Abstract
Background:
The cytochrome P450 enzymes (CYP) play an important role in the metabolism of many therapeutic agents. The activities of different enzymes exhibit variability in different populations, which causes variations in drug response or toxicity. The CYP2B6 and CYP2C8 enzymes are encoded by polymorphic genes characterised by different single nucleotide polymorphisms (SNPs). Several of these CYP variants are often associated with slow metabolism phenotypes. This study aimed to analyse the frequencies of allelic variants of CYP2B6 and CYP2C8 in the Mozambican population.
Methods:
Using a polymerase chain reaction and restriction fragment length polymorphism assay (PCR-RFLP), the frequencies of the allelic variants of CYP2B6 (c.64C>T, c.516G>T, c.777C>A, c.785A>G, c.1459C>T) and CYP2C8 (c.805A>T, c.416G>A, c.1196A>G, c.792C>G) were determined in 360 Mozambican blood donors.
Results:
The frequencies of the allelic variants of the CYP2B6 gene were 0.057, 0.426, 0.0, 0.410, and 0.004. For the CYP2C8 gene, the frequencies of the allelic variants were 0.160, 0.048, 0.0, and 0.005. No significant differences were observed between the gender and geographic distribution of volunteers around the country.
Conclusion:
The frequencies of the allelic variants of the CYP2B6 and CYP2C8 genes were found to be homogeneously distributed in the Mozambican population and were comparable to other African populations. Further studies are required to explore the impact of these variants on the clinical response (efficacy and toxicity) of drugs, including antimalarials.
Keywords: allele frequency, CYP2B6, CYP2C8, polymorphism
Introduction
Cytochrome P450 (CYP) represents a family of isoenzymes responsible for the metabolism of many therapeutic agents. Differences in the activities of these enzymes are thought to be responsible for individual variabilities in drug response or toxicity. Variant allele frequencies of many pharmacogenetically relevant polymorphisms differ greatly among populations of different ethnic groups. However, this information is scarce in some areas of the world with different ethnicities (1). The CYP2B6 and CYP2C8 isoenzymes are both involved in the human biotransformation of a wide variety of drugs (2–4), and several genetic polymorphisms have been found in the genes coding for these enzymes.
Among the drugs metabolised by CYP2B6, the antimalarial drug Artemisinin (3), its derivative artesunate (5), and the antiretroviral drugs Nevirapine and Efavirenz (6) are the most important. CYP2B6 protein is mainly expressed in the liver but can also be expressed in the kidneys, intestines, skin, brain, and lungs (7). Its expression represents approximately 2% to 10% of the total CYP content (8,9) and varies widely (100-fold) between individuals. Such variation has been associated with genetic polymorphism, which can influence the expression and function of the enzyme as reflected in the therapeutic outcomes of CYP2B6-metabolised drugs (10,11).
The CYP2B6 gene has been mapped to chromosome 19 between 19q12 and 19q13.2, and is composed of nine exons (12). To date, 28 allelic variants have been described and characterised (∗1B to ∗29). Some of the allelic variants have been associated with low enzyme activity in vitro (10,13,14). These variants include single nucleotide polymorphisms (SNPs) located in the coding region, such as CYP2B6∗2A (c.64C>T), CYP2B6∗3 (c.777C>A), CYP2B6∗4A (c.785A>G), CYP2B6∗5A (c.1459C>T), CYP2B6∗6A (c.516G>T and c.785A>G) and CYP2B6∗7A (c.516G>T, c.785A> G and c.1459C>T) (10), and some variants in the promoter region, such as 1848C>A, –801G>T, –750T>C, and –82T>C (15). Many of these SNPs have an impact on the enzyme activity. Data from the literature has shown that the c.1459T variant in exon 9 is responsible for 8-fold lower enzyme activity compared to the wild-type protein (10). Regarding the influence of this genetic variability on therapeutic outcomes, the subjects carrying the CYP2B6∗6A allele (c.516G>T, c.785A>G) have an increased risk of Efavirenz toxicity (16–18). The reported frequencies of CYP2B6 polymorphisms in populations of different ethnicities are the following: CYP2B6∗6A (c.516G>T, c.785A>G), 15–40% in Asians and over 50% in African-Americans (10,19,20) and CYP2B6∗5A (c.1459C>T), 14–25% in Caucasians (1).
