Abstract
Circadian clocks are ubiquitous biological oscillators that coordinate an organism’s behavior with the daily cycling of the external environment. To ensure synchronization with the environment, the period of the clock must be maintained near 24 h even as amplitude and phase are altered by input signaling. We show that, in a reconstituted circadian system from cyanobacteria, these conflicting requirements are satisfied by distinct functions for two domains of the central clock protein KaiC: the C-terminal autokinase domain integrates input signals through the ATP/ADP ratio, and the slow N-terminal ATPase acts as an input-independent timer. We find that phosphorylation in the C-terminal domain followed by an ATPase cycle in the N-terminal domain is required to form the inhibitory KaiB•KaiC complexes that drive the dynamics of the clock. We present a mathematical model in which this ATPase-mediated delay in negative feedback gives rise to a compensatory mechanism that allows a tunable phase and amplitude while ensuring a robust circadian period.
Keywords: biological clock, in vitro, robustness, metabolic input processing
Circadian clocks are endogenous oscillatory systems that allow an organism to anticipate daily rhythmic variations in the external environment. Despite having apparently diverse molecular origins, the functional properties of circadian systems are remarkably conserved across most organisms that have been studied. Even when deprived of a rhythmic input, circadian clocks continue to generate self-sustaining oscillations, and these oscillations have a period that is close to 24 h over a wide range of conditions, including varying light levels, nutrient abundance, and temperature (1). The robust period of the oscillator seems to be critical for its physiological role: in multiple organisms, mutants with clocks whose free-running periods are markedly different from 24 h are associated with decreased lifespan and fitness defects (2–4). In contrast, the phase and amplitude of circadian rhythms are generally tunable, and input signals can reset the oscillator to efficiently bring the clock into synchrony with a rhythmic environment.
The competing demands for both a sensitive response to input signaling and a robustly invariant oscillator period place strong constraints on the possible mechanisms underlying the circadian clock. It is currently unclear how the biochemical circuitry that generates circadian rhythms satisfies these constraints in any organism. To study the molecular origins of robust periodicity, we analyzed the biochemically tractable circadian oscillator from the cyanobacterium Synechococcus elongatus.
The Synechococcus core oscillator can be reconstituted in vitro using three purified proteins, KaiA, KaiB, and KaiC, and ATP (Fig. 1A). KaiC is a multifunctional enzyme with slow ATPase, autokinase, and autophosphatase activities. The effector proteins KaiA and KaiB work together to modulate KaiC’s enzymatic activity in a phosphorylation-dependent manner, switching the system between kinase- and phosphatase-dominated modes. The purified oscillator generates coherent, self-sustaining rhythms in the phosphorylation state of KaiC and exhibits many of the conserved properties of the in vivo circadian clock (5). The KaiABC in vitro oscillator is also capable of processing input signals, and the phase of the oscillation can be reset by transiently altering the ratio of ATP to ADP nucleotides in the reaction buffer. This manipulation simulates the metabolic changes that occur in vivo in response to a dark pulse and results in a modulation of KaiC phosphorylation (6).
Fig. 1.
The period of the circadian oscillator is robust to varying input signals. (A) Schematic of the KaiABC oscillator. A purified posttranslational protein circuit generates circadian rhythms in KaiC phosphorylation in the presence of ATP. Active KaiA promotes autophosphorylation on initially unphosphorylated KaiC (U-KaiC) first on Thr432 and then, on Ser431. Ser431-phosphorylated KaiC (S-KaiC) forms KaiB•KaiC complexes that inhibit KaiA and promote system-wide dephosphorylation, driving stable oscillations. The phase of the oscillation can be shifted by metabolic signaling through the ATP/ADP ratio. (B) KaiC autophosphorylation in KaiA-KaiC reactions at various ATP/ADP conditions (colored symbols); all buffers have 10 mM total nucleotide (similar data previously published in ref. 6). (C) KaiABC in vitro reactions in buffers at various ATP/ADP conditions; 10 mM total nucleotide. Oscillations with a robust period persist under the conditions tested. (D) Period of the oscillations shown in C determined by fitting the data after the first trough to a sinusoidal function. Error bars indicate the SE of the least squares fit. Dashed lines indicate linear regressions of the data to show trend. (E) Peak and trough heights of the oscillations shown in C determined by averaging the peaks and troughs after the first trough. Error bars indicate SDs. Dashed lines indicate linear regressions of the data to show trend.
Results
Period of the KaiABC Oscillator Is Robust Against Changes in ATP/ADP Signaling.
