Abstract
Background:
In recent years, DNA methylation as a main epigenetic modification in human cancer is found as a promising biomarker in early detection of breast cancer. Possible applications of numerous hypermethylated genes have been reported in diagnosis of breast cancer but there has been a little comprehensive study on the clinical usefulness of these genes in breast cancer. The aim of the present study was to investigate the promoter methylation status of 14-3-3 sigma gene with the goal of developing a diagnostic application in breast cancer.
Materials and Methods:
Totally 40 cases of cancerous and noncancerous tissues were studied. DNA was extracted from tissue samples, and promoter methylation pattern was determined by using methylation-specific polymerase chain reaction.
Results:
Methylation pattern of 14-3-3 sigma promoter significantly differed between control and malignant breast tissues (P = 0.001), and there was no remarkable correlation between methylation and age (P > 0.05).
Conclusion:
The relationship of promoter methylation of 14-3-3 sigma with development of breast cancer found in this study and confirmed the results of previous reports suggests that we can provide the foundation for possible application of 14-3-3 sigma as a potential biomarker for early detection and monitoring disease status.
Keywords: 14-3-3 Sigma, DNA methylation, early detection
INTRODUCTION
Breast cancer is the most common malignant tumor in women worldwide. According to estimations by the World Health Organization (WHO), breast cancer leads to about 519,000 deaths per year in the world, and is between the top 10 mortal diseases.[1] In Iran, breast cancer is one of the growing and important women's health problems, although its statistics is very similar to that of the regional countries.[2–4] Early detection and treatment of breast cancer increases the chance for survival.[5,6] However, no biomarker has yet proven sufficiently sensitive or specific for routine use in clinical practice.
In recent years, the role of epigenetic change as a distinct and crucial mechanism to silence a variety of methylated tissue-specific and imprinted genes has emerged in many cancer types.[7,8] This epigenetic alteration in DNA is heritable that cannot be explained by changes in the primary structure of DNA. Also in contrast to genetic changes, epigenetic modifications are potentially reversible.[9,10]
DNA methylation is the main epigenetic modification in human cancer and is found early during carcinogenesis.[11] Methylation of promoter CpG islands, CG-rich regions that coincide with the promoters of protein coding genes, is an important mechanism of gene inactivation in human cancers, including breast cancer. These facts have nominated the DNA methylation as a promising marker for clinical applications in cancer management.[12,13] Thus, screening for abnormal methylation patterns seems to provide a significant approach for early cancer diagnosis.[14–16] To this aim, identification of genomic loci whose methylation patterns represent an early diagnostic marker has been a focus of research in the recent years.
14-3-3 Sigma belongs to the 14-3-3 protein family and regulates numerous cellular processes that are important to cancer development. 14-3-3 Sigma is a p53-regulated G2/M inhibitor involved in numerous cellular signaling transduction pathways related to the cell cycle, DNA repair, and apoptosis.[17,18] Recent studies have shown that 14-3-3 sigma promoter is typically hypermethylated in different cancer such as breast.[19–25] However, in previous studies, the methylation of 14-3-3 sigma promoter has been differently reported. In this study, the methylation pattern of 14-3-3 sigma promoter in tumor and normal tissues of breast was investigated, with the goal of determining the clinical significance of its epigenetic silencing in breast cancer.
MATERIALS AND METHODS
Patients and Tissue Specimens
Samples of breast cancer tissues were obtained from 20 women (who had undergone surgery at the Alzahra Hospital of Isfahan University of Medical Sciences. The age of the patients ranged from 44 to 58 years and gave their signed informed consent. Normal breast tissues (n = 20) were also taken from the same patients that underwent partial or total mastectomy, 3 cm away from the site at which the tumor samples were taken. A part of each tumor and representative normal tissue was kept in formalin for histopathologic characterization to confirm the diagnosis. We also studied blood samples from 20 healthy individuals to assess methylation in these genes in normal population.
DNA Isolation
DNA was extracted from 25 mg frozen breast tissues of 20 tumoral and 20 normal samples that were stored at -80°C using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The quality and integrity of the extracted DNA was checked by electrophoresis on 0.8% agarose gel, quantitated spectrophotometrically and stored at -20°C before use.
