Background: The RND efflux transporters of cyanobacteria are largely unknown.
Results: Six RNDs with different functionality and mutant phenotypes exist in Anabaena sp.
Conclusion: Antibiotic export involves a single AcrB-like RND protein.
Significance: The diversity of the RND function in cyanobacteria is initially dissected.
Keywords: Antibiotic Resistance, Cyanobacteria, Membrane, Multidrug Transporters, Nitrosative Stress, RND, Metabolite Transport, TolC
Abstract
The TolC-like protein HgdD of the filamentous, heterocyst-forming cyanobacterium Anabaena sp. PCC 7120 is part of multiple three-component “AB-D” systems spanning the inner and outer membranes and is involved in secretion of various compounds, including lipids, metabolites, antibiotics, and proteins. Several components of HgdD-dependent tripartite transport systems have been identified, but the diversity of inner membrane energizing systems is still unknown. Here we identified six putative resistance-nodulation-cell division (RND) type factors. Four of them are expressed during late exponential and stationary growth phase under normal growth conditions, whereas the other two are induced upon incubation with erythromycin or ethidium bromide. The constitutively expressed RND component Alr4267 has an atypical predicted topology, and a mutant strain (I-alr4267) shows a reduction in the content of monogalactosyldiacylglycerol as well as an altered filament shape. An insertion mutant of the ethidium bromide-induced all7631 did not show any significant phenotypic alteration under the conditions tested. Mutants of the constitutively expressed all3143 and alr1656 exhibited a Fox− phenotype. The phenotype of the insertion mutant I-all3143 parallels that of the I-hgdD mutant with respect to antibiotic sensitivity, lipid profile, and ethidium efflux. In addition, expression of the RND genes all3143 and all3144 partially complements the capability of Escherichia coli ΔacrAB to transport ethidium. We postulate that the RND transporter All3143 and the predicted membrane fusion protein All3144, as homologs of E. coli AcrB and AcrA, respectively, are major players for antibiotic resistance in Anabaena sp. PCC 7120.
Introduction
TolC (tolerance to colicin E1) is a versatile outer membrane protein involved in the secretion of numerous substances across the cell wall of Gram-negative bacteria (1–3). Among these substances are proteins (4), antibiotics (2, 5), siderophores (6–8), and lipids, which in the case of some cyanobacteria are required for the formation of the heterocyst-specific cell wall (e.g. see Ref. 9). To perform these functions, TolC is part of three different secretion systems. (i) TolC complexes with different inner membrane ABC2 transporters involved in the secretion of protein toxins to form the type I secretion system (e.g. see Ref. 10). (ii) TolC was found to be part of cation efflux pumps for extrusion of toxic metal ions (e.g. see Ref. 11). However, in the best understood system (iii), TolC is the outer membrane factor for different superfamilies of multidrug efflux pumps, such as the resistance-nodulation-cell division type (RND), ABC transporter superfamily, or the major facilitator superfamily (MFS) (Fig. 1) (e.g. see Refs. 12 and 13). The best described system is the tripartite multidrug efflux complex AcrAB (acriflavine resistance protein A and B)-TolC in Escherichia coli. The inner membrane transporter confers substrate specificity and recruits TolC upon substrate binding. The contact between TolC and the inner membrane transporter is established by adaptor proteins of the HlyD (hemolysin D) family, also called membrane fusion proteins (MFPs) (4), which stabilize the transient complex and are usually encoded in an operon together with their corresponding inner membrane transporter. After substrate secretion, TolC dissociates from the complex and can be utilized by other transporter systems (e.g. see Ref. 14). However, although a solid description of the TolC-involving complexes has been obtained, it is still not clear how specific TolC recruitment is regulated. The currently favored model suggests that complex formation is triggered by substrate availability in the cytoplasm or periplasm via the transporter or MFP components (15, 16).
FIGURE 1.

Identification of RND and HlyD family proteins of Anabaena sp. PCC 7120. A, a general model of the detoxification efflux pathway in Gram-negative bacteria. Small cytotoxic substances can cross the outer membrane (OM), often facilitated by porins, and accumulate in the periplasm (PP). Upon accumulation in the periplasm, cytotoxic substances can cross the plasma membrane (PM) and enter the cytosol (CYT). Single-component inner membrane transporters (MFS- and SMR-type) catalyze the electrochemical gradient-dependent clearing of the cytoplasm by transporting cytotoxins across the inner membrane into the periplasm, from which these can be extruded across the outer membrane in a RND- and TolC-dependent manner. (Rate constants are indicated; for explanation, see Table 5.) A clustering of the RND (B) or the HlyD (C) family proteins was created with CLANS. Each dot represents a protein. The proteins are grouped by their pairwise sequence similarity determined by all-versus-all BLASTs. The filled red circles highlight proteins with experimentally determined functions. The filled green circles highlight seven proteins of the Anabaena sp. proteome that cluster with RND transporters (B) and 22 that cluster with HlyD family proteins (C) (Table 4). The large, empty circles mark those clusters that contain experimentally characterized proteins. The colors of these circles correspond to the font colors of the RND (B) and HlyD (C) family transporter names given below the respective clustering.
Although global models are proposed for the function of TolC in Gram-negative bacteria, remarkably little is known about these systems in cyanobacteria even with respect to the molecular nature of the components involved. Only recently, the TolC homolog of Anabaena sp. PCC 7120 (hereafter referred to as Anabaena sp.) has been identified experimentally (5, 7, 9, 17). Anabaena sp. is a filamentous, heterocyst-forming cyanobacterium (18, 19). The mutant of the TolC-like gene hgdD (heterocyst glycolipid deposition protein D; alr2887) is impaired in heterocyst development. Based on the comparable phenotype of mutants of hgdD (9) and of the ABC exporter DevBCA (alr3170–alr3172) (20), it was proposed that DevBCA-HgdD is responsible for heterocyst glycolipid (HGL) secretion (9, 17). Furthermore, the mutant of hgdD is impaired in secretion of hydroxamate-type siderophores (7), and initial results suggest that HgdD might also be involved in protein secretion (9). Recently, it could be demonstrated that HgdD is equally essential for secretion of the macrolide antibiotic erythromycin and the fluorescent dye ethidium (5). However, not much is known about other transporter components involved in the HgdD-dependent transport process.
In addition to the HgdD system, two MFS-type transporters, SchE (schizokinen exporter; All4025), which was recently characterized to be essential for schizokinen export (7), and SmsA (secondary metabolite secretion protein A; All2215), have been described as plasma membrane-localized factors involved in secondary metabolite secretion, most likely acting in concert with the HgdD-dependent metabolite secretion system (5).
Here we describe the RND protein family network of Anabaena sp. and discuss their relation to the only TolC-like protein found in this cyanobacterium and the identified MFPs. We analyzed their regulation and involvement in antibiotic resistance as well as the homeostasis of photosystems during fixation of atmospheric nitrogen. Moreover, we present evidence leading to the proposal that All3143 is the major RND component responsible for multidrug efflux in Anabaena sp. We propose its annotation as AcrB, especially because All3143 is able to partially complement the E. coli ΔacrB mutant.
EXPERIMENTAL PROCEDURES
Homology Search for RND Family Proteins
Uniprot IDs assigned to the RND family proteins or HlyD family proteins were downloaded from PFAM (ACR_tran, PF00873; HlyD, PF00529; HlyD_2, PF12700; version 25.0) by a Python script using Biopython. Sequences and taxonomy information were downloaded from UniProt, and bacterial sequences were selected for further processing. Redundant sequences were removed with cd-hit (21). The remaining 7,901 RND and 12,918 HlyD sequences were clustered with CLANS (22, 23). Sequences from functionally characterized RND family proteins were assigned (protein name followed by UniProt ID): CnrA (CNRA_RALME), AcrB (ACRB_ECOLI), CusA (CUSA_ECOLI), CzcA (B1J3Z9_PSEPW), AcrF (ACRF_ECOLI), AcrD (ACRD_ECOLI), ArpB (ARPB_PSEPU), TtgB (TTGB_PSEPU), MdtB (multidrug tolerance B)/YegN (MDTB_ECOLI), MdtF/YhiV (MDTF_ECOLI), MdtC (MDTC_ECOLI), MexB (MEXB_PSEAE), MexD (Q9HVI9_PSEAE), MexF (multidrug efflux transporter F) (Q9I0Y8_PSEAE), MexI (Q9HWH4_PSEAE), EmhB (Q6V6X8_PSEFL), EefB (Q8GC83_ENTAE). Sequences from functionally characterized HlyD family proteins were assigned: AcrA (ACRA_ECOLI), MexA (MEXA_PSEAE), MexC (Q51395_PSEAE), MexE (B7V862_PSEA8), MdtA (MDTA_ECO8A), EefA (B5XTA5_KLEP3), EmhA (C1KA84_PSEFL), HlyD (HLYDC_ECOLX), EmrA (EMRA_ECOLI).
Generation of Anabaena sp. Mutants
Anabaena sp. PCC 7120 and several mutant derivatives used in this work are listed in Table 1. For generation of single recombinant insertion mutants, an internal fragment of the coding region was amplified by PCR on genomic DNA (oligonucleotides listed in Table 2) introducing BglII restriction sites for cloning of the products into pCSV3 (24) containing a SpR/SmR gene cassette yielding the required plasmids for generation of single recombinant mutants (Table 3). Plasmids were transferred from E. coli to Anabaena sp. by conjugations as described (25).
