Abstract
The V617F mutation in the Jak2 pseudokinase domain causes myeloproliferative neoplasms, and the equivalent mutation in Jak1 (V658F) is found in T-cell leukemias. Crystal structures of wild type and V658F mutant human Jak1 pseudokinase reveal a conformational switch that remodels a linker segment encoded by exon 12, which is also a site of mutations in Jak2. This switch is required for V617F-mediated Jak2 activation, and possibly for physiologic Jak activation.
The receptors for diverse cytokines, including erythropoietin (EPO), growth hormone, interleukins and interferons, initiate intracellular signaling via one or more members of the Jak family of non-receptor tyrosine kinases1,2. Jak kinases contain a FERM domain, an SH2-like domain, and a pseudokinase domain (also referred to as the Jak homology 2, or JH2 domain) adjacent to their C-terminal tyrosine kinase domain (Fig. 1a). The pseudokinase domain has a protein kinase fold, but key catalytic residues are not conserved and the domain is thought to inhibit the adjacent tyrosine kinase domain, perhaps via an intramolecular interaction3. Somatic gain-of-function mutations in Jaks underlie a number of hematologic malignancies4, and the pseudokinase domain is the most frequent site of these activating mutations5. In particular, the V617F mutation in the Jak2 pseudokinase is found in ∼95% of patients with polycythemia vera (PV)6-9, a myeloproliferative neoplasm (MPN) characterized by EPO-independent overproduction of red blood cells due to constitutive signaling from Jak2 V617F in complex with the erythropoietin receptor (EPO-R)10. The corresponding mutation in Jak1 (V658F) has been identified in T-cell acute lymphoblastic leukemia11 and also leads to constitutive catalytic activation12. A subset of PV patients who are V617F-negative have mutations in exon 12 of Jak213,14, which encodes a portion of the polypeptide chain that links the SH2-like and pseudokinase domains (SH2-PK linker).
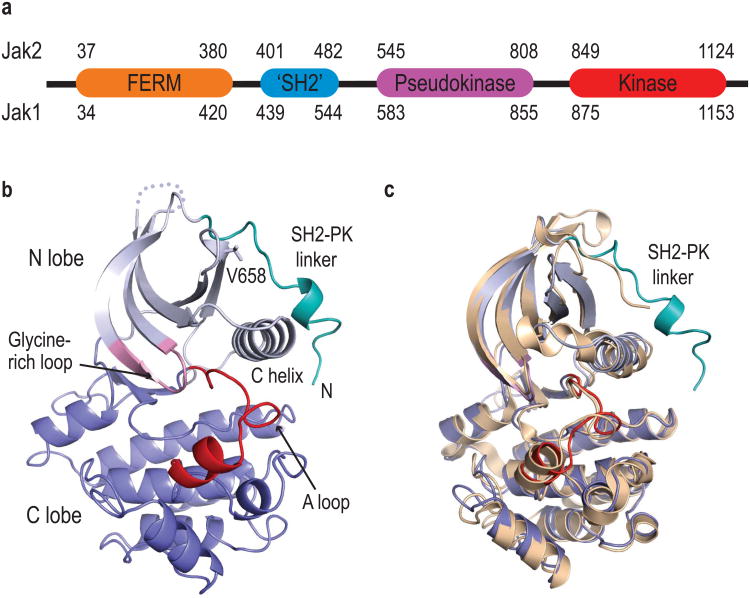
Figure 1.
Structure of the Jak1 pseudokinase domain. (a) The domain structure of Jak family kinases. (b) Structure of the Jak1 pseudokinase domain, with key structural motifs indicated. (c) The Jak1 pseudokinase domain is superimposed on the structure of Jak2 pseudokinase domain (tan).
