Abstract
Posttranscriptional regulatory mechanisms are crucial for protein synthesis during spermatogenesis and are often organized by the chromatoid body. Here, we identify the RNA methyltransferase NSun2 as a novel component of the chromatoid body and, further, show that NSun2 is essential for germ cell differentiation in the mouse testis. In NSun2-depleted testes, genes encoding Ddx4, Miwi, and Tudor domain-containing (Tdr) proteins are repressed, indicating that RNA-processing and posttranscriptional pathways are impaired. Loss of NSun2 specifically blocked meiotic progression of germ cells into the pachytene stage, as spermatogonial and Sertoli cells were unaffected in knockout mice. We observed the same phenotype when we simultaneously deleted NSun2 and Dnmt2, the only other cytosine-5 RNA methyltransferase characterized to date, indicating that Dnmt2 was not functionally redundant with NSun2 in spermatogonial stem cells or Sertoli cells. Specific NSun2- and Dnmt2-methylated tRNAs decreased in abundance when both methyltransferases were deleted, suggesting that RNA methylation pathways play an essential role in male germ cell differentiation.
INTRODUCTION
During spermatogenesis, diploid spermatogonia differentiate into meiotic spermatocytes, which transform into haploid spermatids and then mature into sperm (see Fig. S1 in the supplemental material). All premeiotic cells are called spermatogonia; they include regenerating stem cells, as well as those that have taken the path to terminal differentiation. In the mouse testis, spermatogonia are attached to the basal lamina close to the tubular wall. Spermatogonia undergo several rounds of mitotic divisions before they enter two meiotic divisions as primary spermatocytes, which leads to the production of haploid round spermatids (1). Round spermatids undergo morphological changes and transform into elongated spermatids and finally into spermatozoa. Germ cell differentiation is supported by Sertoli cells, which mature until the second postnatal week, when they lose their proliferative capacity and acquire functions to support spermatogenesis (2).
NSun2 is a posttranscriptional regulator, methylating cytosine-5 in tRNA and possibly other RNA species, and its deletion in mice causes male infertility (3, 4). Especially during mammalian spermatogenesis, transcriptional and posttranscriptional mechanisms need to be tightly controlled, because synthesis and translation of mRNAs are temporally uncoupled twice during the differentiation program (see Fig. S1 in the supplemental material) (5, 6). Transcription is first stalled during the first meiotic prophase to allow DNA repair after homologous recombination at the leptotene and zygotene stages (7). A wave of intense transcription during the pachytene stage then allows the spermatocytes to sustain two consecutive rounds of cell division without a real interphase (8–10). Transcription then continues until the transition from round to elongated spermatids, when a second phase of transcriptional silencing occurs (10, 11). Thus, ongoing protein translation in elongated spermatids at the final steps of spermiogenesis entirely depends on the stability of mRNAs (12, 13).
To compensate for the lack of de novo RNA synthesis, transcriptionally competent pachytene spermatocytes and round spermatids store mRNA in ribonuclear particles that protect the mRNA until translation is required (8, 10, 14). Accordingly, a large number of RNA-binding proteins are highly or uniquely expressed in germ cells, and many knockout mouse models for genes encoding RNA-binding proteins are infertile (15, 16). Many of these RNA-binding proteins localize to the chromatoid body, an RNA-processing organelle in the cytoplasm close to the perinuclear region (17).
The chromatoid body appears for the first time in the cytoplasm of meiotic pachytene spermatocytes as an electron-dense fibrous-granular structure in the interstices of mitochondrial clusters (see Fig. S1 in the supplemental material) (18). The chromatoid body contains proteins involved in RNA processing, such as RNA helicases, decapping enzymes, and Argonaute proteins, as well as mRNAs and microRNAs (19). The proper formation and function of chromatoid bodies is essential for posttranscriptional regulation of gene expression in haploid germ cells (17).
Here, we identify the RNA methyltransferase NSun2 as a novel component of the chromatoid body in male haploid germ cells. Loss of NSun2 leads to reduced expression of genes involved in RNA processing, and we find an essential role for NSun2 in the progression of the first prophase of male meiosis.
MATERIALS AND METHODS
Ethics statement.
All mouse husbandry and experiments were carried out according to the guidelines of the local ethics committee under the terms of a United Kingdom Home Office license.
Mice.
Two lines of NSun2−/− mice were generated using the embryonic stem (ES) cell line D014D11 (German Gene Trap Consortium) and the mouse line Nsun2tm1a(EUCOMM)Wtsi (The Wellcome Trust Sanger Institute). Generation and genotyping were performed as described previously (3). NSun2−/− mice were crossed with Dnmt2−/− mice (Dnmt2tm1Bes/J) to generate NSun2-Dnmt2 double-knockout (DKO) mice (20, 21).
Chromosome spreads.
Meiotic chromosome spreads were performed as described previously (22). Testes were dissected from mice and placed in phosphate-buffered saline (PBS). The tunica albuginea and extracellular material were removed from the seminiferous tubules, which were washed further in PBS. The tubules were then placed in hypotonic extraction buffer, which consisted of 30 mM Tris, 50 mM sucrose, 17 mM trisodium citrate dihydrate, 5 mM EDTA, 0.5 mM dithiothreitol (DTT), and 0.5 mM phenylmethylsulfonyl fluoride (PMSF), pH 8.2, for 1 h. The tubules were then removed from the extraction buffer and placed on a glass slide, on which they were resuspended in 40 μl of 100 mM sucrose, pH 8.2. The tubules were torn using forceps, and the tubule remnants were removed. The remaining suspension was then placed onto one end of a glass slide that had previously been dipped in 1% paraformaldehyde (PFA), pH 9.2, containing 0.15% Triton X-100, and the suspension was spread over the surface of the slide by tilting the slide. The chromosomes were dried for 4 h in a closed box with high humidity before proceeding to immunofluorescence staining.
Immunofluorescence and antibodies.
Freshly dissected testes were placed in 4% PFA for 16 to 20 h and then transferred to 70% ethanol. The testes were then embedded in paraffin, and sections were made on glass slides. The testis sections were then rehydrated in xylene and an ethanol series before antigen retrieval was performed by boiling the slides for 20 min in 10 mM tribasic sodium citrate, pH 6. The slides were washed in PBS and then immersed in 0.5% Triton X-100 for 10 min before being placed in blocking buffer (5% fetal calf serum [FCS] in PBS) for 1 h. Antibody stainings were performed in blocking buffer before washing and mounting of slides in Vectashield. For NSun2 staining, a purified rabbit polyclonal antibody (NSun2 antibody Meth2) (1:500) or 20854-1-AP (1:500) (NSun2p; Proteintech) were used. Mouse monoclonal antibody NPM1 (1:500) was from Sigma and sp56 (1:200) from Sourde Bioscience UK Ltd. Rabbit polyclonal Mili and Miwi antibodies (1:500) were from Cell Signaling Technologies; goat polyclonal Ddx4 (1:100), Ddx25 (1:100), Maelstrom (1:100), and Gata4 (1:100) and rabbit polyclonal Dmrt1 (1:100) were from Santa Cruz. Rabbit polyclonal Sycp3 (1:500) and Ki67 (1:100) were from Abcam, and mouse monoclonal gH2AX antibody (1:200) was from Millipore.
Coimmunoprecipitation and protein expression.