TABLE 1.
Anabaena sp. and Escherichia coli strains used in this study
Sp, spectinomycin; Sm, streptomycin; Cm, chloramphenicol; gi*…, gene interruption; co*…, complementation.
| Strain | Resistance | Genotype | Relevant properties | Souce/Ref. |
|---|---|---|---|---|
| Anabaena sp. PCC 7120 | Wild type | |||
| CSR10 | SpR/SmR | alr4167::SpRSmR | gi* by plasmid pCSV3 derivative | Ref. 57 |
| AFS-I-alr2887 | SpR/SmR | alr2887::SpRSmR | gi* by plasmid pCSV3 derivative | Ref. 9 |
| AFS-I-alr1656 | SpR/SmR | alr1656::SpRSmR | gi* by plasmid pCSV3 derivative | This study |
| AFS-I-all3143 | SpR/SmR | all3143::SpRSmR | gi* by plasmid pCSV3 derivative | This study |
| AFS-I-alr4267 | SpR/SmR | alr4267::SpRSmR | gi* by plasmid pCSV3 derivative | This study |
| AFS-I-all7631 | SpR/SmR | all7631::SpRSmR | gi* by plasmid pCSV3 derivative | This study |
| E.coli BW 25113 | Wild type | This study | ||
| E.coli BW 25113ΔtolC | CmR | ΔtolC/pGR50 | Deletion of tolC | This study |
| E. coli BW 25113ΔacrAB | CmR | ΔacrAB/pGR50 | Deletion of acrA and acrB | This study |
| E. coli BW 25113ΔtolC-tolC | CmR | ΔtolC/ptet:tolC | co* of ΔtolC with tolC | This study |
| E. coli BW 25113ΔtolC-hgdD | CmR | ΔtolC/ptet:hgdD | co* of ΔtolC with hgdD | This study |
| E. coli BW 25113ΔacrAB-acrABHis | CmR | ΔacrAB/ptet:acrABHis | co* of ΔacrAB with acrAB | This study |
| E. coli BW 25113ΔacrAB-all3144–3143 | CmR | ΔacrAB/ptet:all3144-all3143 | co* of ΔacrAB with all3143-all3144 | This study |
TABLE 2.
Primers used for cloning
| Primer name | Oligonucleotide sequence |
|---|---|
| SR1656_F | AGATCTCCATTGAGGTTAACCC |
| SR1656_R | AGATCTCAGAGGTAAAGCAGAACC |
| SR3143_F | AGATCTCGGTGACAAATATCTTGG |
| SR3143_R | AGATCTGGCAAATTCCGTTGCTTCGG |
| SR4267_F | AGATCTCTGCGTTCTTTAGAAGG |
| SR4267_R | AGATCTGGACAAGATGATCGCC |
| SR7631_F | AGATCTGCTCCGCCTCAAGTTG |
| SR7631_R | AGATCTGTTATTGCAGATTTACC |
| 1656_S1F | GTCGAGCAGTTTGGTACC |
| 1656_S1R | CCAAGGCAATAATCAGACC |
| 3143_S1F | GCAATCAGTATACCTACGC |
| 3143_S1R | GTAGGGAATATCCCTATACC |
| 4267_S1F | CCTCTGAAAACTGCCGTG |
| 4267_S1R | GCAAGGATGTAGGAATAGC |
| 7631_S1F | GTTGGATAGTTGTGCTAGG |
| 7631_S1R | CATTCAAACTGCCATCGCC |
| 2887_F | GGTACCAGGAGGCATCATGTGAAAGGACAACACTTATTC |
| 2887_R | CTCGAGCTACCGACTACTGACTAC |
| 3144_F | GGTACCAGGAGGCATCATATGTCATCCTCTGAGCCTCAAAC |
| 3143_R | CTCGAGCCTCTGCCTCATCTATGACTAG |
| AcrA_F | CTCGAGAGGAGGCATCATATGAACAAAAACAGAGGG |
| AcrBHIS_R | CTCGAGTCAGTGATGGTGATGGTGATGTCCACCGCCTCCATGATGATCGACAGTATGG |
| AcrA_R | AACGGATCCTGTTTAAGTTAAGACTTGG |
| 3143_F2 | ATATGGATCCATTCCCCAATGTTTGTTGAC |
| 3143_R2 | ATATGTCGACTCATCTATGACTAGTTG |
| pGRF | ATATGTCGACATGGAAGCCGGCGGCACC |
| tolCdeletion _F | AATTTTACAGTTTGATCGCGCTAAATACTGCTTCACCACAAGGAATGCAAGTGTAGGCTGGAGCTGCTTC |
| tolCdeletion _R | ATCTTTACGTTGCCTTACGTTCAGACGGGGCCGAAGCCCCGTCGTCGTCAATTCCGGGGATCCGTCGACC |
TABLE 3.
Plasmids used in this study
Ap, ampicilin; Sp, spectinomycin; Sm, streptomycin; Cm, chloramphenicol.
| Plasmid | Marker | Properties | Source/Ref. |
|---|---|---|---|
| pCSV3 | SpR/SmR | pRL500 with substituted ApR gene | Ref. 79 |
| pALH12 | SpR/SmR | Internal fragment of alr1656 was cloned in pCSV3 via BamHI | This study |
| pALH13 | SpR/SmR | Internal fragment of all3143 was cloned in pCSV3 via BamHI | This study |
| pALH14 | SpR/SmR | Internal fragment of alr4267 was cloned in pCSV3 via BamHI | This study |
| pALH15 | SpR/SmR | Internal fragment of all7631 was cloned in pCSV3 via BamHI | This study |
| pGR50 | CmR | tet promoter, CmR | Ref. 80 |
| pGR-TolC | CmR | This study | |
| pGR-HgdD | CmR | This study | |
| pGR-All3144-All3143 | CmR | This study | |
| pGR-AcrABHIS | CmR | This study | |
| pGR-AcrA-All3143 | CmR | This study |
E. coli Strains and Generation of Deletion Mutants
E. coli BW25113 (26) and chromosomal deletion derivatives used in this work are listed in Table 1. E. coli knockouts BW25113ΔacrAB and BW25113ΔtolC were constructed according to Ref. 27. E. coli BW25113ΔtolC was generated by PCR-mediated gene replacement (27), introducing the kanamycin resistance cassette using primers listed in Table 2. E. coli BW25113ΔacrAB was created by P1 phage transduction (28) with P1 phage prepared from E. coli MC4100ΔacrAB::Kan strain. Subsequently, plasmid pCP20 was used to eliminate the kanamycin resistance cassette from the generated deletion mutants. Deletions in the resulting mutants were verified by PCR.
Growth of Cyanobacteria
Anabaena sp. wild type and mutants were grown photoautotrophically at 30 °C in BG11 medium (29) with or without (in the case of BG110) 17.6 mm NaNO3, without the source of iron (no ferric ammonium citrate added; BG11−Fe). Mutant strains were grown in the presence of 6 μg ml−1 streptomycin and 6 μg ml−1 spectinomycin. The growth of the strains was analyzed by Equation 1,
 |
For analysis of growth on plates, Anabaena sp. was grown in liquid medium to early exponential phase (2–3 days), and concentration was adjusted to OD750 = 1. Five μl of 1:1, 1:10, and 1:100 dilutions were spotted on plates and incubated under the indicated growth conditions. After 7 days, the 1:10 dilution was used for representation.
Genomic DNA and RNA Isolation
Isolation of genomic DNA, manipulation of plasmid DNA, PCR with the Triple Master PCR System (Eppendorf), and isolation of DNA from Anabaena sp. have been described previously (5, 7, 30). RNA was isolated from cells of a 25-ml culture at OD750 = 1. Cells were collected by centrifugation (5 min, 3200 × g) and frozen in liquid nitrogen. For isolation of total RNA, 1 ml of TRIzol® (Invitrogen) was used. Isolation was performed according to the manufacturer's instructions, and residual DNA was removed by digestion with RNase-free DNase (5). cDNA was synthesized using 1–2 μg of total RNA and RevertAid transcriptase (Thermo Scientific) with random hexamer primers according to the manufacturer's instructions.
Chlorophyll, Phycocyanin, and Lipid Analysis
For chlorophyll a and phycocyanin determination from whole cells, Anabaena sp. was grown in liquid BG11 medium to early exponential growth phase (2–3 days). To obtain comparable results, culture density was adjusted to OD750 = 1 prior to measurement. Chlorophyll a and phycocyanin content of whole cells was determined as described (31, 32). Lipid analysis was performed as described previously (9).
Microscopic Analysis
Microscopic analysis was performed using a Zeiss Axiophot microscope (Zeiss) with a ×63/1.4 numerical aperture plan. Counting of cells and surface estimation for rate calculation were performed as described previously (see Ref. 5).