To better understand the pseudokinase domain and the effects of pathogenic mutations, we determined crystal structures of a wild type and V658F mutant fragment of human Jak1 spanning the pseudokinase domain and a segment of the SH2-PK linker (residues 561-852). The wild type and mutant structures were refined at resolutions of 1.8Å and 1.9Å, respectively (see Supplementary Table 1 and Methods online). The overall architecture of the pseudokinase domain closely resembles that of typical tyrosine kinases, with an N-terminal lobe composed of five β-strands and the C-helix, and a larger C-terminal lobe (Fig. 1b). Several features of the pseudokinase domain appear to be inconsistent with phosphotransfer activity. In particular, the region corresponding to the kinase activation loop adopts a well-defined conformation that is expected to preclude binding of polypeptide substrates (Supplementary Fig. 1). At the N-terminus of the domain, the SH2-PK linker adopts a partially α-helical conformation and extends across the N-lobe, roughly perpendicular to the C-helix. The Jak1 domain superimposes well with a recently reported structure of the Jak2 pseudokinase15 (RMSD 1.26 Å for 261 equivalent residues, Fig. 1c), but the Jak2 structure lacks the SH2-PK linker, which was truncated in the crystallized construct.
Val658 lies within the N-lobe of the domain, at the end of strand β4. In the V658F mutant, the β4-β5 loop folds inward toward the C-helix, and the mutant Phe658 packs in an edge-to-face manner with Phe636 in the C-helix (Fig. 2a). Strikingly, the phenyl ring of Phe658 occupies precisely the same position occupied by that of Phe575 in the wild type structure. Phe575 lies in the SH2-PK linker, and the V658F mutation induces a coordinated rearrangement of this segment. In the mutant, Phe575 is shifted by ∼10Å and becomes solvent-exposed. Both the V658F and wild type crystals contain two molecules in the asymmetric unit, and interestingly, we observe essentially the same rearrangement in molecule “B” in the wild type structure that we observe in the V658F mutant, demonstrating that the wild type pseudokinase domain can also adopt the Phe575-flipped conformation (Fig. 2b). In the V658F mutant structure, both the A and B molecules adopt the “flipped” conformation with Phe575 exposed. The electron density for the linker segments is clear and continuous in both the wild type and V658F structures (Supplementary Fig. 2a, b).
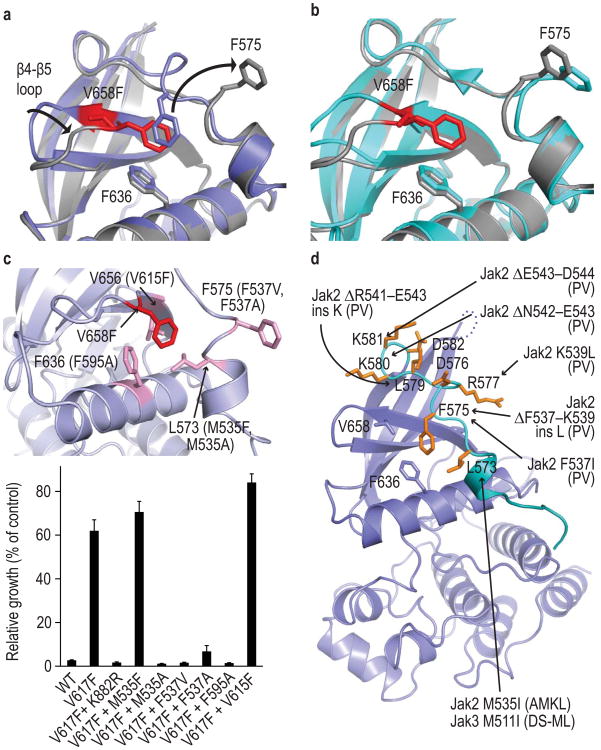
Figure 2.
Crystal structure of the V658F mutant Jak1 pseudokinase domain. (a) Superposition of wild-type Jak1 pseudokinase (blue) on V658F mutant (grey). Side chains of the F-F-V triad residues are shown. The mutant residue V658F displaces Phe575, rearranging the SH2-PK linker. (b) Superposition of crystallographic molecule “B” of the wild type Jak1 pseudokinase (cyan) on the V658F mutant (grey), showing that the wild type pseudokinase can adopt essentially the same conformation observed in the V658F mutant. (c) Mutagenesis of the linker–pseudokinase switch in Ba/F3 cells. Mutations were introduced in mouse V617F Jak2 in residues that appear to support the observed rearrangement of the F-F-V triad (upper panel) and tested for their effect on IL-3-independent proliferation in Ba/F3 cells (lower panel). Locations of mutations are illustrated on the Jak1 structure, with the mutations in the corresponding Jak2 residues in parentheses. Control mutation K882R renders the Jak2 kinase domain catalytically inactive. Error bars indicate standard deviation of triplicate experiments. Corresponding line graphs of daily proliferation measurements are presented in Supplementary Fig. 3. (d) Patient-derived exon 12 mutations in Jak2 or Jak3 mapped on the Jak1 structure. For deletions, the first altered residue is indicated.The clinical presentation associated with each alteration is abbreviated in parentheses (PV, polycythemia vera; AMKL, acute megakaryoblastic leukemia; DS-ML, Down Syndrome associated myeloid leukemia).