Bisulfite Conversion
The treatment of DNA samples performed using EpiTect Bisulfite Kit (Qiagen), which converts unmethylated but not methylated cytosines to uracils and provides efficient DNA deamination and purification.[26]
Methylation-Specific Polymerase Chain Reaction
Methylation-specific polymerase chain reaction (PCR) for 14-3-3 sigma gene promoter was performed with primers specific for methylated DNA [5’-AATCAATAATACGCTTCTTATCGTC-3’ (sense) and 5’-ATAGTTATTTTATTAAGGAGGTCGG-3’ (antisense)], and primers specific for unmethylated DNA [5’-AAATCAATAATACACTTCTTATCATC-3’ (sense) and 5’-GATAGTTATTTTATTAAGGAGGTTGG-3’ (antisense)]. The PCR for methylated primers was 126 bp and for unmethylated primers was 128 bp product. The PCR conditions were as follows: One cycle of 95°C for 12 min; 45 cycles of 95°C for 30 s, 51°C (unmethylated reaction) or 53°C (methylated reaction) for 30 s, 72°C for 30 s; and one cycle of 72°C for 7 mins. Ten mililiters of each 50 mL methylation specific amplified product was loaded directly onto nondenaturing 2% agarose gels, stained with ethidium bromide, and examined under ultraviolet illumination. Samples were scored as methylation positive when methylated alleles were present, visualized as bands in the methylated DNA, and as methylation-negative when bands were seen only in the unmethylated DNA.
We used placental DNA treated in vitro with SssI methyltransferase (New England Biolabs, Beverly, MA, USA) as a positive control for methylated alleles of 14-3-3 sigma, and products of PCR reaction on this gene and DNA from normal breast tissue was used as unmethylated controls.
Statistical Analysis
Statistical analysis of significant methylation of gene was performed with Fisher exact test (2-sided) using SPSS Version 16. P value less than 0.05 was considered significant.
RESULTS
Methylation status of 14-3-3 sigma promoter in tumor and normal samples. Promoter methylation analysis of 14-3-3 sigma gene was carried out in 20 breast cancer tissues and 20 normal breast tissues collected from breast cancer patients. Patient's age ranged from 44 to 58 years with a mean of 51.7 years. Methylation analysis of this gene was also carried out in 20 normal blood collected from healthy individuals. Among breast cancer tissues 70% (14 of 20) were methylated and 30% (6 of 20) were unmethylated. Three of 20 tumor samples showed heterogeneous methylation, that is, both the methylation and unmethylated alleles were detected that are considered as methylated for the analysis.[27] Among normal breast tissues 20% (4 of 20) were methylated and 80% (16 of 20) were unmethylated [Figure 1]. There were significant differences between tumor and normal breast tissues in the methylation pattern (P = 0.001) [Figure 2]. Methylation status in normal population showed that among blood samples 20% (4 of 20) were methylated and 80% (16 of 20) were unmethylated. Also there was no remarkable correlation between methylation and age (P > 0.05).
Figure 1.

MSP analysis of T, cancerous and N normal breast tissue from one patient by U, unmethylated and M, methylated primers. C, control
Figure 2.
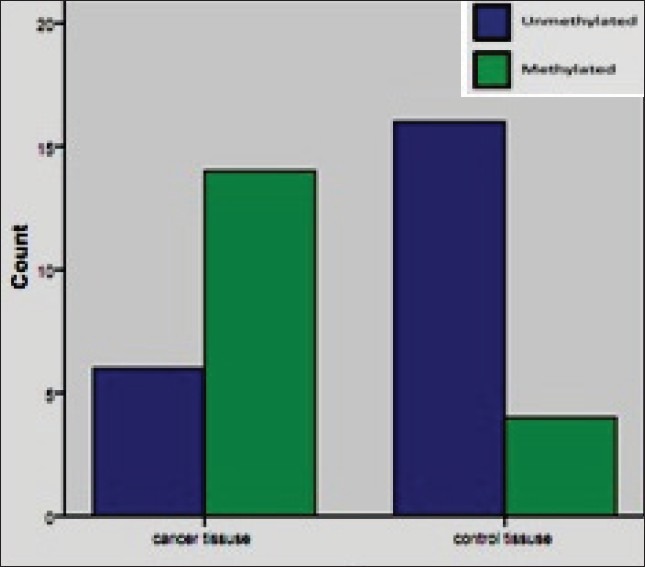
Comparison of methylation pattern of 14-3-3 sigma promoter region in breast cancer and its adjacent normal tissues
DISCUSSION
Use of an unbiased approach and predictive factors for accurate prognosis are necessary for the optimum management of patients with cancer and are especially important in breast cancer.