Ethidium Uptake Measurement by DNA Intercalation
The Intercalation of ethidium into endogenous nucleic acids after uptake was monitored by the increase of fluorescence, as described previously (5). Data points were taken every minute for 30 min (excitation, 316 nm; emission, 620 nm (Tecan Infinite 200, CH)). For protonophore treatment with carbonyl cyanide m-chlorophenylhydrazone (CCCP), CCCP (in DMSO) was added to a final concentration of 200 μm to an E. coli cell suspension. Cells were incubated for 10 min at room temperature prior to measurement. Results were analyzed by a least squares fit analysis (Sigma Plot) as described (5).
RESULTS
Bioinformatic Identification of Putative TolC Partners
The function of TolC depends on transporters of the cytoplasmic membrane energizing the secretion event (Fig. 1A). To identify such transporters in cyanobacteria, we performed a global search for RND-like efflux pumps in Gram-negative bacteria, including Anabaena sp. The results were clustered to assign putative functions to identified genes (e.g. see Ref. 23).
We used different template sequences to search for RND homologs (Fig. 1B), including AcrB, AcrD, AcrF, ArpB, EmhB, EefB, MexB, MexD, MexI, MexF, MdtF/YhiV, and TtgB (33–43) of the RND multidrug efflux family HAE-1; the copper transporter CusA (44); the nickel and cobalt exporter CnrA (45); the zinc, cadmium, and lead exporter CzcA (46); and MdtB/YegN and MdtC (47).
In the set of 7,901 sequences, we identified eight sequences from Anabaena sp. To assign the individual sequences to the various known RND protein families, we have chosen the CLANS representation (22, 23), where each spot represents an individual sequence and the distance between two spots is a reflection of the similarity of the corresponding sequences. This allows the visualization of clusters of similar sequences. We focused on the assignment of sequences to experimentally confirmed sequences, and the clusters containing such sequences are highlighted by circles with different colors (Fig. 1, B and C).
The two proteins encoded by all7618 and all7631 were previously suggested to belong to the copper transport system (7). Here we observed that these two genes cluster with sequences coding for metal transporters of the CnrA and CzcM family (Fig. 1B, violet). Whereas three sequences (Alr1656, Alr4267, and Alr5294) could not be assigned to any cluster with known function, one sequence (All3143) clusters with genes of the MexF family.
RNDs generally consist of 12 transmembrane helices (TMHs). Two extended periplasmic loops (between TMH1 and -2 and TMH7 and -8) serve as drug binding or transport modules and as a TolC docking domain and split the transmembrane domains in three clusters: one TMH at the N terminus, six TMHs in the center of the protein, and five at the C terminus (e.g. see Refs. 2, 33, and 48). As a consequence of the even number of TMHs, the N and C terminus are both localized in the cytoplasm. All7617 and Asr3133 are indeed similar to the C terminus of All7618, but, with 138 and 91 amino acids, respectively, they are too short to represent a full-length RND-type protein. Thus, they were excluded from further analysis.
In addition, taking the deposited coding region of Alr4267 for prediction of transmembrane regions, we realized an exceptional domain organization, because TMH1 could not be predicted (Fig. 2A). However, careful analysis of the genomic context shows an upstream start codon. Using this codon results in a prolonged amino acid sequence at the N terminus, and an additional TMH becomes predictable (Fig. 2B). Thus, Alr4267 has a topology expected for RND, and consequently, we assume that six RND-like proteins are encoded by the genome of Anabaena sp.
FIGURE 2.

Secondary structure prediction of Alr4267. A, the amino acid sequence generated by in silico translation from the upstream ATG (green) and from the start codon according to the annotated reading frame of alr4256 (red). B, the amino acid sequence of Alr4267 as deposited at CyanoBase (78) was used for prediction of TMHs using TMHMM (dashed red line). The amino acid sequence translated from the alternative start codon as shown in A yields the highly conserved transmembrane organization of RND family transporter (green line). The number of the proposed TMH is given. Note that the score for the second predicted TMH at the N terminus is below threshold.
We used a similar approach to identify membrane fusion proteins linking the plasma membrane-localized transporter with TolC (e.g. see Ref. 4) (Fig. 1C). As bait for functional assignment, we used the HlyD family proteins EmrA, MdtA, MexE, AcrA, EefA, EmhA, MexA, MexC, and HlyD (37, 49–56). Among 12,918 sequences from Gram-negative bacteria, we obtained sequences of 22 Anabaena sp. proteins. Except for All5304, all identified HlyD family proteins are in direct genomic proximity to either an identified RND-like efflux pump or a putative ABC transporter (Table 4). Only six sequences (All7314, Alr1200, Alr2675, Alr4240, Alr5148, and Alr5304) cluster with the HlyD sequence from selected baits (Fig. 1C), which might be the first indication of their function.
TABLE 4.
The family of MFPs in Anabaena sp.
| MFP | Upstream/downstream gene | ||
|---|---|---|---|
| RND | |||
| 1 | Alr1655 | alr1656 | |
| 2 | All3144 | all3143 | |
| 3 | Alr5293 | alr5294 | |
| 4 | All7619 | all7618 | |
| 5 | All7632 | all7631 | |
| ABC | |||
| 6 | Alr0445 | alr0446 (putative ABC transporter) | alr0447 (ATP-binding protein) |
| 7 | Alr0451 | alr0452 (putative ABC transporter) | |
| 8 | All0809 | al0808 (devC homolog) | all0807 (ATP-binding protein ) |
| 9 | Alr1200 | alr1201 (ATP-binding protein) | |
| 10 | Alr1501 | alr1502 (putative ABC transporter) | |
| 11 | Alr1505 | alr1506 (ATP-binding protein/permease) | alr1507 (ATP-binding protein) |
| 12 | All2652 | all2651 (putative ABC transporter) | all2650 (ATP-binding protein) |
| 13 | All2675 | all2676 (ATP-binding protein) | |
| 14 | Alr3710 | devB | devA |
| 15 | Alr4240 | alr4239 (ATP-binding protein) | |
| 16 | Alr4280 | alr4281 (devC homolog) | alr4282 (putative ABC transporter) |
| 17 | Alr4973 | alr4973 (devC homolog) | alr4974 (DevA homolog) |
| 18 | Alr5148 | alr5147 (ATP-binding protein) | |
| 19 | Alr5347 | hgdC | hgdA |
| 20 | All7314 | all7315 (HlyB-like ABC transporter) | |
| None | |||
| 21 | All5304 | ||
Expression of Genes Coding for Putative RND-type Proteins
To obtain a first insight into the possible role of the RND-type factors, we analyzed their expression during progressing growth with respect to the expression of hgdD by RT-PCR (Fig. 3A). The constitutively expressed RNase P subunit B (rnpB; Fig. 3A) was used to normalize for total RNA content. In contrast to hgdD, no transcript of any RND was detected during early growth. During increase of culture density, the transcripts of alr1656, all3143, alr4267, and alr5294 became detectable at a similar level, whereas the expression of hgdD remained constant.
FIGURE 3.
Expression analysis of putative RND family genes. A, growth curve of Anabaena sp. to define early (1 day), late exponential (3 days) and stationary growth phase (9 days). Growth is expressed as a natural logarithm of the ratio of the cell density at the indicated times and at time 0. B, RT-PCR analysis of hgdD and RND gene transcript abundance on RNA isolated from wild-type Anabaena sp. at the indicated growth phase (lanes 1–3). Lane 4, RT-PCR in the absence of the reverse transcriptase; lane 5, PCR on isolated genomic DNA (gDNA). The transcript abundance of the constitutive rnpB was analyzed as an internal standard for normalization of total RNA concentration. C, RT-PCR analysis of hgdD and RND transcript abundance (lanes 1–7) on RNA isolated from wild-type cells grown to late exponential phase (3 days) under either deprivation of iron (−Fe) or the addition of 0.25 μm erythromycin (+Ery) or ethidium bromide (+EB). As a control, PCR on isolated genomic DNA is shown in the bottom lane. D, the transcript of the constitutive rnpB was analyzed as an internal standard for normalization of total RNA concentration. Error bars, S.D.
Proteins of the RND family are typically involved in multidrug efflux. In addition, HgdD has been found to be indispensable for efflux of erythromycin, ethidium, and siderophores (5, 7). Thus, we analyzed the expression of the identified RNDs during exponential growth after the removal of iron from the medium or after the addition of erythromycin and ethidium bromide (Fig. 3). Remarkably, alr5294 is repressed in the absence of iron or in the presence of ethidium bromide. Furthermore, all7618 is induced by the addition of either erythromycin or ethidium bromide, whereas all7631 is specifically induced by the addition of ethidium bromide. Thus, in contrast to earlier observations (7), we found that the CusB homologs All7618 and All7631 are expressed as a response to stress treatment. The difference in transcript abundance might suggest that the CusA and CusB homologs of Anabaena sp. are not expressed as an operon and that the recruitment of HgdD for the formation of the tripartite secretion channel for metal extrusion is tightly regulated by the presence of the membrane fusion protein homolog but not the efflux pump itself. Consistently, it has been shown that the affinity and stability for the formation of tripartite secretion complexes depend on the oligomerization kinetics of the MFP (57). Nevertheless, we can conclude that all six RNDs are expressed and thus functionally relevant.
