Abstract
The international project MOSAR was conducted in five rehabilitation centers; patients were screened for rectal carriage of extended-spectrum β-lactamase (ESBL)-producing members of the Enterobacteriaceae. Among 229 Klebsiella pneumoniae isolates, four clonal groups (CG) or complexes (CC) prevailed: CG17 in France, CG101 in Italy, CG15 in Spain, and CC147 in Israel. ESBLs, mainly CTX-Ms, were produced by 226 isolates; three isolates expressed AmpC-like cephalosporinases. High genetic diversity of K. pneumoniae populations was observed, with specific characteristics at each center.
TEXT
Klebsiella pneumoniae is an important human pathogen, causing nosocomial infections (1). Its most common mechanism of resistance to oxyimino-cephalosporins is the production of extended-spectrum β-lactamases (ESBLs) (2, 3), followed by AmpC-like cephalosporinases (4) and some newly emerging carbapenemases, such as KPC (5, 6). Their frequency has been increasing due to clonal spread of β-lactamase-producing strains and transfer of plasmids with β-lactamase genes (6, 7). Lately, multilocus sequence typing (MLST) has opened new perspectives for K. pneumoniae clonality studies (8). Some MLST analyses were carried out in the context of virulence or infection types (9–11); some others targeted ESBL- or AmpC-producing strains (12–18). Recently these were focused mostly on carbapenemase producers (6, 19–23). A number of pandemic clones associated with various β-lactamases were described (6, 19–21, 24). The EU project MOSAR was a prospective study on the spread of resistance in hospital wards across Europe and Israel (www.mosar-sic.org). The aim of this work was to compare ESBL- or AmpC-producing K. pneumoniae strains colonizing patients in five rehabilitation units (RUs) in four countries.
Five RUs, located in the north of France (Berck Maritime Hôpital [BM]) and the areas of Rome, Italy (Fondazione Santa Lucia [FS]), Barcelona, Spain (Guttmann Institute [GI]), and Tel-Aviv, Israel (Loewenstein Hospital [LH] and Tel-Aviv Souraski Medical Center [TA]), participated in the study. From October 2008 until February 2011, rectal swabs were collected from all patients at admission, 2 weeks later, and then once monthly and at discharge. Swabs were plated on the Brilliance ESBL agar (Oxoid, Basingstoke, United Kingdom), and Enterobacteriaceae colonies, identified according to the manufacturer's instructions, were subjected to further analysis (1 colony per morphotype). Species identification was done using the Vitek 2 system (bioMérieux, Marcy l'Etoile, France). ESBLs and AmpCs were detected using the ESBL double-disk test (cefotaxime, ceftazidime, cefepime, and amoxicillin-clavulanate disks) in the absence and presence of 250 μg/ml cloxacillin (25). K. pneumoniae isolates with inhibition zones augmented upon exposure to cloxacillin were tested by PCR for acquired AmpCs (26).
Of the total of 958 K. pneumoniae isolates identified, 229 patient-unique isolates with ESBL or AmpC from 2008-2009 were randomly selected for the molecular study based on their numbers at each center (BM, n = 19; FS, n = 41; GI, n = 25; LH, n = 26; TA, n = 118). Differences in isolate numbers from particular sites resulted from differences in timing of the clinical trials and the overall numbers of K. pneumoniae isolates at each place. All of the isolates were typed first by pulsed-field gel electrophoresis (PFGE) as described previously (27), using the XbaI restriction enzyme (Fermentas, Vilnius, Lithuania). PFGE types and subtypes were discerned visually according to the method of Tenover et al. (28). In order to construct dendrograms, PFGE patterns were compared by using the BioNumerics Fingerprinting software program (version 6.01; Applied Maths, Sint-Martens-Latem, Belgium), as reported previously (29). Then, 100 isolates, including representatives of all PFGE types and of multiple subtypes for each larger PFGE type with subtypes, were typed by MLST (8). The database available at http://www.pasteur.fr/mlst was used for assigning sequence types (STs).
