Background: CD1 antigen-presenting proteins are recycled intracellularly via different pathways to sample diverse lipid antigen pools.
Results: The hydrophobic binding grooves of CD1b-e but not CD1a collapse without a bound lipid antigen.
Conclusion: CD1b-e are likely to be regulated allosterically by lipid transfer proteins not required by CD1a.
Significance: We present a testable hypothesis for the mechanism of lipid antigen recycling by CD1 that is consistent with molecular structure and physiology.
Keywords: Allosteric Regulation, Antigen Presentation, Cellular Immune Response, Lipid Binding Protein, Molecular Dynamics, Adaptive Immunity, Hydrophobic Collapse
Abstract
CD1 proteins mediate the presentation of endogenous and foreign lipids on the cell surface for recognition by T cell receptors. To sample a diverse antigen pool, CD1 proteins are repeatedly internalized and recycled, assisted, in some cases, by lipid transfer proteins such as saposins. The specificity of each CD1 isoform is, therefore, conferred in part by its intracellular pathway but also by distinct structural features of the antigen-binding domain. Crystal structures of CD1-lipid complexes reveal hydrophobic grooves and pockets within these binding domains that appear to be specialized for different lipids. However, the mechanism of lipid loading and release remains to be characterized. Here we gain insights into this mechanism through a meta-analysis of the five human CD1 isoforms, in the lipid-bound and lipid-free states, using all-atom molecular dynamics simulations. Strikingly, for isoforms CD1b through CD1e, our simulations show the near-complete collapse of the hydrophobic cavities in the absence of the antigen. This event results from the spontaneous closure of the binding domain entrance, flanked by two α-helices. Accordingly, we show that the anatomy of the binding cavities is restored if these α-helices are repositioned extrinsically, suggesting that helper proteins encountered during recycling facilitate lipid exchange allosterically. By contrast, we show that the binding cavity of CD1a is largely preserved in the unliganded state because of persistent electrostatic interactions that keep the portal α-helices at a constant separation. The robustness of this binding groove is consistent with the observation that lipid exchange in CD1a is not dependent on cellular internalization.
Introduction
As part of our immune defense, T cell receptors recognize antigen molecules presented by the MHC. These antigens are normally short peptide fragments originating either from foreign proteins (implying pathogenic infection) or from the host. However, it is now apparent that some T cells also recognize lipid antigens presented instead by a class of proteins known as CD1 (1–7). Members of the CD1 family, like MHC class I proteins, have a transmembrane heavy chain comprising three extracellular domains (α1, α2, and α3) that non-covalently bind to β2-microglobulin (β2m)3. The human family consists of five isoforms, named CD1a through CD1e, that differ in their intracellular localization and ligand specificity. CD1 members a-d function as lipid antigen-presenting proteins on cell membranes and are, therefore, recognized by T cell receptors. By contrast, CD1e seems to exist in a soluble form and has been proposed to function as an intracellular chaperone to assist lipid trafficking (1–7).
Crystal structures of CD1 proteins from several species have revealed the architecture of the antigen-binding domain (referred to as α1/α2). This consists of two antiparallel portal helices (helices α1 and α2) flanking the cavity entrance, sitting above a six-stranded antiparallel β-sheet platform (8–18). The α3 and β2m domains are positioned between the binding domain and the membrane (supplemental Fig. S1). The antigen-binding cavity in CD1 proteins is more voluminous and hydrophobic than those in the MHC and seems to be specialized for recognizing lipid hydrocarbon tails from a wide range of antigenic molecules, including mycolates, glycolipids, phospholipids, and lipopeptides (supplemental Fig. S2). Differences in ligand specificity between CD1 isoforms seem to result, at least in part, from the variability in the anatomy of the hydrophobic binding grooves, particularly in the so-called A′ and F′ pockets (supplemental Fig. S2). Additional isoform-characteristic subpockets and portals also seem to contribute to define the specificity of each binding domain (19).
Antigen specificity also results from the particular cellular localization and recycling of each CD1 isoform and their exposure to distinct pools of lipid antigen (20–22, 3, 5). Initially, nascent CD1 molecules fold together with β2m in the endoplasmic reticulum lumen, facilitated by chaperones such as calnexin and calreticulin, and by the presence of stabilizing endogenous lipid (23–27). Subsequently, CD1 molecules traffic to the cell surface before being internalized through the endocytic network, where they may exchange bound and free lipids prior to cycling back to the plasma membrane (20–22, 3, 5). CD1b, CD1c, and CD1d each contain a tyrosine-based targeting motif within their cytoplasmic tail that enables association with specific adaptor proteins and acts as a signal for intracellular sorting. As a result, CD1b and CD1d are routed to late endosomes and lysosomes, whereas CD1c is distributed throughout the endocytic system. In contrast, CD1a does not contain a tyrosine-based targeting motif, and, thus, its trafficking exclusively follows an early endosomal route (28–32).
Despite much progress in our understanding of CD1-mediated antigen processing, little is known about the mechanisms of lipid loading and exchange at the molecular level. The hydrophobic nature of the lipid ligands implies considerable challenges for traditional biophysical assays such as those used to study peptide-MHC interactions. Crystallographic structures have been enormously informative, but these only reveal the end result of the process of lipid recognition, leaving fundamental questions unanswered. For example, is the anatomy of the hydrophobic cavities an intrinsic characteristic of each isoform, or is lipid recognition more akin to an induced-fit process? Are these molecular mechanisms somehow related to the cellular localization of each isoform? It is worth noting that only late compartments contain glycolipid processing enzymes and lipid transfer proteins (LTPs) such as saposins, GM2-A, and apoE (33–36), and that the acidic pH of late endosomes regulates lipid exchange (37–39). By contrast, CD1a is able to exchange lipids even at the plasma membrane surface (40). Therefore, it is plausible that unique mechanisms for lipid exchange may have evolved for specific CD1 members. But how could LTPs or saposins influence the conformational mechanisms of lipid loading and exchange, beyond serving as lipid carriers?