To dissect the mechanism of the oscillator’s response to input signaling through the ATP/ADP ratio, we first isolated its effect on KaiC’s kinase activity by studying nonoscillatory KaiA-KaiC reactions, where the absence of KaiB removes the negative feedback on phosphorylation that permits oscillations. Decreasing the ATP/ADP ratio caused a marked slowing of the phosphorylation rate, a phenomenon that we observed at various absolute concentrations of ATP (Fig. 1B, Fig. S1A, and Table S1), while leaving the phosphatase activity unaffected (6).
If the ∼24-h period of the circadian oscillator is determined by a balance between the KaiC kinase and phosphatase rates, then inhibition of kinase activity through the ATP/ADP ratio would be expected to upset that balance and change the oscillator period. To experimentally test this prediction, we subjected oscillating KaiA-KaiB-KaiC reactions to a sustained rather than transient drop in the ATP/ADP ratio and measured oscillator performance (Fig. 1C and Fig. S1 B–E). Surprisingly, we found that the core oscillator continued to measure time robustly across a range of ATP/ADP ratios, encompassing metabolic conditions measured in living cells under various light levels (6, 7). Rhythms persist with a period that remains close to 24 h (within 5%), although the attenuation of kinase activity caused by lower ATP/ADP ratios does change the amplitude of the oscillator (Fig. 1 D and E).
Thus, the KaiABC protein oscillator is intrinsically capable of robustly adapting to input signaling to maintain circadian rhythmicity. The robustness of the period in the face of perturbations to kinase activity suggests that there may be slow steps in the oscillator dynamics that are critical for appropriate timing but that do not involve phosphorylation. To identify these steps and elucidate the mechanistic basis of how the circadian oscillator maintains a robust period, we undertook a combined experimental and mathematical modeling study of the ability of the biochemical circuit driving the KaiABC in vitro oscillator to process nucleotide input signals.
N-Terminal Domain ATPase Activity Is Required for KaiB•KaiC Complex Assembly and Negative Feedback.
KaiC, the only enzyme in the core oscillator, consists of two homologous catalytic domains that are members of the RecA/DnaB superfamily of P-loop ATPases. Both domains have conserved nucleotide binding motifs, and thus, each is a potential target for the ATP/ADP signals that can reset the oscillator (Fig. 2A). The N-terminal domain (CI) binds ATP with high affinity and plays an important structural role in assembling hexameric KaiC particles (8). Both domains also have conserved putative catalytic glutamates. The C-terminal domain (CII) has both kinase and phosphatase activities that are modulated by KaiA (9, 10). Both activities occur through phosphotransfer reactions at the same CII active site, and dephosphorylation of KaiC involves an ATP synthesis mechanism where a phosphoryl group on KaiC can be transferred back to an ADP acceptor (11, 12). KaiA interacts with the C-terminal tail of KaiC, promoting CII kinase activity by remodeling an inhibitory loop (13) and initiating an ordered pattern of phosphorylation on two adjacent CII residues (Ser431 and Thr432) during the circadian cycle (9, 14). CI also hydrolyzes ATP but does not phosphorylate itself (8). The overall rate of catalytic turnover from both domains of the protein is quite slow (∼15 ATP/KaiC per day), very weakly dependent on temperature, and correlated with the time scale of the circadian rhythm itself (15). Many KaiC mutants with an altered ATP turnover rate show distortion of output signaling and altered control of cell division in vivo (16).
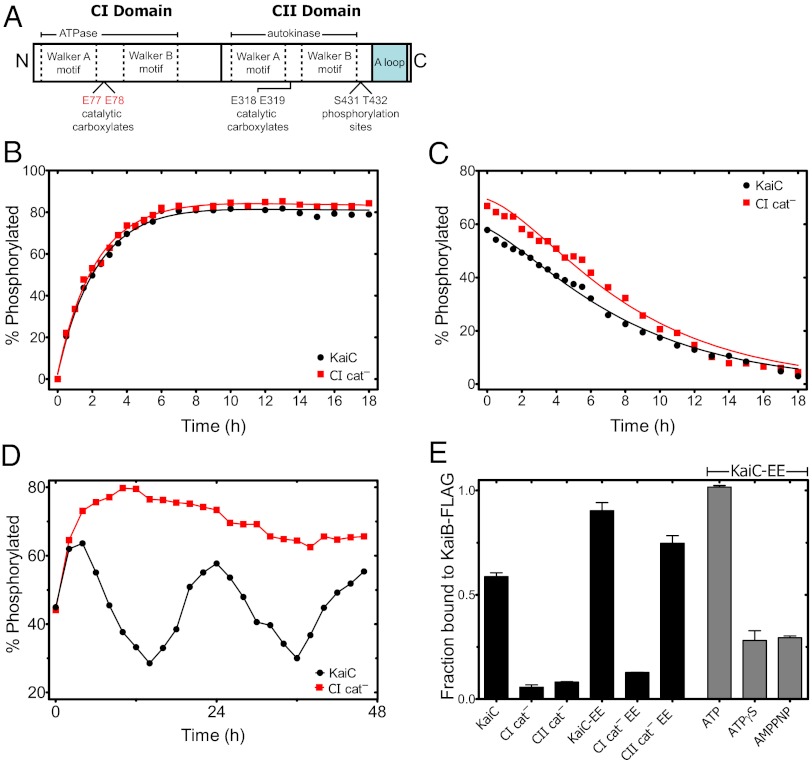
Fig. 2.