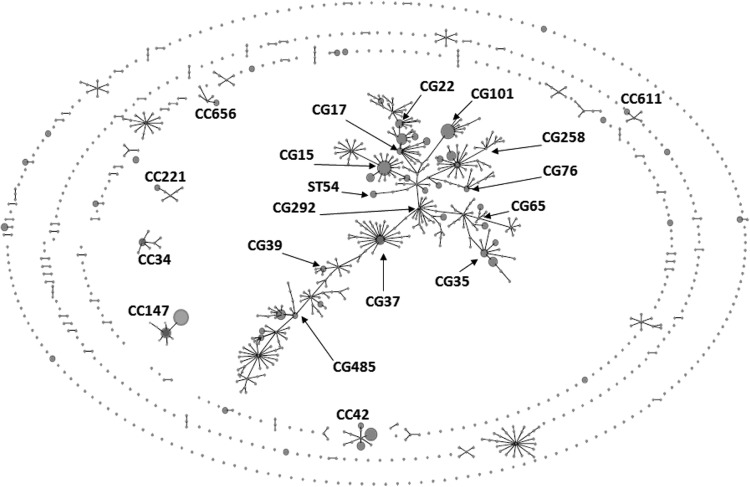
The analysis distinguished 56 STs shown in Tables 1 and 2. All of the STs were analyzed by using eBURST program (eburst.mlst.net) against the MLST database (http://www.pasteur.fr/mlst) with 1,001 STs (as of 2 August 2012) to reveal their relationships with all publicly available STs. As shown in Fig. 1, 30 of our STs, representing 138 isolates, were located within a huge central clonal complex (CC), made of 319 (31.9%) STs known to date. This CC, with a specific branched structure, has been growing proportionally to the total number of STs in the database; e.g., in September 2011, it comprised 139 STs out of 650 STs (21.4%) (18). The accuracy of the eBURST grouping is questionable if the proportion of STs in a single CC exceeds 25% of all STs (30). In addition, this large K. pneumoniae CC was recently described as polyphyletic due to DNA transfer and recombination between related and unrelated STs and thus not representing a true CC as a whole (9, 18). Breurec et al. proposed to subdivide it into clonal groups (CGs), each made of a central ST and its single-locus variants (SLVs), followed by their SLVs, which might reflect clonal evolution of the STs more reliably (18). Using this approach, we split the 30 STs into 12 CGs mapping within the large central “CC.” Six of these matched those discerned by Breurec et al. among their isolates, namely, CG15, CG17, CG39, CG65, CG101, and CG258 (18). Three others—CG35, CG37, and CG485—shared some STs with Breurec et al.'s CGs, CG13, CG394, and CG36, respectively, but had other central genotypes. Finally, three new CGs were defined (CG22, CG76, and CG292), and one ST of this set (ST54) was not assigned to any CG. The remaining 26 STs were not part of the central “CC” but mapped to the eBURST perimeter. Nine STs with 71 isolates (31.0%) were grouped into six CCs, CC34, CC42, CC147, CC221, CC611, and CC656, likely arising from clonal evolution. Seventeen other STs had only single SLVs in the database or were singletons.
Table 1.
K. pneumoniae “major” CGs or CCsa
| CG or CC | ST | No. of isolates (no. of PFGE types) for center |
ESBL or AmpC variant(s) (center[s]) | |||||
|---|---|---|---|---|---|---|---|---|
| BM | FS | GI | LH | TA | Total | |||
| CG15 | ST15 | 4 (3) | 1 (1) | 7 (1) | 2 (2) | 11 (5) | 25 | CTX-M-15 (BM, GI, LH, TA); SHV-5 (BM, FS); SHV-2 (BM) |
| ST277 | 1 (1) | 1 | CTX-M-40 | |||||
| ST326 | 5 (1) | 5 | CTX-M-15 | |||||
| ST709 | 2 (1) | 2 | CTX-M-15 | |||||
| Total (%) | 33 (14.4) | |||||||
| CG17 | ST16 | 1 (1) | 5 (1) | 4 (1) | 2 (1) | 12 | CTX-M-15 | |
| ST17 | 1 (1) | 1 (1) | 2 | CTX-M-15 (LH); SHV-12 (TA) | ||||
| ST20 | 4 (1) | 1 (1) | 5 | CTX-M-15 (BM); SHV-12 (TA) | ||||
| ST630 | 3 (1) | 3 | CTX-M-15 | |||||
| ST676 | 1 (1) | 1 | CTX-M-39 | |||||
| ST845 | 2 (1) | 2 | CTX-M-15 | |||||
| Total (%) | 25 (10.9) | |||||||
| CG35 | ST35 | 4 (1) | 4 | CTX-M-15 | ||||
| ST327 | 3 (1) | 4 (4) | 7 | CTX-M-2 | ||||
| ST835 | 1 (1) | 1 | CTX-M-15 | |||||
| Total (%) | 12 (5.2) | |||||||
| CG37 | ST37 | 7 (1) | 1 (1) | 8 | CTX-M-15 | |||
| Total (%) | 8 (3.5) | |||||||
| CC42 | ST383 | 2 (1) | 15 (2) | 17 | CTX-M-15 | |||
| ST413 | 5 (1) | 5 | CTX-M-15 | |||||
| ST910 | 1 (1) | 1 | CTX-M-14 | |||||