To gain insights into these open mechanistic questions, we carried out a meta-analysis of the complete CD1 family using atomically detailed molecular simulations. We specifically analyzed the structural dynamics of all isoforms, both in the lipid-bound and unbound forms, with particular emphasis on the integrity of the hydrophobic cavities within the binding domain (supplemental Table S1). Our results reveal common patterns and intriguing differences among isoforms that correlate with their particular cellular compartmentalization and the specific amino acid sequence of the portal helices. Importantly, we verify that our results are consistent across a range of methodological choices, such as truncation of the β2m domain, the simulation force field, and the assumed extent of hydration of the empty hydrophobic cavities. The results obtained from this substantial computational effort, totaling over 3 μs of simulation time, permit us to formulate a clear-cut hypothesis for the mechanism of lipid loading by CD1 proteins and how helper proteins might assist this process allosterically.
COMPUTATIONAL PROCEDURES
Software Details
All simulations were performed at constant temperature and pressure, namely 298 K and 1 atm, respectively, using a periodic truncated octahedron box with a side length of 75 Å. A time step of 2 fs was used in all cases. Non-bonded pair lists were generated every 10 steps using a distance cutoff of 14 Å. A Lennard-Jones potential was used to model non-bonded van der Waals interactions, cut off at 12 Å. Electrostatic interactions were computed using the Particle-Mesh-Ewald method with a real space cutoff of 12 Å. The simulations were performed using NAMD (versions 2.6 and 2.7) (41) or GROMACS 4.5.5 (42). The simulations carried out with NAMD used the CHARMM27 protein/lipid force field (43, 44), the TIP3P water model, and a Langevin-based thermostat and barostat. Bonds to hydrogen atoms were constrained rigidly. The force fields used in the simulations carried out with GROMACS were AMBER99SB-ILDN (45) or OPLS-AA/L (46), both with the TIP3P water model. In this case, we used a velocity-rescale thermostat and the Parrinello-Rahman barostat. Bonds to hydrogen atoms were again constrained. The volumetric analysis of the binding cavities employed in-house software.
Simulation Systems
All simulation systems were prepared with CHARMM (47). The initial atomic coordinates for CD1a, CD1b, CD1c, CD1d, and CD1e were obtained from the Protein Data Bank entries 1ONQ (10), 2H26 (14), 3OV6 (17), 2AKR (13), and 3S6C (18), respectively. In CD1e, the missing residues 120–122 (a short β-loop outside of the binding cavity) were modeled using CHARMM. In CD1c, missing residues 87–89 (a loop connecting helix α1 to the β-sheet floor) were modeled and subsequently refined using MODELLER (48). All systems included a charge-neutralizing 150 mm NaCl buffer. The simulations of the truncated α1/α2 binding domains comprise ∼32,000 atoms, including ∼10,000 water molecules. Those of the complete α1–3/β2m include ∼54,000 atoms and ∼16,000 water molecules. The charge state of ionizable residues corresponds to neutral pH. The preparation of each system entailed a series of energy minimizations to remove steric clashes between protein, lipid, and solvent, followed by an equilibration phase during which the positions of atoms in the protein or protein-lipid complex (and in some cases, protein-bound water molecules) were gradually released from their initial configuration.
Lipid-bound Systems
In the lipid-bound systems, the coordinates of cocrystallized lipid and spacer molecules were preserved: namely, 3′ sulfated β-1-d-galactosylceramide in CD1a (10), endogenous phosphatidylcholine plus a tetracosyl-palmitate spacer in CD1b (14), mannosyl-β1-phosphomycoketide and a dodecane spacer in CD1c (17), and cis-tetracosenoyl-sulfatide in CD1d (13). Following a 1.5-ns equilibration phase, production simulations for each system were carried out for 100 ns.
Lipid-free Systems
To prepare the lipid-free systems, a set of constrained simulations was carried out initially to assess the most likely degree of hydration of each of the binding cavities, precluding any significant change in the protein backbone. At the start of these preparatory simulations, the binding cavities were either fully hydrated via superposition of a pre-equilibrated box of water molecules or completely empty. These constrained simulations were carried out for ∼30 ns each, converging to an equivalent degree of hydration irrespective of the starting point. The most frequently observed locations for water within the cavities of each binding domain were then identified, and a new set of simulations was prepared with water molecules at these positions. The total number of water molecules included was equal to the final hydration number of the preparatory simulations. A short constrained simulation of 1 ns was then carried out for each system to further optimize protein-water hydrogen bonds in this hypothetical configuration. This was the starting point for subsequent production simulations, 100 ns in length, with no structural constraints. These simulations were repeated three times for each binding domain.
To test the dependence of the results on the simulation force field, an independent set of 100-ns trajectories was calculated for lipid-free CD1a and CD1b either using AMBER99SB-ILDN or OPLS-AA/L and starting from the same point as the CHARMM27 simulations. In addition, a 100-ns control simulation of the complete α1–3/β2-microglobulin complex was carried out for lipid-free CD1b to evaluate whether the simulation results obtained for the isolated binding domain were representative.
Simulated Recovery of the CD1b Binding Groove
For lipid-free CD1b, a configuration at the end of one of the unconstrained 100-ns trajectories (with CHARMM27) was the starting point for an additional set of simulations in which the backbone of the portal helices was gradually forced to adopt the arrangement in the lipid-bound x-ray structure over the course of 50 ns. This procedure, carried out with the TMD module of NAMD, was repeated three times using force constants of 200, 600, and 1000 kcal mol/Å2. An additional 50-ns simulation was carried out subsequently to assess the structure of the binding groove while preserving the relative distance between the portal helices.
RESULTS
Experimental and Simulated Structures of CD1-Lipid Complexes Are Consistent
Because the main goal in this study was to assess the structural dynamics of the CD1 binding domains in the absence of lipid antigen, it was important to first establish whether our simulation framework produces results that are consistent with the available experimental data for the lipid-bound state. Therefore, we initially carried out a series of 100-ns control simulations of each CD1 binding domain in complex with a lipid antigen.