CI ATPase activity is required for KaiB•KaiC complex formation. (A) KaiC is organized into tandem catalytic domains, CI and CII, both containing conserved Walker A and Walker B motifs that flank pairs of catalytic residues. CII has autokinase activity, which is promoted by interactions between the C-terminal A-loop region and KaiA. In the CI cat– mutant protein, the catalytic carboxylates Glu77 and Glu78 are both mutated to glutamine. (B) Autophosphorylation of KaiC WT (black circles) and CI cat– mutant (red squares) in the presence of KaiA. (C) Autodephosphorylation of KaiC WT and CI cat– mutant. Highly phosphorylated protein was prepared by incubation with KaiA and then subsequently removed by immunoprecipitation to initiate dephosphorylation. Solid lines in B and C are fits to a four-state kinetic model of phosphoform interconversion. (D) Phosphorylation dynamics of KaiC WT and CI cat– mutant protein in the presence of KaiA and KaiB. (E) Levels of WT KaiC, CI cat–, and a CII domain catalytic mutant (CII cat–, E318Q) protein coimmunoprecipitating with KaiB-FLAG in the presence of KaiA after incubation for 10 h. Data are also shown for phosphomimetic (S431E; T432E) KaiC in various backgrounds of catalytic site mutations: WT (KaiC-EE), CI cat– (CI– EE), and CII cat– (CII– EE; black bars). KaiC-EE coimmunoprecipitating with KaiB-FLAG after incubation for 8–10 h in buffers with the nonhydrolyzable ATP analogs ATPγS and adenylyl imidodiphosphate (AMPPNP) (gray bars). Error bars indicate SEs from three replicates.
Clearly, phosphorylation in CII is required to generate a stable circadian rhythm, but the observation that ∼24-h rhythms persist even when CII phosphorylation is partially inhibited by ADP suggests that CII does not solely dictate the oscillator dynamics (Fig. 1C). Indeed, multiple lines of evidence have led to the hypothesis that both KaiC domains are critical for circadian function (15, 17). Because both of these domains interact with nucleotides and both have slow specific activities, we suspected that catalysis in CI might have an unknown function in the oscillator dynamics and that a mathematical description of the functional coupling between CI and CII could account for the robust period of oscillations.
We, therefore, sought to define separate roles for catalysis in each domain by selectively mutating the conserved catalytic residues. We found that, in a mutant where the conserved catalytic glutamates in CI are changed to glutamine (CI cat−), the intact CII domain retains its normal ability to execute posttranslational modifications: KaiA-stimulated autokinase activity, kinetic ordering of multisite phosphorylation, degree of sensitivity to KaiA, and intrinsic phosphatase activity in the CII domain are quantitatively unchanged (rate constants differ by less than 10%) (Fig. 2 B and C, Fig. S2, and Table S2). We interpret these results as an indication that CII function is independent of CI activity.
However, the CI cat– mutant protein is incapable of generating circadian rhythms in vitro. Instead, without CI catalytic activity, the system locks into a highly phosphorylated steady state (Fig. 2D). We traced this behavior to an unexpected defect: the CI cat– mutant is incapable of forming a complex with KaiB and thus incapable of closing the feedback loop that inhibits KaiA (Fig. 2E).
In the WT system, KaiB•KaiC complexes assemble and disassemble rhythmically during the oscillation, peaking in abundance when KaiC phosphatase activity is high (18). The KaiB-KaiC interaction is believed to globally regulate KaiC phosphorylation by then sequestering KaiA in inhibitory KaiA•KaiB•KaiC complexes (9, 19). Previous work has shown that the formation of these inhibitory complexes is closely linked to the phosphorylation state in CII, specifically phosphorylation on Ser431 (9, 14). We now show that there is an additional requirement for catalytic activity in the CI domain that cannot be overcome, even by phosphomimetic mutations in CII (Fig. 2E).
In contrast, catalysis in CII is not required per se to form the KaiB•KaiC complex; when the lack of kinase activity in a CII cat− mutant is bypassed by phosphomimetic mutations (CII cat− EE), KaiB•KaiC complexes assemble normally. As expected from these results, phosphomimetic KaiC with intact catalytic residues (KaiC-EE) incubated with nonhydrolyzable ATP analogs is defective in KaiB•KaiC complex formation, confirming that catalysis in CI is specifically required for this step in the circadian cycle (Fig. 2E). This requirement for CI catalytic activity is consistent with very recent structural data indicating that, although CII phosphorylation promotes KaiB interaction, KaiB makes physical contact with the CI domain of KaiC (20).