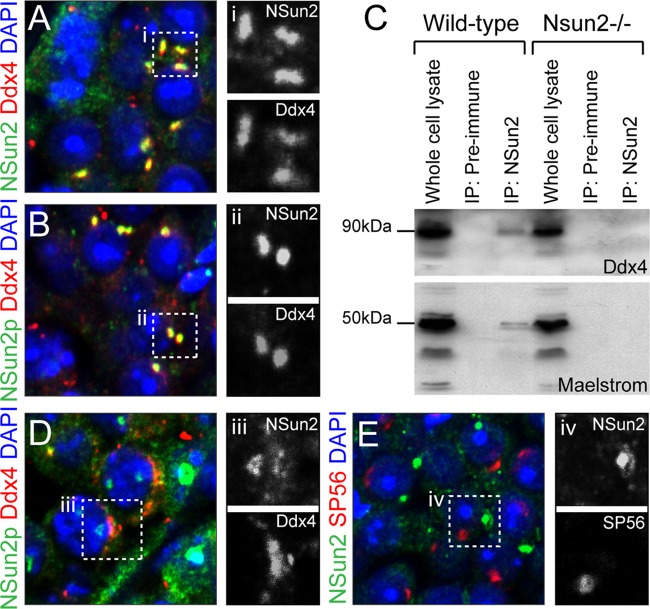
Seminiferous tubules were lysed in lysis buffer (1% NP-40, 150 mM NaCl, 20 mM Tris-HCl, pH 8, 1 mM DTT, and protease inhibitor cocktail) for 1 h on ice. The lysates were cleared by centrifugation at 13,000 rpm, the protein concentration was normalized, and the lysates were diluted 1:10 in immunoprecipitation (IP) buffer (150 mM NaCl, 20 mM Tris-HCl, pH 8, 1 mM DTT, 10% glycerol, and protease inhibitor cocktail) with protein A dynabeads (Invitrogen) and either NSun2p (Proteintech) antibody or rabbit preimmune serum. The IP mixtures were incubated for 16 h at 4°C with gentle mixing. After five 10-ml washes with IP buffer, the beads were resuspended in SDS protein sample buffer, and samples were electrophoresed on a 10% SDS-polyacrylamide gel. The gels were blotted onto nitrocellulose membranes, which were incubated in TBST blocking solution (Tris-buffered saline, pH 8.8, with 5% skim milk powder). The blots were incubated with primary antibodies (Ddx4 or Maelstrom; Santa Cruz) in blocking solution, followed by incubation with the anti-goat horseradish peroxidase (HRP)-conjugated secondary antibody. The chemiluminescent signal was detected using an enhanced-chemiluminescence (ECL) kit (GE Healthcare) according to the manufacturer's instructions. Protein expression of NSun2 (Covalab; 1:1,000) and Dnmt2 (Santa Cruz; 1:500) was analyzed by standard Western blotting using 80 μg of total testis protein extract.
RNA isolation and quantitative reverse transcription-PCR (qRT-PCR).
Expression of Dnmt2 and NSun2 RNA was measured as described previously (21). RNA was isolated from testes using TRIzol (Life Technologies), and cDNA synthesis was performed on 1 μg of RNA using the Superscript III Reverse Transcriptase kit from Invitrogen with random hexamers. Miwi, Tnp2, and Prm1 TaqMan probes were from Applied Biosystems, and quantitative PCR (qPCR) was performed according to the manufacturer's instructions. qPCR for the retrotransposons Line1 5′ untranslated region (UTR), Line1 open reading frame 2 (ORF2), intracisternal A particle (IAP) 3′ long terminal repeat (LTR), and IAP GAG was performed using the SYBR green method, as described previously (23). The primers used were as follows: Line15′UTR-F, GGC GAA AGG CAA ACG TAA GA; Line15′UTR-R, GGA GTG CTG CGT TCT GAT GA; Line1ORF2-F, GGA GGG ACA TTT CAT TCT CAT CA; Line1ORF2-R, GCT GCT CTT GTA TTT GGA GCA TAG A; IAP3′LTR-F, GCA CAT GCG CAG ATT ATT TGT T; IAP3′LTR-R, CCA CAT TCG CCG TTA CAA GAT; IAPGAG-F, AAC CAA TGC TAA TTT CAC CTT GGT; IAPGAG-R, GCC AAT CAG CAG GCG TTA GT. Northern blot analyses for tRNAs were performed as described previously (21).
Gene expression arrays and analyses.
Total RNA (250 ng) was converted to cRNA target using the Illumina TotalPrep-96 kit (Ambion; 4397949). Total RNA was reverse transcribed and converted to double-stranded cDNA using a T7 promoter-oligo(dT) primer and purified with magnetic oligo(dT) beads. This formed the template for an in vitro transcription (IVT) reaction that included a biotinylated nucleotide-ribonucleotide mixture for both cRNA amplification and biotin labeling. After purification, quality control, and quantity normalization, the cRNAs of six samples per genotype and condition were hybridized to arrays. Hybridization, washing, staining, and scanning were performed according to standard Illlumina protocols (Illumina WGGX DirectHyb Assay Guide 11286331 RevA). Microarray hybridization, washing, and scanning were performed at the Genomics Core Facility of Cancer Research United Kingdom (CR-UK) (Cambridge Research Institute [CRI], Cambridge, United Kingdom) (24).
Gene expression analysis was carried out on MouseWG-6 v2.0 Expression BeadChip (Illumina) arrays. All data analyses were carried out in R using Bioconductor packages (25). Raw intensity data from the array scanner were processed using the BASH (26) and HULK algorithms as implemented in the beadarray package (27). Log2 transformation and quantile normalization of the data were performed across all sample groups. Differential expression analysis was carried out using the limma package (28). Differentially expressed genes were selected using a P value cutoff of <0.01 after application of false discovery rate (FDR) correction for multiple testing applied globally to correct for multiple contrasts.
Gene ontology categorizations were performed using DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/).
Microarray data accession number.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO) (29) and are accessible through GEO series accession number GSE39480 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39480).
RESULTS
NSun2 is essential for normal male fertility.
Despite repeated matings between the ages of 6 and 21 weeks, male mice with a homozygous deletion of the NSun2 gene in two independent knockout lines [Nsun2Gt(D014D11)Wrst and Nsun2tm1a(EUCOMM)Wtsi] failed to produce pregnant females (3). To confirm male infertility in the absence of NSun2 (NSun2−/−), we isolated the testes from both knockout lines and found a marked decrease in size compared to those from wild-type (wt) littermates (Fig. 1A and B; see Fig. S2A in the supplemental material). To explore the biological functions of NSun2 during spermatogenesis, we first analyzed the consequences of its deletion for testis morphology in Nsun2tm1a(EUCOMM)Wtsi males at 12 weeks of age (Fig. 1C and D). Lack of NSun2 caused a loss of elongated spermatids in NSun2−/− males, but not spermatogonia or primary spermatocytes (Fig. 1C, arrows, and D). The same morphological defects were observed in Nsun2Gt(D014D11)Wrst mice (see Fig. S2B and C in the supplemental material). Loss of spermatids was confirmed by RNA expression analyses for two markers of round spermatids, transition protein 2 (Tnp2) and Prm1 (30, 31). Both markers were more than 10-fold repressed when NSun2 was deleted (Fig. 1E). Thus, testes of NSun2−/− mice contained spermatocytes but lacked spermatids, indicating that NSun2 is required for successful meiosis during spermatogenesis.
Fig 1.