The epigenetic alterations that initiate and drive tumourigenesis[25,28] are promising targets for early detection because they may precede clinical signs of cancer and can be detected at very low levels. However, preliminary clinical applications of this approach have revealed several shortcomings, including the lack of association between hypermethylation of a given gene promoter and a specific cancer.
There are several reports on methylation profiles of breast cancer patients in various populations. And CpG methylation of 14-3-3 sigma gene is commonly found in breast cancer, including early stages of tumor development.[28,29] Also numerous reports documented CpG methylation of 14-3-3 sigma in several common human malignancies, with a particularly high prevalence in prostate, breast, and ovarian carcinomas.[25]
As described above, 14-3-3 sigma is known as responsible for instituting the G2 cell cycle check point in response to DNA damage in human cell.[30] It was reported that 14-3-3 sigma has been in downexpressed status in breast cancer[19,25,29] and these finding associate with hypermethylation status of sigma gene promoter, which plays an important role in the progression of breast cancer. Therefore, further evaluation of sigma gene promoter methylation in fine-needle biopsies, serum free DNA, tumor tissue, and premalignant lesions, such as carcinoma in situ, can provide the foundation for its development as a biomarker for early detection.
Using methylation-specific PCR techniques, we studied the methylation status of 14-3-3 sigma promoter in 20 breast cancer patients in comparison with 20 controls, including normal breast tissues from same patients. In our study, the comparison of normal and malignant tissues showed a statistically significant (P = 0.001) difference in the methylation pattern. And 14-3-3 sigma was found to be methylated in 70% of breast cancer tissues in contrast with 30% in normal breast tissues. Luo et al[29] found that 14-3-3 sigma gene was methylated in 90% of sporadic breast cancer patients. It has also been demonstrated that losing of 14-3-3 sigma expression is often accompanied by methylation of the CpG island of this gene. Also methylation of 14-3-3 sigma gene and its association with gene silencing have been reported in 91% of primary breast cancer.[19] Various frequencies of 14-3-3 sigma methylation were observed in different studies; whether this variation is due to ethnicity or etiology remains to be determined in a larger study.
Results of recent and previous studies suggest that methylation of 14-3-3 sigma may be a useful biomarker for the diagnosis of breast cancer and for improving the followup of treatment and evaluation of its efficacy. However, the specificity and sensitivity values obtained were inadequate for their use in prospective screening studies. A combination of the several biomarkers may improve prediction of the clinical outcome, but other genes must be investigated to improve the accuracy of molecular diagnosis in cancer.
CONCLUSION
In summary, CpG island promoter methylation is an epigenetic change that is early event and largely responsible for silencing of the sigma gene and occurs in a majority of breast cancers. We analyzed the methylation status of 14-3-3 sigma promoter gene in normal and cancerous tissues. Our study showed that there is a significant difference in the methylation pattern, indicating the role of promoter hypermethylation of 14-3-3 sigma in the development of breast cancer.
It seems that measurement of circulating methylated DNA is a promising and noninvasive approach to cancer risk assessment. Therefore, further research is required to establish the link between 14-3-3 sigma hypermethylated gene promoter measured in the serum of breast cancer patients and grade and stage of tumor and response to chemotherapy, including control of symptoms and improvement in quality of life. This biomarker may be potentially useful to monitor disease status and treatment.
