Abstract
PURPOSE
The ETS2 transcription factor is an evolutionarily conserved gene that is deregulated in cancer. We analyzed the transcriptome of lung adenocarcinomas and normal lung tissue by expression profiling and found that ETS2 was significantly down-regulated in adenocarcinomas. In this study, we probed the yet unknown functional role of ETS2 in lung cancer pathogenesis.
EXPERIMENTAL DESIGN
Lung adenocarcinomas (n=80) and normal lung tissues (n=30) were profiled using the Affymetrix Human Gene 1.0 ST platform. Immunohistochemical (IHC) analysis was performed to determine ETS2 protein expression in NSCLC histological tissue specimens (n=201). Patient clinical outcome, based on ETS2 IHC expression, was statistically assessed using the log-rank and Kaplan-Meier tests. RNA interference and over-expression strategies were employed to assess effects of ETS2 expression on the transcriptome and on various malignant phenotypes.
RESULTS
ETS2 expression was significantly reduced in lung adenocarcinomas compared to normal lung (p<0.001). Low ETS2 IHC expression was a significant predictor of shorter time to recurrence in NSCLC (p=0.009, HR=1.89) and adenocarcinoma (p=0.03, HR=1.86). Moreover, ETS2 was found to significantly inhibit lung cancer cell growth, migration and invasion (p<0.05), and microarray and pathways analysis revealed significant (p<0.001) activation of the HGF pathway following ETS2 knockdown. In addition, ETS2 was found to suppress MET phosphorylation and knockdown of MET expression significantly attenuated (p<0.05) cell invasion mediated by ETS2-specific siRNA. Furthermore, knockdown of ETS2 augmented HGF-induced MET phosphorylation, cell migration and invasion.
CONCLUSION(S)
Our findings point to a tumor suppressor role for ETS2 in human NSCLC pathogenesis through inhibition of the MET proto-oncogene.
Keywords: NSCLC, ETS2, tumor suppressor, MET, HGF
Introduction
Lung cancer remains the leading cause of cancer-related deaths in the United States and worldwide (1). Non-small cell lung cancer (NSCLC) represents the majority of diagnosed lung cancer cases and is associated with a relatively poor 15% overall five-year survival rate (2). Understanding the molecular profiles of NSCLC as well as elucidating the roles oncogenes and tumor suppressors elicit in the development of this malignancy is expected to identify aberrant signaling pathways and molecular targets for therapy.
V-ets erythroblastosis virus E26 oncogene homolog 2 (ETS2) belongs to the ETS family of transcription factors and controls gene expression by binding to numerous genes with GGA(A/T) ETS response elements (EREs) thus impacting a broad spectrum of cellular functions including proliferation, differentiation, migration, transformation and apoptosis (3-5). During embryonic development, ETS2 was shown to insure proper development of the trophoblast (6) and to regulate endothelial cell survival during embryonic angiogenesis (7). Moreover, ETS2 was reported to be activated in response to extra-cellular mitogenic signaling mediated by the Ras oncogene (8) in murine fibroblasts, to maintain telomerase gene expression in breast cancer cells (9) and, notably, to exhibit both tumor promoting and suppressive effects in different types of carcinomas (10-15). However, ETS2 expression and function in human lung cancer is still unknown.
Our current lung adenocarcinoma expression profiling efforts revealed marked down-regulation of ETS2 transcript expression in lung tumors compared to paired normal lung tissues prompting us to examine ETS2 function in lung cancer pathogenesis. In this study, we found that low ETS2 immunohistochemical protein expression was significantly associated with shorter time to recurrence in NSCLC. Moreover, we found that ETS2 negatively regulates cellular growth, migration and invasion, MET oncogene phosphorylation and activation as well as HGF-mediated signaling. Our findings reveal for the first time a potential tumor suppressive role for ETS2 in lung cancer pathogenesis that is, in part, mediated by inhibition of MET oncogenic signaling.
Methods
NSCLC frozen tissue specimens and tissue microarrays
All human tissues were obtained from the MD Anderson Cancer Center Lung Cancer Specialized Program of Research Excellence (SPORE) tissue bank (Houston, TX) and had been classified using the 2004 World Health Organization (WHO) classification system as described before (16). All specimens were obtained from patients, who underwent surgery at the same institution from 2003 to 2005, under a protocol that was approved by the MD Anderson Cancer Center institutional review board. Detailed clinical and pathologic information was available for most of these cases and included patients’ demographic data, smoking history (never smokers or ever smokers, patients who had smoked at least 100 cigarettes in their lifetime), and pathologic tumor-node-metastasis (TNM) staging.
A collection of 80 lung adenocarcinomas and 30 non-tumoral paired tissues were snap-frozen and preserved in liquid nitrogen for total RNA extraction and microarray profiling. For each tissue sample, the percentage of malignant tissue was calculated and the cellular composition of specimens was determined by histological examination (J.F.) following Hematoxylin-Eosin (H&E) staining. All malignant samples retained contained more than 40% tumor cells.
For NSCLC tissue microarray analysis, we obtained archived FFPE samples from surgically resected lung cancer specimens from the lung cancer tissue bank. The tissue microarray analyzed in this study was composed of 201 NSCLC tumor specimens (135 lung adenocarcinomas and 66 SCCs) (Supplementary Table 1). After histological examination of NSCLC specimens, the NSCLC TMAs were constructed by obtaining three 1 mm in diameter cores from each tumor at 3 different sites (periphery, intermediate, and central tumor sites). The TMAs were prepared with a manual tissue arrayer (Advanced Tissue Arrayer ATA100, Chemicon International) as described previously (16).
Cell culture
Lung cancer cell lines were either originally purchased from the ATCC (H441, H2291, H3255, H1299, H1693, H522, H1792, H23 and H2009) or were obtained from Dr. Adi Gazdar (University of Texas Southwestern, Dallas, TX, USA) (DFC1024, HCC4006, H2228, H1650 and H1944) and were grown in DMEM-F12 low glucose medium supplemented with 10% fetal bovine serum (FBS) and maintained in humidified 5% CO2 incubator. Information on the smoking status of patients from whom the cell lines were isolated was obtained from the ATCC or from Dr. Adi Gazdar. All cell lines used in the study were authenticated by short tandem repeat (STR) DNA fingerprinting using the PowerPlex 16 HS System (Promega, Madison, WI). For MET inhibition studies, lung cancer cells were treated with the MET tyrosine kinase inhibitor, PHA-665752 (SelleckBio, Houston, TX) at a 100 nM final concentration. For treatment with hepatocyte growth factor (HGF) and activation of MET, cells were washed twice with phosphate-buffered saline (PBS), serum-deprived overnight and then treated with HGF (EMD Millipore, Darmstadt, Germany), at a final concentration of 50 ng/ml in serum-free cell culture medium.