Mutants of the Putative RND-type Components Are Viable
We generated insertion mutants of the constitutively expressed alr1656, all3143, and alr4267 and of the ethidium bromide-induced all7631, the latter to compare the importance of the global and specifically expressed genes (Table 1). All generated strains were fully segregated because no wild-type gene was detectable by PCR (Fig. 4A).
FIGURE 4.

Generation of RND mutants and growth analysis. A, derivative plasmids of pCSV3, which contain a homologous region of alr1656, all3143, alr4267, or all7631 (left), were used to generate single insertion mutants. Segregation was confirmed by PCR analysis on genomic DNA from wild-type (lane 1) or mutant strains (lanes 2–4) using the indicated primers with depicted binding site and orientation. B, RT-PCR analysis of hgdD (alr2887), rnpB, and RND gene transcript abundance on cDNA isolated from the indicated strains at exponential growth phase (day 3). Lane 7, PCR on genomic DNA isolated from wild-type Anabaena sp. C, antibiotic resistance of Anabaena sp. (strain CSR10) and deletion strains. Growth in the presence of ethidium bromide (1 μg ml−1), erythromycin (10 ng ml−1), roxithromycin (30 ng ml−1), tylosin (100 ng ml−1), neomycin (1 μg ml−1), FeACi (0.1 mm), and CuSO4 (5 μm) and in the absence of iron and copper (−Fe/−Cu) was analyzed on solid BG11 medium containing streptomycin (Sp) and spectinomycin (Sm) (6 μg ml−1 each) as selective antibiotic. Cells of an early exponential growth phase culture were adjusted to OD750 and diluted 1-, 10-, and 100-fold, and 5 μl of the cell suspensions were spotted onto the BG11 agar plates with the indicated additives. Shown is the 10-fold dilution after 7 days of incubation at constant light (30 μmol cm−2 s−1).
We first analyzed whether the disruption of an individual RND-like gene results in an altered expression pattern of the remaining RND network. To this end, the expression of the six RND-like genes and hgdD was analyzed by RT-PCR in the individual insertion mutants (Fig. 4B). Most notably, we observed an enhanced expression of alr1656 in AFS-I-hgdD and of alr4267 in AFS-I-all3143. In addition, we could observe a slight induction of alr7631 in AFS-I-alr4267, but otherwise we did not find a noteworthy enhanced or reduced expression of the RND genes or hgdD. Next, we performed a phenotype screening by analyzing growth on plates. We substituted the wild-type strain by CSR10 (58) to retain the selective antibiotic pressure. Similar to the ASF-I-hgdD, none of the RND mutant strains shows any growth impairment in BG11 medium (Fig. 4C, panel 1), which confirms that the analyzed RND-like genes (alr1656, all3143, alr4267, and all7631) are not essential because the mutant strains are fully segregated. Therefore, we analyzed the growth of the mutants under conditions at which the TolC function might be required. As documented previously, AFS-I-hgdD shows a Fox− phenotype (panel 2) (9) and is highly sensitive to ethidium bromide and erythromycin (panels 3 and 4) (5). The antibiotic sensitivity seems to be somewhat specific for macrolides, because the presence of roxithromycin and tylosin results in a similar growth inhibition as compared with erythromycin (panels 3–6), whereas the aminoglycoside antibiotic neomycin does not induce a growth inhibition under the conditions tested (panel 7). Interestingly, AFS-I-all3143 is the only mutant that appears to be Fox− and shows a similar sensitivity toward erythromycin and tylosin. Its growth on roxithromycin is slightly affected as judged by the pale phenotype of the spotted culture (panel 5). All other mutant strains did not show a significant alteration in growth under the conditions tested. Also, the deprivation of iron and copper or the excess of each of these metals had no influence on growth.
Two RND Proteins Are Involved in Heterocyst Viability during Nitrogen Fixation
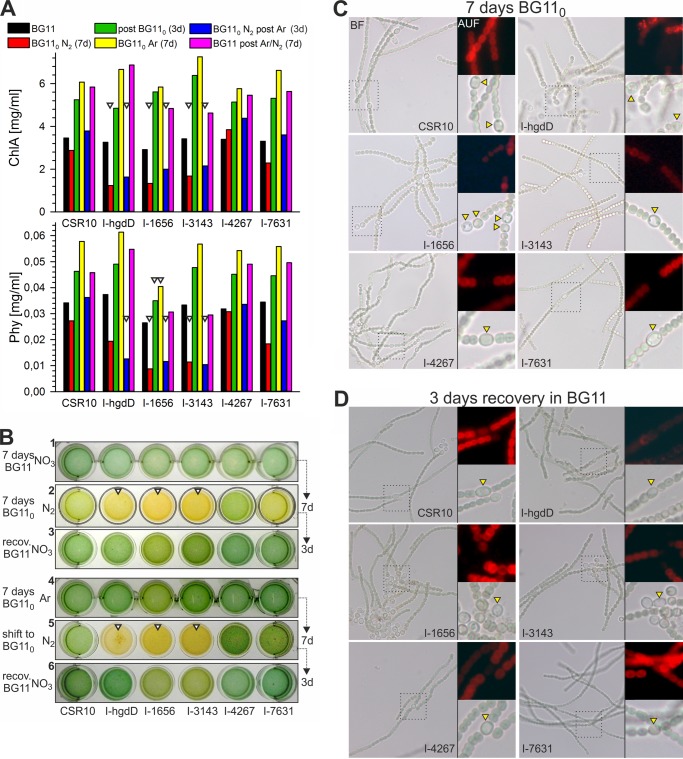
The initial growth analysis pointed to a possible specific relation between HgdD and All3143 in heterocyst formation or function. To further analyze a possible function of RND proteins in heterocysts, we performed microscopic analyses of liquid cultures after nitrogen step-down and determined the chlorophyll a (Chl) and phycocyanin (Phy) concentration of whole cell cultures. Similar to growth on solid plates (Fig. 4), there was no growth retardation or any significant difference in Chl or Phy content after 7 days in BG11 medium (Fig. 5, A and B). After a shift to nitrogen-deprived BG110 medium, AFS-I-hgdD and AFS-I-all3143 showed the expected pale Fox− phenotype. Surprisingly, also AFS-I-alr1656 appeared to be Fox− under these conditions. All three mutants showed a drastic reduction in Chl and Phy content under these conditions. AFS-I-hgdD has been described to be essential for heterocyst development (9), and consistently, heterocyst differentiation was arrested at an early stage in this mutant (Fig. 5C). In contrast, both AFS-I-alr1656 and AFS-I-all3143 developed heterocysts, but their cytoplasmic content appeared lysed or retracted to the poles of the cell (Fig. 5, C and D). Furthermore, AFS-I-alr1656 shows a high tendency to form double heterocysts (Fig. 6). Apparently, this phenotype is not a consequence of nitrogen starvation but of active fixation of atmospheric nitrogen, because deprivation of nitrogen under a pure argon atmosphere does not induce the Fox− phenotype (Fig. 5B, panel 4). Also, heterocyst differentiation in wild-type or mutant strains could not be observed under the argon atmosphere. Upon shift from argon to air, the Fox− phenotype became evident again (Fig. 5B, panel 5). After transfer to BG11 medium, all strains recovered the Chl and Phy content (Fig. 5A), but AFS-I-alr1656 and AFS-I-all3143 did so to a low level.
FIGURE 5.
Analysis of RND mutants during nitrogen step down. A, measurement of Chl and Phy content of whole cells after the indicated growth period and condition (top panel). Arrows, those strains and conditions that exhibit substantial differences in chlorophyll a and phycocyanin content with respect to CSR10 (57) used as control strain. B, 1 ml of cells with OD750 = 1 were placed in a culture dish for visual inspection of filament color. Arrows indicate those strains and conditions with differences in filament color with respect to CSR10. The indicated differences are representative of three independent biological replicas. Shown are bright field light microscopic (BF) and autofluorescence (AUF) representations of Anabaena filaments after 7 days of nitrogen deprivation (BG110) (C) and 3 days of recovery in nitrate-containing (BG11) media under oxic conditions (D). The position of (in the case of I-hgdD partially) differentiated heterocysts can be easily recognized by the gap in the autofluorescence due to disassembly of photosystem II.
FIGURE 6.

Quantitative analysis of double and triple heterocyst formation. CSR10, AFS-I-alr1656, and AFS-I-all3143 were grown in BG110, and images were taken as described in the legend to Fig. 4. Heterocysts were counted (100%) and classified as being single standing in the filament (1), forming a cluster of two heterocysts (2), or forming an even larger cluster (3). The average and S.D. values (error bars) of multiple analyzed images and cultures are shown.