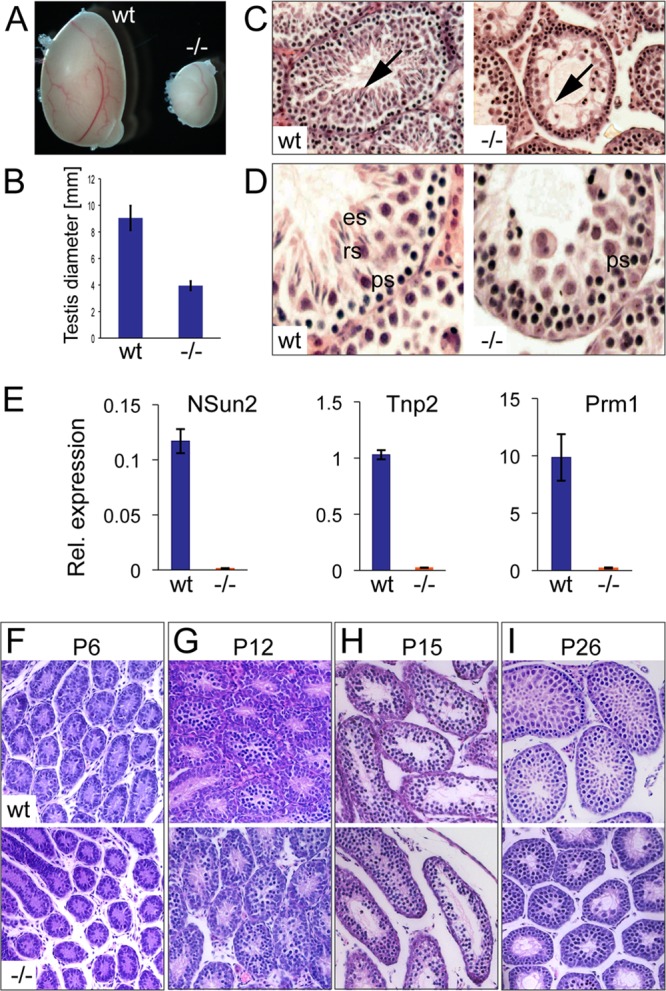
Testis size is reduced and sperm differentiation is blocked in NSun2−/− mice [Nsun2tm1a(EUCOMM)Wtsi]. (A) A testis from an NSun2−/− mouse (−/−) is smaller than one from a wild-type littermate (wt). (B) Quantification of data from panel A. Error bars, standard deviations (SD). (C and D) Hematoxylin and eosin staining of testis sections from wild-type and NSun2−/− mice. The arrows indicate the absence of elongated sperm in the NSun2−/− testis. ps, primary spermatocytes; rs, round spermatids; es, elongated spermatids. (E) qPCR confirming repression of NSun2, Tnp2, and Prm1 RNAs in the NSun2−/− testis. (F to I) Hematoxylin and eosin staining of sections from wt and NSun2−/− testes at the indicated time points.
To determine the precise developmental stage at which the morphological changes became apparent, we analyzed histological sections from wild-type and NSun2−/− testes at postnatal days 6 (P6), 12, 15, and 26 (Fig. 1F to I). We observed morphological differences between NSun2−/− and wild-type testes at P26 only by the lack of round spermatids in NSun2−/− testes (Fig. 1I). We therefore focused our further studies on adult mice 3 months of age, if not otherwise indicated.
To identify the defective meiotic stage in the absence of NSun2, we immunolabeled surface-spread testicular cells for Sycp3, a marker of the lateral component of the synaptonemal complex, and γH2AX, which marks double-strand breaks and the sex body (Fig. 2A to E). The dynamic localization of both markers during meiosis has been well described (32). The vast majority of germ cells in NSun2−/− testes failed to progress beyond early spermatocyte differentiation and arrested at the leptotene and zygotene stages (Fig. 2A, B, and F). We observed a 6-fold reduction of cells at the pachytene stage in the absence of NSun2 (Fig. 2C, D, and F). The vast reduction of pachytene and lack of diplotene spermatocytes indicate that spermatogenesis is aborted during the pachytene stage, which is also confirmed by the presence of spermatocytes with a normal sex body (Fig. 2D; see Fig. S3A and B in the supplemental material). NSun2−/− testes lacked diplotene germ cells (Fig. 2E and F). The increase in germ cells at leptotene and zygotene stages further indicated that spermatocyte differentiation is blocked at the entry to the pachytene stage in NSun2−/− testes (Fig. 2F), an effect that was not due to increased apoptosis (see Fig. S3C and D in the supplemental material). We further confirmed the lack of pachytene cells as early as P15 (see Fig. S4 in the supplemental material). Although wild-type testis was morphologically indistinguishable from knockout testis at P15 (Fig. 1H), in rare cases we did observe the appearance of pachytene spermatocytes in wild-type, but not in knockout tubules (see Fig. S4A to C in the supplemental material).
Fig 2.
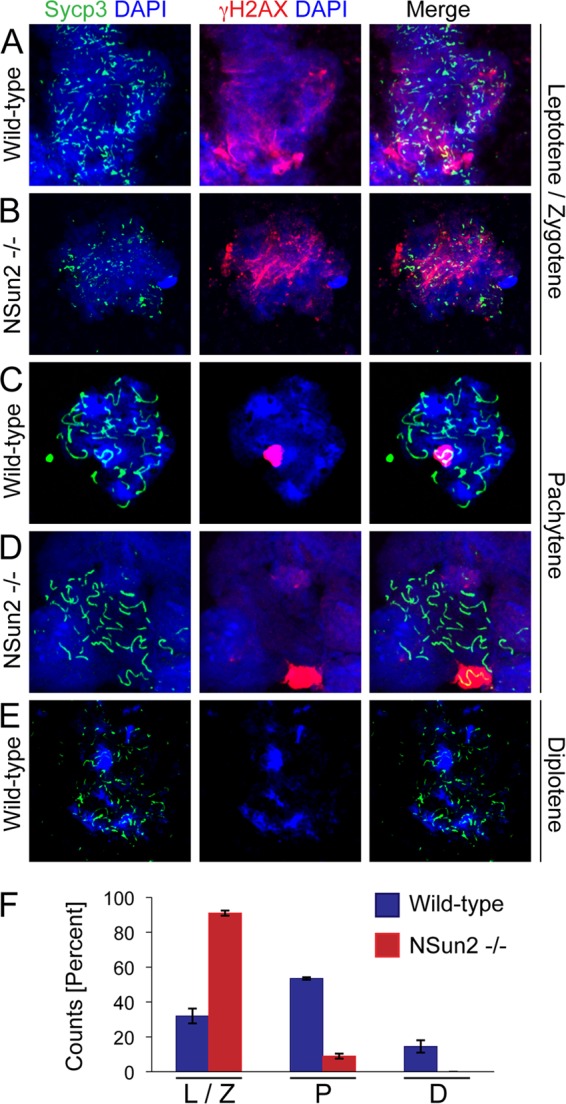
Germ cells arrest at the leptotene and zygotene stages of prophase I at 6 weeks of age. (A to E) Surface-spread germ cells isolated from wild-type (A, C, and E) and NSun2−/− (B and D) testes labeled for Sycp3 (green) and γH2X (red). DAPI (4′,6-diamidino-2-phenylindole) (blue) served as a counterstain. (F) Quantification of germ cells at the indicated stages of early prophase. The error bars indicate SD.
Since transcription is stalled before but highly increased during the pachytene stage (see Fig. S1 in the supplemental material), we asked whether lack of the posttranscriptional modification of cytosine-5 methylation in RNAs might hinder transcriptional processes at the pachytene stage.
Transcriptional and posttranscriptional pathways are repressed in the absence of NSun2.