Total RNA isolation
Total RNA was isolated from cells using the RNeasy kit from Qiagen according to the manufacturer's instructions. Lung adenocarcinoma and normal lung tissue samples were homogenized using Omni plastic disposable probes and an Omni (TH-115) homogenizer (Kennesaw, GA) for 1 min on dry ice after which total RNA was isolated using Trizol reagent according to the manufacturer's instructions. Total RNA was quantified using the Nanodrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA). RNA quality was assessed based on RNA integrity numbers (RINs) generated by the Agilent Bioanalyzer 2000 (Agilent, SantaClara, CA) according to the manufacturer's instructions.
Microarray processing and analysis
Total RNA isolated from lung adenocarcinomas (n=80) and adjacent normal lung tissues (n=30). RNA was also isolated from lung cancer cells transfected with scrambled siRNA and ETS2-specific siRNA (three replicates each, n=6). RNA samples were analyzed by microarray expression profiling using the Affymetrix Human Gene 1.0 ST platform (Affymetrix, Santaclara, CA) according to the manufacturer's instructions. Briefly, in vitro transcription and cDNA target preparation were performed using the WT expression kit (Life Technologies, Grand Island, NY). 2.5 μg of fragmented and labeled cDNA, was generated using the Affymetrix GeneChip® WT Terminal Labeling and Controls Kit and hybridized onto Human Gene 1.0 ST arrays according to the manufacturer's instructions (Affymetrix). Arrays were washed, stained and processed using Affymetrix GeneChip Fluidics Station 450 systems after which they were imaged using Affymetrix GeneChip Scanner 3000 7G for subsequent generation of raw data (*CEL files). Microarray data were submitted to the Gene Expression Omnibus (GEO) and were MIAME compliant. Lung tumors and normal lung tissue were deposited under series GSE43458 (samples GSM1062765-GSM1062874) and array data of transfected cells were submitted under series GSE43459 (samples GSM1062875-GSM1062880). Raw data were normalized using Robust Multichip Array (RMA) and log2 transformed using BRB-ArrayTools v 4.3.0 developed by Dr. Richard Simon and the BRB-ArrayTools Development Team (17). Genes significantly differentially expressed between lung adenocarcinomas and normal lung tissues were selected based on a false discovery rate (FDR) threshold of <0.001 and a 1.5 fold-change between the two groups (n=2665 transcripts). Genes differentially expressed between H441 lung cancer cells transfected with ETS2-specific siRNA compared to cells transfected with scrambled siRNA were selected based on a p<0.01 (n=1816 transcripts) (Supplementary Table 2). Functional pathways analysis was performed using the commercially available software Ingenuity Pathways Analysis according to the manufacturer's instructions.
Transfection of siRNA and expression vectors
siRNAs against ETS2 and MET as well as scrambled (control) siRNA were synthesized by a proprietary design as SMARTpool siRNA (Dharmacon, Thermoscientific). Knockdown of ETS2 or MET expression was performed using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer's instructions. ETS2 over-expression was achieved using an ETS2 cDNA clone (Origene, Rockville, MD) inserted into a Cytomegalovirus plasmid pCMV-XL5 vector. Cells were transfected with the expression vectors using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. For HGF treatment experiments, transfections were performed one day before overnight incubation with serum-free cell culture medium.
Migration and invasion assays
Transwell cell migration was quantified by seeding cells (n=8 × 104) in serum-free medium onto the upper layer of 24-well BD BioCoat™ 8.0 μm PET membrane inserts (BD Biosciences, Bedford, MA). Cells (n=8 × 104) in serum-free medium were seeded onto the upper layer of 24-well BD BioCoat Matrigel invasion chambers (BD Biosciences) for quantitative assessment of cell invasion. After 48 hours, migrating or invading cells were washed with PBS, fixed with 10% formalin and stained with 0.5% crystal violet then counted using bright-field microscopy. For HGF treatment experiments, a concentration of 50 ng/ml of HGF in serum-free medium was supplied to the upper layer. Qualitative assessment of cell migration was performed by the wound healing assay in which cells were scratched with a 200 μl pipette tip after which the wound was monitored for closure. All conditions were performed in three replicates.
Trypan blue exclusion and cell count
Cells were seeded in three replicate wells for each experimental condition at a density of 3 × 104 cells/well in 12-well plates and then transfected the following day with siRNAs or over-expression vectors. 72 hours following transfection, cells were washed with 1x PBS, trypsinized, mixed with 0.4% Trypan blue solution (Sigma Aldrich) then counted using the Reichert Bright-Line Hemacytometer (Hausser Scientific, Horsham, PA, USA) by trypan blue exclusion principle.
Anchorage-dependent and –independent colony formation assays
For anchorage-dependent colony formation on plastic, cells were seeded at a density of 150 cells/well in six-well plates. Plates were monitored for colony development for two to three weeks under a phase-contrast microscope after which cells were washed with 1x PBS, fixed with 10% formalin and stained with 0.5% crystal violet before counting. All experiments were done in triplicates and student's t-test was used to test for statistical significance. Cells were assessed for anchorage-independent colony formation by growth on soft agar. Cells were seeded (1 × 104/well) in 2 ml culture medium containing of 0.3% (w/v) of granulated agar (Difco, BD Biosciences). Cell-agar suspensions were plated on a 2 ml lower layer comprised of 0.6% (w/v) of agar. Both layers of soft agar were supplemented with 20% FBS. All conditions were performed in triplicates and colonies were scored two to three weeks after cell seeding.
Western blot analysis
Cell monolayers were washed twice with PBS, harvested and lysed with ice-cold RIPA buffer (Sigma-Aldrich, St. Louis, MO) after which protein lysates (20 micrograms) were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and western blotting. The antibodies used for immunoblotting included those raised against ERK1/2, phosphorylated-ERK1/2 (THR202/TYR204), MET, phosphorylated-MET (Y1234/Y1235), (Cell Signaling Technology, Danvers, MA), ETS2 (Santa Cruz Biotechnology, Santa Cruz, CA) and β-ACTIN (loading control) (Sigma-Aldrich). Antibody binding was detected by enhanced chemiluminescence (Amersham Biosciences Corp.). Band intensities were quantified relative to intensity of β-ACTIN using Image J software.
Phospho-MET enzyme-linked immunosorbent assay (ELISA)
Levels of phosphorylated MET (Y1234/Y1235) were quantified 48 h following knockdown of ETS2 expression using the phospho-MET(Y1234/Y1235) PathScan sandwich ELISA (Cell signaling Technology) according to the manufacturer's instructions.
Quantitative real-time PCR
A total of 1 μg of RNA was reverse-transcribed using the High Capacity RNA-to-cDNA kit (Life Technologies) according to the manufacturer's instructions and diluted in nuclease-free water. Real-time quantitative-PCR was performed using pre-designed TaqMan expression assays for ETS2 (Hs00232009_m1), β-ACTIN (Hs99999903_m1), MET (Hs01565584_m1), RAB3B (Hs01001137_m1), SERPINB5 (Hs00985285_m1), ZEB1 (Hs00232783_m1), SNAI2 (Hs00950344_m1) and SPDEF (Hs01026050_m1) (Life Technologies) on an ABI 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA). Relative quantification was calculated using the 2−ΔΔCT relative quantification method.