To quantify the similarity between crystallographic and simulated structures, we calculated the displacement of the protein backbone as a function of the simulation time. In all cases, the overall domain fold and the configuration of the main structural components, namely the β-sheet floor of the binding cavity and the two portal α-helices flanking its entrance, were highly consistent with the experimental structures. Histograms of the root mean square displacement time series typically peak at < 1 Å (supplemental Fig. S3). Such small deviations are simply due to entropic factors at room temperature (versus the cryogenic conditions in the crystals) and the inexact nature of the simulation framework. The only notable exception is helix α1 in CD1c, which drifts by 1.2 Å, most likely because of the fact that the adjacent loop was not resolved in the reference crystal structure and had to be modeled prior to the simulation (see “Computational Procedures”).
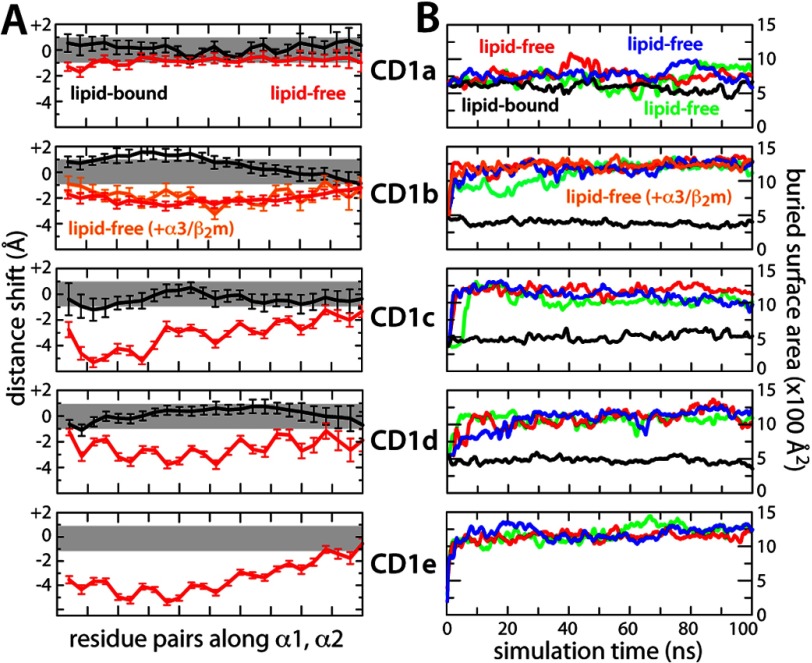
Consistent with the overall similarity of the protein backbone in the simulated and experimental structures, the openness of the binding cavity was also well preserved in the calculated trajectories. The change in openness relative to the crystal structures may be monitored by computing the distances between residue pairs along the two portal helices, α1 and α2. These distances did not change significantly, with a maximum shift of ± 1 Å (Fig. 1A). An alternative metric is the protein surface area buried between the two portal helices (excluding the lipid from the computation). In none of the simulations did the calculated area show a substantial drift over time. Instead, it fluctuated around ∼500 Å2, a value comparable with that for the x-ray structures (Fig. 1B).
FIGURE 1.
Displacement of the portal helices (α1 and α2) flanking the entrance of the antigen binding grooves in CD1a through CD1e, relative to the experimental crystal structures, in simulations of the lipid-bound (black lines) and lipid-free (red lines) states of each binding domain. A, the displacement of the helices is quantified by monitoring the distance between pairs of residues facing each other along the length of the helices. For CD1b, data are also shown for the complete α1–3/β2m complex in the lipid-free state. For CD1e, only the lipid-free state was simulated because no lipids were resolved in the available crystal structure. The values plotted are time averages over the final 50 ns of the simulations. For the lipid-free states, global averages are obtained from three independent simulations. Error bars show mean ± S.D. The gray band indicates variations of ±1 Å to be expected at room temperature. B, protein surface area buried between α1 and α2, as a function of simulation time, for the lipid-bound (black) and lipid-free states (colors).
Finally, and importantly, we observed that the distinct binding cavity anatomies of each of the CD1 isoforms were largely preserved during the simulations of the lipid-bound forms (Fig. 2). It should be noted that the distinct size and shape of these hydrophobic cavities are conferred by the precise arrangement of key amino acid side chains lining the pocket and tunnels therein and not only by the overall protein fold. We show, therefore, that this precise arrangement is also largely sustained on the simulation time scale in the presence of the lipid. In sum, none of these control calculations reveal a systematic deficiency of our simulation framework, suggesting that it may also provide meaningful insights into the characteristics of the lipid-free state.
FIGURE 2.
Persistence or collapse of the antigen binding grooves in simulations of lipid-bound and lipid-free CD1 isoforms relative to the experimental crystal structures. The anatomy of the binding grooves are depicted at 20-ns intervals throughout the simulations. This anatomy is represented graphically by the surface encompassing the volume of the various pockets and tunnels, as calculated with a grid-based search method. The simulation systems analyzed are the same as in Fig. 1.
Collapse of the Binding Cavities in the Lipid-free State with a Notable Exception
As mentioned above, the presentation of non-self antigens mediated by CD1 in vivo eventually entails the removal of endogenous lipids carried by these proteins from the endoplasmic reticulum lumen. At the molecular scale, any realistic mechanism of lipid exchange necessarily implies a transient evacuation of the hydrophobic grooves within the binding domain. On the basis of the available x-ray structures of CD1-lipid complexes, the shape and size of the binding cavities appear to be perfectly complementary to the bound lipids. However, an open question is whether the anatomy of these binding cavities is preserved in the lipid-free state.
To address this question in the most conservative way, we first designed a set of simulations under the assumed premise that the removal of the lipid ligand does not result in significant changes in the conformation of the proteins. In addition, we assumed that water molecules would occupy the binding cavities. To determine the idealized hydration number for each CD1 isoform, we carried out two sets of simulations in which the binding cavities were initially prepared either entirely empty or entirely full of solvent molecules, whereas the protein backbone was constrained to the lipid-bound configuration. These simulations showed a dynamic flux of water in and out of the hydrophobic cavities, converging to a stable hydration number after ∼15 ns irrespective of the hydration state imposed at the start (supplemental Fig. S4). The number of bound water molecules after 30 ns of simulation was ∼30 for CD1a, b, c, and e and ∼15 for CD1d (supplemental Fig. S4). These water molecules were distributed primarily within the F′ pocket. In CD1d, this pocket is rather shallow (∼1000 Å3) compared with the other CD1 isoforms (∼1200–1600 Å3) (19), consistent with the smaller hydration number.