Three residues appear to be centrally important for the observed rearrangement; Phe575 in the SH2-PK linker, Phe636 in the C-helix, and V658F, the site of the MPN mutation (V617F-site). We refer to these three residues as the F-F-V triad. This triad is highly conserved in Jak-family kinases, and these residues appear to be evolutionarily “coupled”; that is, substitutions in one position are almost invariably accompanied by changes in one of the other residues in the triad (Supplementary file ConSurf_Alignment.pdf). Interestingly, human Jak3 has a methionine at the V617F-site, and is the only human Jak kinase that is not activated by phenylalanine mutations at this site12. This pattern of conservation of residues comprising the F-F-V triad, combined with our observation that the wild type pseudokinase domain can also adopt the flipped conformation, leads us to speculate that the observed rearrangement is part of a structural switch controlling catalytic activation both in wild type Jak kinases and in the context of the V617F-site mutation. Consistent with a model in which the mutation promotes a rearrangement related to that induced by cytokine, cells bearing Jak2 V617F are hyper-responsive to EPO and even absent EPO stimulation, they proliferate at a rate equivalent to cells bearing wild type Jak2 with cytokine stimulation8. In contrast, pseudokinase domain deletions result in constitutive but diminished catalytic activation, and deletion mutants are not further stimulated by cytokine3.
To examine the requirement for the observed phenylalanine-flip rearrangement in Jak activation, we introduced point mutations in the SH2-PK linker and F-F-V triad and tested their effect on the ability of the V617F mutation in Jak2 to promote IL-3 independent growth of Ba/F3 cells. We carried out this structure-function analysis in Jak2 because of the availability of the well-characterized Ba/F3 cell system (Ba/F3.EPO-R) in which the EPO receptor is co-expressed with mutant Jak2 to allow its proper folding and maturation16. In the SH2-PK linker, mutation of the flipped phenylalanine (Phe537 in Jak2) to either valine or alanine abrogated V617F-driven proliferation (Fig. 2c, Supplementary Fig. 3). Residue Leu573 forms hydrophobic interactions that appear to stabilize the helical segment of the SH2-PK linker, and mutation of the corresponding residue in Jak2 from methionine to alanine (M535A) also reversed V617F-driven proliferation. In contrast, a conservative M535F substitution at this position was without effect (Fig. 2c). Phe636 in the C-helix forms the platform for the observed rearrangement, and we found that mutation of the corresponding residue in Jak2 (Phe595) to alanine also abrogated the transforming effect of V617F (Fig. 2c), as previously observed17,18. The V615F mutation in Jak2 modestly enhanced proliferation as compared with V617F alone, consistent with a role in promoting the SH2-PK linker rearrangement, but was not activating on its own (Fig. 2c and Supplementary Fig. 4f). Collectively, the effects of these mutations support a role for the phenylalanine-flip rearrangement in V617F-driven Jak2 activation. Analysis of these mutations in the context of wild type Jak2 reconstituted in Jak2-deficient cells will be required to establish whether the rearrangement is also required for EPO-driven activation of Jak2.
We also probed a crystal packing interaction between the two molecules in the asymmetric unit. This non-crystallographic dimer buries a large surface area (∼2080 Å2) and involves both V658F and Phe575 in the SH2-PK linker, but we found no role for this interface in Jak regulation (Supplementary Fig. 4).