Immunohistochemistry
IHC analysis was performed on 4-μm formalin-fixed paraffin-embedded (FFPE) histological sections performed using purified rabbit polyclonal primary antibodies raised against ETS2 (1:1000 dilution, clone RB24588, AP9846c, Abgent, San Diego, CA) and phospho-MET (Y1234/Y1235) (1:250 dilution, catalog number AF2480; R&D systems, Minneapolis, MN). Antigen retrieval was carried out using the Dako Target Retrieval System at a pH of 6 (Dako, Carpinteria, CA). Intrinsic peroxidase activity was blocked by 3% methanol and hydrogen peroxide for 15 minutes and serum-free protein block from Dako was used for 30 minutes for blocking non-specific antibody binding. Slides were then incubated with antibodies against ETS2 and phospho-MET (Y1234/Y1235) at room temperature for 65 and 90 minutes, respectively. After 3 washes in TBS, slides were then incubated for 30 minutes with Dako Envision+ Dual Link at room temperature. Following three additional washes, slides were incubated with Dako chromogen substrate for 5 minutes and were counterstained with hematoxylin for another 5 minutes. FFPE pellets from lung cancer cell lines displaying positive ETS2 and phospho-MET (Y1234/Y1235) expression were used as positive controls for antibody optimization, whereas samples and whole-section tissue specimens processed similarly, except for the omission of the primary antibodies, were used as negative controls. Immunostaining intensity and reactivity were examined by an experienced pathologist (J.F.) using a light microscope under a ×20 magnification objective. ETS2 expression was quantified using a 4-value intensity score (0, none; 1, weak; 2, moderate; and 3, strong) and the percentage (0%–100%) of the extent of reactivity. A final expression score was obtained by multiplying the intensity and reactivity extension values (range, 0–300) (16). For analysis of phosphorylated MET expression, sections with membranous immunoreactivity were considered positive for phospho-MET expression as previously described (18, 19) and phospho-MET staining was quantified using a score range of 0 to 3+ (20).
Statistical analysis
ANOVA and Student's t-test were utilized to test for statistical significance among different groups in the in vitro experiments. Statistical analysis of the IHC data was first summarized using standard descriptive statistics and frequency tabulations. Associations of ETS2 IHC protein expression alone or in combination phospho-MET (Y1234/Y1235) with patient outcome (time to recurrence) was estimated using the Kaplan-Meier method and compared among groups by log-rank statistical tests. Multivariate Cox proportional hazard models were applied to assess the effects of ETS2 and/or phospho-MET IHC expression on time to recurrence and adjusted for tumor stage. All computations were carried out in SAS 9.3 and S-plus 8.2.
Results
ETS2 expression is significantly lower in lung adenocarcinomas relative to normal lung
We performed gene expression profiling of lung adenocarcinomas (N=80) and matched normal lung (N=30) and identified, using an FDR threshold of 0.1% and a 1.5 fold-change cut-off, 2665 transcripts that were significantly differentially expressed between lung adenocarcinomas and normal lung. Further analysis from our current expression profiling efforts revealed significantly decreased expression of the ETS2 transcription factor transcript in lung adenocarcinomas compared to normal lung tissues (p<10−7) (Figure 1A). In silico analysis of a publicly available microarray dataset (21) corroborated our findings and similarly showed a significant decrease in ETS2 expression in NSCLC tumors compared to adjacent normal lung. QRTPCR analysis confirmed the ETS2 decreased expression (p=0.003) (Figure 1B). Moreover, ETS2 expression was highly correlated between gene microarray and QRTPCR in a panel of normal lung and lung adenocarcinoma tissues (R=0.72, p<0.001) (Figure 1C). Analysis of ETS2 expression in our gene expression microarray dataset in the context of various clinicopathological features revealed that ETS2 was significantly lower in smoker compared to never-smoker adenocarcinomas (p<0.001) (Supplementary Figure 1A) which was confirmed in a subset of adenocarcinoma tumors by QRTPCR (p<0.001) (Supplementary Figure 1B). Similarly, ETS2 expression, assessed by QRTPCR, was significantly lower in established NSCLC cell lines previously derived from smokers compared to those isolated from never-smoker patients (p<0.05) (Supplementary Figure 2). These findings demonstrate that ETS2 mRNA expression is reduced in lung adenocarcinomas relative to normal lung, in particular, in smoker lung tumors.
Figure 1. ETS2 transcript is significantly decreased in lung adenocarcinomas compared to normal lung tissue.
Lung adenocarcinomas (n=80) and normal lung tissues (n=30) were profiled using the Human Gene 1.0 ST platform as detailed in the Methods section. A. ETS2 normalized expression in adenocarcinomas (ADC) and normal lung. B. Relative ETS2 mRNA expression was assessed by QRTPCR, normalized to that of β-ACTIN and quantified using the 2−ΔΔCT relative quantification method as detailed in the Methods section. P-values indicate statistical significance, by the Student's t-test, of difference in ETS2 expression between ADCs and normal lung. C. Statistical correlation of ETS2 expression quantified by microarray profiling and QRTPCR; R, Pearson's correlation coefficient.
Decreased ETS2 immunohistochemical expression is associated with shorter time to recurrence in NSCLC patients
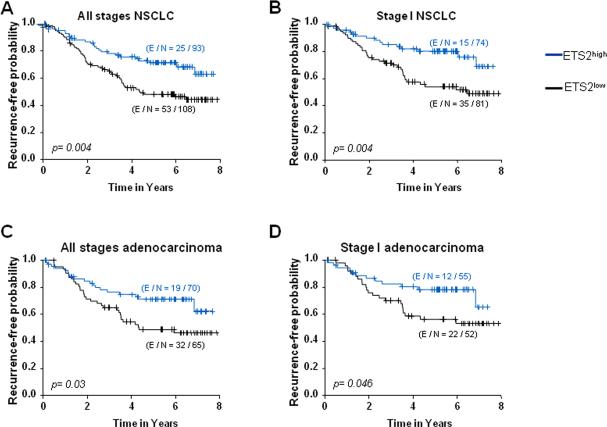
ETS2 is a canonical transcription factor that affects a broad spectrum of genes by binding to ERE sequences in the genome thereby affecting various molecular and cellular functions (3, 5). Our findings on reduced ETS2 transcript expression in lung tumors prompted us to examine the IHC expression of ETS2 protein in association with patient outcome which has not been characterized before. We analyzed ETS2 expression in a NSCLC (n=201) tissue microarray (135 adenocarcinomas and 66 SCCs) derived from patients who did not receive neoadjuvant or adjuvant treatment. Representative photomicrographs of low (left) and high (right) ETS2 IHC expression are depicted in Supplementary Figure 3. Association of ETS2 IHC expression with patient clinical outcome was performed following dichotomization of patients into groups with relatively higher or lower ETS2 expression than the IHC score of 55 which was determined by the martingale residual. Low ETS2 expression predicted shorter time to recurrence in all-stages (Figure 2A) and stage-I (Figure 2B) NSCLC (both p=0.004 of the log-rank test). Moreover, relatively low ETS2 expression predicted shorter time to recurrence in all-stages (p=0.03) (Figure 2C) and stage-I (Figure 2D) (p=0.046) lung adenocarcinoma but not in lung SCC. Importantly, multivariate Cox proportional hazard models demonstrated that, after adjusting for stage, relatively lower ETS2 IHC protein expression was an independent predictor of shorter time to recurrence in NSCLC (p=0.009, HR=1.89, 95% confidence interval 1.17-3.06) and in lung adenocarcinoma (p=0.03, HR=1.86, 95% confidence interval 1.05-3.31). Our findings on the association of low ETS2 immunohistochemical expression with shorter time to recurrence in NSCLC points to a potential tumor suppressor function for ETS2 in human lung cancer.