The Mutants of RND Proteins Modulate the Lipid Composition
Because HgdD has been described to be important for the secretion of metabolites involved in the formation of the heterocyst-specific glycolipid HGL2 (9, 17), we analyzed the lipid content of mutant and wild-type cells after growth in nitrate-containing BG11 or nitrate-deprived BG110 growth medium. Although heterocyst development is negligible in the presents of nitrate (Anabaena sp. BG11; lane 1), nitrate deprivation induces heterocyst development and the subsequent accumulation of heterocyst-specific glycolipids (HGL1 and HGL2) (Anabaena sp. BG110; lane 2). In contrast, the amounts of the galactolipids (monogalactosyldiacylglycerol, digalactosyldiacylglycerol, and sulfoquinovosyldiacylglycerol) and phospholipids (phosphatidylglycerol) are not affected by nitrate deprivation (Anabaena sp. BG110; lane 2). Subsequently, we compared the lipid profile of wild type and mutants grown in BG110. We observed an altered profile for the analyzed mutants with respect to wild type, but also with respect to the AFS-I-hgdD strain (Fig. 7). Whereas the all3143, alr7631, and alr4267 mutants showed a reduction but not loss of HGL1 and HGL2, AFS-I-alr1656 exhibited an enhanced level of these lipids, which most likely reflects the multiple-heterocyst phenotype (Figs. 5 and 6). In addition, we noticed the presence of an enhanced level of MGDG in AFS-I-all3143, whereas this lipid was reduced in AFS-I-alr4267. Thus, none of the RND components is exclusively involved in secretion of HGL precursors, but a partial function in this process can be assumed at least for All3143, Alr7631, and Alr4267.
FIGURE 7.

Analysis of the lipid content in RND mutants. Thin layer chromatography analysis of the lipid content from whole filaments of the indicated strains is shown. Arrows indicate significant changes in the abundance of individual lipids of mutant strains with respect to the wild type. PG, phosphatidylglycerol; SQDG, sulfoquinovosyl diacylglycerol; DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; HGL1, heterocyst-specific glycolipid 1; HGL2, heterocyst-specific glycolipid 2.
Secondary Metabolite Transport by the Different RND Mutants
Recently, we have established a method to analyze uptake and secretion by Anabaena sp. using the model substrate ethidium bromide (5). The transport cycle can be described analytically, which enables us to determine the process that is affected by a mutant (5). Here we analyzed uptake by measuring the intercalation of ethidium into intracellular DNA (5), which can be described by a two-state model (59). At a low concentration of ethidium bromide (2.5 μm), we observed a significant intercalation for AFS-I-hgdD and a slightly lower one for AFS-I-all3143 (Fig. 8). At elevated concentrations (10 μm), the phenotype of ASF-I-hgdD and AFSI-all3143 became comparable, whereas AFS-I-alr4267 and AFS-I-all7631 still behaved like the wild type. This suggests that All3143 is the major RND homolog energizing the HgdD (TolC)-dependent export of ethidium.
FIGURE 8.
Analysis of ethidium efflux by RND mutants. The transport of ethidium at low (2.5 μm) and high (10 μm) concentrations of ethidium bromide (EB) was analyzed by monitoring the intercalation of ethidium into nucleic acids in whole cells over time. Wild type and I-hgdD are shown as controls. Kinetic parameters (Table 5) were calculated by consecutive reaction kinetics, as established previously (5).
We analyzed the intercalation by the equation presented previously (5) and were able to describe the data observed at ethidium bromide concentrations of 2.5, 5.0, 7.5, 10, 12.5, 25, and 125 μm by a combined least squares fit analysis with open parameters with a confidence of more than 95% (Table 5). As expected, the rate for uptake across the outer membrane was not significantly affected in any of the mutants (k1), whereas the import across the plasma membrane (k2) was reduced by 3-fold in AFS-I-all7631 when compared with the wild type. For the export rate across the plasma membrane (k−2), slight variations by a factor of 2 were found (Table 5).
TABLE 5.
Kinetic parameters determined in this study
Boldface type indicates significance of change compared with the wild type.
| Strain | k1 | k−1 | k2 | k−2 |
|---|---|---|---|---|
| s−1 | s−1 | s−1 | s−1 | |
| Wild type | (12 ± 5) × 10−2 | (13 ± 6) × 10−2 | (30 ± 4) × 10−4 | (24 ± 5) × 10−4 |
| AFS-I-hgdD | (20 ± 2) × 10−2 | (2 ± 1) × 10−7a | (39 ± 9) × 10−4 | (46 ± 9) × 10−4 |
| AFS-I-all1656 | (10 ± 3) × 10−2 | (6 ± 3) × 10−2 | (42 ± 8) × 10−4 | (23 ± 6) × 10−4 |
| AFS-I-all3143 | (18 ± 4) × 10−2 | (15 ± 3) × 10−7a | (40 ± 7) × 10−4 | (46 ± 6) × 10−4 |
| AFS-I-alr4267 | (5 ± 2) × 10−2 | (11 ± 3) × 10−3b | (42 ± 5) × 10−4 | (31 ± 3) × 10−4 |
| AFS-I-alr7631 | (16 ± 4) × 10−2 | (8 ± 5) × 10−2 | (9 ± 6) × 10−4b | (11 ± 7) × 10−4 |
a p < 0.005.
b p < 0.01.
The major variations were observed for the export across the outer membrane. The rate for the export by AFS-I-hgdD is reduced by 6 orders of magnitude, and the rate for the export of AFS-I-all3143 is reduced by 5 orders of magnitude when compared with wild type. Interestingly, the export rate across the outer membrane is reduced by a factor of 10 for AFS-I-alr4267 as well, whereas no reduction was observed for AFS-I-all1656 or AFS-I-alr7631. Thus, we can conclude that All1656 and Alr7631 are not involved in energizing the ethidium secretion and that Alr4267 influences somewhat the secretion behavior, whereas All3143 is the major RND transporter for this process.
Alr3143-All3144 Partially Complements the E. coli ΔacrAB Mutant
After demonstrating that All3143 is the only RND transporter whose mutation shows a similar ethidium efflux phenotype as mutation of hgdD, we propose that All3143 may represent the functional homolog of AcrB in Anabaena sp. To test this hypothesis, we amplified the ORF of hgdD and the successive ORFs all3143-all3144 and cloned them into pGR50 plasmid (Table 3) to generate the strains listed in Table 1. In addition, E. coli ΔtolC-tolC and ΔacrAB-acrABHis were generated to control the complementation efficiency of the transient plasmids (Table 1). The E. coli tripartite efflux system composed of ArcAB-TolC is efficient in secretion of ethidium, because no intercalation can be observed in the E. coli BW25113 strain (Fig. 9A). In contrast, the ΔtolC and ΔacrAB strains showed strong intercalation (Fig. 9, B and C). Interestingly, when preincubated with a 20–200 μm concentration of the uncoupler of the protein gradient CCCP typically used in experiments (60, 61), intercalation was significantly stronger than in the ΔtolC and ΔacrAB strains (Fig. 9, A and D). These observations suggest that a residual membrane potential-dependent secretion activity exists in the strains with impaired function of the AcrAB-TolC system.
FIGURE 9.
Complementation of E. coli ΔtolC and ΔacrAB by hgdD and all3144-all3143. A, intercalation of ethidium in E. coli wild-type cells (BW25113) and derived deletion mutant strains ΔtolC and ΔacrAB is shown in arbitrary fluorescence units (AFU) under constant settings for direct comparison. CCCP was used as uncoupler of the plasma membrane potential of E. coli wild-type strain. B, intercalation of ethidium in E. coli ΔtolC strain with transiently expressed E. coli tolC or Anabaena sp. tolC-like hgdD under the control of the tetracycline promoter. C, intercalation of ethidium in E. coli ΔacrAB strain with transiently expressed E. coli acrABHis or Anabaena sp. all3144-all3143 gene cluster under the control of the tetracycline promoter. D, intercalation of ethidium in E. coli wild-type cells (BW25113) in the presence of indicated concentrations of CCCP. The signal given on the y axis in A–D is represented in arbitrary fluorescence units. E, immunoblot of AcrB expression in ΔtolC (top) and ΔarcAB (bottom) strains.
We analyzed whether ΔtolC can be complemented by expression of hgdD under the control of a tetracyclin promoter. Although we could observe the complementation of ΔtolC by E. coli TolC, expression of hgdD did not complement the observed secretion phenotype (Fig. 9B). This might be explained by the different size of the two proteins, especially by the distinct N terminus of HgdD. Interestingly, in the ΔtolC background, the expression of endogenous acrB was up-regulated (Fig. 9E, lane 1 versus lane 2). The same effect could be observed in the strain complemented with hgdD, but not for the tolC complementation (lanes 3 and 4), consistent with the notion that HgdD cannot substitute the function of E. coli TolC.
Next, we aimed the complement the ΔacrAB phenotype by expression of all3144-all3143 (Fig. 9C). Here we observed a significant reduction of the ethidium intercalation rate when compared with the ΔacrAB mutant. Thus, All3144-All3143 partially complements the function of AcrAB. The lack of AcrAB in the All3144-All3143 complementation strain was confirmed by immunodecoration with αAcrB antibodies (Fig. 9E). This observation suggests that All3144-All3143 exhibits an AcrAB-like function.
DISCUSSION
The Complexity of the RND/MFP System in Anabaena
We dissected the network of RND family and MFP proteins in Anabaena sp. likely to be involved in the formation of tripartite efflux pumps with the TolC-like outer membrane channel HgdD. We identified six RND-like efflux pumps, of which all but Alr4267 are encoded downstream of a gene coding for an MFP (Fig. 1 and Table 4). In turn, all identified MFPs except All5304 can be assigned to either an identified RND-like protein or a putative ABC-transporter (Fig. 1 and Table 4).