The results of this preparatory set of calculations were the basis for a second series of simulations where, instead, we investigated the response of the protein structure to the removal of the lipid antigen. Specifically, we calculated triplicates of 100-ns trajectories for each of the CD1 isoforms without any restriction on the protein dynamics. At the start of these simulations, the hydrophobic cavities were not empty but, in each case, included the number of bound water molecules derived from the previous analysis, located at the positions observed most frequently.
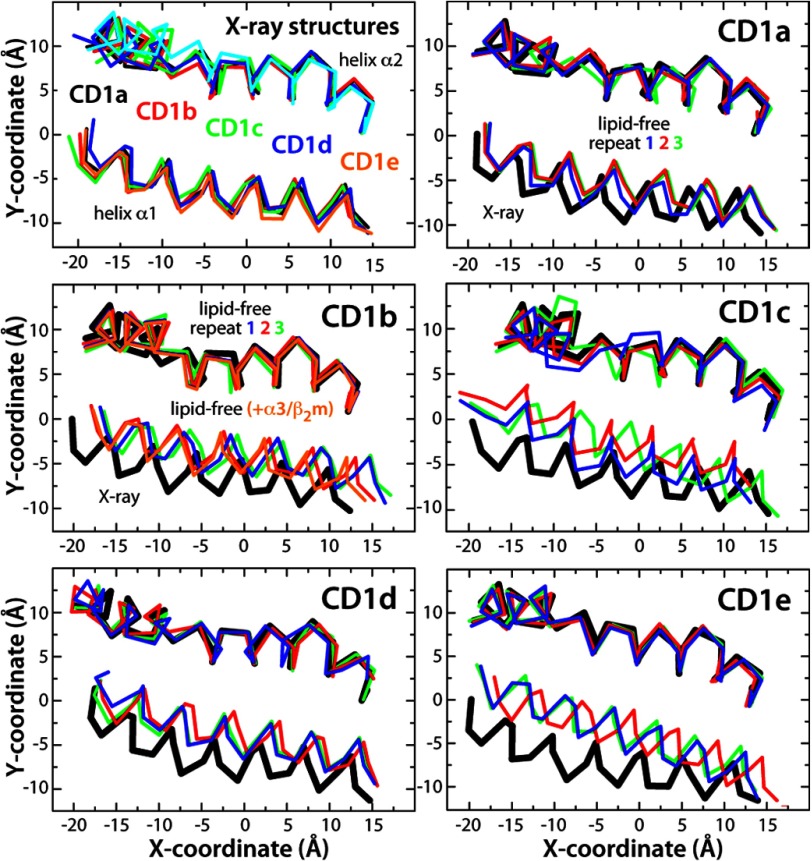
This conservative procedure notwithstanding, these unconstrained simulations revealed pronounced structural changes in the binding domains of CD1b through CD1e compared with the equivalent calculations for the lipid-bound state (supplemental Fig. S3). These changes reflect primarily the relative drift of the portal helices α1 and α2, which led to varying degrees of closure of the cavity entrance (Figs. 1A and 3). The helices closed by up to ∼3 Å in CD1b and by up to ∼4 Å along most of the length of CD1d. In CD1c, most of the space between helices α1 and α2 also closed by ∼4 Å and by up to ∼6 Å at the entrance of the F′ pocket, indicating that this pocket might be occupied by unresolved ligands in the crystal structure of the lipid-bound state (17). Similarly, for CD1e, we observed that much of the binding domain entrance closed by ∼4–6 Å. Consistent with these results, in all cases we calculated an additional ∼750–1000 Å2 of buried protein surface between helices α1 and α2 compared with the lipid-bound states (Fig. 1B).
FIGURE 3.
Configuration of the portal helices α1 and α2 in the x-ray structures of CD1a through CD1e compared with the lipid-free simulations. The plots show a two-dimensional projection of the Cα-trace of the helices viewed perpendicularly to the floor of the binding cavity. In each case, the atomic coordinates plotted are time averages over the last 50 ns of each repeat simulation. The S.D. around these averages are omitted because these are typically smaller than the differences between repeats.
Strikingly, these changes in CD1b, CD1c, CD1d, and CD1e were associated with a near-complete collapse of their internal hydrophobic cavity (Fig. 2). The features of the majority of the F′ pocket and connecting tunnels were lost in the absence of bound lipid. Only disconnected portions of the A′ pocket were partially maintained, apparently because of a conserved disulfide bridge within this pocket. Concurrent with the collapse of the binding cavities, most of the water molecules that were initially bound returned to the bulk solvent (supplemental Figs. S5 and S6).