| Total (%) | 23 (10.0) | |||||||
| CG101 | ST101 | 24 (1) | 1 (1) | 25 | CTX-M-15 | |||
| Total (%) | 25 (10.9) | |||||||
| CC147 | ST147 | 1 (1) | 1 (1) | 14 (1) | 16 | CTX-M-15 (FS, TA); SHV-12 (BM) | ||
| ST392 | 7 (1) | 19 (1) | 26 | CTX-M-15 (LH, TA); SHV-12 (TA) | ||||
| Total (%) | 42 (18.3) | |||||||
| CG258 | ST11 | 2 (2) | 2 | CTX-M-1 | ||||
| ST340 | 1 (1) | 1 (1) | 2 | SHV-2a (GI); CMY-2 (LH) | ||||
| ST833 | 3 (1) | 5 (1) | 8 | CTX-M-15 (LH, TA); SHV-12 (TA) | ||||
| Total (%) | 12 (5.2) | |||||||
| CG485 | ST26 | 1 (1) | 1 | CTX-M-9 | ||||
| ST45 | 2 (2) | 1 (1) | 1 (1) | 5 (2) | 9 | CTX-M-15 (LH, TA); CTX-M-1 (FS); TEM-3 or CMY-2 (BM) | ||
| ST323 | 1 (1) | 1 | CTX-M-2 | |||||
| ST485 | 1 (1) | 1 | CTX-M-2 | |||||
| ST834 | 1 (1) | 1 | CTX-M-2 | |||||
| Total (%) | 13 (5.7) | |||||||
| Total, all (%) | 193 (84.3) | |||||||
STs, geographic and quantitative distribution, PFGE types, and ESBL or AmpC types. New STs identified in this study are in bold. MLST was performed for 100 isolates, representing all of the PFGE types in the entire collection; for the PFGE types that were split into subtypes, the analysis was carried out for several subtypes each.
Table 2.
K. pneumoniae clones of lower incidence in the studya
| STb | Clonal status | Center(s) | No. of isolates (no. of PFGE types) | ESBL or AmpC variant(s) (center) |
|---|---|---|---|---|
| ST1 | Member of CC656 | GI | 1 (1) | CTX-M-15 |
| ST34 | Member of CC34 | GI, TA | 3 (3) | SHV-2 (GI); CTX-M-15 or SHV-5 (TA) |
| ST39 | Member of CG39 | BM | 1 (1) | CTX-M-15 |
| ST54c | TA | 2 (1) | CTX-M-15 | |
| ST76 | Member of CG76 | TA | 1 (1) | SHV-5 |
| ST105 | Member of CG22 | TA | 1 (1) | CTX-M-2 |
| ST252 | Member of CC221 | GI | 1 (1) | CTX-M-9 |
| ST403 | Member of CG65 | TA | 1 (1) | CTX-M-39 |
| ST483 | Member of CC611 | GI | 1 (1) | CTX-M-3 |
| ST869 | Member of CG65 | TA | 2 (1) | CTX-M-14 |
| ST904 | Member of CG292 | TA | 2 (1) | SHV-2a |
| Total | 16 | |||
| ST6 | SLV of ST908 | TA | 1 (1) | SHV-5 |
| ST225 | SLV of ST633 | TA | 1 (1) | SHV-12 |
| ST378 | SLV of ST183 | FS | 1 (1) | CTX-M-15 |
| ST484 | SLV of ST849 | GI | 1 (1) | DHA-1 |
| ST836 | SLV of ST198 | GI | 3 (1) | CTX-M-15 |
| ST843 | SLV of ST440 | TA | 1 (1) | CTX-M-15 |
| Total | 8 | |||
| ST104 | Singleton | BM | 1 (1) | CTX-M-1 |
| ST244 | Singleton | TA | 1 (1) | SHV-5 |
| ST261 | Singleton | TA | 1 (1) | CTX-M-14 |
| ST278 | Singleton | TA | 2 (1) | SHV-27 |
| ST299 | Singleton | LH | 1 (1) | CTX-M-39 |
| ST307 | Singleton | GI | 1 (1) | CTX-M-15 |
| ST321 | Singleton | TA | 1 (1) | CTX-M-14 |
| ST348 | Singleton | BM | 1 (1) | TEM-3 |
| ST844 | Singleton | TA | 1 (1) | CTX-M-15 |
| ST846 | Singleton | TA | 1 (1) | SHV-5 |
| ST906 | Singleton | TA | 1 (1) | SHV-12 |
| Total | 12 |
Clonal status in the global MLST database, frequency, PFGE types, and ESBL or AmpC types. STs shown in this table have been split into three subgroups according to their position in the global MLST database (http://www.pasteur.fr/mlst; searched by eBURST on 2 August 2012): (i) members of the CGs or CCs that were incidentally identified in the study; (ii) STs which had only single SLVs; and (iii) singletons. In all these subgroups, the STs have been ordered according to their ascending number symbols.