Figure 2. Reduced ETS2 immunohistochemical expression is associated with shorter time to recurrence in NSCLC patients.
ETS2 protein expression was analyzed by IHC in a NSCLC TMA comprised of 201 FFPE histological tissue specimens (135 lung adenocarcinomas and 66 SCCs). Differences in time to recurrence between patients stratified by ETS2 IHC expression (ETS2high: ≥55, blue; ETS2low: <55, black) were statistically assessed by the log-rank test and Kaplan-Meier survival probability method and depicted for all-stages (A) and stage-I (B) NSCLC patients as well as in all-stages (C) and stage-I (D) adenocarcinoma patients. E; number of censored patients in each subgroup; N; number of patients in each group based on relatively higher or lower ETS2 IHC expression.
ETS2 inhibits lung cancer cell growth, migration and invasion
We then sought to test the role of ETS2 in NSCLC pathogenesis. H441 lung cancer cells transfected with small interfering RNA (siRNA) specific to ETS2 exhibited significantly reduced ETS2 transcript compared to cells transfected with control scrambled siRNA (p<0.05) (Figure 3A). Expression of ETS2 protein was also reduced by 0.6-fold, albeit less than that observed for the transcript, following RNA interference (Figure 3A). Moreover, ETS2 knockdown significantly increased cell growth (1.55-fold) evidenced by trypan blue exclusion (Figure 3B, right) count as well as augmented cell migration (Figure 3B, left) and invasion (Figure 3B, middle) by 4.4-fold and 6.2-fold, respectively (all p<0.05). Conversely, ETS2 over-expression in H1299 lung cancer cells, with a relatively low basal expression of the gene (Figure 3C), significantly decreased cell migration (Figure 3D, left) and invasion (Figure 3D, middle) by 2.6 fold and proliferation by 1.5 fold (Figure 3D, right) (all p<0.05). Moreover, over-expression of ETS2 in H1299 cells significantly suppressed anchorage-dependent (1.8-fold) and –independent (2.1-fold) colony formation evidenced by growth on plastic and soft agar, respectively (p<0.05) (Supplementary Figures 4A and 4B). Importantly, siRNA-mediated knockdown of ETS2 expression nearly completely rescued cells from cell growth inhibition mediated by over-expression of the gene suggesting that the observed cell growth inhibition was ETS2-mediated (Supplementary Figure 4C). It is worthwhile to mention that the fold-changes in cellular migration and invasion following knockdown or over-expression of ETS2 were greater than the noted differences in cell growth. These findings suggest a tumor suppressor function for the ETS2 gene in vitro in human lung cancer.
Figure 3. ETS2 expression is a negative regulator of cellular migration, invasion and proliferation.
QRTPCR (upper panels) and western blotting analyses (lower panels) of ETS2 mRNA and protein expression, respectively, were performed 48 hours after transfection of H441 cells with scrambled (control) siRNA and siRNA targeting ETS2 (A) and of H1299 cells with control and ETS2 over-expression vectors (C). ETS2 expression was normalized to that of the β-ACTIN housekeeping gene and quantified relative to the expression in cells transfected with control siRNA. For western blotting, 20 μg of total protein from samples were analyzed by SDS-PAGE as detailed in the Methods section. Membranes were stained with polyclonal antibody against ETS2 and with a β-ACTIN monoclonal antibody to ensure equal protein loading. Analysis of Transwell cell migration (left panels) and invasion (middle panels) was performed in the transfected H441 (B) and H1299 (D) cells using BD BioCoat transwell inserts or inserts with Matrigel, respectively. Cells that migrated or invaded after 48 hours to the lower side of the inserts were fixed, stained and quantified, as described in the Methods section, relative to number of cells transfected with control siRNA. Growth of cells (right panels, B and D) was assessed by the trypan blue exclusion method as mentioned before. All assays are representative of three independent experiments. * indicate p-values <0.05 assessed by the Student's t-test.
Global gene expression analysis downstream of ETS2 knockdown reveals modulation of key pathways in lung cancer cells
To gain insights into the mechanisms of ETS2 tumor suppressor function, we sought to compare and contrast the transcriptome of cells transfected with scrambled siRNA and ETS2-specific siRNA. Gene expression profiling, using the Affymetrix Human Gene 1.0 ST platform, identified 1816 transcripts that were significantly differentially expressed, based on a p<0.01 threshold, in H441 lung cancer cells with ETS2 knockdown compared to their control counterparts (Figure 4A, Supplementary Table 2). Moreover, functional analysis of the genes using Ingenuity Pathways Analysis (IPA), revealed that ETS2 knockdown modulated key pathways typically activated in cancer, in particular, HGF, integrin, tissue factor, semaphorin and mitogen activated protein kinase (MAPK) signaling (all p<0.001) (Figure 4B). Notably, the pathways analysis revealed that HGF signaling was the top modulated canonical pathway following ETS2 knockdown (p<10-5) (Figure 4B) and that an HGF-mediated gene-interaction network was among the top significant gene networks following topological arrangement of the differentially expressed genes by IPA (Supplementary Figure 5). In addition, we observed significant induction of genes involved in epithelial mesenchymal transition (EMT) (22) (e.g., zinc finger E-box binding homeobox 1/ZEB1 and snail homolog 2/SNAI2), GTPases (e.g., RAB3B), known ETS2 targets (23) (e.g., serpin peptidase inhibitor clade B member 5/SERPINB5) and members of the ETS family (5) (e.g., SAM pointed domain containing ets transcription factor/SPDEF) all of which were confirmed by QRTPCR analysis following selection by a combination of statistical criteria and pathways analysis (Figure 4C). Our global transcriptome analysis further points to a tumor suppressor function by ETS2 in human lung cancer cells.
Figure 4. Global changes in lung cancer cell transcriptome following knockdown of ETS2 expression.
H441 lung cancer cells were transfected with control siRNA and ETS2-specific siRNA (three replicates each). Total RNA was isolated 48 hours after transfection and was analyzed using the Human Gene 1.0 ST platform, as detailed in the Methods section, to compare and contrast the transcriptome of H441 cells transfected with control siRNA and ETS2-specific siRNA. A. Heat map depicting 1,816 transcripts that were significantly differentially expressed between cells transfected with scrambled (blue) and ETS2-specific (red) siRNA based on a p<0.01. Rows and columns represent transcripts and samples, respectively. Up-regulated and down-regulated gene expression is indicated by red and green colors, respectively. B. Functional pathways analysis of the differentially expressed genes was performed using the IPA commercially available software. Statistically significant modulation (indicated by the inverse log of p-values) of the top five over-represented canonical pathways following ETS2 knockdown and predicted by IPA is depicted. C. Confirmation of microarray profiling by QRTPCR analysis of RAB3B, SPDEF, ZEB1, SNAI2 and SERPINB5 genes in H441 lung cancer cells transfected with control and ETS2-specific siRNA. Expression changes are depicted relative to cells transfected with control siRNA. * indicate p-values <0.05 assessed by the Student's t-test.