CI ATPase Activity Is Insensitive to the ATP/ADP Input.
Having established that the CI and CII catalytic domains play distinct and essential roles in the core oscillator, we sought to assess how each domain responds to the ATP/ADP input signal and the functional consequences for the circadian rhythm. To isolate the activity of each KaiC domain, we incubated the CI cat– mutant (which has intact CII kinase activity) or an N-terminal fragment consisting only of the CI domain in mixtures of ATP and ADP. Consistent with previous work, we found that ADP strongly inhibited phosphorylation in CII in a manner consistent with competitive inhibition (6). However, we found that the rate of ATP hydrolysis catalyzed by the CI fragment was only weakly affected by the presence of ADP (Fig. 3 A and B) and that mutations to the catalytic residues of phosphomimetic KaiC-EE gave a similar specific activity to the CI fragment alone (Fig. S3C).
Fig. 3.
CI ATPase and CII autokinase activities are differentially regulated by ADP. (A) KaiA-stimulated CII domain autophosphorylation in the CI cat– mutant protein at various ATP/ADP conditions (colored symbols); all buffers have 1 mM ATP. (B) Kinase rate constants with error bars indicating SEs of fits (△, kCII kinase) obtained from data in A. CI ATPase rate constants (●, kCI ATPase) measured as total ADP production by the CI fragment (KaiC residues 1–247) in buffers at various ATP/ADP levels; all buffers have 1 mM ATP. Error bars indicate SEMs of at least three measurements. Dashed lines indicate fits to a competitive inhibition model, k{[ATP]/([ATP] + KI[ADP])}: KI/CII kinase = 0.96 ± 0.18 and KI/CI ATPase = 0.10 ± 0.03. (C) Amount of phosphomimetic KaiC-EE coimmunoprecipitating with KaiB-FLAG as a function of time under various total protein concentrations with equimolar amounts of KaiB-FLAG and KaiC-EE (0.5× = 1.75 μM, 1× = 3.5 µM, 2× = 7 μM, and 4× = 14 μM). Dashed lines indicate predicted assembly curves for KaiB•KaiC complex assembly rates set by bimolecular collisions for each concentration. (D) Amount of phosphomimetic KaiC-EE coimmunoprecipitating with KaiB-FLAG as a function of time under various ATP/ADP conditions. Solid lines in C and D indicate fits to a pseudofirst-order reversible binding reaction (SI Text). (E) Schematic of requirements for KaiB•KaiC complex assembly. U-KaiC first autophosphorylates in the CII domain on Thr432 and then Ser431. Autokinase activity in CII depends on KaiA and the ATP/ADP input signal. After Ser431 phosphorylation is achieved, KaiC undergoes a catalytic cycle in the CI domain, causing a hypothetical structural rearrangement to occur and permitting a stable KaiB•KaiC complex to form. ATPase activity in CI is insensitive to the ATP/ADP input signal.
From these data, we conclude that KaiC consists of two catalytic domains, one of which changes activity markedly in response to KaiA and the ATP/ADP metabolic input signal and one of which maintains a slow, environment-independent activity (17). Redox-dependent input signals have also been shown to modulate KaiA activity and thus, also control activity in the CII domain (21). Taking these observations together, we propose here that CII activity is generally responsive to the environment and its autophosphorylation reflects input signals to the clock, whereas the CI ATPase acts as an invariant timer that mediates negative feedback through KaiB binding after CII phosphorylation.
KaiB•KaiC Complex Assembly Rates Are Controlled by the CI ATPase and Insensitive to ATP/ADP.
The KaiABC protein oscillator is perfectly robust against increases in total protein concentration: the dynamics of the system are quantitatively unchanged if the concentrations of the components are scaled up together. This property is presumably important for insulating circadian timing from changes in cell volume and gene expression in vivo (18). A simple class of mechanisms that can generate this robustness property relies on all on- and off-rates for protein complexes being fast relative to the timescale of oscillation and concentrations being much higher than the relevant binding affinities. Under these approximations, the dynamics are set purely by the KaiC enzymatic rates, which are first order in protein concentration (9).
However, it is clear from multiple studies that the formation of the inhibitory KaiB•KaiC complex is, in fact, slow and that this complex plays a critical role in switching the oscillator from a kinase-dominated mode to a phosphatase-dominated mode (9, 14, 19). If the slow rate of KaiB•KaiC complex assembly followed a mass-action dependence on the product of the KaiB and KaiC concentrations, the robustness of the oscillator to total protein concentration would be threatened. Given that catalysis in CI is required for formation of the KaiB•KaiC complex and that the previously measured rate of CI-catalyzed ATP hydrolysis is also slow (∼10/KaiC per day), we speculated that catalytic turnover in CI might provide a rate-limiting step that dictates the assembly rate of KaiB•KaiC complexes during the circadian cycle (15, 19).