New STs identified in this study are in bold. MLST was performed for 100 isolates representing all of the PFGE types in the entire collection.
ST54 is positioned by eBURST within the large heterogeneous complex; however, it cannot be unambiguously classified into any CG.
Fig 1.
Population structure of ESBL- or AmpC-producing K. pneumoniae isolates identified in the five rehabilitation centers (BM, FS, GI, LH, and TA), shown in the context of all of the 1,001 STs present in the global MLST database (http://www.pasteur.fr/mlst; as of2 August 2012). The scheme was constructed using eBURST analysis. STs are symbolized by dots; the STs identified in the study are represented by dots in circles. The size of a dot in circle corresponds to the number of isolates belonging to an ST. Single locus variants (SLVs) are linked by solid lines. Numbers indicate all of the CGs/CCs (and ST54) to which the STs identified in the study were classified. The STs which had only single SLVs in the database and singletons are not numbered.
As shown in Table 1, 11 STs were represented by at least five isolates from at least two centers each, grouping 158 (69.0%) isolates altogether. They belonged to nine “major” CGs or CCs, and were: ST15 (CG15), ST16 & ST20 (CG17), ST37 (CG37), ST45 (CG485), ST101 (CG101), ST147 & ST392 (CC147), ST327 (CG35), ST383 (CC42), and ST833 (CG258). Most of these 11 STs have been reported globally (http://www.pasteur.fr/mlst). On the other hand, 42 STs with 64 isolates (27.9%) occurred only in single hospitals (Tables 1 and 2), and these also included some globally spread clones, e.g., ST11 (CG258). Their contribution to local K. pneumoniae populations varied from 4.9% in FS to 64.0% in GI, defining the population in GI as the relatively most specific. New STs (n = 17) were identified in all of the centers.
The data shown in Table 1 revealed that four of the “major” CGs/CCs were more prevalent than others, especially in particular areas. CG15 was found in all of the clinical sites and was one of the most frequent groups (n = 33; 14.4%). Of four STs in this CG, ST15 was most prominent (n = 25) and was the only ST present in each hospital. ST15 together with ST326 caused the CG15 predominance among the rectal colonizers in GI in Barcelona (48.0%), in contrast to ESBL- or AmpC-producing K. pneumoniae clinical and carrier isolates from other Spanish centers (13, 31). CG15, usually ST15 and/or ST14 (not found here), has been commonly observed with ESBLs or other β-lactamases worldwide (12, 16, 18, 32–34). CC147 was identified in four hospitals and was the largest CG/CC overall (n = 42; 18.3%). It consisted of ST147 and ST392, found mostly in the Israeli sites, where CC147 was the main clonal structure (26.9% in LH and 28.0% in TA). Recently ST147 has been associated with carbapenemases in many countries (24, 34–37) but not in Israel so far (19). CG17, comprising six STs, occurred in four centers and dominated in BM, France (42.1%), due to ST16, ST20, and ST630. Previously, CG17 was reported as the key clonal group among ESBL producers in Canada (17) and was notable in other studies worldwide (13, 16, 31). CG101 was represented only by ST101 in two sites, highly prevailing in FS in Rome (58.5%). ST101 has been observed with various β-lactamases worldwide (14–16, 33, 38, 39); e.g., it was the main carbapenem-nonsusceptible strain in another Italian hospital recently (40).