ETS2 suppresses MET activation and HGF signaling in human lung cancer cells
Our expression profiling analysis demonstrated that HGF signaling was the top significant modulated pathway following siRNA-mediated ETS2 knockdown in lung cancer cells. Since HGF is known to function as ligand for the MET receptor oncogene (24, 25), we sought to determine the effects of ETS2 knockdown on the latter tyrosine kinase receptor. Western blotting analysis demonstrated that siRNA-mediated knockdown of ETS2 expression increased phosphorylated levels of MET by 1.2-fold (Figure 5A). Enzyme-linked immunosorbent assay for phosphorylated MET levels similarly revealed an increase in MET phosphorylation at Y1234/Y1235 following ETS2 knockdown (Supplementary Figure 6). Conversely, ETS2 over-expression suppressed MET phosphorylation which was rescued by co-transfection of ETS2-targeting siRNA (Supplementary Figure 7). Importantly, co-transfection of H441 lung cancer cells with siRNA targeting MET (Figure 5B) abrogated cell migration (Figure 5C, upper panel), invasion (Figure 5C, middle panel) and, to a less pronounced effect, cell growth (Figure 5C lower panel) (p<0.05) mediated by knockdown of ETS2 expression alone (Figures 5B and 5C). Moreover, we examined the expression of genes we had identified to be induced by ETS2 knockdown (Figure 4C) in cells transfected with siRNAs targeting both ETS2 and MET. Co-transfection of cells with MET-specific siRNA significantly attenuated the induction of SNAI2, RAB3B, SPDEF and SERPINB5 mediated by knockdown of ETS2 alone (all p<0.05) as well as reduced, albeit not significant, expression of the ZEB1 gene (Figure 5D). These data demonstrate that ETS2 suppresses key features of the lung cell malignant phenotype, at least in part, through inhibition of the MET oncogene.
Figure 5. ETS2 inhibits cell migration and invasion through suppression of MET phosphorylation and activation.
A. H441 lung cancer cells were transfected with control and ETS2-specific siRNA and analyzed by western blotting analysis, as described earlier, for ETS2 and total and phosphorylated (P-MET) MET levels 48 hours following transfection. B. QRTPCR analysis of ETS2 and MET transcript expression (left panel) and western blotting analysis of total and phosphorylated MET protein levels (right panel) was performed 48 hours following transfection of H441 cells with control siRNA, siRNA against ETS2 or MET alone, and with both ETS2- and MET-specific siRNAs. * indicate p-values <0.05 assessed by the Student's t-test. C. H441 cells, transfected as described in B were analyzed for differences in migration (upper panel), invasion (middle panel) and cell growth (lower panel) 48 hours following transfection. Analysis of cellular migration, invasion and growth, relative to control cells transfected with scrambled siRNA only, was performed as described earlier in Figure 3. D. QRTPCR analysis of the expression of RAB3B, SPDEF, ZEB1, SNAI2 and SERPINB5 in cells transfected with ETS2-specific siRNA, MET-specific siRNA and with both ETS2- and MET-specific siRNAs compared to cells transfected with scrambled siRNA only. * indicate p-values (<0.05) indicating statistically significant pairwise (indicated by the dotted connectors) differences. All assays are representative of three independent experiments.
ETS2 suppresses HGF-mediated signaling in lung cancer cells
Since we found that ETS2 suppresses cellular migration and invasion through inhibition of MET phosphorylation and activation, we sought to examine the role of ETS2 in HGF-mediated signaling. Following an overnight serum starvation, H441 lung cancer cells were treated with HGF (50 ng/ml final concentration) for one hour. HGF treatment increased ETS2 protein expression by 1.3-fold in H441 cells (Figure 6A). The increase in ETS2 protein was also noted in additional lung cancer cell lines (Supplementary Figure 8). Moreover, and as shown in previous studies (26, 27), HGF treatment also increased phosphorylation of both MET and extracellular-regulated kinase (ERK1/2) MAPK (Figure 6A) in H441 lung cancer cells which was alleviated at later treatment time points (Supplementary Figure 8A) as previously reported for earlier HGF treatment and ligand stimulation studies (28, 29). Furthermore, expression of ETS2 protein and of phospho-ERK1/2 and phospho-MET was attenuated by co-treatment of cells with the MET tyrosine kinase inhibitor PHA-665752 (100 nM final concentration) (Figure 6A and Supplementary Figure 8B).
Figure 6. ETS2 inhibits signaling downstream of HGF.
A. Western blotting analysis was performed 1 hour after treatment of serum-starved H441 cells with HGF, 100 nM PHA-665752, and the combination of both HGF and PHA-665752. Protein lysates were immunoblotted with antibodies against ETS2, total and phosphorylated levels of MET and ERK1/2 as well as with an antibody against β-ACTIN to ensure equal protein loading. B. H441 cells were transfected with scrambled and ETS2-specific siRNA and serum-starved overnight the following day. 24 hours after serum-starvation, transfected cells were incubated in medium with and without 50ng/ml HGF for 1 hour for subsequent western blotting (left panel) for total and phosphorylated MET protein levels or were transferred onto the upper layer of 24-well BD BioCoat transwell inserts or inserts with Matrigel, in the presence or absence of 50 ng/ml HGF, for analysis of cell migration (upper right) and invasion (lower right), respectively. C. QRTPCR analysis of the expression of the indicated genes in cells transfected with control and ETS2-specific siRNA and incubated in cell culture medium with and without 50 ng/ml HGF for 48 hours. * indicate p-values (<0.05) indicating statistically significant pairwise (indicated by the connectors) differences. All assays were performed in triplicates and representative of three independent experiments.