In this scenario, although the encounter frequency between KaiB and KaiC molecules would depend on protein concentration, KaiC would be unable to form a stable complex except after a CI ATPase reaction. We would then expect the kinetics of complex assembly to be approximately independent of the frequency of bimolecular collisions and therefore, independent of increasing protein concentration. To test this prediction, we used an immunoprecipitation assay to measure the kinetics of KaiB•KaiC complex formation at a variety of total protein concentrations using the KaiC-EE phosphomimetic mutant that is constitutively capable of forming KaiB•KaiC complexes. Indeed, assembly occurs on a timescale of several hours, and the assembly rate is only weakly dependent on concentration (less than a 20% increase in the rate constant is observed for an eightfold increase in protein concentration as opposed to the eight times acceleration expected from the mass-action increase in collision frequency) (Fig. 3C and Table S3). We interpret these results to indicate that the reason KaiB•KaiC complexes form slowly is not because it is intrinsically difficult for KaiB and phosphorylated KaiC to engage each other in solution, but because each KaiC molecule must first become activated by ATP hydrolysis in CI, resulting in a short-lived, binding-competent state.
This ATPase requirement ensures that KaiB•KaiC complexes form on an appropriate circadian timescale and maintains the robustness of the oscillator to protein concentration. The precise structural nature of this CI-dependent activation is currently unclear. Because ATP binding in CI is known to be important for stabilizing the KaiC hexamer (8), ATP hydrolysis in CI may allow KaiB•KaiC complex formation by transiently depleting ATP from the monomer–monomer interface and disrupting hexamer structure, possibly exposing an otherwise hidden binding surface for KaiB on CI.
If ATPase activity in CI is unaffected by the ATP/ADP input signal and this catalytic step is permitting the formation of KaiB•KaiC complexes, we would expect the kinetics of assembly of phosphomimetic KaiC with KaiB to also be independent of the ATP/ADP ratio. Indeed, kinetic analysis of KaiB•KaiC complex formation in the presence of various amounts of ADP did not show systematic variation in the rate, consistent with our direct measurement of the ATPase activity of the isolated CI domain (Fig. 3D and Table S3).
We can now describe the sequence of events that lead to the formation of a KaiB•KaiC complex in the circadian oscillator (Fig. 3E). First, a KaiC molecule must achieve phosphorylation on the critical Ser431 residue. Because of the ordered nature of KaiC phosphorylation, multiple reactions in CII are required: phosphorylation on Thr432 followed by phosphorylation on Ser431. Each of these steps in CII can be controlled by KaiA activity and external input signals. Our data suggest that, after KaiC has achieved an appropriate phosphorylation state, it must go through a cycle of ATP hydrolysis in CI to bind stably to KaiB. This CI catalytic event is not modulated by the ATP/ADP ratio. Thus, it represents a constant, slow-timing step in the circadian mechanism.
Mathematical Models with Sequential Catalytic Steps in CII and CI Support a Robust Period.
KaiB•KaiC inhibitory complexes close a negative feedback loop by sequestering the activator KaiA (9, 19). Thus, the CI-dependent step required to form the KaiB•KaiC complex can serve as a delay between phosphorylation and negative feedback, and we expect its presence and dependence on input signals to have important consequences for the robustness of the oscillator period. To analyze the functional implications of the distinct roles for CI and CII catalytic activity, we developed simple mathematical models with various reaction topologies based on measured rates of KaiC enzymatic activity and assessed the impact of the CI-dependent KaiB binding step on their ability to generate robust circadian rhythms under varying ATP/ADP input signals (SI Text discusses differential equations). In these models, collective oscillations arise from global changes in KaiA activity driven by sequestration of KaiA in phosphorylation-dependent KaiA•KaiB•KaiC complexes. We assume that, when the required KaiC phosphorylation is lost, the inhibitory complexes dissociate immediately (9).
A model that explicitly incorporates the requirement for slow, invariant CI catalysis in forming the KaiB•KaiC inhibitory complex is capable of generating robust circadian oscillations over a wide range of ATP/ADP conditions (Fig. 4A), while retaining an ability to be phase-shifted in response to transient changes in ATP/ADP ratio (Fig. 4D). We set the rate constant for CI-dependent KaiB•KaiC complex formation from experimental data (compare Fig. 3); the model requires that this rate be similar to the phosphorylation rates (Fig. S4A). In general, robustness does not depend on highly fine-tuned choices of the kinetic parameters in the model; the period remains robust when these parameters are randomly perturbed about their experimentally estimated original values (Fig. S4B). In our model, the effective KaiB•KaiC complex formation rate constant is set by the CI ATPase rate and thus, first order in protein concentration. This scheme displays perfect robustness to total protein concentration (Fig. S4C). If the complex formation rate instead has a mass-action dependence on protein concentration, robust scaling of the oscillator dynamics is lost (Fig. S4D).