A good correlation between PFGE and MLST results, with higher resolution of PFGE, was reported for K. pneumoniae previously (8, 41). Consistently, as shown in Tables 1 and 2, several STs in this study were split into multiple PFGE types each, while for larger PFGE types, isolates of different subtypes usually had identical MLST allelic profiles. An interesting exception was the Israeli CC147 isolates, split by MLST into ST147 and ST392 (SLVs differing by tonB alleles) but clustered by PFGE into 1 type with 23 subtypes. The detailed analysis (sequencing tonB) revealed a good correlation between the two STs and all PFGE subtypes. These results showed that in some K. pneumoniae CGs/CCs, isolates of two related STs may be clustered into a single PFGE type, as also reported by others (17, 42).
β-Lactamase profiling was done by isoelectric focusing as reported elsewhere (43); identification of the blaCTX-M-, blaSHV-, blaTEM-, blaCMY-2-, and blaDHA-like genes was done by PCRs (44–47). Sequencing of the genes was performed as reported previously (44). As shown in Table 3, 226 isolates (98.7%) produced ESBLs and 3 had AmpCs (1.3%). The ESBLs belonged to the CTX-M (n = 202; 88.2%), SHV (n = 22; 9.6%), and TEM (n = 2) families, while the AmpCs were CMY-2 (n = 2) and DHA-1 (n = 1). The prevalence of CTX-Ms ranged from 63.2% in BM to 97.5% in FS, and they represented all of the five subgroups known: CTX-M-1, -2, -8, -9, and -25. The CTX-M-1 group was predominant (n = 180; 78.6%), mostly due to CTX-M-15, produced by 175 isolates (76.4%; varying from 52.6% in BM to 92.7% in FS). SHVs, mainly SHV-5 and -12, were identified widely, while TEM-3 occurred only in BM. As shown in Tables 1 to 3, most of the β-lactamases were produced by at least two clones, also locally. CTX-M-15 was expressed by 27 clones overall. Similarly, isolates of most of the STs of wider prevalence and/or spread (ST15, ST17, ST20, ST45, ST147, ST340, ST392, and ST833) produced alternatively 2 to 4 different enzymes. The data were consistent with the recent CTX-M predominance, and CTX-M-15 in particular (3, 48), often assigned to the spread of blaCTX-M-15-carrying plasmids (7) and specific Escherichia coli clones (6). However, blaCTX-M-15 was more prevalent in K. pneumoniae than in E. coli in the same RUs at the same time (76.4% versus 40.6% of isolates, respectively) (29). The lack of strict correlation between β-lactamases and the clones showed that the clones have been spreading largely as such, locally acquiring β-lactamase genes. This has often been observed for E. coli (6) but much less for K. pneumoniae (14, 15, 17, 18).
Table 3.
β-Lactamase variants in the study isolates; geographic and quantitative distribution
| β-Lactamase | CTX-M group | Center(s) | No. (%) of isolates | No. of clones |
|---|---|---|---|---|
| CTX-M-15 | 1 | BM, FS, GI, LH, TA | 175 (76.4) | 27 |
| CTX-M-2 | 2 | FS, LH, TA | 11 (4.8) | 5 |
| CTX-M-14a | 9 | TA | 5 (2.1) | 4 |
| CTX-M-1 | 1 | BM, FS, GI | 4 (1.7) | 3 |
| CTX-M-39 | 25 | LH, TA | 3 (1.3) | 3 |
| CTX-M-9 | 9 | GI | 2 (0.9) | 2 |
| CTX-M-3 | 1 | GI | 1 (0.4) | 1 |
| CTX-M-40 | 8 | BM | 1 (0.4) | 1 |
| SHV-5 | BM, FS, TA | 7 (3.1) | 6 | |
| SHV-12 | BM, TA | 7 (3.1) | 7 | |
| SHV-2 | BM, GI | 3 (1.3) | 2 | |
| SHV-2a | GI, TA | 3 (1.3) | 2 | |
| SHV-27 | TA | 2 (0.9) | 1 | |
| TEM-3 | BM | 2 (0.9) | 2 | |
| CMY-2 | BM, LH | 2 (0.9) | 2 | |
| DHA-1 | GI | 1 (0.4) | 1 | |
| Total | 229 |
CTX-M-14 was encoded by the blaCTX-M-14a (n = 4) or blaCTX-M-14b (n = 1) gene.
This study has been one of the hitherto few extensive MLST analyses of ESBL-producing K. pneumoniae (12–18), and like even fewer such works (13), it was performed on rectal colonizers and not clinical isolates, which might explain some differences observed. Although several clonal groups and the CTX-M-15 enzyme dominated in the entire collection, a high genetic diversity was observed, combined with unique features at each center.