We then sought to assess the relevance of ETS2 to HGF signaling in lung cancer cells. RNA interference-mediated knockdown of ETS2 expression significantly augmented HGF-induced cell migration (1.9-fold) and invasion (1.7-fold) (p<0.05) (Figure 6B, right panel). These effects were concomitant with increased MET phosphorylation in HGF-treated cells transfected with ETS2-specific siRNA (1.8-fold with respect to control cells) which was higher than that observed in treated cells transfected with control siRNA (1.5-fold with respect to control cells) (Figure 6B, left panel). Furthermore, we also found that, similar to knockdown of ETS2 alone (p<0.05), HGF treatment also significantly increased the expression of the SNAI2, RAB3B, SPDEF, ZEB1 and SERPINB5 genes (all p<0.05) (Figure 6C). It is worthwhile to mention, that HGF treatment also significantly increased ETS2 mRNA levels (p<0.05) (Figure 6C). Importantly, the expression of the aforementioned genes was significantly higher in HGF-treated cells transfected with ETS2-specific siRNA compared to similarly treated cells that were transfected with control siRNA (p<0.05) (Figure 6C). The observed additive effect of the combination of HGF treatment and ETS2 knockdown on MET phosphorylation was also confirmed in an additional lung cancer cell line (H1944) (Supplementary Figures 9A and 9B). Whereas HGF treatment alone increased phospho-MET protein, evidenced by western blotting, by 61-fold, phosphorylated MET protein was increased by 98-fold following HGF treatment in cells with knockdown of ETS2 (Supplementary Figure 9B). This effect was concomitant with increased cell growth (1.6-fold), rather than invasion, compared to HGF-treated cells with control siRNA (Supplementary Figure 9C). Moreover and conversely, over-expression of ETS2 in H1299 lung cancer cells decreased HGF-induced phosphorylated MET levels and decreased HGF-induced cell migration evidenced by the wound healing assay (Supplementary Figure 9D). Our findings suggest that ETS2 may impact dissimilar phenotypic and anti-malignant effects downstream of HGF in different lung cancer cell lines. Interestingly, differential anti-malignant phenotypic effects by the MET inhibitor, PHA-665752, in different cancer cell lines were also previously noted in the original report by Christensen et al (26). Our findings suggest that ETS2 inhibits HGF-mediated oncogenic signaling in a negative-feedback manner.
It is worthwhile to mention that we also analyzed phosphorylated MET IHC expression (Supplementary Figure 10) along with that of ETS2 in association with patient time to recurrence. Interestingly, NSCLC (p=0.01) or adenocarcinoma (p=0.02) patients with relatively lower ETS2 expression and higher phosphorylated MET membrane immunoreactivity exhibited significantly shortest time to recurrence compared to other patient subgroups, in particular those with higher ETS2 and higher phosphorylated MET expression (Supplementary Figures 111A-D). These data further suggest that ETS2 suppression of MET oncogene signaling is operative in human lung cancer.
Discussion
Our expression profiling efforts revealed significant down-regulation of the ETS2 transcription factor in lung adenocarcinomas compared to normal lung tissues. We questioned the relevance of ETS2 down-regulation to NSCLC biology, given the important cellular and molecular roles this transcription factor plays (3, 5), and found that low ETS2 protein was predictive of shorter time to recurrence and the gene was a negative regulator of lung cancer cell growth, migration and invasion and of HGF/MET oncogenic signaling. It is worthwhile to note that we had found significant reduced expression of ETS2 in smoker compared to never-smoker lung adenocarcinomas (Supplementary Figure 1) and, similarly, NSCLC cell lines originally derived from smokers exhibited reduced ETS2 expression compared to lung cancer cell lines isolated from never-smokers. It is worthwhile to mention that in silico analysis of publicly available expression datasets also revealed reduced ETS2 transcript expression in smoker adenocarcinomas as well as marked down-regulation of expression of the gene in airways of phenotypically healthy smokers compared to disease-free non-smokers (data not shown). In addition and notably, ETS2 mRNA expression, part of a genomic signature of phosphatidylinositol 3-kinase (PI3K) pathway activation, was decreased in the normal bronchial airways of smokers with lung cancer compared to airways of disease-free smokers (30). It is plausible to suggest that cigarette smoking reduces expression of the ETS2 gene in the course of NSCLC pathogenesis.
Microarray profiling coupled with functional pathways analysis revealed that ETS2 expression is linked to various canonical cancer-associated pathways in lung cancer cells, in particular the HGF pathway. Moreover, QRTPCR analysis confirmed the array findings and demonstrated up-regulation of the ZEB1, SNAI2, RAB3B, SERPINB5 and SPDEF genes following siRNA-mediated knockdown of ETS2 expression concomitant with elevated cell migration and invasion. ZEB1 and SNAI2 (also known as SLUG) are transcription factors with well-established roles in promotion of EMT in various physiological and pathological conditions (22). RAB3B is a relatively under-studied RAS family member and GTPase that was found to be an important positive regulator of epithelial cell polarity (31) and of the stemness of adult human mesenchymal stem cells (32). SERPINB5, a serpin peptidase inhibitor, was demonstrated to be a target repressed by murine Ets2 (23) and was shown to be up-regulated in human lung tumors compared to normal lung tissues (33) and hypomethylated and elevated in lung SCCs (34). SPDEF (also known as PDEF), a member of the ETS-domain protein family (5), was shown to be up-regulated in mammary tumors (35) and to positively control cancer cell migration and invasion (36). Notably, SPDEF was found to promote airway epithelial cell (e.g., goblet cell) hyperplasia, inflammation and mucus hyper secretion (37). It is also noteworthy, that HGF signaling, known for its pro-migratory and -invasive effects (27), was the top pathway significantly modulated following ETS2 knockdown in vitro. In addition, ETS2 knockdown significantly augmented HGF-mediated induction of all five genes. It is reasonable to suggest, given the increased expression of the aforementioned genes following ETS2 knockdown, along with their given previous reported promalignant roles, that ETS2 functions as a lung tumor suppressor in lung cancer cells, at least in part, by regulation of the migration- and invasion-promoting transcriptome.
In this study we found that ETS2 suppressed HGF-mediated MET phosphorylation and cell migration and invasion in a negative feedback manner. Our findings are in accordance with previous reports demonstrating ETS2 negative feedback regulation of other signaling networks, namely FGF signaling (38) and phorbol ester-induced MAPK signaling (39) by up-regulation of MAPK phosphatases (38, 39). Moreover, a previous study by Cao et al showed modulation of ETS2 expression and intracellular localization downstream of HGF/MET pathway activation in mesothelioma cancer cell lines, albeit without functional assessment of the relevance of increased ETS2 expression (40). Since we also demonstrated that ETS2 basally suppressed MET phosphorylation and activation, it is plausible to suggest that ETS2 functions both upstream and downstream of MET oncogene to inhibit cell migration and invasion. In an attempt to further probe the relevance of the identified MET/ETS2 signaling to human lung cancer, we studied the association of both ETS2 and phosphorylated MET IHC protein expression with NSCLC patient outcome. The combination of both decreased ETS2 and increased phosphorylated MET IHC expression identified a subgroup of patients with further reduced time to recurrence compared to patients with low ETS2 and high phosphorylated-MET protein or when compared to analysis of ETS2 expression alone. Further studies to confirm the combinatorial effect of low ETS2 protein and high phosphorylated MET immunohistochemical expression on reduced time to recurrence in lung cancer or other malignancies are warranted. It is worthwhile to note that MET function was shown to be deregulated in different types of solid tumors and hematological malignancies by increased transcript expression or copy number gain or amplification (27, 41, 42). It is intriguing to suggest that ETS2 may represent an additional endogenous level of pathway control of MET activation in NSCLC.