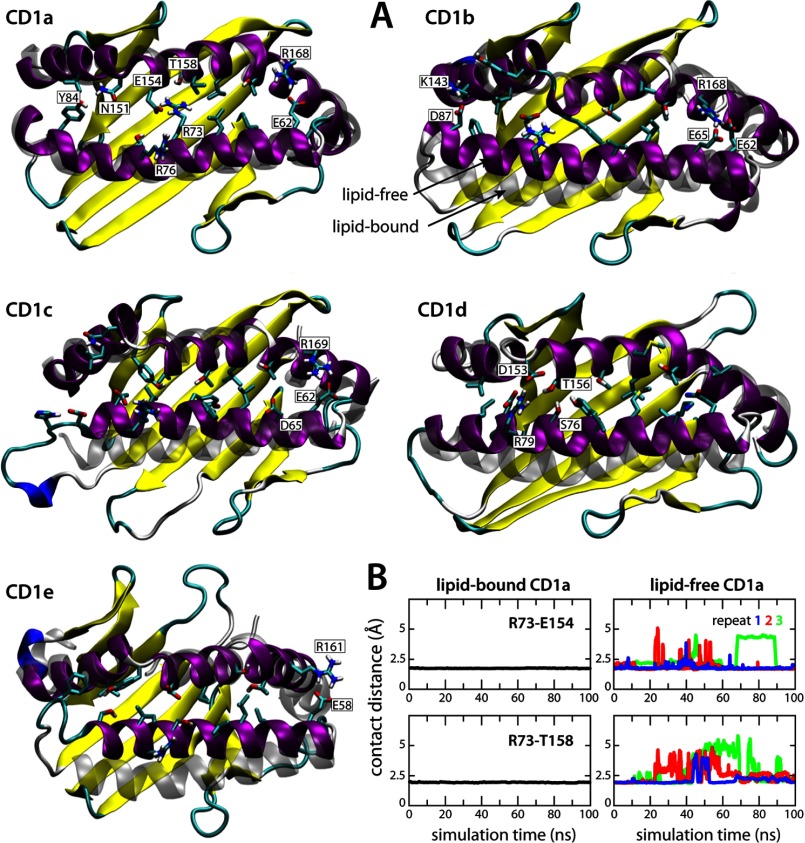
Interestingly, very limited changes were observed for CD1a. The entrance of its binding cavity closed only slightly in the lipid-free simulations, by about 1 Å (Figs. 1A and 3), whereas the overall shape and interconnectivity of the hydrophobic pockets was largely maintained (Fig. 2) and hydrated significantly (supplemental Figs. S5 and S6). The distinct structural stability of CD1a seems to result principally from the interaction between Arg-73 and Glu-154, positioned midway along the length of the portal helices α1 and α2, which tethers them at a constant separation (Fig. 4). Stable helix-helix interactions were also observed in other isoforms, e.g. above the edge of the A′ pocket of CD1b, CD1c, and CD1e, but these appear to be too peripheral to prevent the closure of the cavity entrance (Fig. 4).
FIGURE 4.
Side chain interactions sustain the entrance of CD1a but not CD1b–e in the absence of a lipid antigen. A, final simulation snapshot of each CD1 binding domain in the lipid-free state. Each domain is represented as a schematic and colored according to the secondary structure. The configurations of helices α1 and α2 in the respective crystal structures are overlaid (fitted to the α2 helix backbone) and shown in transparent gray. Side chains of key residues in α1 and α2 are labeled. The surrounding solvent molecules included in the simulation are omitted for clarity. B, persistence of the electrostatic interactions across the center of the entrance to the binding groove in CD1a in the lipid-bound and lipid-free simulations.
Simulation Results Are Robust to Choice of Force Field and Molecular Construct
To assess the reproducibility of our observations, we repeated the lipid-free simulations of CD1a and CD1b using two different protein force fields, namely the all-atom AMBER99SB-ILDN (45) and OPLS-AA/L (46) parameter sets. Although these force fields are based upon the same principles as CHARMM27, the specific parameterization of bonded and non-bonded interactions is significantly different. Nevertheless, we obtained comparable results for all force fields. The portal mouth and cavity of CD1a were stable in the absence of lipid, whereas the simulation of CD1b exhibited a drastic collapse of the binding cavity and near-complete dehydration (supplemental Fig. S7).
For CD1b, which reveals the most dramatic changes, we also simulated the complete soluble portion of the complex, i.e. including the α3 domain and β2-microglobulin in addition to the α1/α2 binding domain. β2-microglobulin mostly interacts with the β-sheet floor of the binding domain, but the α3 domain is connected to the portal helix α2 (supplemental Fig. S1), and therefore it is reasonable to ask whether the results for our truncated construct are representative of the entire complex. Nevertheless, the results for the complete CD1b complex were again comparable with those from the simulations of the binding domain only (Figs. 1–3 and supplemental Fig. S5). We thus conclude that the collapse of the binding cavity upon removal of the lipid antigen is not due to either the protein construct considered in the simulations or the particular force field used.
The Anatomy of Hydrophobic Cavities Is Restored by Repositioning of the Portal Helices
The irreversible collapse of the binding cavities in the lipid-free state implies that CD1 proteins, once folded, are by themselves largely unable to load lipid antigens (except CD1a). Our results also reveal a clear correlation between the displacement of the portal helices α1 and α2 and the collapse of the hydrophobic grooves. Because the portal helices are likely to participate in interactions with helper proteins such as saposins or LTPs, we hypothesized that these protein partners might not only deliver a lipid ligand but also induce or regulate the opening of the binding cavity entrance, as has been proposed for MHC proteins (49–51). Even so, the key question is whether this hypothetical reopening of the cavity entrance would be sufficient to restore the size and shape of the hydrophobic pockets. It should be noted that the anatomy of the binding grooves is defined by the specific conformations of the lining amino acid side chains and not only by arrangements of the protein backbone.
To assess this hypothesis, we carried out an additional series of simulations of lipid-free CD1b in which the portal helices α1 and α2 are gradually repositioned at a distance similar to that observed in the lipid-bound state over a 50-ns time-window (see “Computational Procedures”) without directly interfering with the dynamics of the protein side chains. The simulations were then extended by a further 50 ns, keeping the distance between the portal helices constant (supplemental Fig. S8). As mentioned, CD1b features the most intricate network of binding pockets among all CD1 isoforms, and thus we reasoned that this would be the most challenging case. Nonetheless, we observed that significant stretches of the F′ and A′ pockets, and intervening connecting tunnels, were recovered rapidly (Fig. 5). By the end of the calculated trajectory, much of the anatomy of the binding cavity was restored, except for a slight contraction, comparable with what we observed for lipid-free CD1a (Fig. 2). Thus, we conclude that by regulating the displacement of the portal helices flanking the entrance of binding groove, helper proteins found in the recycling pathway may restore the ability of lipid-free CD1 proteins to load lipid antigens with specific features.
FIGURE 5.