Most RND-type proteins in E. coli are constitutively expressed at a very low level and do not contribute to general multidrug resistance (62). In E. coli, only acrB is constitutively expressed at a higher level than other RNDs and confers multidrug resistance, whereas mdtF expression increases during progression of growth and contributes to drug tolerance (63). MdtF is involved in the secretion of indole, which is an important signal molecule in stationary phase, biofilm formation, and intestinal growth (63, 64). The expression of the RND-type genes of Anabaena sp. was not detectable by RT-PCR at very early exponential growth with the experimental condition applied (Fig. 3). In contrast to E. coli, not one but four of them show a gradually increased expression in a growth phase-dependent manner (Fig. 3). This could suggest that RNDs in Anabaena sp. have a function in adaptation to stationary growth phase. In addition, the difference of expression when compared with E. coli might result from the different habitats of both bacteria. The freshwater-living Anabaena sp. does not form biofilms, nor is it known to be involved in infectious diseases. Furthermore, Anabaena sp. does not have to adapt to high bile salt concentrations present in the intestine. Therefore, a global response system might be sufficient for adaptation at the late growth phase in Anabaena sp.
The Relation between RND Function and Heterocyst Development
When grown in liquid BG110 culture medium and under a constant supply of 1% CO2, the insertion mutants of alr1656 and all3143, but not of alr4267 or all7631, seem to be impaired in fixation of nitrogen under oxygenic conditions (Figs. 4–6). In contrast to the phenotype of AFS-I-hgdD (9), the heterocysts are developed to a mature state but show a condensation of the cytoplasmic content. Additionally, we observed a highly frequent formation of double heterocysts in AFS-I-alr1656 (Figs. 5 and 6). A similar observation has been reported for a hetN-null mutant and a deletion mutant of patS (65). HetN is involved in maintaining the heterocyst periodic pattern subsequent to the action of PatS. PatS is a small peptide that is expressed in heterocysts and represses the initiation of heterocyst development in adjacent vegetative cells (e.g. see Refs. 19 and 66). The small peptide binds to HetR, the master regulator of heterocyst development, and inhibits its function (e.g. see Ref. 67). The pattern formation is thereby defined by the distribution of PatS between two developing heterocysts (e.g. see Ref. 19). Curiously, the transporter for PatS is not yet known. There are reports where RNDs are involved in secretion of small peptides of non-ribosomal origin, such as the syringomycins of Pseudomonas syringae (68). Moreover, PatS (13–17 amino acids) is of a similar size as syringomycin (69). Contradicting the hypothesis that Alr1656 is involved in PatS export, however, is the observation that alr1656 has an upstream gene encoding an MFP, which would imply a direct interaction with HgdD (Fig. 1 and Table 4). In the absence of further evidence, it is difficult to imagine that extracellular PatS diffusion regulates heterocyst spacing, and thus, considering the hypothesis formulated, one would have to argue that Alr1656 releases PatS into the periplasm.
Because MexF of Pseudomonas aeruginosa and MdtF of E. coli are involved in the efflux of nitrosyl indole derivatives (70, 71), Alr1656 and All3143 could also be involved in the detoxification of nitrosatives during diazotrophic growth. Nitrosatives, such as nitrosyl indole, are reactive nitrogen species that are generated in the presence of nitric oxide under anaerobic conditions. Nitric oxide damages the photosystems and is an inhibitor of nitrogenase activity (e.g. see Ref. 72). The microoxic environment of heterocysts might be especially prone to generation of nitrosatives. This notion is supported by the exclusive expression of two flavodiiron proteins known to protect cells from nitrosative or oxidative stress (Flv1B and Flv3B) in heterocysts (73). An accumulation of nitric oxide and nitrosatives during diazotrophic growth in AFS-I-alr1656 and AFS-I-all3143 could explain the Fox− phenotype and the reduction in chlorophyll a and phycocyanin content in vegetative cells (Fig. 5). This possibility is further supported by the growth of the two mutants under a protective argon atmosphere, which is depleted of dinitrogen, oxygen, and carbon dioxide and does not lead to an impaired photosystem. In contrast, the protection from oxidative stress even leads to an increase in chlorophyll a and phycocyanin content (Fig. 5, A and B).
The Ethidium Efflux from Anabaena sp. Cells Involves at Least Two RNDs
HgdD is essential for ethidium efflux in Anabaena sp. (5). The efflux of the corresponding insertion mutant has a k0.5 of 40 m, most likely as a result of passive diffusion. Thus, HgdD is an essential part of a non-redundant major efflux system, at least under standard conditions (5). So far, the nature of the plasma membrane factor that confers drug resistance was unknown, although recent experiments using CCCP suggested that the complex is energized by the proton gradient of the plasma membrane (5). The mutant of all3143 shows a striking similarity in susceptibility toward macrolide antibiotics (Fig. 4), with respect to the Fox− phenotype (Fig. 5) and, to some extent, also in lipid content (Fig. 7) as compared with the hgdD mutant. More importantly, AFS-I-all3143 is the only mutant that has an efflux rate for ethidium, which is comparable with AFS-I-hgdD (Fig. 8 and Table 5). Therefore, All3143 (and most likely in concert with the MFP All3144) is part of the tripartite secretion complex for ethidium.
However, a second RND has to be involved in the efflux of ethidium, because the accumulation of ethidium in AFS-I-all3143 is still somewhat lower than in AFS-I-hgdD (Table 5). At higher concentrations, this alternative capacity is saturated, and the specific action of All3143 becomes indispensable. A possible candidate for a second ethidium exporting RND might be the constitutively expressed alr4267 (Figs. 3 and 4), the mRNA of which is somewhat enhanced in AFS-I-all3143 when compared with wild-type cells (Fig. 4) and the mutant of which shows a slightly reduced ethidium efflux rate (Table 5). The mutant of alr4267 does not show an HgdD-related phenotype but displays a crimped filament shape independent of the nitrogen source (Figs. 4–6). In addition, the mutant of alr4267 has a slightly reduced content of the galactolipid MGDG (Fig. 7), which is the most abundant lipid of the plasma membrane (74–76). Thus, one might speculate that Alr4267 has a function in cell wall integrity or cell division by transport of the galactolipid MGDG (Table 5).
Our observations suggest that Alr4267 has a broad substrate compatibility but with low affinity. The substrate specificity might be defined by the associated MFP because Alr4267 is the only RND in Anabaena sp. without this adaptor encoded in the same genomic context. Thus, it is tempting to speculate that Alr4267 is a general RND that can utilize the MFP of different RNDs to provide a low affinity background efflux independent of expression level of other, more specialized, RNDs. According to this hypothesis, Alr4267 would associate with All3144 still expressed in the AFS-I-all3143 and would provide a low level of ethidium resistance, as seen in Fig. 8 and Table 5. This notion, however, has to be further addressed in the future.
All3143 Is the Functional Homolog of AcrB in Anabaena sp.
Consistent with the function of All3143 in metabolite secretion, All3143 and All3144 partially complement the AcrAB mutant of E. coli (Fig. 9). In contrast, HgdD is not able to complement the function of TolC, most likely due to its large N-terminal domain of about 30 kDa with a predicted unstructured conformation, which is not present in TolC of E. coli. In turn HgdD lacks the C-terminal domain (∼7 kDa) of E. coli TolC. Both the N and C terminus form the equatorial domain that contributes to the specific interaction with MFPs (1–3). Furthermore, Anabaena sp. PCC 7120, like other cyanobacteria, has a much larger periplasmic space of up to 30–40 nm (77) with a much thicker peptidoglycan layer when compared with E. coli. Thus, the N-terminal domain of HgdD might be an adjustment to the cyanobacterial periplasmic space in addition to its function in interacting with the peptidoglycan layer and thus disturb a correct RND-MFP-HgdD assembly in E. coli, where the distance between plasma and outer membrane only measures up to 20 nm.
We conclude that the RND proteins from Anabaena sp. fulfill multiple physiological roles and exhibit a wide variety of phenotypes concerning resistance to toxins and cell wall and heterocyst formation. Further, the partial complementation of the E. coli AcrAB mutant by All3143-All3144 with respect to ethidium efflux confirms a functional conservation of the bacterial toxin efflux systems in general. In addition, a partial functional redundancy of Alr4267 and All3143 exists because both mutations influence the ethidium efflux, and Alr4267 is up-regulated in the all3143 insertion mutant. Consistently, in contrast to the hgdD mutant, the all3143 mutant still confers resistance to roxithromycin (and in part to macrolides), which might be due to the function of Alr4267 and which might form a tripartite complex with HgdD and All3144 (MFP). Nevertheless, based on the phenotype of the two mutants, it can be suggested that All3143 is the major RND, whereas Alr4267 might act as a backup system with perhaps a specialized function. Starting from a bioinformatic identification of six putative RNDs, we have provided evidence of their possible functions, which calls for their detailed dissection in future work.
Acknowledgment
We thank Enrique Flores (Sevilla) for constant support and helpful discussions.