Somatic gain of function mutations in exon 12 of Jak2 map to the SH2-PK linker, and an activating mutation in Jak3 is also found in this region (Fig. 2d). These alterations are expected to disrupt the observed conformation of the linker and may promote repositioning of the β4-β5 loop as we observe in the Jak1 V658F mutant (Supplementary Fig. 2c, d). Such a shift in the β4-β5 loop would in turn require rearrangement of the linker segment. Interestingly, an in vitro screen with Jak2 identified similar activating point mutations in the SH2-PK linker, leading to the conclusion that the SH2-PK linker is critical for mediating autoinhibition and cytokine-induced activation of Jak219.
While this manuscript was under review, a structure of the wild type Tyk2 pseudokinase domain was released in the Protein Data Bank (PDB ID 3ZON). The Tyk2 structure does include the SH2-PK linker and is very similar overall to the wild type Jak1 structure described here (RMSD 1.15Å for 258 aligned residues), however the linker conformation differs in detail (Supplementary Fig. 5). In Tyk2, the linker takes a similar path across the N-lobe, but does not adopt a helical conformation. Despite this difference, Phe581 (the equivalent of Phe575 in Jak1) is also in contact with the phenylalanine in the C-helix and would also be expected to rearrange in the context of the V617F-site mutation. It is unclear whether the difference in linker conformation represents a genuine difference between Jak1 and Tyk2, or whether it may arise from crystal packing interactions in this region in one or both structures.
In the Jak2 pseudokinase domain, stabilization of the C-helix is proposed to mediate in part the activating effect of the V617F mutation15. The C-helix is three residues longer at its N-terminus in the V617F mutant, and a break in regular hydrogen bonding near the center of the helix is also “repaired” in the V617F structure. This break in the C-helix appears to be unique to the Jak2 wild type structure, as the C-helix is continuous in all of our Jak1 structures and in Tyk2. However, we do observe a corresponding difference in the length of the C-helix in molecule “A” as compared with molecule “B” in the Jak1 pseudokinase structures, irrespective of the presence of the V658F mutation. In the Tyk2 pseudokinase domain, the longer C-helix is also observed in the wild-type context. Thus this is a conformationally labile region of the pseudokinase domain, but further work will be required to understand how alternate conformations of this segment may relate to Jak regulation.
Despite the lack of conserved active site residues, the Jak2 pseudokinase domain is reported to have catalytic activity and to autophosphorylate itself on both Ser523 and Tyr57020. These are autoinhibitory phosphorylation sites, and the V617F mutation has been proposed to promote Jak2 activation in part by abrogating pseudokinase catalytic activity and therefore phosphorylation of these sites15,20. The Ser523 and Tyr570 phosphorylation sites are not conserved in Jak1 or other family members, and we do not observe autophosphorylation of the Jak1 pseudokinase domain at alternate sites (Supplementary Fig. 6). Furthermore, the pseudokinase activation loop in Jak1 and Jak2, in both wild type and V617F-site mutants, adopts a conformation that blocks the expected position of a phosphoacceptor (Supplementary Fig. 1e). While we do not dismiss the possibility that catalytic activity of the pseudokinase domain plays a role in regulation of Jak2 and perhaps other family members, we favor a model in which the V617F mutation promotes activation primarily via rearrangement of the pseudokinase domain and SH2-PK linker, as described here. The F-F-V triad, which is central to this rearrangement, is highly conserved among all Jak kinases. Furthermore, this model provides a unifying explanation for the activating effects of both the V617F and exon 12 mutations in myeloproliferative disorders.
An obvious outstanding question is how the rearrangement we observe here leads to activation of the adjacent tyrosine kinase domain. The remodeled surface of the linker/pseudokinase unit could destabilize autoinhibitory interactions within the intact Jak kinase and also potentially promote activation by favoring dimerization or via intramolecular interaction with the kinase domain proper. Structural studies of longer fragments of Jak including at a minimum both the pseudokinase and kinase domains will be required to definitively address this question at a structural level.