Murine spermatogenesis initiates a few days after birth and takes approximately 35 days (Fig. 3A). To determine gene transcription levels in testes that lacked NSun2, we performed gene expression profiling in wild-type and NSun2−/− mice at postnatal days 15 and 49. Postnatal day 15 was chosen because pachytene is initiated at this age, and we did not observe any morphological differences in wild-type and knockout mouse models. At postnatal day 49, spermatogenesis is completed, and NSun2−/− mice lack spermatids and sperm (Fig. 3A, dotted lines).
Fig 3.
Factors of the RNA-processing machinery are repressed in the NSun2−/− testis. (A) Overview of murine spermatogenesis in days (d). The blue lines indicate the appearance of the indicated germ cells, and the dotted lines mark germ cells missing in the NSun2−/− testis. The red dotted lines with asterisks mark the time points when samples for RNA expression arrays were taken. (B) Heat map showing significantly changed genes (probes) at an FDR of <0.01 in wild-type versus NSun2−/− testes at P15 in six biological replicates (A to F). (C) Fold change in expression of RNAs (log2FC) in NSun2−/− versus wild-type (wt) testes at postnatal day 15 (15d). (D) Fold change in expression of Miwi, Trd5, and Trd6 RNAs in NSun−/− versus wt testes at postnatal day 15 (−/− 15d), NSun2−/− versus wt testes at postnatal day 49 (−/− 49d), and testes of wt mice at 49 days versus 15 days (Wt 49d). (E and F) qRT-PCR determining RNA expression of NSun2 and Miwi RNAs (E) and selected transposons (F) in NSun2−/− and wt testes. The error bars indicate SD. (G) Venn diagram showing the overlap of significantly repressed genes in NSun2−/− versus wt testes at 49 days (blue) and significantly upregulated genes in wt testes at 49 versus 15 days (orange). (H) Heat map of the 602 genes (probes) shown in panel G in wild-type and NSun2−/− testes at P15 and P49 in six biological replicates (A to F). (I) Fold change of the 27 most repressed RNAs in the NSun2−/− testis at 49 compared to 15 days.
Despite the lack of morphological differences at P15, RNA microarray analyses distinguished gene expression profiles according to genotype (see Fig. S5 in the supplemental material). A total of 3,155 genes (probes) were found to be differentially expressed in wild-type versus NSun2−/− testes (FDR < 0.01) (Fig. 3B; see Table S1 in the supplemental material). To analyze whether inhibition of transcriptional processes might hinder entry into the pachytene stage, we determined the gene ontology categories of the 1,347 repressed genes in NSun2−/− testes (Table 1). Genes encoding proteins involved in RNA processing, such as nucleotide and RNA-binding proteins, as well as ATP-dependent helicases, were significantly underrepresented in NSun2−/− testes at P15 (Table 1; see Table S2 in the supplemental material).
Table 1.
Gene ontology categorization of significantly (FDR < 0.01) underrepresented genes (probes) in NSun2−/− testes at P15 and P49
| Term | Count | % | P value |
|---|---|---|---|
| Underrepresented in NSun2−/− at 15 days (1,186 probes) | |||
| DAVID biological processes (GOTERM_BP_FAT) | |||
| Spermatogenesis | 46 | 3.88 | 5.50E−12 |
| Cell cycle | 78 | 6.57 | 1.44E−11 |
| Multicellular-organism reproduction | 54 | 4.55 | 9.83E−09 |
| Protein localization | 79 | 6.66 | 9.21E−08 |
| RNA processing | 53 | 4.47 | 2.26E−07 |
| DNA metabolic process | 51 | 4.30 | 4.10E−07 |
| DAVID molecular processes (GOTERM_MF_FAT) | |||
| Nucleotide binding | 214 | 18.03 | 1.00E−19 |
| Helicase activity | 28 | 2.36 | 1.13E−09 |
| ATPase activity | 43 | 3.62 | 2.19E−09 |
| RNA binding | 74 | 6.23 | 9.35E−09 |
| ATP-dependent helicase activity | 19 | 1.60 | 6.71E−07 |
| Serine/threonine kinase activity | 46 | 3.88 | 1.12E−05 |
| tRNA binding | 7 | 0.59 | 5.14E−04 |
| KEGG database pathways | |||
| Aminoacyl-tRNA biosynthesis | 8 | 0.67 | 0.002 |
| Cell cycle | 13 | 1.10 | 0.007 |
| RNA degradation | 8 | 0.67 | 0.012 |
| Ubiquitin-mediated proteolysis | 12 | 1.01 | 0.026 |
| Nucleotide excision repair | 6 | 0.51 | 0.032 |
| Spliceosome | 11 | 0.93 | 0.034 |
| Underrepresented in NSun2−/− at 49 days (602 probes) | |||
| DAVID biological processes (GOTERM_BP_FAT) | |||
| Protein transport | 36 | 7.83 | 1.78E−05 |
| Chromosome organization | 25 | 5.43 | 8.68E−05 |
| Translation | 20 | 4.35 | 4.53E−04 |
| Cell cycle | 30 | 6.52 | 7.60E−04 |
| DAVID molecular processes (GOTERM_MF_FAT) | |||
| Nucleotide binding | 92 | 20 | 1.80E−08 |
| RNA binding | 34 | 7.39 | 6.25E−05 |
| GTP binding | 20 | 4.35 | 8.30E−04 |
| Translation initiation factor activity | 9 | 1.96 | 0.002 |
| ATP binding | 50 | 10.87 | 0.006 |
| Nucleoside binding | 52 | 11.30 | 0.011 |
| KEGG database pathways | |||
| Basal transcription factors | 6 | 1.30 | 5.90E−04 |
| Pathways in cancer | 12 | 2.61 | 0.081 |
To identify potential candidate genes causing the block in germ cell development, we asked which genes showed the highest changes in expression in NSun2−/− testes. We found a total of 118 genes more than 2-fold downregulated and very few (33) genes 2-fold or more upregulated in NSun2−/− versus wild-type testis (Fig. 3C; see Table S3 in the supplemental material). One of the most repressed genes in NSun2−/− testes was Miwi (Fig. 3D and E). Miwi is specifically expressed in spermatocytes and spermatids, and lack of Miwi causes spermatogenic arrest at the round spermatid stage (34). Interestingly, Miwi localizes to chromatoid bodies, which function in RNA storage and processing, and we found two more chromatoid body components (Tdrd5 and Tdrd6) downregulated in NSun2−/− testes (Fig. 3D). Miwi directly interacts with Tdrd6, and also in Tdrd6−/− mice, the development from round to elongated spermatids is abrogated (35). In wild-type mice, expression of Miwi, Tdrd5, and Tdrd6 were upregulated during spermatogenesis (Fig. 3D, Wt 49d). Miwi has been shown to regulate retrotransposon silencing in mouse testes (36). However, we did not observe increased expression of transposons, such as Line1 and IAP (Fig. 3F). Thus, male infertility in NSun2−/− mice is not due to deleterious transposon activation (36–39).
RNA microarray analyses of testes from adult mice (P49) revealed that 6,590 genes (probes) were significantly (FDR < 0.01) downregulated in NSun2−/− testes (see Table S4 in the supplemental material). To exclude genes that were repressed because of the specific loss of spermatids and sperm in NSun2−/− testes (Fig. 3A), we compared this gene list with transcripts that are significantly upregulated during normal spermatogenesis by comparing the gene expression profiles of wild-type mice at P49 versus P15 (see Table S4 in the supplemental material). Only repression of 602 genes (probes) in NSun2−/− testes at P49 was due to lack of NSUN2 rather than loss of spermatids and sperm (Fig. 3G and H; see Table S5 in the supplemental material). Gene ontology categorization using the 602 probes confirmed that genes encoding proteins involved in transcription and RNA processing were again overrepresented (Table 1). When we plotted the top 27 genes showing the highest fold change in expression in NSun2−/− testes at P49 but less than 1.5-fold change in expression in wild-type controls (P49 versus P15), we found three ATP-dependent RNA helicases (Ddx4, Ddx5, and Eif4a2) within this group (Fig. 3I).
