Abstract
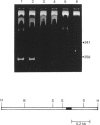
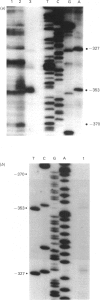
The pyrimidine/purine-biased region located upstream of the EGF (epidermal growth factor) receptor gene transcription initiation sites was sensitive to S1 nuclease when under superhelical tension. The structural basis of this specific reactivity to S1 nuclease was probed by the use of diethyl pyrocarbonate. The patterns of modification suggested that the H-form proposed by Mirkin, Lyamichev, Drushlyak, Dobrynin, Filippov & Frank-Kamenetskii [Nature (London) (1987) 330, 495-497], which includes an intramolecular triplex and a single-stranded region, was the most plausible model for the sequence tested. The results of dimethyl sulphate modification also supported this model.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C. Heteronomous DNA. Nucleic Acids Res. 1983 Jun 25;11(12):4141–4155. doi: 10.1093/nar/11.12.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder G. D., Latimer L. J., Lee J. S. Immunofluorescent staining of mammalian nuclei and chromosomes with a monoclonal antibody to triplex DNA. Chromosoma. 1988 Nov;97(3):185–192. doi: 10.1007/BF00292959. [DOI] [PubMed] [Google Scholar]
- Cantor C. R., Efstratiadis A. Possible structures of homopurine-homopyrimidine S1-hypersensitive sites. Nucleic Acids Res. 1984 Nov 12;12(21):8059–8072. doi: 10.1093/nar/12.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T., Efstratiadis A. Sequence-dependent S1 nuclease hypersensitivity of a heteronomous DNA duplex. J Biol Chem. 1986 Nov 5;261(31):14771–14780. [PubMed] [Google Scholar]
- Futcher B. Supercoiling and transcription, or vice versa? Trends Genet. 1988 Oct;4(10):271–272. doi: 10.1016/0168-9525(88)90166-7. [DOI] [PubMed] [Google Scholar]
- Gralla J. D. Rapid "footprinting" on supercoiled DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3078–3081. doi: 10.1073/pnas.82.10.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Hidaka S., Sakaki Y. Sequence analysis of a KpnI family member near the 3' end of human beta-globin gene. Nucleic Acids Res. 1985 Nov 11;13(21):7813–7827. doi: 10.1093/nar/13.21.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgpeth J., Goodman H. M., Boyer H. W. DNA nucleotide sequence restricted by the RI endonuclease. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3448–3452. doi: 10.1073/pnas.69.11.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr W. Diethyl pyrocarbonate: a chemical probe for secondary structure in negatively supercoiled DNA. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8009–8013. doi: 10.1073/pnas.82.23.8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Single strands, triple strands, and kinks in H-DNA. Science. 1988 Sep 30;241(4874):1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Topology and formation of triple-stranded H-DNA. Science. 1989 Mar 24;243(4898):1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- Htun H., Lund E., Dahlberg J. E. Human U1 RNA genes contain an unusually sensitive nuclease S1 cleavage site within the conserved 3' flanking region. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7288–7292. doi: 10.1073/pnas.81.23.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H., Lund E., Westin G., Pettersson U., Dahlberg J. E. Nuclease S1-sensitive sites in multigene families: human U2 small nuclear RNA genes. EMBO J. 1985 Jul;4(7):1839–1845. doi: 10.1002/j.1460-2075.1985.tb03858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S., Xu Y. H., Stratton R. H., Roe B. A., Merlino G. T., Pastan I. Characterization and sequence of the promoter region of the human epidermal growth factor receptor gene. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4920–4924. doi: 10.1073/pnas.82.15.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. C., Ishii S., Jinno Y., Pastan I., Merlino G. T. Epidermal growth factor receptor gene promoter. Deletion analysis and identification of nuclear protein binding sites. J Biol Chem. 1988 Apr 25;263(12):5693–5699. [PubMed] [Google Scholar]
- Johnson A. C., Jinno Y., Merlino G. T. Modulation of epidermal growth factor receptor proto-oncogene transcription by a promoter site sensitive to S1 nuclease. Mol Cell Biol. 1988 Oct;8(10):4174–4184. doi: 10.1128/mcb.8.10.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R., Merlino G. T., Pastan I. A transcription factor active on the epidermal growth factor receptor gene. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5016–5020. doi: 10.1073/pnas.85.14.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R., Merlino G. T., Pastan I. Epidermal growth factor (EGF) receptor gene transcription. Requirement for Sp1 and an EGF receptor-specific factor. J Biol Chem. 1988 May 5;263(13):6329–6336. [PubMed] [Google Scholar]
- Kawasaki K., Kudoh J., Omoto K., Shimizu N. Mega base map of the epidermal growth factor (EGF) receptor gene flanking regions and structure of the amplification units in EGF receptor-hyperproducing squamous carcinoma cells. Jpn J Cancer Res. 1988 Nov;79(11):1174–1183. doi: 10.1111/j.1349-7006.1988.tb01542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Gelinas R., Weintraub H. Detection of an altered DNA conformation at specific sites in chromatin and supercoiled DNA. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4389–4393. doi: 10.1073/pnas.80.14.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAWLEY P. D., BROOKES P. FURTHER STUDIES ON THE ALKYLATION OF NUCLEIC ACIDS AND THEIR CONSTITUENT NUCLEOTIDES. Biochem J. 1963 Oct;89:127–138. doi: 10.1042/bj0890127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Honda B. M., Mills A. D., Finch J. T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978 Oct 5;275(5679):416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon C., Schmidt A., de Crombrugghe B. A sequence conserved in both the chicken and mouse alpha 2(I) collagen promoter contains sites sensitive to S1 nuclease. J Biol Chem. 1984 May 25;259(10):6636–6640. [PubMed] [Google Scholar]
- Mirkin S. M., Lyamichev V. I., Drushlyak K. N., Dobrynin V. N., Filippov S. A., Frank-Kamenetskii M. D. DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature. 1987 Dec 3;330(6147):495–497. doi: 10.1038/330495a0. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Purvis I. J., Moseley B. E. Isolation and characterisation of DraI, a type II restriction endonuclease recognising a sequence containing only A:T basepairs, and inhibition of its activity by uv irradiation of substrate DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5467–5474. doi: 10.1093/nar/11.16.5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu H. L., Michot B., Bachellerie J. P. Improved methods for structure probing in large RNAs: a rapid 'heterologous' sequencing approach is coupled to the direct mapping of nuclease accessible sites. Application to the 5' terminal domain of eukaryotic 28S rRNA. Nucleic Acids Res. 1983 Sep 10;11(17):5903–5920. doi: 10.1093/nar/11.17.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Skuzeski J. M., Lund E., Murphy J. T., Steinberg T. H., Burgess R. R., Dahlberg J. E. Synthesis of human U1 RNA. II. Identification of two regions of the promoter essential for transcription initiation at position +1. J Biol Chem. 1984 Jul 10;259(13):8345–8352. [PubMed] [Google Scholar]
- Tsao Y. P., Wu H. Y., Liu L. F. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989 Jan 13;56(1):111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- Voloshin O. N., Mirkin S. M., Lyamichev V. I., Belotserkovskii B. P., Frank-Kamenetskii M. D. Chemical probing of homopurine-homopyrimidine mirror repeats in supercoiled DNA. Nature. 1988 Jun 2;333(6172):475–476. doi: 10.1038/333475a0. [DOI] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Wells R. D. Unusual DNA structures. J Biol Chem. 1988 Jan 25;263(3):1095–1098. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]