Online Methods
Protein expression and purification
Constructs spanning residues 561-852 of human Jak1 bearing the wild-type sequence or V658F mutation were subcloned into a modified pTriEx vector (Novagen) for expression as N-terminal 6xHis plus glutathione-S-transferase (GST) fusion proteins with a tobacco etch virus (TEV) protease recognition sequence. The recombinant baculoviruses were generated using the BacVector3000 system (Invitrogen) and subsequently plaque purified. For induction of protein expression the BIICs/TIPS method was used21. Hi5 cells were cultured in shaker flasks using Express Five serum free medium and at a cell density of ∼1.4 million/mL, cultures were infected with recombinant baculovirus. Infection was allowed to proceed for 60-66 hr to achieve optimal expression of target protein prior to harvest by centrifugation. The cells were washed once in 20mM Tris pH 8.0, 150 mM NaCl, 5% glycerol and flash-frozen in liquid nitrogen and stored at −80°C.
For lysis, cells were thawed, resuspended in lysis buffer (100mM Tris pH8, 300 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, 1% Nonidet-P40 and Roche Complete EDTA-free Protease Inhibitor Cocktail) and incubated on ice for 45 minutes. Cell debris was removed by centrifugation and the supernatant was incubated for 45 minutes at 4°C with Ni-NTA agarose (Qiagen). Beads were washed with a step gradient of Ni wash buffer (20 mM Tris pH 8, 500 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol and 0.02% CHAPS) containing 0 mM, 10 mM, 20 mM and 40 mM imidazole and then eluted with elution buffer (Ni wash buffer with 250 mM imidazole). The eluate was supplemented with 5 mM DTT and then incubated with glutathione sepharose (GE Healthcare) beads for 2 hours at 4°C. The beads were washed and protein eluted by GST elution buffer (20 mM Tris pH 8.0, 500 mM NaCl, 10% glycerol, 2.5 mM TCEP, 5 mM glutathione and 0.02% CHAPS). TEV protease was then added and digestion was carried out at 4°C overnight to remove the 6xHis-GST tag. The digested protein was supplemented with 15 mM imidazole and passed over a Ni-NTA agarose (Qiagen) column to remove 6xHis-GST tag and 6xHis-tagged TEV protease. The flow-through was concentrated and run on a Superdex 200 gel filtration column equilibrated in final buffer (20 mM Tris pH 8.0, 500 mM NaCl, 10% glycerol, 4 mM DTT and 0.02% CHAPS). The eluted peak was concentrated to 4 mg/ml, flash frozen in liquid nitrogen, and stored at −80°C.
Crystallization and structure determination
Crystals of both the wild type and V658F Jak1 pseudokinase were grown by hanging drop vapor diffusion at 20°C by mixing equal volumes of protein solution with a well solution consisting of 1.2 – 1.5 M sodium citrate (pH 6.3-6.6), 6 mM DTT and 0-30 mM NDSB 256. Prior to data collection crystals were coated in NVH oil and then high-pressure cryocooled22 or directly plunge-frozen in liquid nitrogen. Data for Jak1 WT were collected at CHESS on beamline A1. Data for Jak1 V658F were collected at the Advanced Photon Source on beamline 24-C. Diffraction data were processed with the HKL suite of programs23. The structure was determined by molecular replacement using the C-lobe of focal adhesion kinase (PDB ID 1MP824 as a search model. The model was fit to the crystallographic data via manual model building with Coot25 and refinement with CNS26,27 and REFMAC28,29, and for the Jak1 WT structure was ultimately refined to a crystallographic R-value of 18.2 (Rfree=20.9). Analogous procedures were used to obtain the structure of the V658F mutant.
Mass spectrometry
The Jak1 pseudokinase domain (residues 561-852) was incubated with 300 units/mL calf intestinal phosphatase (CIP) (New England Biosciences) at 37°C for 30 minutes or 1 mM ATP/10 mM MgCl2 at 37°C for 30 minutes followed by 300 units/mL CIP at 37°C for 30 minutes. Treated and untreated Jak1 pseudokinase (5 μg) were injected onto a self-packed reversed phased column (1/32” O.D. × 500 μm I.D. PEEK tubing with 5 cm of POROS 10R2 resin). After desalting, protein was eluted with an HPLC gradient (0-100% B in 1 minute, A=0.2M acetic acid in water, B=0.2 M acetic acid in acetonitrile, flow rate = 10 μL/min) into an LTQ ion trap mass spectrometer (ThermoFisher Scientific, San Jose, CA). Data were acquired in profile mode scanning m/z 300-2000. Mass spectra were deconvoluted using MagTran1.03b2 software30.