The CYP2C8 enzyme is part of the four member of CYP2C subfamily (CYP2C9, CYP2C18, and CYP2C19) and is involved in the metabolism of several therapeutically important drugs and endogenous compounds (21). This enzyme is mainly expressed in the liver but also in various extra-hepatic tissues, such as the kidney, heart, mammary gland, and duodenum (22,23). The CYP2C8 gene locus is polymorphic and several variants have been reported, resulting in more than 16 different alleles Polymorphisms in the CYP2C8 gene have been implicated in the variabilities of CYP2C8 activity with different phenotypes (24,25). The main variants are CYP2C8∗2, CYP2C8∗3, CYP2C8∗4, and CYP2C8∗5. The reported frequencies of the CYP2C8 polymorphisms in populations of different ethnicities are the following: CYP2C8∗2 (c.805A>T), 11–17% in African-Americans (26–28); CYP2C8∗3 (c.416 G>A), 0.4–2.1% in African-Americans (28,29) and > 15% in Caucasians (30); CYP2C8∗4 (c.792C>G), 0–5.8% in Caucasians and very rare in African and Asian populations; and CYP2C8∗5 (c.475delA), 0–0.2% in Asians and absent in Caucasian and African populations (24,31).
Malaria poses a significant health threat to individuals in Africa, including Mozambique. The use of artemisinin derivative-based combination therapy (ACT) drugs has been approved in many countries in Africa to respond to the increasing parasite resistance to chloroquine and sulfadoxineperimethamine. Today, it is known that genetic variability in the human genes coding for drugmetabolising enzymes, including antimalarials, may contribute to differences in drug response or toxicity and may be responsible for treatment failure and adverse drug reactions. Unfortunately, in several populations, only limited information is available regarding the functionally relevant SNPs in genes potentially involved in the elimination of the major antimalarial drugs. Here, for the first time, we present the frequencies of the most commonly observed CYP2B6 (c.64C>T, c.516G>T, c.777C>A, c.785A>G, c.1459C>T) and CYP2C8 (c.805A>T, c.416G>A, c.1196A>G, c.792C>G) allelic variants in the Mozambican population. This study can guide future studies to elucidate the importance of these polymorphisms to the therapeutic efficacy and in adverse drug events.
Materials and Methods
Study population
The study population was composed of Mozambican blood donor volunteers from all three regions of the country. Written informed consent before enrolment was obtained from each subject. Ethical approval for this study was obtained from the Ethics Committees of the Ministry of Health, Mozambique and the National Committee for Research Ethics (CONEP), Instituto Oswaldo Cruz, Oswaldo Cruz Foundation-FIOCRUZ, Brazil.
Blood sample collection and DNA extraction
Blood samples were collected by venepuncture from the consenting volunteers, and approximately 200 μL were spotted onto FTA Classic Cards (Whatman®) and allowed to air-dry. The cards were placed in plastic bags and shipped to the Instituto Oswaldo Cruz, FIOCRUZ (Brazil), for analysis. For use in PCR, discs were punched out of the FTA blood spot using a clean 1.2 mm diameter Harris Micropunch™ (Whatman®). The discs were washed three times with 200 μL of FTA purification reagent (Whatman® ) and twice with 200 μL of 1 mM Tris Ethylenediaminetetraacetic acid (TE) (10 mM Tris, 1 mM ethylenediaminetetraacetic acid (EDTA) buffer, pH 8.0, with a 5 min incubation with each wash per the manufacturer’s instructions. The washed discs were dried in a heat block at 56 °C for 10 min and then used in the PCR reactions.