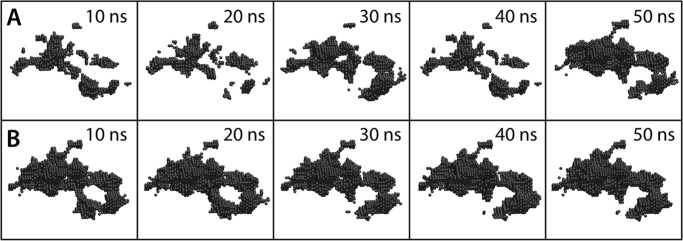
Near-complete recovery of the anatomy of the CD1b binding groove upon repositioning of the portal helices α1 and α2 to the lipid-bound configuration, starting from the collapsed state depicted in Fig. 4. The displacement of the helices is induced progressively over a 50-ns targeted simulation (A) in which biasing forces act exclusively on the backbone atoms of the helices (k = 600 kcal mol/Å2). The resulting configuration was then sustained for another 50 ns (B). The hydrophobic groove is represented graphically by the surface encompassing the volume of the various pockets and tunnels therein, calculated as in Fig. 2. The recovery is shown in 10-ns intervals. The anatomy of the binding groove is largely restored, despite the fact that the simulation does not impose a specific configuration on the protein side chains.
DISCUSSION
The findings presented in this study underscore the notion that the lipid antigen specificity of each CD1 isoform is in part conferred by the anatomy of the hydrophobic cavities within their binding domain (5). This is not to say, however, that lipid recognition proceeds according to a simple lock-and-key mechanism, as could be inferred from inspection of available structural data. On the contrary, we have shown that in the lipid-free state, which is a necessary intermediate during antigen processing in vivo, the binding cavities collapse irreversibly (except for CD1a). Thus, reloading of lipid antigens, which are ultimately presented on the cell surface, must proceed according to an induced-fit mechanism. Nevertheless, we cannot envisage that it is the lipid ligand itself that induces the formation of a suitable binding cavity. The observation that the entrance of these cavities is almost entirely closed in the lipid-free state makes such a mechanism unlikely. Rather, we propose that helper proteins involved in lipid trafficking, such as LTPs or saposins, will not only function as carriers for potential lipid antigens but also regulate the arrangement of the two α-helices flanking the portal of the binding cavity. Our simulations indicate that, by inducing a sufficient separation between them, much of the anatomy of hydrophobic pockets and interconnecting tunnels within the binding domain can be restored prior to lipid binding. Conditions of low pH in late endosomes or lysosomes may also facilitate this process of lipid exchange, e.g. for CD1c (39), possibly by weakening ionic interactions that lock the protein in the lipid-bound form.
In contrast to other isoforms, CD1a is not exposed to acidic pH, glycolipid processing enzymes, or LTPs, and, hence, is likely to have evolved a distinctive mechanism for exchanging lipid ligands. Indeed, CD1a is able to exchange lipids while on the cell surface, without internalization (40). It was proposed previously that the shallow, exposed F′ pocket of CD1a may facilitate this process (2). Our simulation data indeed indicate that the CD1a binding domain is distinct in that it is constrained to a bound-like conformation even in the absence of a bound lipid. Several salt bridges, and particularly that between Arg-73 and Glu-154, help to tether the portal helices and prevent the collapse of the hydrophobic binding cavity, which remains significantly hydrated. It should be noted that both Arg-73 and Glu-154 can be expected to be ionized under the neutral pH conditions in the plasma membrane or the early endocytic pathway in which CD1a is localized. Although, evidently, our simulations do not reach physiological time scales, the reproducibility of our observations in independent trajectories and using different simulation force fields leads us to conclude that CD1a is indeed more conformationally stable than the other isoforms. Lipid exchange in CD1a may, therefore, proceed unassisted by helper proteins or acidic pH conditions. Indeed, the key salt bridge within the roof of the F′ pocket contributes to coordinate the head group of the sulfated galactose lipid bound in the x-ray structure of CD1a (10).
It is worth noting that our results lend support to the view that existing crystal structures of putatively empty CD1 isoforms (11, 18), which resemble those in the lipid-bound state, may be either influenced by crystallographic contacts and/or include bound hydrophobic molecules that are too disordered to be resolved clearly by x-ray diffraction. The latter possibility would be consistent with the weak electron densities observed within the binding grooves of CD1e (18) and mouse CD1d (11) as well as in the F′ pocket of CD1c (17). Contacts mediated by both portal helices α1 and α2 are also apparent in the crystal lattices of CD1e and mouse CD1d. According to our simulations, these structures are unlikely to represent the unliganded conformation.
Finally, we note that previous simulation studies of MHC proteins have indicated some interesting parallels to this work (49–51). In particular, a tapasin-dependent MHC class I protein was observed to adopt a non-receptive form in the peptide-free state induced by a distortion in the arrangement of the helices flanking the binding site equivalent to α1 and α2 in CD1. Conversely, a tapasin-independent MHC allele was found to be more prone to sustain the peptide-bound configuration in the absence of the peptide antigen. Thus, the allosteric regulation of conformationally dynamic ligand-binding pockets appears to be a common feature among antigen-presenting immunological proteins.
Supplementary Material
Acknowledgments
We thank the Rechenzentrum Garching of the Max Planck Society for computational resources, as well as the Jülich Supercomputing Center, the Darwin Supercomputer of the University of Cambridge, and the Swiss National Supercomputing Center via DECI/PRACE-2IP.
This work was supported in part by German Research Foundation Cluster of Excellence Grant EXC115 (to J. D. F. G.) and by an IMPReS fellowship of the Max Planck Institute of Biophysics (to D. G.).

This article contains supplemental Figs. S1–S8 and Table S1.
- β2m
- β2-microglobulin
- LTP
- lipid transfer protein.