It is important to note that ETS2 has been shown to exhibit both oncogenic and tumor suppressor roles in various malignancies. For example, ETS2 was demonstrated to have mitogenic activity downstream of Ras in murine fibroblasts (8), mediate transformation and tumorigenicity of prostate and breast epithelial cells (13, 14) and to be up-regulated in breast, thyroid, prostate and pancreatic tumors as well as in hematologic malignancies such as leukemias (4, 43-45). Moreover, loss of murine Ets2 expression in tumor associated macrophages was demonstrated to decrease breast cancer metastasis in mice (23). On the other hand, various studies have revealed tumor suppressor functions and effects mediated by the ETS2 gene. Foos et al demonstrated that over-expression of full length Ets2, with intact transactivator function, reversed Ras-induced NIH3T3 cell transformation (46) and, in a separate study, Hever and colleagues showed that Ets2 was not required for ErbB2-mediated cell transformation in vitro (47). In addition, mice modeling Down's syndrome and colon cancer exhibited reduced intestinal tumor incidence which was attributed to increased gene dosage of Ets2, located in the 21q22.2 chromosomal locus, in Down's syndrome with trisomy of chromosome 21 (15). In this context and similarly, mice with Ets2-deficient intestinal cells were shown to develop more colon tumors in response to treatment with carcinogens (12). Additional potential mechanisms of ETS2-mediated tumor suppression have been attributed to suppression of miR-196b-induced invasion in gastric cancer cells (11) and promotion of cell cycle arrest by negative feedback inhibition of MAPK signaling following treatment with the phorbol ester PMA (39). Interestingly, over-expression in lung cancer cells of the frequently epigenetically inactivated lung tumor suppressor RASSF1A was demonstrated to increase ETS2 transcript expression concomitant with arrest of cell cycle progression (48). Furthermore, ETS2 expression was shown to be down-regulated in human lung invasive adenocarcinomas compared to non-invasive bronchioalveolar carcinomas (49). The opposing tumor repressive and supportive functions attributable to ETS2 in different tumors may be, in part, related to the dissimilar roles ETS2 may exhibit in tumors of different cell lineages.
In conclusion, we demonstrated, in this study, reduced expression of ETS2 transcript expression in human lung tumors relative to normal lung tissues, association of low ETS2 protein expression with shorter time to recurrence in NSCLC patients and inhibition of human lung cancer cell invasion, migration and growth by ETS2, at least in part, through suppression of MET oncogene phosphorylation and HGF signaling. Our findings provide collective testament to a potential tumor suppressor role of the ETS2 gene in human NSCLC pathogenesis, in part, through inhibition of MET oncogene activation.
Supplementary Material
Statement of Translational Relevance.
The expression and role of ETS2, a transcription factor with various important cellular and molecular functions, in NSCLC biology and pathogenesis is still unknown. In this study, we found that the ETS2 gene exhibited down-regulated expression in lung adenocarcinomas compared to normal lung following microarray profiling and reduced expression of its coding protein product was an independent predictor of shorter time to recurrence in human NSCLC. Moreover, we found that ETS2 negatively regulated cell growth, migration and invasion, in part, through suppression of MET phosphorylation and activation and inhibited HGF-mediated oncogenic signaling. Our findings point to an additional level of intracellular pathway control of aberrant MET oncogene activation, through ETS2 tumor suppressor function, in human NSCLC and that may possibly be used as a marker for selection of patients for MET-targeted therapy.
Acknowledgments
Grant support
Supported in part by a Lung Cancer Research Foundation grant (H.K.), Department of Defense (DoD) (W81XWH-04-1-0142 to I.I.W.) and by UT Lung SPORE P50CA70907 (to I.I.W.) and Cancer Center Support Grant CA-16672 from the National Cancer Institute.
Footnotes
Conflict of interest disclosure: The authors declare no conflict of interest
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu T, Trojanowska M, Watson DK. Ets proteins in biological control and cancer. J Cell Biochem. 2004;91:896–903. doi: 10.1002/jcb.20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–78. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–37. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, et al. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12:1315–26. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei G, Srinivasan R, Cantemir-Stone CZ, Sharma SM, Santhanam R, Weinstein M, et al. Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood. 2009;114:1123–30. doi: 10.1182/blood-2009-03-211391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang BS, Hauser CA, Henkel G, Colman MS, Van Beveren C, Stacey KJ, et al. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol Cell Biol. 1996;16:538–47. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu D, Dwyer J, Li H, Duan W, Liu JP. Ets2 maintains hTERT gene expression and breast cancer cell proliferation by interacting with c-Myc. J Biol Chem. 2008;283:23567–80. doi: 10.1074/jbc.M800790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Nigris F, Mega T, Berger N, Barone MV, Santoro M, Viglietto G, et al. Induction of ETS-1 and ETS-2 transcription factors is required for thyroid cell transformation. Cancer Res. 2001;61:2267–75. [PubMed] [Google Scholar]
- 11.Liao YL, Hu LY, Tsai KW, Wu CW, Chan WC, Li SC, et al. Transcriptional regulation of miR-196b by ETS2 in gastric cancer cells. Carcinogenesis. 2012;33:760–9. doi: 10.1093/carcin/bgs023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munera J, Cecena G, Jedlicka P, Wankell M, Oshima RG. Ets2 regulates colonic stem cells and sensitivity to tumorigenesis. Stem Cells. 2011;29:430–9. doi: 10.1002/stem.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neznanov N, Man AK, Yamamoto H, Hauser CA, Cardiff RD, Oshima RG. A single targeted Ets2 allele restricts development of mammary tumors in transgenic mice. Cancer Res. 1999;59:4242–6. [PubMed] [Google Scholar]
- 14.Sementchenko VI, Schweinfest CW, Papas TS, Watson DK. ETS2 function is required to maintain the transformed state of human prostate cancer cells. Oncogene. 1998;17:2883–8. doi: 10.1038/sj.onc.1202220. [DOI] [PubMed] [Google Scholar]
- 15.Sussan TE, Yang A, Li F, Ostrowski MC, Reeves RH. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down's syndrome. Nature. 2008;451:73–5. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- 16.Tang X, Kadara H, Behrens C, Liu DD, Xiao Y, Rice D, et al. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: implications in lung cancer pathogenesis and prognosis. Clin Cancer Res. 2011;17:2434–43. doi: 10.1158/1078-0432.CCR-10-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–7. [PMC free article] [PubMed] [Google Scholar]