CYP2B6 and CYP2C8 Genotyping
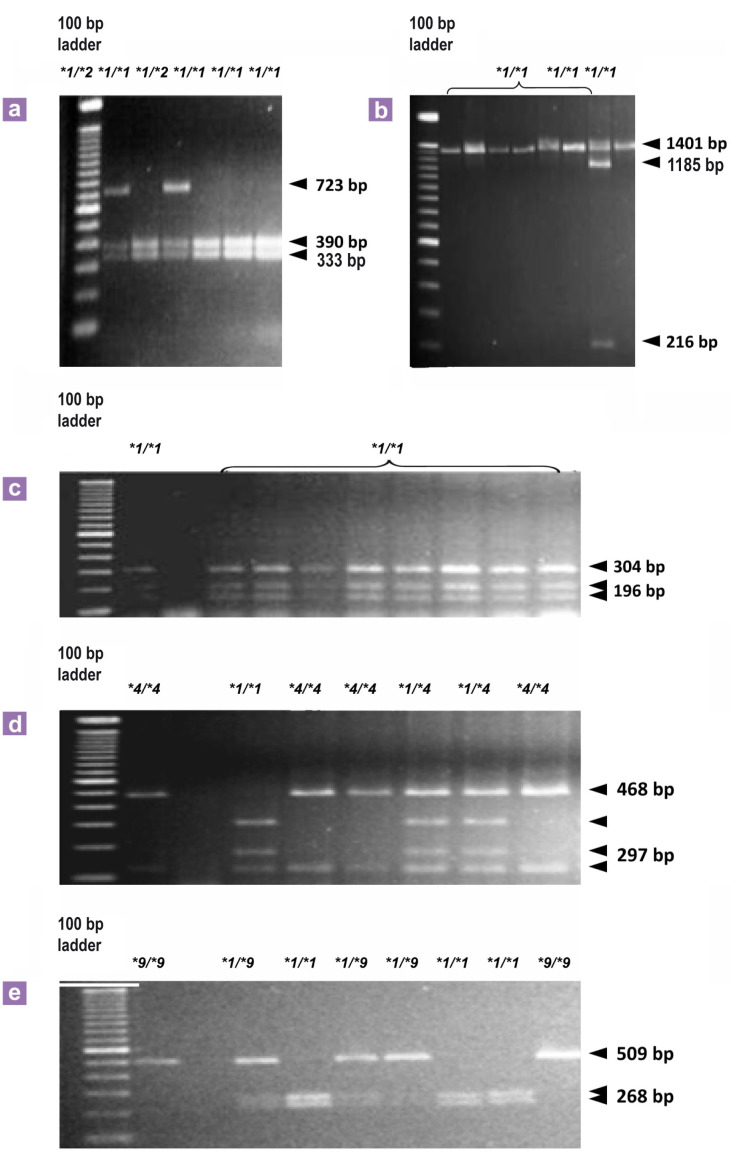
Genotyping of the CYP2B6 coding region, c.64C>T (CYP2B6∗2A), c.777 C>A (CYP2B6∗3), c.785A>G (CYP2B6∗4A,∗6A or ∗7A), c.1459C>T (CYP2B6∗5A or ∗7A) and c.516G>T (CYP2B6∗9A), and the CYP2C8 coding region, c.805A>T (CYP2C8∗2), c.416G>A (CYP2C8∗3), c.792C>G (CYP2C8∗4) and c.1196A>G (CYP2C8∗3), was achieved by PCR-RFLP according to previously described methods with pre-designed primers (10,25,32). For the SNPs in the CYP2B6 gene, the PCR reactions were conducted using a reaction mixture of six FTA card discs of 1.2 mm diameter, 10x PCR buffer, 0.2 mM dNTPs, 1.5 to 3.0 mM MgCl2, 1 to 2.5 U of Taq polymerase (Invitrogen, Applied Biosystems, Foster City, CA, USA) and 20 pmol of each primer. The PCR fragments were produced under the following optimal conditions: an initial denaturation step of 5 min at 95 °C followed by 30 cycles of denaturation at 95 °C for 30 s, annealing for 30 s at 50 °C to 60 °C, and extending for 60 s at 72 °C using a Veriti Thermal Cycler (Applied Biosystems, Foster City, CA). The PCR products were digested with the following: Hae II which digests the wild-type sequence into fragments of 390 bp, 333 bp, and 304 bp for c.64C>T and 196 bp and 1401 bp for c.777 C>A; Sty I for c.785A>G, which digests the wild-type sequence into fragments of 297 bp, 171 bp and 116 bp; Bgl II for c.1459C>T, which leaves the wildtype sequence of 1401 bp undigested; and Bsr I for c.516G>T, which digests the wild-type sequence into fragments of 268 bp and 241 bp. The digestion products were analysed by electrophoresis on a 2% agarose gel (Figure 1) (Table 1).