REFERENCES
- 1. Porcelli S. A., Segelke B. W., Sugita M., Wilson I. A., Brenner M. B. (1998) The CD1 family of lipid antigen-presenting molecules. Immunol. Today 19, 362–368 [DOI] [PubMed] [Google Scholar]
- 2. Moody D. B., Zajonc D. M., Wilson I. A. (2005) Anatomy of CD1-lipid antigen complexes. Nat. Rev. Immunol. 5, 387–399 [DOI] [PubMed] [Google Scholar]
- 3. Barral D. C., Brenner M. B. (2007) CD1 antigen presentation. How it works. Nat. Rev. Immunol. 7, 929–941 [DOI] [PubMed] [Google Scholar]
- 4. Zajonc D. M., Wilson I. A. (2007) Architecture of CD1 proteins. Curr. Top. Microbiol. Immunol. 314, 27–50 [DOI] [PubMed] [Google Scholar]
- 5. Silk J. D., Salio M., Brown J., Jones E. Y., Cerundolo V. (2008) Structural and functional aspects of lipid binding by CD1 molecules. Annu. Rev. Cell Dev. Biol. 24, 369–395 [DOI] [PubMed] [Google Scholar]
- 6. Salio M., Silk J. D., Cerundolo V. (2010) Recent advances in processing and presentation of CD1 bound lipid antigens. Curr. Opin. Immunol. 22, 81–88 [DOI] [PubMed] [Google Scholar]
- 7. De Libero G., Mori L. (2012) Novel insights into lipid antigen presentation. Trends Immunol. 33, 103–111 [DOI] [PubMed] [Google Scholar]
- 8. Zeng Z., Castaño A. R., Segelke B. W., Stura E. A., Peterson P. A., Wilson I. A. (1997) Crystal structure of mouse CD1. An MHC-like fold with a large hydrophobic binding groove. Science 277, 339–345 [DOI] [PubMed] [Google Scholar]
- 9. Gadola S. D., Zaccai N. R., Harlos K., Shepherd D., Castro-Palomino J. C., Ritter G., Schmidt R. R., Jones E. Y., Cerundolo V. (2002) Structure of human CD1b with bound ligands at 2.3 Å, a maze for alkyl chains. Nat. Immunol. 3, 721–726 [DOI] [PubMed] [Google Scholar]
- 10. Zajonc D. M., Elsliger M. A., Teyton L., Wilson I. A. (2003) Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 Å. Nat. Immunol. 4, 808–815 [DOI] [PubMed] [Google Scholar]
- 11. Koch M., Stronge V. S., Shepherd D., Gadola S. D., Mathew B., Ritter G., Fersht A. R., Besra G. S., Schmidt R. R., Jones E. Y., Cerundolo V. (2005) The crystal structure of human CD1d with and without α-galactosylceramide. Nat. Immunol. 6, 819–826 [DOI] [PubMed] [Google Scholar]
- 12. Zajonc D. M., Crispin M. D., Bowden T. A., Young D. C., Cheng T. Y., Hu J., Costello C. E., Rudd P. M., Dwek R. A., Miller M. J., Brenner M. B., Moody D. B., Wilson I. A. (2005) Molecular mechanism of lipopeptide presentation by CD1a. Immunity 22, 209–219 [DOI] [PubMed] [Google Scholar]
- 13. Zajonc D. M., Maricic I., Wu D., Halder R., Roy K., Wong C. H., Kumar V., Wilson I. A. (2005) Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J. Exp. Med. 202, 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia-Alles L. F., Versluis K., Maveyraud L., Vallina A. T., Sansano S., Bello N. F., Gober H. J., Guillet V., de la Salle H., Puzo G., Mori L., Heck A. J., De Libero G., Mourey L. (2006) Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. EMBO J. 25, 3684–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zajonc D. M., Striegl H., Dascher C. C., Wilson I. A. (2008) The crystal structure of avian CD1 reveals a smaller, more primordial antigen-binding pocket compared to mammalian CD1. Proc. Natl. Acad. Sci. U.S.A. 105, 17925–17930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Girardi E., Wang J., Mac T. T., Versluis C., Bhowruth V., Besra G., Heck A. J., Van Rhijn I., Zajonc D. M. (2010) Crystal structure of bovine CD1b3 with endogenously bound ligands. J. Immunol. 185, 376–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scharf L., Li N. S., Hawk A. J., Garzón D., Zhang T., Fox L. M., Kazen A. R., Shah S., Haddadian E. J., Gumperz J. E., Saghatelian A., Faraldo-Gómez J. D., Meredith S. C., Piccirilli J. A., Adams E. J. (2010) The 2.5 Å structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity 33, 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garcia-Alles L. F., Giacometti G., Versluis C., Maveyraud L., de Paepe D., Guiard J., Tranier S., Gilleron M., Prandi J., Hanau D., Heck A. J., Mori L., De Libero G., Puzo G., Mourey L., de la Salle H. (2011) Crystal structure of human CD1e reveals a groove suited for lipid-exchange processes. Proc. Natl. Acad. Sci. U.S.A. 108, 13230–13235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garzón D., Bond P. J., Faraldo-Gómez J. D. (2009) Predicted structural basis for CD1c presentation of mycobacterial branched polyketides and long lipopeptide antigens. Mol. Immunol. 47, 253–260 [DOI] [PubMed] [Google Scholar]
- 20. Moody D. B., Porcelli S. A. (2001) CD1 trafficking: invariant chain gives a new twist to the tale. Immunity 15, 861–865 [DOI] [PubMed] [Google Scholar]
- 21. Briken V., Jackman R. M., Dasgupta S., Hoening S., Porcelli S. A. (2002) Intracellular trafficking pathway of newly synthesized CD1b molecules. EMBO J. 21, 825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Libero G., Mori L. (2005) Recognition of lipid antigens by T cells. Nat. Rev. Immunol. 5, 485–496 [DOI] [PubMed] [Google Scholar]
- 23. Brutkiewicz R. R., Bennink J. R., Yewdell J. W., Bendelac A. (1995) TAP-independent, β 2-microglobulin-dependent surface expression of functional mouse CD1.1. J. Exp. Med. 182, 1913–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bauer A., Hüttinger R., Staffler G., Hansmann C., Schmidt W., Majdic O., Knapp W., Stockinger H. (1997) Analysis of the requirement for β 2-microglobulin for expression and formation of human CD1 antigens. Eur. J. Immunol. 27, 1366–1373 [DOI] [PubMed] [Google Scholar]
- 25. Sugita M., Porcelli S. A., Brenner M. B. (1997) Assembly and retention of CD1b heavy chains in the endoplasmic reticulum. J. Immunol. 159, 2358–2365 [PubMed] [Google Scholar]