Fig. 4.
Mathematical model integrating roles of CI and CII predicts a robust period. (A) Model A consists of a modified reaction network including a slow binding step between Ser431-phosphorylated KaiC and KaiB that is invariant to ATP/ADP (red arrow) in addition to kinase reactions sensitive to ATP/ADP (black arrows) and phosphatase reactions (gray arrows). Numerical integration of this model reproduces oscillations of KaiC phosphorylation with a robust period over a range of ATP/ADP conditions. (B) Model B neglects the role of CI and assumes that KaiB binding occurs much faster than changes in CII phosphorylation. Numerical integration of this model shows that the period elongates and oscillations lose stability at low ATP/ADP conditions. (C) Period of the oscillations at various ATP/ADP for models A (red symbols) and B (blue symbols) determined by fitting the simulated trajectory to a sinusoidal function. Periods under conditions where oscillations are unstable were estimated by fitting to initial transient oscillations and are represented by dashed lines. All periods have been scaled relative to an assumed 24-h period for the 100% ATP condition for each model. (D) Phase response curve showing relationship between time of 50% ADP pulse and the resulting phase advance or delay (model A, red symbols) compared with the phase response from model B (blue symbols) and from in vitro (orange triangles) and in vivo (green squares) experimental data. Comparisons were made by scaling the period of the models to 24 h and shifting the data so that peak phosphorylation occurs at CT 12.
We used our model to estimate the total production rate of ADP from both domains during the circadian cycle, assuming that the CI ATPase is activated by CII phosphorylation during the KaiB binding steps and ignoring the possibility that some ADP could be converted back to ATP during dephosphorylation (11, 12). This calculation predicts that ADP production should be highest near peak phosphorylation, which has been observed experimentally, but overestimates the magnitude of ATP consumption (15) (Fig. S5).
In contrast, if we assume that the enzymatic reactions in CII are the only slow processes and remove the CI-dependent step in KaiB•KaiC complex formation, the model is unable to sustain rhythms with an appropriate period at ATP/ADP ratios across the physiological range (6, 9) (Fig. 4B). Thus, the addition of a slow formation rate for KaiB•KaiC complexes, set by the CI ATPase activity, stabilizes the oscillatory behavior. The inclusion of the CI ATPase step also makes the model behavior much less sensitive to the choice of the inhibitory strengths assigned to phosphorylated forms of KaiC (Fig. S6 A and B).
In the model that accounts for the dynamical roles of both CI and CII, the oscillator period remains within 20% of 24 h as the nucleotide input signal changes from 100% to 40% ATP (Fig. 4C and Table S4), whereas the peak height of the oscillation decreases in qualitative agreement with the experimental data. However, this model does not correctly predict the quantitative extent of phosphorylation at the trough of the oscillation. (Fig. S6 C and D). The inclusion of the CI-dependent delay in negative feedback improves robustness relative to models where KaiB binding is fast, even if the CI ATPase rate is made sensitive to the ATP/ADP ratio contrary to our experimental data (compare Fig. 3 B and D). However, the period of this alternative model elongates inappropriately at low ATP/ADP relative to the model where the CI rate is invariant (Fig. S7 A and B).
The models that we analyzed here are simplified representations of the KaiABC system that neglects some of the known biochemistry of the system, including the facts that KaiC functions as a hexameric ring and that monomers are exchanged between KaiC particles at specific clock times (22). We chose models with a simple structure to minimize the number of parameters that were not constrained by experimental measurements (Table S5), allowing us to search exhaustively across the undetermined parameters and make general statements about the properties conferred by the addition of a CI-dependent KaiB binding step to model topology. For example, we treat the microscopic collisions between KaiB and KaiC implicitly and assume that KaiB is always present in excess, so that the concentration of KaiB does not appear in the dynamics. If the model is made more detailed by explicitly describing the association and dissociation of KaiB from KaiC after ATP hydrolysis in CI, we find that the dynamics are nearly unchanged as long as the affinity constant for the KaiB-KaiC interaction is in the low nanomolar range or below (Fig. S7 C and D). Because more biochemically detailed models of the KaiABC oscillator typically share the core feature of phosphorylation-dependent negative feedback mediated by KaiB, we expect that the inclusion of a slow CI-dependent delay will improve robustness in any model with this feedback loop structure.