ACKNOWLEDGMENTS
We thank Karol Diestra for her excellent support.
This work was part of the activities of the MOSAR integrated project (LSHP-CT-2007-037941), supported by the European Commission under the Life Science Health priority of the 6th Framework Program (WP2 and WP5 study teams). A.B., R. I., J.F., W.H., and M.G. were also financed by complementary grant no. 934/6.PR UE/2009/7 from the Polish Ministry of Science and Higher Education.
Apart from the authors of this work, the MOSAR WP2 and WP5 study groups included A. Grabowska, M. Herda, and E. Nikonorow, National Medicines Institute, Warsaw, Poland; M. J. Schwaber, E. Bilavsky, M. Elenbogen, A. Klein, S. Navon-Venezia, M. Shklyar, L. Keren, R. Glick, S. Klarfeld-Lidji, E. Mordechai, S. Cohen, R. Fachima, Y. Zdonevsky, and B. Knubovets, Division of Epidemiology and Preventive Medicine, Tel-Aviv Sourasky Medical Center, Tel Aviv, Israel; J. Lasley, I. Bertucci, M.-L. Delaby, C. Colmant, and C. Sacleux, Hôpital Maritime de Berck, Berck, France; R. Formisano, M. P. Balice, and E. Guaglianone, Fondazione Santa Lucia IRCCS, Rome, Italy; S. Camps, Institute Guttmann, Barcelona, Spain; J. Hart, E. Isakov, A. Friedman, A. Rachman, G. Franco, and I. Or, Loewenstein Hospital, Ra'anana, Israel; and A. Rabinovich, Geriatric Division, Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel.
Footnotes
Published ahead of print 12 February 2013
REFERENCES
- 1. Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11:589–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bradford PA. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F, Poirel L, Woodford N. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165–174 [DOI] [PubMed] [Google Scholar]
- 4. Jacoby GA. 2009. AmpC beta-lactamases. Clin. Microbiol. Rev. 22:161–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35:736–755 [DOI] [PubMed] [Google Scholar]
- 7. Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982 doi:10.1371/journal.pone.0004982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jung SW, Chae HJ, Park YJ, Yu JK, Kim SY, Lee HK, Lee JH, Kahng JM, Lee SO, Lee MK, Lim JH, Lee CH, Chang SJ, Ahn JY, Lee JW, Park YG. 2012. Microbiological and clinical characteristics of bacteraemia caused by the hypermucoviscosity phenotype of Klebsiella pneumoniae in Korea. Epidemiol. Infect. 14:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chung DR, Lee H, Park MH, Jung SI, Chang HH, Kim YS, Son JS, Moon C, Kwon KT, Ryu SY, Shin SY, Ko KS, Kang CI, Peck KR, Song JH. 2012. Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 31:481–486 [DOI] [PubMed] [Google Scholar]
- 12. Damjanova I, Toth A, Paszti J, Hajbel-Vekony G, Jakab M, Berta J, Milch H, Fuzi M. 2008. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type beta-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005—the new ‘MRSAs’? J. Antimicrob. Chemother. 62:978–985 [DOI] [PubMed] [Google Scholar]
- 13. Oteo J, Cuevas O, Lopez-Rodriguez I, Banderas-Florido A, Vindel A, Perez-Vazquez M, Bautista V, Arroyo M, Garcia-Caballero J, Marin-Casanova P, Gonzalez-Sanz R, Fuentes-Gomez V, Ona-Compan S, Garcia-Cobos S, Campos J. 2009. Emergence of CTX-M-15-producing Klebsiella pneumoniae of multilocus sequence types 1, 11, 14, 17, 20, 35 and 36 as pathogens and colonizers in newborns and adults. J. Antimicrob. Chemother. 64:524–528 [DOI] [PubMed] [Google Scholar]
- 14. Elhani D, Bakir L, Aouni M, Passet V, Arlet G, Brisse S, Weill FX. 2010. Molecular epidemiology of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae strains in a university hospital in Tunis, Tunisia, 1999–2005. Clin. Microbiol. Infect. 16:157–164 [DOI] [PubMed] [Google Scholar]
- 15. Ko KS, Lee JY, Baek JY, Suh JY, Lee MY, Choi JY, Yeom JS, Kim YS, Jung SI, Shin SY, Heo ST, Kwon KT, Son JS, Kim SW, Chang HH, Ki HK, Chung DR, Peck KR, Song JH. 2010. Predominance of an ST11 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J. Med. Microbiol. 59:822–828 [DOI] [PubMed] [Google Scholar]