- 26. Hüttinger R., Staffler G., Majdic O., Stockinger H. (1999) Analysis of the early biogenesis of CD1b. Involvement of the chaperones calnexin and calreticulin, the proteasome and β(2)-microglobulin. Int. Immunol. 11, 1615–1623 [DOI] [PubMed] [Google Scholar]
- 27. Kang S. J., Cresswell P. (2002) Calnexin, calreticulin, and ERp57 cooperate in disulfide bond formation in human CD1d heavy chain. J. Biol. Chem. 277, 44838–44844 [DOI] [PubMed] [Google Scholar]
- 28. Sugita M., Jackman R. M., van Donselaar E., Behar S. M., Rogers R. A., Peters P. J., Brenner M. B., Porcelli S. A. (1996) Cytoplasmic tail-dependent localization of CD1b antigen-presenting molecules to MIICs. Science 273, 349–352 [DOI] [PubMed] [Google Scholar]
- 29. Sugita M., Peters P. J., Brenner M. B. (2000) Pathways for lipid antigen presentation by CD1 molecules. Nowhere for intracellular pathogens to hide. Traffic 1, 295–300 [DOI] [PubMed] [Google Scholar]
- 30. Salamero J., Bausinger H., Mommaas A. M., Lipsker D., Proamer F., Cazenave J. P., Goud B., de la Salle H., Hanau D. (2001) CD1a molecules traffic through the early recycling endosomal pathway in human Langerhans cells. J. Invest. Dermatol. 116, 401–408 [DOI] [PubMed] [Google Scholar]
- 31. Sugita M., Cernadas M., Brenner M. B. (2004) New insights into pathways for CD1-mediated antigen presentation. Curr. Opin. Immunol. 16, 90–95 [DOI] [PubMed] [Google Scholar]
- 32. Cernadas M., Cavallari M., Watts G., Mori L., De Libero G., Brenner M. B. (2010) Early recycling compartment trafficking of CD1a is essential for its intersection and presentation of lipid antigens. J. Immunol. 184, 1235–1241 [DOI] [PubMed] [Google Scholar]
- 33. Kang S. J., Cresswell P. (2004) Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat. Immunol. 5, 175–181 [DOI] [PubMed] [Google Scholar]
- 34. Winau F., Schwierzeck V., Hurwitz R., Remmel N., Sieling P. A., Modlin R. L., Porcelli S. A., Brinkmann V., Sugita M., Sandhoff K., Kaufmann S. H., Schaible U. E. (2004) Saposin C is required for lipid presentation by human CD1b. Nat. Immunol. 5, 169–174 [DOI] [PubMed] [Google Scholar]
- 35. Zhou D., Cantu C., 3rd, Sagiv Y., Schrantz N., Kulkarni A. B., Qi X., Mahuran D. J., Morales C. R., Grabowski G. A., Benlagha K., Savage P., Bendelac A., Teyton L. (2004) Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science 303, 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan W., Qi X., Tsang P., Kang S. J., Illarionov P. A., Illaniorov P. A., Besra G. S., Gumperz J., Cresswell P. (2007) Saposin B is the dominant saposin that facilitates lipid binding to human CD1d molecules. Proc. Natl. Acad. Sci. U.S.A. 104, 5551–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ernst W. A., Maher J., Cho S., Niazi K. R., Chatterjee D., Moody D. B., Besra G. S., Watanabe Y., Jensen P. E., Porcelli S. A., Kronenberg M., Modlin R. L. (1998) Molecular interaction of CD1b with lipoglycan antigens. Immunity 8, 331–340 [DOI] [PubMed] [Google Scholar]
- 38. Cheng T. Y., Relloso M., Van Rhijn I., Young D. C., Besra G. S., Briken V., Zajonc D. M., Wilson I. A., Porcelli S., Moody D. B. (2006) Role of lipid trimming and CD1 groove size in cellular antigen presentation. EMBO J. 25, 2989–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Relloso M., Cheng T. Y., Im J. S., Parisini E., Roura-Mir C., DeBono C., Zajonc D. M., Murga L. F., Ondrechen M. J., Wilson I. A., Porcelli S. A., Moody D. B. (2008) pH-dependent interdomain tethers of CD1b regulate its antigen capture. Immunity 28, 774–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Manolova V., Kistowska M., Paoletti S., Baltariu G. M., Bausinger H., Hanau D., Mori L., De Libero G. (2006) Functional CD1a is stabilized by exogenous lipids. Eur. J. Immunol. 36, 1083–1092 [DOI] [PubMed] [Google Scholar]
- 41. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hess B., Kutzner C., Van der Spoel D., Lindahl E. (2008) GROMACS 4. Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 43. Feller S. E., Yin D., Pastor R. W., MacKerell A. D., Jr. (1997) Molecular dynamics simulation of unsaturated lipid bilayers at low hydration: parameterization and comparison with diffraction studies. Biophys. J. 73, 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. MacKerell A. D., Bashford D., Bellott, Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 45. Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J. L., Dror R. O., Shaw D. E. (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kaminski G. A., Friesner R. A., Tirado-Rives J., Jorgensen W. L. (2001) Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B 105, 6474–6487 [Google Scholar]
- 47. Brooks B. R., Brooks C. L., Mackerell A. D., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) CHARMM: The biomolecular simulation program. J. Comp. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2006) Comparative protein structure modeling using MODELLER. Curr. Protoc. Prot. Sci. 50, 2.9.1–2.9.31 [DOI] [PubMed] [Google Scholar]
- 49. Sieker F., Springer S., Zacharias M. (2007) Comparative molecular dynamics analysis of tapasin-dependent and -independent MHC class I alleles. Protein Sci. 16, 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sieker F., Straatsma T. P., Springer S., Zacharias M. (2008) Differential tapasin dependence of MHC class I molecules correlates with conformational changes upon peptide dissociation. A molecular dynamics simulation study. Mol. Immunol. 45, 3714–3722 [DOI] [PubMed] [Google Scholar]
- 51. Garstka M. A., Fritzsche S., Lenart I., Hein Z., Jankevicius G., Boyle L. H., Elliott T., Trowsdale J., Antoniou A. N., Zacharias M., Springer S. (2011) Tapasin dependence of major histocompatibility complex class I molecules correlates with their conformational flexibility. FASEB J. 25, 3989–3998 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.