- 104.Bell CG, Beck S. The epigenomic interface between genome and environment in common complex diseases. Brief Funct Genomics. 2010;9:477–485. doi: 10.1093/bfgp/elq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Perry C, Mackay-Sim A, Feron F, McGrath J. Olfactory neural cells: an untapped diagnostic and therapeutic resource. The 2000 Ogura Lecture. Laryngoscope. 2002;112(4):603–607. doi: 10.1097/00005537-200204000-00002. [DOI] [PubMed] [Google Scholar]
- 106.Merikangas AK, Corvin AP, Gallagher L. Copy-number variants in neurodevelopmental disorders: promises and challenges. Trends Genet. 2009;25(12):536–544. doi: 10.1016/j.tig.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 107.Need AC, Ge D, Weale ME, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5(2):e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jung SH, Yim SH, Hu HJ, et al. Copy number alterations and expression profles of candidate genes in a pulmonary infammatory myofbroblastic tumor. Lung Cancer. 2010;70(2):152–157. doi: 10.1016/j.lungcan.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 109▪.Lu Y, Wu X, Dong Y, Xu Z, Zhang Y, Xie Z. Anesthetic sevofurane causes neurotoxicity differently in neonatal naive and Alzheimer disease transgenic mice. Anesthesiology. 2010;112(6):1404–1416. doi: 10.1097/ALN.0b013e3181d94de1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lahiri DK, Maloney B. Genes are not our destiny: the somatic epitype bridges between the genotype and the phenotype. Nat Rev Neurosci. 2006;7 www.nature.com/nrn/journal/v7/n12/full/nrn2022-c1.html. [Google Scholar]
- 202.ADNI 2 Study. www.adcs.org/Studies/ImagineADNI2.aspx.
- 203.Candy Dynamics Expands Recall to all Toxic Waste®Brand Nuclear Sludge®Products all Flavors and all Sizes. www.fda.gov/Safety/Recalls/ucm241359.htm.