- 16. Lee MY, Ko KS, Kang CI, Chung DR, Peck KR, Song JH. 2011. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int. J. Antimicrob. Agents. 38:160–163 [DOI] [PubMed] [Google Scholar]
- 17. Peirano G, Sang JH, Pitondo-Silva A, Laupland KB, Pitout JD. 2012. Molecular epidemiology of extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae over a 10 year period in Calgary, Canada. J. Antimicrob. Chemother. 67:1114–1120 [DOI] [PubMed] [Google Scholar]
- 18. Breurec S, Guessennd N, Timinouni M, Le TA, Cao V, Ngandjio A, Randrianirina F, Thiberge JM, Kinana A, Dufougeray A, Perrier-Gros-Claude JD, Boisier P, Garin B, Brisse S. 15 February 2012. Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin. Microbiol. Infect. [Epub ahead of print.] doi:10.1111/j.1469-0691.2012.03805.x [DOI] [PubMed] [Google Scholar]
- 19. Leavitt A, Carmeli Y, Chmelnitsky I, Goren MG, Ofek I, Navon-Venezia S. 2010. Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob. Agents Chemother. 54:3002–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baraniak A, Grabowska A, Izdebski R, Fiett J, Herda M, Bojarska K, Zabicka D, Kania-Pudlo M, Mlynarczyk G, Zak-Pulawska Z, Hryniewicz W, Gniadkowski M. 2011. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008-2009. Antimicrob. Agents Chemother. 55:5493–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. 2011. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66:307–312 [DOI] [PubMed] [Google Scholar]
- 23. Potron A, Kalpoe J, Poirel L, Nordmann P. 2011. European dissemination of a single OXA-48-producing Klebsiella pneumoniae clone. Clin. Microbiol. Infect. 17:E24–E26 [DOI] [PubMed] [Google Scholar]
- 24. Samuelsen O, Toleman MA, Hasseltvedt V, Fuursted K, Leegaard TM, Walsh TR, Sundsfjord A, Giske CG. 2011. Molecular characterization of VIM-producing Klebsiella pneumoniae from Scandinavia reveals genetic relatedness with international clonal complexes encoding transferable multidrug resistance. Clin. Microbiol. Infect. 17:1811–1816 [DOI] [PubMed] [Google Scholar]
- 25. Drieux L, Brossier F, Sougakoff W, Jarlier V. 2008. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: review and bench guide. Clin. Microbiol. Infect. 14(Suppl 1):90–103 [DOI] [PubMed] [Google Scholar]
- 26. Perez-Perez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43:4328–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Izdebski R, Baraniak A, Fiett J, Adler A, Kazma M, Salomon J, Lawrence C, Rossini A, Salvia A, Vidal Samso J, Fierro J, Paul M, Lerman Y, Malhotra-Kumar S, Lammens C, Goossens H, Hryniewicz W, Brun-Buisson C, Carmeli Y, Gniadkowski M. 2013. Clonal structure, extended-spectrum β-lactamases and acquired AmpC-type cephalosporinases of Escherichia coli populations colonizing patients in rehabilitation centers in four countries. Antimicrob. Agents Chemother. 57:309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Turner KM, Hanage WP, Fraser C, Connor TR, Spratt BG. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 7:30 doi:10.1186/1471-2180-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Diestra K, Miro E, Marti C, Navarro D, Cuquet J, Coll P, Navarro F. 2011. Multiclonal epidemic of Klebsiella pneumoniae isolates producing DHA-1 in a Spanish hospital. Clin. Microbiol. Infect. 17:1032–1036 [DOI] [PubMed] [Google Scholar]
- 32. Nielsen JB, Skov MN, Jorgensen RL, Heltberg O, Hansen DS, Schonning K. 2011. Identification of CTX-M15-, SHV-28-producing Klebsiella pneumoniae ST15 as an epidemic clone in the Copenhagen area using a semi-automated Rep.-PCR typing assay. Eur. J. Clin. Microbiol. Infect. Dis. 30:773–778 [DOI] [PubMed] [Google Scholar]