This work was supported by the Centre of Membrane Proteomics (Goethe University Frankfurt) (to A. H.), Deutsche Forschungsgemeinschaft-EXC115 (Cluster of Excellence Macromolecular Complexes) (to E. S. and K. M. P.), Deutsche Forschungsgemeinschaft Grants DFG SCHL 585/2 (to E. S.) and SFB 807 (to K. M. P. and E. S.), and Innovative Medicine Initiative (IMI) Project TRANSLOCATION (to K. M. P.).
- ABC
- ATP-binding cassette
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- Chl
- chlorophyll a
- HGL
- heterocyst-specific glycolipid
- HlyD
- hemolysin D
- MFP
- membrane fusion protein
- MFS
- major facilitator superfamily
- Phy
- phycocyanin
- RND
- resistance-nodulation-cell division
- TMH
- transmembrane helix.
REFERENCES
- 1. Piddock L. J. (2006) Multidrug-resistance efflux pumps. Not just for resistance. Nat. Rev. Microbiol. 4, 629–636 [DOI] [PubMed] [Google Scholar]
- 2. Pos K. M. (2009) Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta 1794, 782–793 [DOI] [PubMed] [Google Scholar]
- 3. Mirus O., Hahn A., Schleiff E. (2010) Outer Membrane Proteins. in Prokaryotic Cell Wall Compounds: Structure and Biochemistry (König H., Claus H., Varma A., eds) pp. 175–230, Springer-Verlag, Berlin [Google Scholar]
- 4. Delepelaire P. (2004) Type I secretion in Gram-negative bacteria. Biochim. Biophys. Acta. 1694, 149–161 [DOI] [PubMed] [Google Scholar]
- 5. Hahn A., Stevanovic M., Mirus O., Schleiff E. (2012) The TolC-like protein HgdD of the cyanobacterium Anabaena sp. PCC 7120 is involved in secondary metabolite export and antibiotic resistance. J. Biol. Chem. 287, 41126–41138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bleuel C., Grosse C., Taudte N., Scherer J., Wesenberg D., Krauss G. J., Nies D. H., Grass G. (2005) TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J. Bacteriol. 187, 6701–6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nicolaisen K., Hahn A., Valdebenito M., Moslavac S., Samborski A., Maldener I., Wilken C., Valladares A., Flores E., Hantke K., Schleiff E. (2010) The interplay between siderophore secretion and coupled iron and copper transport in the heterocyst-forming cyanobacterium Anabaena sp. PCC 7120. Biochim. Biophys. Acta 1798, 2131–2140 [DOI] [PubMed] [Google Scholar]
- 8. Newton S. M., Trinh V., Pi H., Klebba P. E. (2010) Direct measurements of the outer membrane stage of ferric enterobactin transport. Postuptake binding. J. Biol. Chem. 285, 17488–17497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moslavac S., Nicolaisen K., Mirus O., Al Dehni F., Pernil R., Flores E., Maldener I., Schleiff E. (2007) A TolC-like protein is required for heterocyst development in Anabaena sp. strain PCC 7120. J. Bacteriol. 189, 7887–7895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Awram P., Smit J. (1998) The Caulobacter crescentus paracrystalline S-layer protein is secreted by an ABC transporter (type I) secretion apparatus. J. Bacteriol. 180, 3062–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goldberg M., Pribyl T., Juhnke S., Nies D. H. (1999) Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J. Biol. Chem. 274, 26065–26070 [DOI] [PubMed] [Google Scholar]
- 12. Bolhuis H., van Veen H. W., Poolman B., Driessen A. J., Konings W. N. (1997) Mechanisms of multidrug transporters. FEMS Microbiol. Rev. 21, 55–84 [DOI] [PubMed] [Google Scholar]
- 13. Nikaido H. (1996) Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178, 5853–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thanabalu T., Koronakis E., Hughes C., Koronakis V. (1998) Substrate-induced assembly of a contiguous channel for protein export from E. coli. Reversible bridging of an inner membrane translocase to an outer membrane exit pore. EMBO J. 17, 6487–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blair J. M., Piddock L. J. (2009) Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria. An update. Curr. Opin. Microbiol. 12, 512–519 [DOI] [PubMed] [Google Scholar]
- 16. Nikaido H., Takatsuka Y. (2009) Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta 1794, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Staron P., Forchhammer K., Maldener I. (2011) Novel ATP-driven pathway of glycolipid export involving TolC protein. J. Biol. Chem. 286, 38202–38210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nicolaisen K., Hahn A., Schleiff E. (2009) The cell wall in heterocyst formation by Anabaena sp. PCC 7120. J. Basic Microbiol. 49, 5–24 [DOI] [PubMed] [Google Scholar]
- 19. Flores E., Herrero A. (2010) Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 8, 39–50 [DOI] [PubMed] [Google Scholar]
- 20. Fiedler G., Arnold M., Hannus S., Maldener I. (1998) The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 27, 1193–1202 [DOI] [PubMed] [Google Scholar]
- 21. Li W., Godzik A. (2006) Cd-hit. A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 [DOI] [PubMed] [Google Scholar]
- 22. Frickey T., Lupas A. (2004) CLANS. A Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20, 3702–3704 [DOI] [PubMed] [Google Scholar]
- 23. Mirus O., Strauss S., Nicolaisen K., von Haeseler A., Schleiff E. (2009) TonB-dependent transporters and their occurrence in cyanobacteria. BMC Biol. 7, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Valladares A., Rodríguez V., Camargo S., Martínez-Noël G. M., Herrero A., Luque I. (2011) Specific role of the cyanobacterial PipX factor in the heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 193, 1172–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Elhai J., Vepritskiy A., Muro-Pastor A. M., Flores E., Wolk C. P. (1997) Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179, 1998–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants. The Keio collection. Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Datsenko K. A., Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Provence D. L., Curtiss R., III. (1994) Gene transfer in Gram-negative bacteria. in Methods for General and Molecular Bacteriology (Gerhardt P., Murray R. G. E., Wood W. A., Krieg N. R., eds) pp. 317–347 American Society for Microbiology Press, Washington, D. C [Google Scholar]
- 29. Rippka R., Deruelies J., Waterbury J. B., Herdman M., Stanier R. Y. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111, 1–61 [Google Scholar]
- 30. Nicolaisen K., Mariscal V., Bredemeier R., Pernil R., Moslavac S., López-Igual R., Maldener I., Herrero A., Schleiff E., Flores E. (2009) The outer membrane of a heterocyst-forming cyanobacterium is a permeability barrier for uptake of metabolites that are exchanged between cells. Mol. Microbiol. 74, 58–70 [DOI] [PubMed] [Google Scholar]
- 31. Arnon R. I. (1949) Copper enzymes in isolated chloroplasts. Polyphenolpxidase in Beta vulgatis. Plant Physiol. 24, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Porra R. J., Thompson W. A., Kriedemann P. E. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents. Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 [Google Scholar]
- 33. Murakami S., Nakashima R., Yamashita E., Matsumoto T., Yamaguchi A. (2006) Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443, 173–179 [DOI] [PubMed] [Google Scholar]
- 34. Rosenberg E. Y., Ma D., Nikaido H. (2000) AcrD of Escherichia coli is an aminoglycoside efflux pump. J. Bacteriol. 182, 1754–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Daley D. O., Rapp M., Granseth E., Melén K., Drew D., von Heijne G. (2005) Global topology analysis of the Escherichia coli inner membrane proteome. Science 308, 1321–1323 [DOI] [PubMed] [Google Scholar]
- 36. Kieboom J., de Bont J. (2001) Identification and molecular characterization of an efflux system involved in Pseudomonas putida S12 multidrug resistance. Microbiology 147, 43–51 [DOI] [PubMed] [Google Scholar]
- 37. Tian T., Wu X. G., Duan H. M., Zhang L. Q. (2010) The resistance-nodulation-division efflux pump EmhABC influences the production of 2,4-diacetylphloroglucinol in Pseudomonas fluorescens 2P24. Microbiology 156, 39–48 [DOI] [PubMed] [Google Scholar]
- 38. Masi M., Pagès J. M., Villard C., Pradel E. (2005) The eefABC multidrug efflux pump operon is repressed by H-NS in Enterobacter aerogenes. J. Bacteriol. 187, 3894–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X. Z., Zhang L., Poole K. (1998) Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J. Bacteriol. 180, 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stover C. K., Pham X. Q., Erwin A. L., Mizoguchi S. D., Warrener P., Hickey M. J., Brinkman F. S., Hufnagle W. O., Kowalik D. J., Lagrou M., Garber R. L., Goltry L., Tolentino E., Westbrock-Wadman S., Yuan Y., Brody L. L., Coulter S. N., Folger K. R., Kas A., Larbig K., Lim R., Smith K., Spencer D., Wong G. K., Wu Z., Paulsen I. T., Reizer J., Saier M. H., Hancock R. E., Lory S., Olson M. V. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964 [DOI] [PubMed] [Google Scholar]
- 41. Köhler T., Michéa-Hamzehpour M., Henze U., Gotoh N., Curty L. K., Pechère J. C. (1997) Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23, 345–354 [DOI] [PubMed] [Google Scholar]