Together, the RNA expression assays demonstrated that male infertility of NSun2−/− mice may be caused by misregulation of the transcriptional process during the pachytene stage, as the genes showing the highest fold change in repression were involved in RNA processing.
Proteins of the RNA-processing machinery are reduced in NSun2−/− testes.
We next asked whether NSun2 might be part of the RNA-processing machinery in testes and determined the localization of the NSun2 protein during spermatogenesis (Fig. 4). The NSun2 protein was present in round spermatids localized to cytoplasmic granules close to the nuclei (Fig. 4A and C, arrows). The specificity of the NSun2 staining was confirmed using two different antibodies (NSun2p and NSun2) (see Materials and Methods) (Fig. 4A to D). Although NSun2 was also expressed in nucleoli of Sertoli cells, neither the formation of the nucleoli nor the number of Sertoli cells was affected when NSun2 was deleted (see Fig. S6A to C in the supplemental material). Both signals in the nucleoli of Sertoli cells and in round spermatids were lost in NSun2−/− testes (Fig. 4B and D; see Fig. S6B in the supplemental material).
Fig 4.
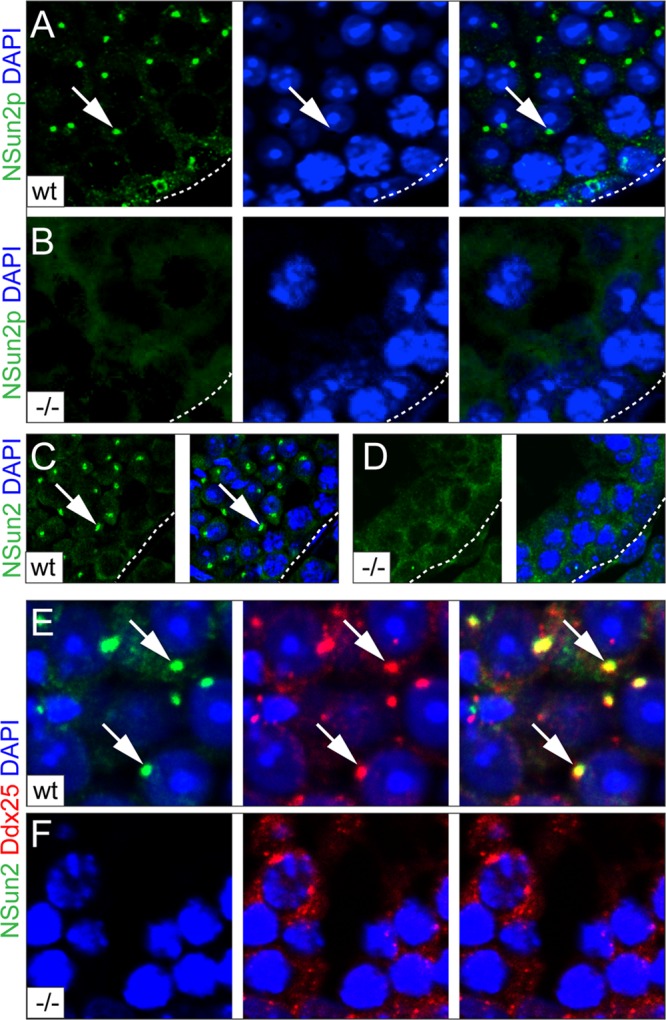
Localization of NSun2 in adult testis. (A and B) Immunofluorescence labeling of NSun2 (NSun2p; green) in wild-type (wt) (A) and NSun2−/− (−/−) (B) testes. (C and D) Higher magnification of wt (C) and NSun2−/− (D) testes showing that NSun2 protein is lost in round spermatids. (E and F) Colocalization of NSun2 (green) and Ddx25 (red) in wt (E) and NSun2−/− (F) testes. Nuclei were counterstained in blue (DAPI) (A to F). The arrows (A, C, and E) point to NSun2 staining in round spermatids.
We next asked whether the NSun2-positive granules were chromatoid bodies. We detected colocalization of NSun2 with Ddx25 and Ddx4 only in wild-type testes (Fig. 4E and F, arrows, and 5A and B; see Fig. S7 in the supplemental material). Ddx4 and Ddx25 are ATP-dependent RNA helicases that localize to the chromatoid body and are required for germ cell development (40–42). We further confirmed localization of NSun2 to chromatoid bodies using two different antibodies (NSun2p and NSun2) (see Materials and Methods) (Fig. 4E and F and 5A and B; see Fig. S7 in the supplemental material) and by coimmunoprecipitation with Ddx4 and Maelstrom (Fig. 5C) (33). The small amount of NSun2 protein in the coimmunoprecipitations with Ddx4 and Maelstrom might be due to the fact that NSun2 colocalized with both markers only in round spermatids, but not the intermitochondrial cement of spermatocytes, where Nsun2 was found in nucleolar structures (Fig. 5D; see Fig. S8A in the supplemental material). NSun2 showed no overlap with the acrosomal protein sp56 in spermatids (Fig. 5E) or Ddx4 in prospermatogonia at embryonic day 16.5 (E16.5) (see Fig. S8B in the supplemental material).
Fig 5.
NSun2 localizes to the chromatoid body. (A and B) NSun2 (green) colocalizes with Ddx4 (red) in round spermatids using two different antibodies for NSun2 (Meth2) (A) and 20854-AP (NSun2p) (B). (C) Coimmunoprecipitation of NSun2 with Ddx4 (top) and Maelstrom (bottom) in wild-type and NSun2−/− testes. Preimmune serum served as the negative control. (D and E) NSun2 (green) does not colocalize with Ddx4 (red) at the intermitochondrial cement in spermatocytes (D) or sp56 (red) at the acrosomal matrix in spermatids (E). (i to iv) Higher magnifications of the respective boxed areas in panels A, B, D, and E). (A, B, D, and E) Nuclei were counterstained in blue (DAPI).
Cytosine-5 tRNA methyltransferases are dispensable for the spermatogonium and early spermatocytes.
Methylation of tRNA at cytosine-5 is catalyzed by Nsun2 and Dnmt2 (20, 43, 44). tRNAs are uniquely methylated by NSun2 and Dnmt2, since tRNAs isolated from testes lacking both enzymes are not methylated (21). Therefore, we considered that the unaffected development of spermatogonia and early spermatocytes up to pachytene stage in Nsun2−/− testes might be due to complementation of NSun2 deficiency by Dnmt2. Dnmt2−/− mice are viable and fertile and do not exhibit any gross phenotype (20).
To confirm that both RNA methyltransferases were coexpressed in the same cell types, we measured RNA and protein levels of NSun2 and Dnmt2 during germ cell differentiation (Fig. 6A and B). Until P15, NSun2 RNA was weakly expressed, but it was 6-fold upregulated from P20, which coincides with the appearance of spermatids (Fig. 3A and 6A, left). We observed a similar expression pattern for Dnmt2 RNA, although Dnmt2 increased slightly earlier during germ cell development (P15) (Fig. 6A, right). We observed upregulation of the NSun2 and Dnmt2 proteins with similar kinetics (Fig. 6B).
Fig 6.