- 33. Hrabak J, Empel J, Bergerova T, Fajfrlik K, Urbaskova P, Kern-Zdanowicz I, Hryniewicz W, Gniadkowski M. 2009. International clones of Klebsiella pneumoniae and Escherichia coli with extended-spectrum beta-lactamases in a Czech hospital. J. Clin. Microbiol. 47:3353–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giske CG, Froding I, Hasan CM, Turlej-Rogacka A, Toleman M, Livermore D, Woodford N, Walsh TR. 2012. Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. Antimicrob. Agents Chemother. 56:2735–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Papagiannitsis CC, Kotsakis SD, Petinaki E, Vatopoulos AC, Tzelepi E, Miriagou V, Tzouvelekis LS. 2011. Characterization of metallo-beta-lactamase VIM-27, an A57S mutant of VIM-1 associated with Klebsiella pneumoniae ST147. Antimicrob. Agents Chemother. 55:3570–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Giakkoupi P, Papagiannitsis CC, Miriagou V, Pappa O, Polemis M, Tryfinopoulou K, Tzouvelekis LS, Vatopoulos AC. 2011. An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009-10). J. Antimicrob. Chemother. 66:1510–1513 [DOI] [PubMed] [Google Scholar]
- 37. Peirano G, Pillai DR, Pitondo-Silva A, Richardson D, Pitout JD. 2011. The characteristics of NDM-producing Klebsiella pneumoniae from Canada. Diagn. Microbiol. Infect. Dis. 71:106–109 [DOI] [PubMed] [Google Scholar]
- 38. Seki LM, Pereira PS, de Souza MDPAH, Conceicao MDS, Marques EA, Porto CO, Colnago EM, Alves CDFM, Gomes D, Assef AP, Samuelsen O, Asensi MD. 2011. Molecular epidemiology of KPC-2-producing Klebsiella pneumoniae isolates in Brazil: the predominance of sequence type 437. Diagn. Microbiol. Infect. Dis. 70:274–277 [DOI] [PubMed] [Google Scholar]
- 39. Pitart C, Sole M, Roca I, Fabrega A, Vila J, Marco F. 2011. First outbreak of a plasmid-mediated carbapenem-hydrolyzing OXA-48 beta-lactamase in Klebsiella pneumoniae in Spain. Antimicrob. Agents Chemother. 55:4398–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mammina C, Bonura C, Aleo A, Fasciana T, Brunelli T, Pesavento G, Degl'Innocenti R, Nastasi A. 2012. Sequence type 101 (ST101) as the predominant carbapenem-non-susceptible Klebsiella pneumoniae clone in an acute general hospital in Italy. Int. J. Antimicrob. Agents 39:543–545 [DOI] [PubMed] [Google Scholar]
- 41. Vimont S, Mnif B, Fevre C, Brisse S. 2008. Comparison of PFGE and multilocus sequence typing for analysis of Klebsiella pneumoniae isolates. J. Med. Microbiol. 57:1308–1310 [DOI] [PubMed] [Google Scholar]
- 42. Andrade LN, Curiao T, Ferreira JC, Longo JM, Climaco EC, Martinez R, Bellissimo-Rodrigues F, Basile-Filho A, Evaristo MA, Del Peloso PF, Ribeiro VB, Barth AL, Paula MC, Baquero F, Canton R, Darini AL, Coque TM. 2011. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob. Agents Chemother. 55:3579–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bauernfeind A, Grimm H, Schweighart S. 1990. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection. 18:294–298 [DOI] [PubMed] [Google Scholar]
- 44. Empel J, Baraniak A, Literacka E, Mrowka A, Fiett J, Sadowy E, Hryniewicz W, Gniadkowski M. 2008. Molecular survey of beta-lactamases conferring resistance to newer beta-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob. Agents Chemother. 52:2449–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Empel J, Hrabak J, Kozinska A, Bergerova T, Urbaskova P, Kern-Zdanowicz I, Gniadkowski M. 2010. DHA-1-producing Klebsiella pneumoniae in a teaching hospital in the Czech Republic. Microb. Drug Resist. 16:291–295 [DOI] [PubMed] [Google Scholar]
- 46. Literacka E, Empel J, Baraniak A, Sadowy E, Hryniewicz W, Gniadkowski M. 2004. Four variants of the Citrobacter freundii AmpC-type cephalosporinases, including novel enzymes CMY-14 and CMY-15, in a Proteus mirabilis clone widespread in Poland. Antimicrob. Agents Chemother. 48:4136–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Woodford N, Fagan EJ, Ellington MJ. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J. Antimicrob. Chemother. 57:154–155 [DOI] [PubMed] [Google Scholar]
- 48. Rossolini GM, D'Andrea MM, Mugnaioli C. 2008. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14(Suppl 1):33–41 [DOI] [PubMed] [Google Scholar]