- 42. Bohnert J. A., Schuster S., Fähnrich E., Trittler R., Kern W. V. (2007) Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF). J. Antimicrob. Chemother. 59, 1216–1222 [DOI] [PubMed] [Google Scholar]
- 43. Ramos J. L., Duque E., Godoy P., Segura A. (1998) Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 180, 3323–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Outten F. W., Huffman D. L., Hale J. A., O'Halloran T. V. (2001) The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276, 30670–30677 [DOI] [PubMed] [Google Scholar]
- 45. Liesegang H., Lemke K., Siddiqui R. A., Schlegel H. G. (1993) Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J. Bacteriol. 175, 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rensing C., Pribyl T., Nies D. H. (1997) New functions for the three subunits of the CzcCBA cation-proton antiporter. J. Bacteriol. 179, 6871–6879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baranova N., Nikaido H. (2002) The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184, 4168–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paulsen I. T., Brown M. H., Skurray R. A. (1996) Proton-dependent multidrug efflux systems. Microbiol. Rev. 60, 575–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lomovskaya O., Lewis K. (1992) Emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. U.S.A. 89, 8938–8942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Touchon M., Hoede C., Tenaillon O., Barbe V., Baeriswyl S., Bidet P., Bingen E., Bonacorsi S., Bouchier C., Bouvet O., Calteau A., Chiapello H., Clermont O., Cruveiller S., Danchin A., Diard M., Dossat C., Karoui M. E., Frapy E., Garry L., Ghigo J. M., Gilles A. M., Johnson J., Le Bouguénec C., Lescat M., Mangenot S., Martinez-Jéhanne V., Matic I., Nassif X., Oztas S., Petit M. A., Pichon C., Rouy Z., Ruf C. S., Schneider D., Tourret J., Vacherie B., Vallenet D., Médigue C., Rocha E. P., Denamur E. (2009) Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5, e1000344–e100034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Winstanley C., Langille M. G., Fothergill J. L., Kukavica-Ibrulj I., Paradis-Bleau C., Sanschagrin F., Thomson N. R., Winsor G. L., Quail M. A., Lennard N., Bignell A., Clarke L., Seeger K., Saunders D., Harris D., Parkhill J., Hancock R. E., Brinkman F. S., Levesque R. C. (2009) Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool epidemic strain of Pseudomonas aeruginosa. Genome Res. 19, 12–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ma D., Cook D. N., Alberti M., Pon N. G., Nikaido H., Hearst J. E. (1993) Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175, 6299–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Coudeyras S., Nakusi L., Charbonnel N., Forestier C. (2008) A tripartite efflux pump involved in gastrointestinal colonization by Klebsiella pneumoniae confers a tolerance response to inorganic acid. Infect. Immun. 76, 4633–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li X. Z., Nikaido H., Poole K. (1995) Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39, 1948–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Poole K., Gotoh N., Tsujimoto H., Zhao Q., Wada A., Yamasaki T., Neshat S., Yamagishi J., Li X. Z., Nishino T. (1996) Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21, 713–724 [DOI] [PubMed] [Google Scholar]
- 56. Felmlee T., Pellett S., Welch R. A. (1985) Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J. Bacteriol. 163, 94–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tikhonova E. B., Dastidar V., Rybenkov V. V., Zgurskaya H. I. (2009) Kinetic control of TolC recruitment by multidrug efflux complexes. Proc. Natl. Acad. Sci. U.S.A. 106, 16416–16421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pernil R., Picossi S., Mariscal V., Herrero A., Flores E. (2008) ABC-type amino acid uptake transporters Bgt and N-II of Anabaena sp. strain PCC 7120 share an ATPase subunit and are expressed in vegetative cells and heterocysts. Mol. Microbiol. 67, 1067–1080 [DOI] [PubMed] [Google Scholar]
- 59. Chrastil J. (1993) Determination of the first-order consecutive reversible reaction kinetics. Comp. Chem. 17, 103–106 [Google Scholar]
- 60. Ocaktan A., Yoneyama H., Nakae T. (1997) Use of fluorescence probes to monitor function of the subunit proteins of the MexA-MexB-oprM drug extrusion machinery in Pseudomonas aeruginosa. J. Biol. Chem. 272, 21964–21969 [DOI] [PubMed] [Google Scholar]
- 61. Murakami S., Tamura N., Saito A., Hirata T., Yamaguchi A. (2004) Extramembrane central pore of multidrug exporter AcrB in Escherichia coli plays an important role in drug transport. J. Biol. Chem. 279, 3743–3748 [DOI] [PubMed] [Google Scholar]
- 62. Sulavik M. C., Houseweart C., Cramer C., Jiwani N., Murgolo N., Greene J., DiDomenico B., Shaw K. J., Miller G. H., Hare R., Shimer G. (2001) Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45, 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kobayashi A., Hirakawa H., Hirata T., Nishino K., Yamaguchi A. (2006) Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J. Bacteriol. 188, 5693–5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bansal T., Alaniz R. C., Wood T. K., Jayaraman A. (2010) The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Borthakur P. B., Orozco C. C., Young-Robbins S. S., Haselkorn R., Callahan S. M. (2005) Inactivation of patS and hetN causes lethal levels of heterocyst differentiation in the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 57, 111–123 [DOI] [PubMed] [Google Scholar]
- 66. Corrales-Guerrero L., Mariscal V., Flores E., Herrero A. (2013) Functional dissection and evidence for intercellular transfer of the heterocyst-differentiation PatS morphogen. Mol. Microbiol. 88, 1093–1105 [DOI] [PubMed] [Google Scholar]
- 67. Feldmann E. A., Ni S., Sahu I. D., Mishler C. H., Levengood J. D., Kushnir Y., McCarrick R. M., Lorigan G. A., Tolbert B. S., Callahan S. M., Kennedy M. A. (2012) Differential binding between PatS C-terminal peptide fragments and HetR from Anabaena sp. PCC 7120. Biochemistry 51, 2436–2442 [DOI] [PubMed] [Google Scholar]
- 68. Kang H., Gross D. C. (2005) Characterization of a resistance-nodulation-cell division transporter system associated with the syr-syp genomic island of Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 71, 5056–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bender C. L., Alarcón-Chaidez F., Gross D. C. (1999) Pseudomonas syringae phytotoxins. Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63, 266–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fetar H., Gilmour C., Klinoski R., Daigle D. M., Dean C. R., Poole K. (2011) mexEF-oprN multidrug efflux operon of Pseudomonas aeruginosa. Regulation by the MexT activator in response to nitrosative stress and chloramphenicol. Antimicrob. Agents Chemother. 55, 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang Y., Xiao M., Horiyama T., Zhang Y., Li X., Nishino K., Yan A. (2011) The multidrug efflux pump MdtEF protects against nitrosative damage during the anaerobic respiration in Escherichia coli. J. Biol. Chem. 286, 26576–26584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xue L., Li S., Zhang B., Shi X., Chang S. (2011) Counteractive action of nitric oxide on the decrease of nitrogenase activity induced by enhanced ultraviolet-B radiation in cyanobacterium. Curr. Microbiol. 62, 1253–1259 [DOI] [PubMed] [Google Scholar]
- 73. Ermakova M., Battchikova N., Allahverdiyeva Y., Aro E. M. (2013) Novel heterocyst-specific flavodiiron proteins in Anabaena sp. PCC 7120. FEBS Lett. 587, 82–87 [DOI] [PubMed] [Google Scholar]
- 74. Omata T., Murata N. (1983) Isolation and characterization of the cytoplasmic membranes from the blue-green alga (cyanobacterium) Anacystis nidulans. Plant Cell Physiol. 24, 1101–1112 [Google Scholar]
- 75. Huang H., Zhong Z. P., Wang K. B., Bai K. Z., Li L. B., Kuang T. Y. (2004) Isolation and characterization of the cytoplasmic membrane from the terrestrial cyanobacterium–Nostoc flagelliforme. Acta Bot. Sin. 46, 1186–1191 [Google Scholar]
- 76. Huang F., Hedman E., Funk C., Kieselbach T., Schröder W. P., Norling B. (2004) Isolation of outer membrane of Synechocystis sp. PCC 6803 and its proteomic characterization. Mol. Cell Proteomics 3, 586–595 [DOI] [PubMed] [Google Scholar]
- 77. Wilk L., Strauss M., Rudolf M., Nicolaisen K., Flores E., Kühlbrandt W., Schleiff E. (2011) Outer membrane continuity and septosome formation between vegetative cells in the filaments of Anabaena sp. PCC 7120. Cell Microbiol. 13, 1744–1754 [DOI] [PubMed] [Google Scholar]
- 78. Nakao M., Okamoto S., Kohara M., Fujishiro T., Fujisawa T., Sato S., Tabata S., Kaneko T., Nakamura Y. (2010) CyanoBase. The cyanobacteria genome database update 2010. Nucleic Acids Res. 38, D379–D381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Black T. A., Cai Y., Wolk C. P. (1993) Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9, 77–84 [DOI] [PubMed] [Google Scholar]
- 80. Bundschuh F. A., Hannappel A., Anderka O., Ludwig B. (2009) Surf1, associated with Leigh syndrome in humans, is a heme-binding protein in bacterial oxidase biogenesis. J. Biol. Chem. 284, 25735–25741 [DOI] [PMC free article] [PubMed] [Google Scholar]