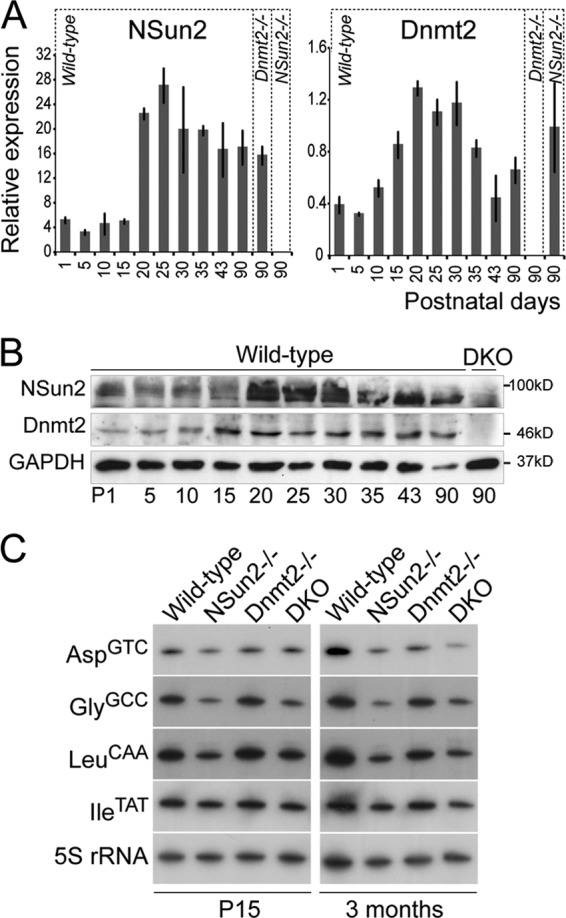
Deletion of NSun2 and Dnmt2 reduced tRNA stability in testes at P15 and 3 months of age. (A) RNA expression levels of NSun2 (left) and Dnmt2 (right) at the indicated postnatal days. The error bars indicate SD. (B) Protein expression levels of NSun2 (top) and Dnmt2 (middle) at the indicated postnatal days (P) in wild-type and NSun2-Dnmt2 double knockouts (DKO). GAPDH served as a loading control (bottom). (C) Northern blot analyses of NSun2- and Dnmt2-methylated tRNAs at P15 and 3 months of age. 5S RNA served as a loading control.
The low abundances of both proteins from P1 to P15 indicated that they might not be functionally active in early stages of germ cell differentiation. Although recent studies suggest that cytosine-5 methylation also occurs in mRNA (4), we were unable to detect any significant overlap between putative methylated mRNAs and differentially abundant mRNAs in NSun2−/− testes (data not shown). The confirmed target substrates of NSun2 and Dnmt2 are tRNAs, and loss of cytosine-5 methylation in tRNAs decreases their overall stability (21). In line with these data, we find that the abundances of the NSun2 and Dnmt2 target substrate tRNAs AspGTC, GlyGCC, and LeuCAA decreased after deletion of Nsun2 (NSun2−/−) or NSun2 and Dnmt2 (DKO) both at P15 and in adult testes, whereas the negative-control tRNA IleTAT remained unchanged (Fig. 6C). Thus, although only weakly expressed at P15, deletion of NSun2 and Dnmt2 already decreased tRNA stability early in germ cell differentiation, even before the pachytene stage.
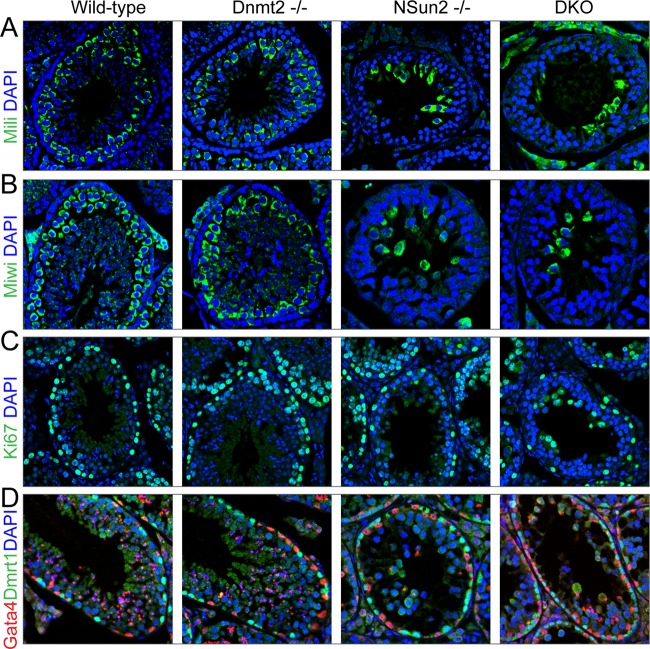
Comparable to NSun2 protein localization in wild-type testes, we found Mili to be absent in the spermatogonium, but then both localized to the cytoplasm of primary spermatocytes of double-knockout testes (Fig. 4A and 7A, wild type) (45). Whereas localization of Mili in Dnmt2−/− testes was comparable to that in the wild type (Fig. 7A, Dnmt2−/−), the number of Mili-positive cells was dramatically reduced in the absence of NSun2 (Fig. 7A, NSun2−/−). Similarly, Miwi, which normally localizes to spermatocytes and the chromatoid bodies of spermatids, was reduced only upon deletion of NSun2 (Fig. 7B, NSun2−/−). We observed the same reduction in Mili- and Miwi-positive cells in the second independent NSun2 knockout line (Nsun2Gt(D014D11)Wrst) (see Fig. S9 in the supplemental material). Double deletion of NSun2 and Dnmt2 (DKO) resulted in a comparable loss of Mili- and Miwi-positive germ cells in NSun2−/− testes, whereas Dnmt2−/− testes showed normal levels of both proteins (Fig. 7A and B).
Fig 7.
Cytosine-5 methylation is dispensable in spermatogonial and Sertoli cells. (A to D) Immunofluorescence labeling of Mili (green) (A), Miwi (green) (B), Ki67 (green) (C), and Gata4 (sc-1237) (red) and Dmrt1 (green) to mark Sertoli and spermatogonial stem cells, respectively (D), in wild-type testis or upon deletion of Dnmt2 (Dnmt2−/−), NSun2 (NSun2−/−), or both Dnmt2 and NSun2 (DKO). DAPI served as a nuclear counterstain.
To confirm that spermatogonia were not affected by deletion of NSun2 and Dnmt2, we stained testis sections for Ki67, a marker for dividing cells. The number of Ki67-positive spermatogonial cells remained unchanged in NSun2−/− testes (Fig. 7C). Although we observed a slight reduction in proliferating Ki67-positive cells in DKO testes (Fig. 7C; see Fig. S10A in the supplemental material), the number and distribution of Sertoli cells labeled by two different antibodies to Gata4 and spermatogonial stem cells marked by Dmrt1 were unchanged in the absence of both NSun2 and Dnmt2 proteins (Fig. 7D; see Fig. S10B in the supplemental material) (46). Gata4 labeled a dot-like structure in wild-type round spermatids, which were not representative of chromatoid bodies and are likely to be nonspecific (see Fig. S10C in the supplemental material).
In conclusion, our data suggested that in mouse testes, posttranscriptional cytosine-5 modifications are dispensable in spermatogonial stem cells and early spermatocytes but are specifically required for the meiotic progression from the leptotene/zygotene stages to pachytene.