Intuitively, how does the combination of input-sensitive phosphorylation and a slow, invariant timer step produce an oscillator period that is robust to changing input signals? Consider a pool of weakly phosphorylated KaiC near the trough of the circadian rhythm. KaiC will begin to autophosphorylate in response to KaiA until it has accumulated enough Ser431 phosphorylation to inhibit KaiA through KaiB•KaiC complexes. The time that it takes to achieve this threshold phosphorylation will depend inversely on the CII kinase rate and thus, the ATP/ADP input signal. However, before inhibition can occur, KaiC must first go through a slow catalytic cycle in CI, providing a fixed time delay in the negative feedback loop in the oscillator. While waiting for a sufficient concentration of inhibitory KaiB•KaiC complexes to assemble, the pool of KaiC will continue to autophosphorylate, overshooting the inhibitory threshold of phosphorylation by an amount proportional to the CII kinase rate.
If the ATP/ADP ratio were lowered KaiC would need more time to reach a phosphorylation state capable of triggering inhibition, but phosphorylation is also slower during the CI-mediated KaiB binding phase, so that the peak level of phosphorylation is depressed. Here, CII acts as an integrator, accumulating information about the level of input signaling and encoding it in the amplitude of overshoot phosphorylation (23).
After CI catalysis and KaiB•KaiC complex formation, the system enters the dephosphorylation phase, and the overshoot phosphorylation must then be removed. Under low ATP/ADP conditions, slowed phosphorylation is compensated by less time spent dephosphorylating before the cycle can repeat (SI Text has a quantitative version of this argument) (Fig. S8 A and B).
By this mechanism, the period of the oscillator can be held approximately constant at the expense of a variable amplitude (compare Fig. 1 D and E), a general phenomenon seen throughout circadian biology (1, 24). This argument applies to perturbations that change only the kinase activity of KaiC. If both the kinase and phosphatase rates are slowed or accelerated together, which may be the case in some of the period mutants caused by point mutations in CII, the model predicts that the oscillator period will change more sensitively (Fig. S8C). Kinase activity can also be modulated by increasing the concentration of KaiA, but this manipulation presumably changes the dynamics in other ways, because increased KaiC phosphorylation is needed to inactivate the excess KaiA. Indeed, experimental studies show that the period of the in vitro oscillator is more sensitive to changes in the [KaiA]/[KaiC] ratio than the nucleotide signals studied here (25).
Discussion
Oscillators play crucial roles in a variety of biological contexts, including development, the cell cycle, and circadian rhythms. These oscillating systems must be coupled to the rest of the organism to receive input signals and transduce output signals, but this coupling must be achieved through mechanisms that do not compromise the functional properties of the oscillator. In the case of circadian rhythms, utility for the organism depends on an agreement between the free-running period of the oscillator and the periodicity of the external environment (2).
The core of the cyanobacterial circadian clock is a posttranslational reaction network composed of interaction between three protein components: KaiA, KaiB, and KaiC. Recent work has shown that metabolic signals, including the ATP/ADP ratio and the redox state of the quinone pool, impinge directly on oscillator components (6, 26). When these parameters are perturbed during the phosphorylation phase of the circadian rhythm, phase shifts can be induced, but the system then adapts to the altered metabolic conditions to allow oscillations to continue. We have shown that an oscillator design composed of sequential sensitive and insensitive enzymatic steps is capable of receiving input signals while maintaining a robust period and that this design is implemented by the biochemical properties of the two catalytic domains of KaiC in the cyanobacterial clock. This architecture, where CII receives variable signals and CI then allows negative feedback after a slow, insensitive catalytic step, may be a general scheme that applies beyond nucleotide sensing. Temperature compensation may also be explicable in this framework. We expect that other biological clocks may use similar combinations of linked input-sensitive and -independent processes to achieve a balance between tunability and robust timing.
Materials and Methods
Recombinant Protein Purification and in Vitro Protein Reactions.
Procedures and reaction conditions are provided in SI Text.
ATPase Assay.
The purified KaiC CI fragment was incubated at 3.5 μM in buffers containing 1 mM ATP and various amounts of ADP at 30 °C for 48 h. ATP and ADP in the reaction mixtures were separated on a MonoQ 5/50 GL anion exchange column (GE Healthcare) at room temperature at a flow rate of 1.0 mL/min in a mobile phase of 0.05 M NH4H2PO4 (pH 4.8) and eluted with a salt gradient up to 0.75 M NH4H2PO4 (pH 4.8). ATP and ADP amounts were determined as the area under the A280 absorbance for each peak on the chromatograph using Unicorn software (GE Healthcare). ATPase activity was determined as ADP produced per hour per KaiC molecule.
Immunoprecipitation of KaiB•KaiC Complexes.