Autoradiography
Jak1 pseudokinase at a concentration of 1 mg/mL was incubated at room temperature with 1 μCi [γ-32P]-ATP in a reaction buffer containing 20 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 4 mM DTT, 1 mM ATP, and 10 mM MgCl2 or MnCl2, and quenched with the addition of 100 mM EDTA. The autophosphorylation of EGFR kinase domain was measured under identical conditions as a positive control for detection of phosphorylation. Phosphorylation was monitored by SDS-PAGE followed by autoradiography.
Analysis of Jak2 mutants in Ba/F3 cells
Structure-function studies of Jak2 mutants in Ba/F3 cells bearing the Epo receptor were carried out essentially as previously described31. Briefly, point mutations were introduced in the pMSCV.JAK2V617F expression vector by site-directed mutagenesis using the QuickChange II XL Mutagenesis Kit (Agilent technologies) and specific primers constructed. The sequence of the entire mutagenized cDNA was verified by DNA sequencing. Mouse Ba/F3 cells expressing EPO-R (Ba/F3.EPO-R) were maintained in RPMI 1640 containing 10% fetal bovine serum and WEHI-3B conditioned medium. Ba/F3.EPO-R cells were transduced with the mutagenized Jak2 V617F using retroviral supernatant (293T cell method) by spin infection. Cells were grown for 2 days and sorted for GFP positive cells. Proliferation was measured by growing cells in 1 mL cultures with or without IL-3 for three days. The number of viable cells was determined daily using trypan blue exclusion.
Supplementary Material
Acknowledgments
We thank beamline personnel at the Macromolecular Crystallography Resource at the Cornell High-Energy Synchrotron Source (MacCHESS) and the Northeast Collaborative Access Team at the Advanced Photon Source, Argonne National Laboratory (NE-CAT) for assistance with data collection and processing. We thank K. Arnett for generous help with Jak1 autophosphorylation experiments. MacCHESS and NE-CAT are supported by grants from the U.S. National Institutes of Health (N.I.H). This work was supported in part by N.I.H. training grants GM008313 (J.M.R.) and CA936132 (R.M.), and by funding from Novartis Institutes for Biomedical Research (M.J.E).
Footnotes
Accession Codes: Crystallographic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 4L00 and 4L01 and can be accessed at www.pdb.org.
Author Contributions: A.V.T., A.D., R.M., and J.M.R. designed and performed experiments and analyzed data. Y.J. carried out experiments. S.M.G. and C.U.K contributed pressure cryo-cooling, and S.B.F. and J.A.M. contributed mass spectrometry analysis. A.V.T., R.M., M.S., J.D.G., and M.J.E. designed experiments, analyzed data, and wrote the manuscript.
Competing financial interests: J.D.G. and M.J.E. are consultants for and receive research support from Novartis Institutes for Biomedical Research.