DISCUSSION
Our finding that the RNA methyltransferase NSun2 is required for the progression of the first prophase of male meiosis highlights the essential roles of posttranscriptional mechanisms during spermatogenesis. Spermatids and sperm cannot be formed in the absence of NSun2. Although the lack of NSun2 and Dnmt2 can cause differentiation defects in somatic tissues, such as brain and skin, the complete lack of a specific differentiated lineage seems to be unique to the male testis (3, 21, 43).
In somatic cells, the NSun2 protein localizes to the nucleoli in interphase of the cell cycle (47–49). The nucleolus is a distinct nuclear domain in which RNA processing and maturation, as well as tRNA methylation, take place (50). Also in the testis, we find NSun2-positive nucleolar structures in Sertoli cells and primary spermatocytes at interphase of the cell cycle (51). The deletion of NSun2 in Sertoli cells may contribute to the impaired spermatogenesis in NSun2 knockout mice, because the correct proliferation and maturation of Sertoli cells are essential for germ cells to progress through differentiation and meiosis (2). However, two observations argue against a contribution by impaired Sertoli cells to the observed NSun2−/− phenotype. First, the number and location of Sertoli cells were unaffected by deletion of NSun2. Second, in the absence of NSun2, Sertoli cells still showed the characteristic nucleolar tripartite structure typical of mature, adult mouse Sertoli cells, indicating that the maturation of Sertoli cells was unaffected (52).
During cell divisions at the beginning of both mitosis and meiosis, nucleolar structures disassemble, but the components are stored at various cellular locations throughout the cell cycle (48, 51, 53, 54). Nucleolar reassembly starts at telophase in somatic cells and depends on the activation of RNA-processing complexes (55). During spermatogenesis, the nucleolus becomes fragmented at the zygotene-pachytene stage and migrates to the cytoplasm, where it forms a structure that has been suggested to be the origin of the chromatoid body (51, 56, 57).
Lack of NSun2 causes a block of progression of the first prophase of male meiosis at the zygotene-pachytene stage before the chromatoid bodies first appear in the cytoplasm at the late pachytene stage (18), indicating that NSun2's functions are essential for meiotic prophase progression before the chromatoid body is formed. Since NSun2−/− testis lacks germ cells containing a chromatoid body, it is difficult to determine whether NSun2 is also required for chromatoid body assembly, or even RNA processing at the chromatoid body. However, RNA expression profiling revealed that genes encoding proteins involved in transcriptional and posttranscriptional processes are already reduced at P15 before any chromatoid body is formed, indicating that male infertility in NSun2−/− mice is not simply due to the lack of chromatoid bodies. However, we did find downregulation of a number of mRNAs encoding proteins linked to functions of the chromatoid body. Dissecting whether deletion of NSun2 is directly linked to decay of those specific mRNAs or indirectly prevents the formation of functional chromatoid bodies is hampered by the fact that these processes are intertwined (35).
NSun2 generally localizes to cellular RNA-processing centers, and while NSun2 is the first RNA methyltransferase identified as a component of the chromatoid body, Drosophila NSun2 is also a component of ribonuclear particles involved in RNA silencing and germ cell development (58). How NSun2 mechanistically blocks the progression of the first prophase of male meiosis before the pachytene stage remains to be determined but may, at least in part, be dependent on its tRNA methyltransferase activity.
NSun2 is the orthologue of Saccharomyces cerevisiae Trm4 tRNA methylase (47, 59, 60). NSun2 catalyzes the formation of cytosine-5 methylation (m5C) in several tRNAs in vivo in tissues, including skin, liver, and testis (3, 21). Functionally, the m5C posttranscriptional modification influences translation rates, as well as correct RNA folding and stability (21, 61–64). In the absence of NSun2, tRNAs lack specific m5C modifications, which can cause reduced protein translation rates (3, 21). Thus, the methylation of tRNAs by NSun2 at ribonuclear particles might allow translation of stalled mRNAs. In this scenario, global transcription should be unaffected, and reduced expression of specific mRNAs might reflect a delay in germ cell development before the first meiotic prophase is blocked.
Supplementary Material
ACKNOWLEDGMENTS
We are most grateful to everybody who provided us with reagents. In particular, we thank Shinseog Kim, Noora Kotaja, and Duncan Odom for advice and helpful comments on the manuscript and Tanja Musch for technical assistance. We also thank the CRI Genomics and Bioinformatics Core Facilities. We gratefully acknowledge the support of the Cambridge Stem Cell Initiative and Stephen Evans-Freke.
This work was funded by the Deutsche Forschungsgemeinschaft (FOR1082), CR-UK, and the Medical Research Council. F.T. is supported by the Institute of Genetics and Biophysics A. Buzzati-Traverso, CNR, Italy.
Footnotes
Published ahead of print 11 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01523-12.
REFERENCES
- 1. de Rooij DG. 2001. Proliferation and differentiation of spermatogonial stem cells. Reproduction 121:347–354 [DOI] [PubMed] [Google Scholar]
- 2. Sharpe RM, McKinnell C, Kivlin C, Fisher JS. 2003. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction 125:769–784 [DOI] [PubMed] [Google Scholar]
- 3. Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. 2011. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 7:e1002403 doi:10.1371/journal.pgen.1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. 2012. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40:5023–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braun RE. 1998. Post-transcriptional control of gene expression during spermatogenesis. Semin. Cell Dev. Biol. 9:483–489 [DOI] [PubMed] [Google Scholar]
- 6. Paronetto MP, Messina V, Barchi M, Geremia R, Richard S, Sette C. 2011. Sam68 marks the transcriptionally active stages of spermatogenesis and modulates alternative splicing in male germ cells. Nucleic Acids Res. 39:4961–4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS. 2005. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 37:41–47 [DOI] [PubMed] [Google Scholar]
- 8. Geremia R, Boitani C, Conti M, Monesi V. 1977. RNA synthesis in spermatocytes and spermatids and preservation of meiotic RNA during spermiogenesis in the mouse. Cell Differ. 5:343–355 [DOI] [PubMed] [Google Scholar]
- 9. Monesi V, Geremia R, D'Agostino A, Boitani C. 1978. Biochemistry of male germ cell differentiation in mammals: RNA synthesis in meiotic and postmeiotic cells. Curr. Top. Dev. Biol. 12:11–36 [DOI] [PubMed] [Google Scholar]
- 10. Paronetto MP, Sette C. 2010. Role of RNA-binding proteins in mammalian spermatogenesis. Int. J. Androl. 33:2–12 [DOI] [PubMed] [Google Scholar]
- 11. Monesi V. 1964. Ribonucleic acid synthesis during mitosis and meiosis in the mouse testis. J. Cell Biol. 22:521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang J, Medvedev S, Reddi PP, Schultz RM, Hecht NB. 2005. The DNA/RNA-binding protein MSY2 marks specific transcripts for cytoplasmic storage in mouse male germ cells. Proc. Natl. Acad. Sci. U. S. A. 102:1513–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhong J, Peters AH, Lee K, Braun RE. 1999. A double-stranded RNA-binding protein required for activation of repressed messages in mammalian germ cells. Nat. Genet. 22:171–174 [DOI] [PubMed] [Google Scholar]
- 14. Kleene KC. 2001. A possible meiotic function of the peculiar patterns of gene expression in mammalian spermatogenic cells. Mech. Dev. 106:3–23 [DOI] [PubMed] [Google Scholar]
- 15. Venables JP, Cooke HJ. 2000. Lessons from knockout and transgenic mice for infertility in men. J. Endocrinol. Invest. 23:584–591 [DOI] [PubMed] [Google Scholar]