Reaction samples were collected after incubation at 30 °C, mixed with monoclonal anti-FLAG (anti-DYKDDDDK) M2 antibody-coupled Protein G Dynabeads (Invitrogen), eluted with 3xFLAG peptide (Sigma-Aldrich) or nonreducing SDS/PAGE sample buffer, and analyzed by SDS/PAGE. Where appropriate, all proteins were first exchanged into buffer containing an ATP analog using Micro Bio-Spin P-30 polyacrylamide columns (Bio-Rad) and then incubated at 30 °C for 96 h before the addition of KaiB-FLAG.
Mathematical Modeling and Kinetic Analysis.
Governing differential equations, kinetic fitting schemes, and numerical analyses performed are described in SI Text.
Supplementary Material
Acknowledgments
We thank K. Cook, D. A. Drummond, E. Ferguson, and L. Rothman-Denes for valuable discussions and comments on the manuscript. This work was supported by a Burroughs-Wellcome Career Award at the Scientific Interface and a Chicago Biomedical Consortium Junior Investigator Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212113110/-/DCSupplemental.
References
- 1.Winfree AT. The Geometry of Biological Time. 2nd Ed. New York: Springer; 2000. [Google Scholar]
- 2.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95(15):8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106(11):4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martino TA, et al. Circadian rhythm disorganization produces profound cardiovascular and renal disease in hamsters. Am J Physiol Regul Integr Comp Physiol. 2008;294(5):R1675–R1683. doi: 10.1152/ajpregu.00829.2007. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308(5720):414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 6.Rust MJ, Golden SS, O’Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331(6014):220–223. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kallas T, Castenholz RW. Internal pH and ATP-ADP pools in the cyanobacterium Synechococcus sp. during exposure to growth-inhibiting low pH. J Bacteriol. 1982;149(1):229–236. doi: 10.1128/jb.149.1.229-236.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi F, et al. Roles of two ATPase-motif-containing domains in cyanobacterial circadian clock protein KaiC. J Biol Chem. 2004;279(50):52331–52337. doi: 10.1074/jbc.M406604200. [DOI] [PubMed] [Google Scholar]
- 9.Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318(5851):809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99(24):15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egli M, et al. Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism. Biochemistry. 2012;51(8):1547–1558. doi: 10.1021/bi201525n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishiwaki T, Kondo T. Circadian autodephosphorylation of cyanobacterial clock protein KaiC occurs via formation of ATP as intermediate. J Biol Chem. 2012;287(22):18030–18035. doi: 10.1074/jbc.M112.350660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YI, Dong G, Carruthers CW, Jr, Golden SS, LiWang A. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2008;105(35):12825–12830. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishiwaki T, et al. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26(17):4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terauchi K, et al. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2007;104(41):16377–16381. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong G, et al. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 2010;140(4):529–539. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami R, et al. ATPase activity and its temperature compensation of the cyanobacterial clock protein KaiC. Genes Cells. 2008;13(4):387–395. doi: 10.1111/j.1365-2443.2008.01174.x. [DOI] [PubMed] [Google Scholar]
- 18.Kageyama H, et al. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell. 2006;23(2):161–171. doi: 10.1016/j.molcel.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 19.Qin X, et al. Intermolecular associations determine the dynamics of the circadian KaiABC oscillator. Proc Natl Acad Sci USA. 2010;107(33):14805–14810. doi: 10.1073/pnas.1002119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang YG, Tseng R, Kuo NW, Liwang A. Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc Natl Acad Sci USA. 2012;109(42):16847–16851. doi: 10.1073/pnas.1211508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood TL, et al. The KaiA protein of the cyanobacterial circadian oscillator is modulated by a redox-active cofactor. Proc Natl Acad Sci USA. 2010;107(13):5804–5809. doi: 10.1073/pnas.0910141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito H, et al. Autonomous synchronization of the circadian KaiC phosphorylation rhythm. Nat Struct Mol Biol. 2007;14(11):1084–1088. doi: 10.1038/nsmb1312. [DOI] [PubMed] [Google Scholar]
- 23.Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387(6636):913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 24.Lakin-Thomas PL, Brody S, Coté GG. Amplitude model for the effects of mutations and temperature on period and phase resetting of the Neurospora circadian oscillator. J Biol Rhythms. 1991;6(4):281–297. doi: 10.1177/074873049100600401. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima M, Ito H, Kondo T. In vitro regulation of circadian phosphorylation rhythm of cyanobacterial clock protein KaiC by KaiA and KaiB. FEBS Lett. 2010;584(5):898–902. doi: 10.1016/j.febslet.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Kim YI, Vinyard DJ, Ananyev GM, Dismukes GC, Golden SS. Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc Natl Acad Sci USA. 2012;109(44):17765–17769. doi: 10.1073/pnas.1216401109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.