- 18.Arriola E, Canadas I, Arumi-Uria M, Domine M, Lopez-Vilarino JA, Arpi O, et al. MET phosphorylation predicts poor outcome in small cell lung carcinoma and its inhibition blocks HGF-induced effects in MET mutant cell lines. Br J Cancer. 2011;105:814–23. doi: 10.1038/bjc.2011.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuta K, Kozu Y, Mimae T, Yoshida A, Kohno T, Sekine I, et al. c-MET/phospho-MET protein expression and MET gene copy number in non-small cell lung carcinomas. J Thorac Oncol. 2012;7:331–9. doi: 10.1097/JTO.0b013e318241655f. [DOI] [PubMed] [Google Scholar]
- 20.Birner P, Oberhuber G, Stani J, Reithofer C, Samonigg H, Hausmaninger H, et al. Evaluation of the United States Food and Drug Administration-approved scoring and test system of HER-2 protein expression in breast cancer. Clin Cancer Res. 2001;7:1669–75. [PubMed] [Google Scholar]
- 21.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 23.Zabuawala T, Taffany DA, Sharma SM, Merchant A, Adair B, Srinivasan R, et al. An ets2-driven transcriptional program in tumor-associated macrophages promotes tumor metastasis. Cancer Res. 2010;70:1323–33. doi: 10.1158/0008-5472.CAN-09-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–25. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 25.Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991;251:802–4. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- 26.Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 2003;63:7345–55. [PubMed] [Google Scholar]
- 27.Liu X, Newton RC, Scherle PA. Developing c-MET pathway inhibitors for cancer therapy: progress and challenges. Trends Mol Med. 2010;16:37–45. doi: 10.1016/j.molmed.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Dulak AM, Gubish CT, Stabile LP, Henry C, Siegfried JM. HGF-independent potentiation of EGFR action by c-Met. Oncogene. 2011;30:3625–35. doi: 10.1038/onc.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radtke S, Milanovic M, Rosse C, de Rycker M, Lachmann S, Hibbert A, et al. ERK2 but not ERK1 mediates HGF-induced motility in non small cell lung carcinoma cell lines. J Cell Sci. 2013 doi: 10.1242/jcs.115832. Epub ahead of print doi: 10.1242/jcs.115832. [DOI] [PubMed] [Google Scholar]
- 30.Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med. 2010;2:26ra5. doi: 10.1126/scitranslmed.3000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto Y, Nishimura N, Morimoto S, Kitamura H, Manabe S, Kanayama HO, et al. Distinct roles of Rab3B and Rab13 in the polarized transport of apical, basolateral, and tight junctional membrane proteins to the plasma membrane. Biochem Biophys Res Commun. 2003;308:270–5. doi: 10.1016/s0006-291x(03)01358-5. [DOI] [PubMed] [Google Scholar]
- 32.Song L, Webb NE, Song Y, Tuan RS. Identification and functional analysis of candidate genes regulating mesenchymal stem cell self-renewal and multipotency. Stem Cells. 2006;24:1707–18. doi: 10.1634/stemcells.2005-0604. [DOI] [PubMed] [Google Scholar]
- 33.Smith SL, Bowers NL, Betticher DC, Gautschi O, Ratschiller D, Hoban PR, et al. Overexpression of aurora B kinase (AURKB) in primary non-small cell lung carcinoma is frequent, generally driven from one allele, and correlates with the level of genetic instability. Br J Cancer. 2005;93:719–29. doi: 10.1038/sj.bjc.6602779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon YJ, Lee SJ, Koh JS, Kim SH, Lee HW, Kang MC, et al. Genome-wide analysis of DNA methylation and the gene expression change in lung cancer. J Thorac Oncol. 2012;7:20–33. doi: 10.1097/JTO.0b013e3182307f62. [DOI] [PubMed] [Google Scholar]
- 35.Feldman RJ, Sementchenko VI, Gayed M, Fraig MM, Watson DK. Pdef expression in human breast cancer is correlated with invasive potential and altered gene expression. Cancer Res. 2003;63:4626–31. [PubMed] [Google Scholar]
- 36.Gunawardane RN, Sgroi DC, Wrobel CN, Koh E, Daley GQ, Brugge JS. Novel role for PDEF in epithelial cell migration and invasion. Cancer Res. 2005;65:11572–80. doi: 10.1158/0008-5472.CAN-05-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park KS, Korfhagen TR, Bruno MD, Kitzmiller JA, Wan H, Wert SE, et al. SPDEF regulates goblet cell hyperplasia in the airway epithelium. J Clin Invest. 2007;117:978–88. doi: 10.1172/JCI29176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ekerot M, Stavridis MP, Delavaine L, Mitchell MP, Staples C, Owens DM, et al. Negative-feedback regulation of FGF signalling by DUSP6/MKP-3 is driven by ERK1/2 and mediated by Ets factor binding to a conserved site within the DUSP6/MKP-3 gene promoter. Biochem J. 2008;412:287–98. doi: 10.1042/BJ20071512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunes-Xavier CE, Tarrega C, Cejudo-Marin R, Frijhoff J, Sandin A, Ostman A, et al. Differential up-regulation of MAP kinase phosphatases MKP3/DUSP6 and DUSP5 by Ets2 and c-Jun converge in the control of the growth arrest versus proliferation response of MCF-7 breast cancer cells to phorbol ester. J Biol Chem. 2010;285:26417–30. doi: 10.1074/jbc.M110.121830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao X, Littlejohn J, Rodarte C, Zhang L, Martino B, Rascoe P, et al. Up-regulation of Bcl-xl by hepatocyte growth factor in human mesothelioma cells involves ETS transcription factors. Am J Pathol. 2009;175:2207–16. doi: 10.2353/ajpath.2009.090070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008;7:504–16. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- 42.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-azawi D, Ilroy MM, Kelly G, Redmond AM, Bane FT, Cocchiglia S, et al. Ets-2 and p160 proteins collaborate to regulate c-Myc in endocrine resistant breast cancer. Oncogene. 2008;27:3021–31. doi: 10.1038/sj.onc.1210964. [DOI] [PubMed] [Google Scholar]
- 44.Ge Y, LaFiura KM, Dombkowski AA, Chen Q, Payton SG, Buck SA, et al. The role of the proto-oncogene ETS2 in acute megakaryocytic leukemia biology and therapy. Leukemia. 2008;22:521–9. doi: 10.1038/sj.leu.2405066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito Y, Miyoshi E, Takeda T, Sakon M, Ihara S, Tsujimoto M, et al. Ets-2 overexpression contributes to progression of pancreatic adenocarcinoma. Oncol Rep. 2002;9:853–7. [PubMed] [Google Scholar]
- 46.Foos G, Garcia-Ramirez JJ, Galang CK, Hauser CA. Elevated expression of Ets2 or distinct portions of Ets2 can reverse Ras-mediated cellular transformation. J Biol Chem. 1998;273:18871–80. doi: 10.1074/jbc.273.30.18871. [DOI] [PubMed] [Google Scholar]
- 47.Hever A, Oshima RG, Hauser CA. Ets2 is not required for Ras or Neu/ErbB-2 mediated cellular transformation in vitro. Exp Cell Res. 2003;290:132–43. doi: 10.1016/s0014-4827(03)00315-x. [DOI] [PubMed] [Google Scholar]
- 48.Agathanggelou A, Bieche I, Ahmed-Choudhury J, Nicke B, Dammann R, Baksh S, et al. Identification of novel gene expression targets for the Ras association domain family 1 (RASSF1A) tumor suppressor gene in non-small cell lung cancer and neuroblastoma. Cancer Res. 2003;63:5344–51. [PMC free article] [PubMed] [Google Scholar]
- 49.Borczuk AC, Kim HK, Yegen HA, Friedman RA, Powell CA. Lung adenocarcinoma global profiling identifies type II transforming growth factor-beta receptor as a repressor of invasiveness. Am J Respir Crit Care Med. 2005;172:729–37. doi: 10.1164/rccm.200504-615OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.