Figure 1:
Genotyping of CYP2B6 variants by PCR-RFLP analysed with 2% agarose gel electrophoresis: (a) show the fragments after digestion with Hae II of the PCR product for identification of CYP2B68∗2 (c. 64C>T) variant. (b) show the fragments after digestion with Bgl II of the PCR product for identification of CYP2B6∗5 (c.1459C>T). (c) show the fragments after digestion with Hae II of the PCR product for identification of CYP2B6∗3 (c.777C>A). (d) show the fragments after digestion with StyI of the PCR product for identification of CYP2B6∗4 (c.785A>G) and (e) fragments of the PCR product after digestion with Bsr I for identification of CYP2B6∗9 (c.516G>T). ∗1/∗1- represent a typical profile for subject genotyped as homozygous wild type; ∗1/∗2, ∗1/∗5, ∗1/∗4, ∗1/∗9- represent a profile for a single subject genotyped as heterozygous; ∗4/∗4, ∗9/∗9- represent a typical profile for subject genotyped as homozygous mutant.
Table 1.
Genotyping method for CYP2B6 and CYP2C8 polymorphisms by PCR-RFLP
| Polymorphisms | Primer Set | Cycling Pattern | Restriction enzyme digestion pattern | Fragment Length (bp) |
|---|---|---|---|---|
| c.1196A>G (CYP2C8∗3) | F- 5’-CTTCCGTGCTACATGATGACG-3’ | 95 °C–5min (1×), 94 °C–20s | 10 μL PCR Product | wt: 390, 333 |
| R- 5´-GTAAATACCACTTGACCA-3’ | 50 °C–30 s, 72 °C–60 s (30×) | 1 U of Hae II | mt: 723 | |
| 72 °C–10 s (1×) | 4 hours at 37 °C | |||
| c. 777 C>A (CYP2B6∗3) | F- 5´-GACAGAAGGATGAGGGAGGAA-3´ | 95 °C–5 min (1×), 95 °C–30 s | 10 μL PCR Product | wt: 304, 196, 140 |
| R- 5´-CTCCCTCTGTCTTTCATTCTGT-3´ | 59 °C–30 s, 72 °C–60 s (30×) | 1U of Hae II | mt: 500, 140 | |
| 72 °C–10 s (1×) | 4 hours at 37 °C | |||
| c.785A>G (CYP2B6∗ 4A,∗6A or ∗7A) | F- 5´-GACAGAAGGATGAGGGAGGAA-3´ | 95 °C–5 min (1×), 95 °C–30s | 10 μL PCR Product | wt: 297, 171, 116 |
| R- 5´-CTCCCTCTGTCTTTCATTCTGT-3´ | 59 °C–30 s, 72 °C–60s (30×) | 1 U of Sty I | mt: 468, 116 | |
| 72 °C–10 s (1×) | 4 hours at 37 °C | |||
| c.1459.C>T (CYP2B6∗ 5A or ∗7A) | F- 5´-TGAGAATCAGTGGAAGCCATAGA-3 | 95 °C–5 min (1×), 95 °C–30 s | 10 μL PCR Product | wt: 1401 |
| R-5´-TAATTTTCGATAATCTCACTCCTGC-3´ | 59 °C–30 s, 72 °C–60s (30×) | 1 U of Bgl II | mt: 1185, 216 | |
| 72 °C–10 s (1×) | 4 hours at 37 °C | |||
| c.516G>T (CYP2B6∗9A) | 5´-GACAGAAGGATGAGGGAGGAA-3´ | 95 °C–5min (1×), 95 °C–30 s | 10 μL PCR Product | wt: 268, 241 |
| 5´-CTCCCTCTGTCTTTCATTCTGT- 3´ | 56 vC–30 s, 72 °C–40 s (30×) | 1 U of Bsr I | mt: 509, 17 | |
| 72 °C–10 s (1×) | 16 hours at 65 °C | |||
| c.805A>T (CYP2C8∗2) | F-5’-AAGATACATATATCTTATGACATG-3’ | 95 °C–5 min (1×), 94 °C–20 s | 10 μL PCR Product | wt: 214, 98 |
| - 5’-ATCCTTAGTAAATTACAGAAGG-3’ | 55 °C–20 s, 72 °C–10 s (38×) | 1 U of Bcl I | mt: 312 | |
| 72 °C–5 s (1×) | 16 hours at 37 °C | |||
| c.416G>A (CYP2C8∗3) | F- 5’-AGGCAATTCCCCAATATCTC -3’ | 95 °C–5 min (1×), 94 °C–20 s | 10 μL PCR Product | wt: 310, 37 |