References
- 1.Baker SJ, Rane SG, Reddy EP. Hematopoietic cytokine receptor signaling. Oncogene. 2007;26:6724–37. doi: 10.1038/sj.onc.1210757. [DOI] [PubMed] [Google Scholar]
- 2.Ghoreschi K, Laurence A, O'Shea JJ. Janus kinases in immune cell signaling. Immunol Rev. 2009;228:273–87. doi: 10.1111/j.1600-065X.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol. 2000;20:3387–95. doi: 10.1128/mcb.20.10.3387-3395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ihle JN, Gilliland DG. Jak2: normal function and role in hematopoietic disorders. Curr Opin Genet Dev. 2007;17:8–14. doi: 10.1016/j.gde.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Haan C, Behrmann I, Haan S. Perspectives for the use of structural information and chemical genetics to develop inhibitors of Janus kinases. J Cell Mol Med. 2010;14:504–27. doi: 10.1111/j.1582-4934.2010.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–8. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 7.Kralovics R, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–90. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 8.Levine RL, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–97. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 9.Baxter EJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–61. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 10.Tefferi A, Vainchenker W. Myeloproliferative neoplasms: molecular pathophysiology, essential clinical understanding, and treatment strategies. J Clin Oncol. 2011;29:573–82. doi: 10.1200/JCO.2010.29.8711. [DOI] [PubMed] [Google Scholar]
- 11.Jeong EG, et al. Somatic mutations of JAK1 and JAK3 in acute leukemias and solid cancers. Clin Cancer Res. 2008;14:3716–21. doi: 10.1158/1078-0432.CCR-07-4839. [DOI] [PubMed] [Google Scholar]
- 12.Staerk J, Kallin A, Demoulin JB, Vainchenker W, Constantinescu SN. JAK1 and Tyk2 activation by the homologous polycythemia vera JAK2 V617F mutation: cross-talk with IGF1 receptor. J Biol Chem. 2005;280:41893–9. doi: 10.1074/jbc.C500358200. [DOI] [PubMed] [Google Scholar]
- 13.Scott LM. The JAK2 exon 12 mutations: a comprehensive review. Am J Hematol. 2011;86:668–76. doi: 10.1002/ajh.22063. [DOI] [PubMed] [Google Scholar]
- 14.Scott LM, et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356:459–68. doi: 10.1056/NEJMoa065202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bandaranayake RM, et al. Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat Struct Mol Biol. 2012;19:754–9. doi: 10.1038/nsmb.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wernig G, et al. The Jak2V617F oncogene associated with myeloproliferative diseases requires a functional FERM domain for transformation and for expression of the Myc and Pim proto-oncogenes. Blood. 2008;111:3751–9. doi: 10.1182/blood-2007-07-102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gnanasambandan K, Magis A, Sayeski PP. The constitutive activation of Jak2-V617F is mediated by a pi stacking mechanism involving phenylalanines 595 and 617. Biochemistry. 2010;49:9972–84. doi: 10.1021/bi1014858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dusa A, Mouton C, Pecquet C, Herman M, Constantinescu SN. JAK2 V617F constitutive activation requires JH2 residue F595: a pseudokinase domain target for specific inhibitors. PLoS One. 2010;5:e11157. doi: 10.1371/journal.pone.0011157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao L, et al. A JAK2 interdomain linker relays Epo receptor engagement signals to kinase activation. J Biol Chem. 2009;284:26988–98. doi: 10.1074/jbc.M109.011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ungureanu D, et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol. 2011;18:971–6. doi: 10.1038/nsmb.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valiev M, Yang J, Adams JA, Taylor SS, Weare JH. Phosphorylation reaction in cAPK protein kinase-free energy quantum mechanical/molecular mechanics simulations. J Phys Chem B. 2007;111:13455–64. doi: 10.1021/jp074853q. [DOI] [PubMed] [Google Scholar]
- 22.Flex E, et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J Exp Med. 2008;205:751–8. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bercovich D, et al. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down's syndrome. Lancet. 2008;372:1484–92. doi: 10.1016/S0140-6736(08)61341-0. [DOI] [PubMed] [Google Scholar]
- 24.Mullighan CG, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2009;106:9414–8. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearney L, et al. Specific JAK2 mutation (JAK2R683) and multiple gene deletions in Down syndrome acute lymphoblastic leukemia. Blood. 2009;113:646–8. doi: 10.1182/blood-2008-08-170928. [DOI] [PubMed] [Google Scholar]
- 26.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–33. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin J, Pawson T. Modular evolution of phosphorylation-based signalling systems. Philos Trans R Soc Lond B Biol Sci. 2012;367:2540–55. doi: 10.1098/rstb.2012.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pawson T, Kofler M. Kinome signaling through regulated protein-protein interactions in normal and cancer cells. Curr Opin Cell Biol. 2009;21:147–53. doi: 10.1016/j.ceb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z, Marshall AG. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. Journal of the American Society for Mass Spectrometry. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 31.Deshpande A, et al. Kinase domain mutations confer resistance to novel inhibitors targeting JAK2V617F in myeloproliferative neoplasms. Leukemia. 2011 doi: 10.1038/leu.2011.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.