ACKNOWLEDGMENTS
We are grateful to department of biology. genetic ward of Isfahan university of medical sciences for providing financial support to conduct this study
Footnotes
Source of Support: Nil
Conflict of Interest: None declared
REFERENCES
- 1.Brooks M. Breast cancer screening and biomarkers. Methods Mol Biol. 2009;472:307–21. doi: 10.1007/978-1-60327-492-0_13. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Bray F, Pisani P, Parkin DM. Cancer incidence, mortality and prevalence worldwide 2000. Int J Cancer. 2000. [Last accessed on 2012 Feb 23]. Version 1.0. Available from: http://www.dep.iarc.fr/globocan/globocan.html .
- 3.Pisani P, Bray F, Parkin DM. Estimates of the world-wide prevalence of cancer for 25 sites in the adult population. Int J Cancer. 2002;97:72–81. doi: 10.1002/ijc.1571. [DOI] [PubMed] [Google Scholar]
- 4.Montazeri A, Ebrahimi M, Mehrdad N, Ansari M, Sajadian A. Delayed presentation in breast cancer: A study in Iranian women. BMC Womens Health. 2003;3:4. doi: 10.1186/1472-6874-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagel GC, Schmidt S, Strauss BM, Katenkamp D. Quality of life in breast cancer patients: A cluster analytic approach.Empirically derived subgroups of the EORTC-QLQ BR 23-a clinically oriented assessment. Breast Cancer Res Treat. 2001;68:75–87. doi: 10.1023/a:1017975609835. [DOI] [PubMed] [Google Scholar]
- 6.Efficace F, Therasse P, Piccart MJ, Coens C, van Steen K, Welnicka-Jaskiewicz M, et al. Health-related quality of life parameters as prognostic factors in a nonmetastatic breast cancer population: An international multicenter study. J Clin Oncol. 2004;22:3381–8. doi: 10.1200/JCO.2004.02.060. [DOI] [PubMed] [Google Scholar]
- 7.Yang X, Yan L, Davidson NE. DNA methylation in breast cancer. Endocr Relat Cancer. 2001;8:115–27. doi: 10.1677/erc.0.0080115. [DOI] [PubMed] [Google Scholar]
- 8.Franklin TB, Mansuy IM. Epigenetic inheritance in mammals: Evidence for the impact of adverse environmental effects. Neurobiol Dis. 2010;39:61–5. doi: 10.1016/j.nbd.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg AP. Cancer epigenetics takes center stage. Proc Natl Acad Sci U S A. 2001;98:392–4. doi: 10.1073/pnas.98.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10:687–92. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 11.Barekati Z, Radpour R, Kohler C, Zhong XY. Specificity of methylation assays in cancer research: A guideline for designing primers and probes. Obstet Gynecol Int. 2010;2010:pii: 870865. doi: 10.1155/2010/870865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Widschwendter M, Jones PA. The potential prognostic, predictive, and therapeutic values of DNA methylation in cancer. Clin Cancer Res. 2002;8:17–21. [PubMed] [Google Scholar]
- 13.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–66. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 14.Tlsty TD, Crawford YG, Holst CR, Fordyce CA, Zhang J, McDermott K, et al. Genetic and epigenetic changes in mammary epithelial cells may mimic early events in carcinogenesis. J Mammary Gland Biol Neoplasia. 2004;9:263–74. doi: 10.1023/B:JOMG.0000048773.95897.5f. [DOI] [PubMed] [Google Scholar]
- 15.Evron E, Dooley WC, Umbricht CB, Rosenthal D, Sacchi N, Gabrielson E, et al. Detection of breast cancer cells in ductal lavage fluid by methylation-specific PCR. Lancet. 2001;357:1335–6. doi: 10.1016/s0140-6736(00)04501-3. [DOI] [PubMed] [Google Scholar]
- 16.Krassenstein R, Sauter E, Dulaimi E, Battagli C, Ehya H, Klein-Szanto A, et al. Detection of breast cancer in nipple aspirate fluid by CpG island hypermethylation. Clin Cancer Res. 2004;10:28–32. doi: 10.1158/1078-0432.ccr-0410-3. [DOI] [PubMed] [Google Scholar]