| R- 5`- ACTCCTCCACAAGGCAGTGA-3 | 55 °C–20 s, 72 °C–10 s (38×) | 1 U of Bse RI | mt: 347 | |
| 72 °C–5s (1×) | 16 hours at 37 °C | |||
| c.1196A>G (CYP2C8∗3) | F- 5’-CTTCCGTGCTACATGATGACG-3’ | 95 °C–5min (1×), 94 °C–20s | 10 μL PCR Product | wt: 92, 25 |
| R- 5`-CTGCTGAGAAAGGCATGAAG-3 | 55 °C–20 s, 72 °C–10 s (38×) | 1 U of Xmn I | mt: 117 | |
| 72 °C–5 s (1×) | 16 hours at 37 °C | |||
| c.792C>G (CYP2C8∗4) | F- 5’- AAAGTAAAAGAACACCAAGC-3’ | 94 °C–5 min (1×), 94 °C–30 s | 10 μL PCR Product | wt: 83,53, 31 |
| R- 5’-AAACATCCTTAGTAAATTACA-3’ | 48 °C–30 s, 72 °C–30 s (30×) | 1 U of Taq I | mt: 136, 31 | |
| 72 °C–5 s (1×) | 4 hours at 65 °C |
Abbreviations: bp = base pairs, wt = wild type, mt = variant type.
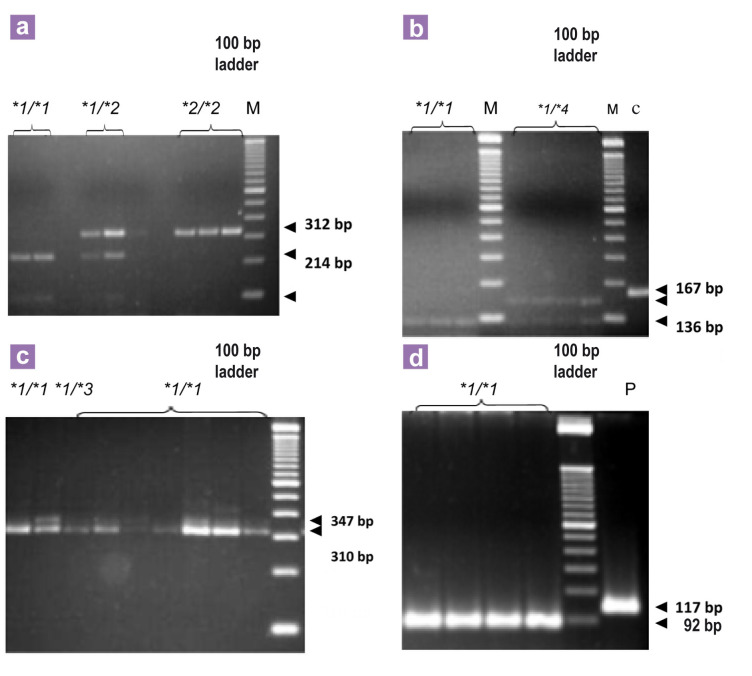
The amplification of c.805A>T (CYP2C8∗2), c.416G>A (CYP2C8∗3) and c.1196A>G (CYP2C8∗3) was performed by the addition of 5 FTA card discs of 1.2 mm diameter to a reaction mixture containing 1× buffer, 2.5 mM MgCl2, 0.2 mM dNTPs, 0.25 mM of each primer and Taq polymerase 1 U/μL, 10% Betaine. For the c.792C>G (CYP2C8∗4) variant, the reaction mixture contained 2.5 mM MgCl2, 1× buffer, 250 μM of dNTPs, 0.4 mM of each oligonucleotide, 10% glycerol, 1 U Taq polymerase and 3 FTA card discs of 1.2 mm diameter. The products were digested with the following: Bcl I for c.805A>T, which digests the wild-type sequence into fragments of 214 bp and 98 bp; Bse RI for c.416G>A, which digests the wild-type sequence into fragments of 310 bp and 37 bp; Xmn I for c.1196A>G, which digests the wild-type sequence into fragments of 92 bp and 25 bp; and Taq I for c.792C>G, which digests the wild-type sequence into fragments of 83 bp, 53 bp, and 31 bp. The digestion products were analysed by electrophoresis on a 4% agarose gel (Figure 2), stained with 0.5 μg/mL ethidium bromide and visualised under a ultraviolet UV light. The primer sets and annealing temperatures are summarised in Table 1
Figure 2:
Genotyping of CYP2C8 variants by PCR-RFLP analysed with 4% agarose gel electrophoresis: (a) show the fragments after digestion with Bcl I of the PCR product for identification of CYP2C8∗2 (c.805A>T) variant. (b) show the fragments after digestion with Taq I of the PCR product for identification of CYP2C8∗4 (c. 792C>G). (c) and (d) show the fragments after digestion of the PCR product with Bse RI and Xmn I, respectively for identification of CYP2C8∗3 (c.416G>A and c.1196A>G). ∗1/∗1- represent a typical profile for subject genotyped as homozygous wild type; ∗1/∗2, ∗1/∗3, ∗1/∗4- represent a profile for a single subject genotyped as heterozygous; ∗2/∗2- represent a typical profile for subject genotyped as homozygous mutant.