- 16. Venables JP, Eperon I. 1999. The roles of RNA-binding proteins in spermatogenesis and male infertility. Curr. Opin. Genet. Dev. 9:346–354 [DOI] [PubMed] [Google Scholar]
- 17. Kotaja N, Lin H, Parvinen M, Sassone-Corsi P. 2006. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J. Cell Sci. 119:2819–2825 [DOI] [PubMed] [Google Scholar]
- 18. Fawcett DW, Eddy EM, Phillips DM. 1970. Observations on the fine structure and relationships of the chromatoid body in mammalian spermatogenesis. Biol. Reprod. 2:129–153 [DOI] [PubMed] [Google Scholar]
- 19. Kotaja N, Sassone-Corsi P. 2007. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat. Rev. Mol. Cell Biol. 8:85–90 [DOI] [PubMed] [Google Scholar]
- 20. Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311:395–398 [DOI] [PubMed] [Google Scholar]
- 21. Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. 2012. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 19:900–905 [DOI] [PubMed] [Google Scholar]
- 22. Peters AH, Plug AW, van Vugt MJ, de Boer P. 1997. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5:66–68 [DOI] [PubMed] [Google Scholar]
- 23. Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental Cell 12:503–514 [DOI] [PubMed] [Google Scholar]
- 24. Nascimento EM, Cox CL, MacArthur S, Hussain S, Trotter M, Blanco S, Suraj M, Nichols J, Kubler B, Benitah SA, Hendrich B, Odom DT, Frye M. 2011. The opposing transcriptional functions of Sin3a and c-Myc are required to maintain tissue homeostasis. Nat. Cell Biol. 13:1395–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80 doi:10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cairns JM, Dunning MJ, Ritchie ME, Russell R, Lynch AG. 2008. BASH: a tool for managing BeadArray spatial artefacts. Bioinformatics 24:2921–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dunning MJ, Smith ML, Ritchie ME, Tavare S. 2007. Beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 23:2183–2184 [DOI] [PubMed] [Google Scholar]
- 28. Smyth G. 2005. Limma: linear models for microarray data, p 397–420. In Gentleman VCR, Dudoit S, Huber W, Irizarry R. (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY [Google Scholar]
- 29. Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marret C, Avallet O, Perrard-Sapori MH, Durand P. 1998. Localization and quantitative expression of mRNAs encoding the testis-specific histone TH2B, the phosphoprotein p19, the transition proteins 1 and 2 during pubertal development and throughout the spermatogenic cycle of the rat. Mol. Reprod. Dev. 51:22–35 [DOI] [PubMed] [Google Scholar]
- 31. Wykes SM, Nelson JE, Visscher DW, Djakiew D, Krawetz SA. 1995. Coordinate expression of the PRM1, PRM2, and TNP2 multigene locus in human testis. DNA Cell Biol. 14:155–161 [DOI] [PubMed] [Google Scholar]
- 32. Turner JM, Burgoyne PS, Singh PB. 2001. M31 and macroH2A1.2 colocalise at the pseudoautosomal region during mouse meiosis. J. Cell Sci. 114:3367–3375 [DOI] [PubMed] [Google Scholar]
- 33. Costa Y, Speed RM, Gautier P, Semple CA, Maratou K, Turner JM, Cooke HJ. 2006. Mouse MAELSTROM: the link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum. Mol. Genet. 15:2324–2334 [DOI] [PubMed] [Google Scholar]
- 34. Deng W, Lin H. 2002. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Developmental Cell 2:819–830 [DOI] [PubMed] [Google Scholar]
- 35. Vasileva A, Tiedau D, Firooznia A, Muller-Reichert T, Jessberger R. 2009. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr. Biol. 19:630–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480:264–267 [DOI] [PubMed] [Google Scholar]
- 37. Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316:744–747 [DOI] [PubMed] [Google Scholar]
- 38. De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D. 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480:259–263 [DOI] [PubMed] [Google Scholar]
- 39. Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22:908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. 2006. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc. Natl. Acad. Sci. U. S. A. 103:2647–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. 2000. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 14:841–853 [PMC free article] [PubMed] [Google Scholar]
- 42. Tsai-Morris CH, Sheng Y, Lee E, Lei KJ, Dufau ML. 2004. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is essential for spermatid development and completion of spermatogenesis. Proc. Natl. Acad. Sci. U. S. A. 101:6373–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR. 2007. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 21:261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. 2010. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24:1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131:839–849 [DOI] [PubMed] [Google Scholar]
- 46. Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, Zarkower D. 2010. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Developmental Cell 19:612–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Frye M, Watt FM. 2006. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 16:971–981 [DOI] [PubMed] [Google Scholar]
- 48. Hussain S, Benavente SB, Nascimento E, Dragoni I, Kurowski A, Gillich A, Humphreys P, Frye M. 2009. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J. Cell Biol. 186:27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, Vincent AK, Malli R, Ali G, Khan FS, Ishak GE, Doherty D, Weksberg R, Ayub M, Windpassinger C, Ibrahim S, Frye M, Ansar M, Vincent JB. 2012. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 90:856–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Colonna A, Kerr SJ. 1980. The nucleus as the site of tRNA methylation. J. Cell. Physiol. 103:29–33 [DOI] [PubMed] [Google Scholar]
- 51. Paniagua R, Nistal M, Amat P, Rodriguez MC. 1986. Ultrastructural observations on nucleoli and related structures during human spermatogenesis. Anat Embryol. 174:301–306 [DOI] [PubMed] [Google Scholar]
- 52. Myers M, Ebling FJ, Nwagwu M, Boulton R, Wadhwa K, Stewart J, Kerr JB. 2005. Atypical development of Sertoli cells and impairment of spermatogenesis in the hypogonadal (hpg) mouse. J. Anat. 207:797–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hernandez-Verdun D. 2011. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2:189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takeuchi IK, Takeuchi YK. 1990. Ethanol-phosphotungstic acid and bismuth staining of spermatid nucleoli in mouse spermiogenesis. J. Struct. Biol. 103:104–112 [DOI] [PubMed] [Google Scholar]
- 55. Hernandez-Verdun D, Roussel P, Gebrane-Younes J. 2002. Emerging concepts of nucleolar assembly. J. Cell Sci. 115:2265–2270 [DOI] [PubMed] [Google Scholar]
- 56. Andonov MD, Chaldakov GN. 1989. Morphological evidence for calcium storage in the chromatoid body of rat spermatids. Experientia 45:377–378 [DOI] [PubMed] [Google Scholar]
- 57. Comings DE, Okada TA. 1972. The chromatoid body in mouse spermatogenesis: evidence that it may be formed by the extrusion of nucleolar components. J. Ultrastruct. Res. 39:15–23 [DOI] [PubMed] [Google Scholar]
- 58. Gerbasi VR, Preall JB, Golden DE, Powell DW, Cummins TD, Sontheimer EJ. 2011. Blanks, a nuclear siRNA/dsRNA-binding complex component, is required for Drosophila spermiogenesis. Proc. Natl. Acad. Sci. U. S. A. 108:3204–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. 2006. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res. 34:6034–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Motorin Y, Grosjean H. 1999. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA 5:1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Agris PF. 2004. Decoding the genome: a modified view. Nucleic Acids Res. 32:223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grosjean B, Ha R. 2005. Modification and editing of RNA. Springer, Berlin, Germany [Google Scholar]
- 63. Motorin Y, Helm M. 2010. tRNA stabilization by modified nucleotides. Biochemistry 49:4934–4944 [DOI] [PubMed] [Google Scholar]
- 64. Motorin Y, Lyko F, Helm M. 2010. 5-Methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 38:1415–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.