- 17.Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14-3-3 Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–20. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 18.Pulukuri SM, Rao JS. CpG island promoter methylation and silencing of 14-3-3 sigma gene expression in LNCaP and Tramp-C1 prostate cancer cell lines is associated with methyl-CpG-binding protein MBD2. Oncogene. 2006;25:4559–72. doi: 10.1038/sj.onc.1209462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson AT, Evron E, Umbricht CB, Pandita TK, Chan TA, Hermeking H, et al. High frequency of hypermethylation at the 14-3-3 sigma locus leads to gene silencing in breast cancer. Proc Natl Acad Sci U S A. 2000;97:6049–54. doi: 10.1073/pnas.100566997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaneuchi M, Sasaki M, Tanaka Y, Shiina H, Verma M, Ebina Y, et al. Expression and methylation status of 14-3-3 sigma gene can characterize the different histological features of ovarian cancer. Biochem Biophys Res Commun. 2004;316:1156–62. doi: 10.1016/j.bbrc.2004.02.171. [DOI] [PubMed] [Google Scholar]
- 21.Mhawech P, Benz A, Cerato C, Greloz V, Assaly M, Desmond JC, et al. Downregulation of 14-3-3 sigma in ovary, prostate and endometrial carcinomas is associated with CpG island methylation. Mod Pathol. 2005;18:340–8. doi: 10.1038/modpathol.3800240. [DOI] [PubMed] [Google Scholar]
- 22.Lodygin D, Yazdi AS, Sander CA, Herzinger T, Hermeking H. Analysis of 14-3-3 sigma expression in hyperproliferative skin diseases reveals selective loss associated with CpG-methylation in basal cell carcinoma. Oncogene. 2003;22:5519–24. doi: 10.1038/sj.onc.1206854. [DOI] [PubMed] [Google Scholar]
- 23.Osada H, Tatematsu Y, Yatabe Y, Nakagawa T, Konishi H, Harano T, et al. Frequent and histological type-specific inactivation of 14-3-3 sigma in human lung cancers. Oncogene. 2002;21:2418–24. doi: 10.1038/sj.onc.1205303. [DOI] [PubMed] [Google Scholar]
- 24.Iwata N, Yamamoto H, Sasaki S, Itoh F, Suzuki H, Kikuchi T, et al. Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 sigma gene in human hepatocellular carcinoma. Oncogene. 2000;19:5298–302. doi: 10.1038/sj.onc.1203898. [DOI] [PubMed] [Google Scholar]
- 25.Lodygin D, Hermeking H. The role of epigenetic inactivation of 14-3-3 sigma in human cancer. Cell Res. 2005;15:237–46. doi: 10.1038/sj.cr.7290292. [DOI] [PubMed] [Google Scholar]
- 26.Korshunova Y, Maloney RK, Lakey N, Citek RW, Bacher B, Budiman A, et al. Massively parallel bisulphate pyrosequencing reveals the molecular complexity of breast cancer-associated cytosine-methylation patterns obtained from tissue and serum DNA. Genome Res. 2008;18:19–29. doi: 10.1101/gr.6883307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirza S, Sharma G, Prasad CP, Parshad R, Srivastava A, Gupta SD, et al. Promoter hypermethylation of TMS1, BRCA1, ERalpha and PRB in serum and tumor DNA of invasive ductal breast carcinoma patients. Life Sci. 2007;81:280–7. doi: 10.1016/j.lfs.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 28.Zurita M, Lara PC, del Moral R, Torres B, Linares-Fernández JL, Arrabal SR, et al. Hypermethylated 14-3-3-sigma and ESR1 gene promoters in serum as candidate biomarkers for the diagnosis and treatment efficacy of breast cancer metastasis. BMC Cancer. 2010;10:217. doi: 10.1186/1471-2407-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo J, Feng J, Lu J, Wang Y, Tang X, Xie F, et al. Aberrant methylation profile of 14-3-3 sigma and its reduced transcription/expression levels in Chinese sporadic female breast carcinogenesis. Med Oncol. 2010;27:791–7. doi: 10.1007/s12032-009-9287-8. [DOI] [PubMed] [Google Scholar]
- 30.Roma AA, Goldblum JR, Fazio V, Yang B. Expression of 14-3-3 sigma, p16 and p53 proteins in anal squamous intraepithelial neoplasm and squamous cell carcinoma. Int J Clin Exp Pathol. 2008;1:419–25. [PMC free article] [PubMed] [Google Scholar]