Confirmation of the heterozygous patterns was achieved by direct DNA sequencing. The amplicons were purified using the ChargeSwitch® PCR Clean-Up commercial kit (Invitrogen, Applied Biosystems, Foster City, CA. USA). The sequencing reactions were performed using the ABI PRISM Big Dye Terminator v. 3.1 Kit (PE Applied BioSystems), according to the manufacturer’s recommendations, on an ABI PRISM 3730 DNA Analyser (PE Applied BioSystems). Analyses of the obtained sequences were performed using the SeqScape v.s2.6 software (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
The frequencies of the allelic variants in the CYP2B6 and CYP2C8 genes were analysed by the Statistical Package for the Social Sciences (SPSS) 11.5 for Windows software. Genotype deviations from the Hardy-Weinberg equilibrium were also determined. The expected genotype frequencies for all of the tested SNPs were calculated from the respective single allelic frequencies and were consistent with the Hardy Weinberg equilibrium using the chi-square (χ2) test. When the tested variants were not found, the individuals were considered to have the wild-type alleles CYP2B6 ∗1A and CYP2C8 ∗1A for the genes CYP2B6 and CYP2C8, respectively. The allelic variant frequencies were compared within gender and region using the chi-square test or Fisher's exact test. A P value < 0.05 was considered significant.
Results
Frequencies of the allelic variants of the CYP2B6 and CYP2C8 genes
Genotyping data were available from a total of 360 Mozambican blood donor volunteers of African descent (mean age of 30.2 years (SD 9.74); 261 male and 99 female subjects) from the provinces of Cabo Delgado (n = 47), Nampula (n = 49), Niassa (n = 2), and Zambézia (n = 24) from the Northern region, Sofala (n = 30) and Tete (n = 6) from the Central region, and Inhambane (n = 24), Gaza (n = 86), and Maputo (n = 92) from the Southern region. The frequencies of the allelic variants commonly reported in the CYP2B6 and CYP2C8 genes evaluated in this study are listed in Table 2. In the CYP2B6 gene, the allelic variants c.516 T (42.6%) and c.785 G (41%) were predominantly observed in our study population compared to the allelic variants c.64 T (5.7%) and c.1459 T (0.4%). For the SNP at position c.777C>A, all of the tested individuals were genotyped as wild-type. For the analysis of the CYP2C8 gene, the most frequent allelic variant was c.805 T, compared to c.416 A (4.8%) and c.792 G (0.5%). Samples from all of the individuals tested for the SNP at position c.1196A>G presented the wild-type allele. Table 3 shows the distribution of the CYP2B6 and CYP2C8 allelic variants by gender in the studied populations. There was no significant difference in the frequency between the two groups. Intra-population variations were determined between the three regions of Mozambique. Our data were similar for all of the SNPs analysed. The study population was found to a large extent to be genetically homogenous.
Table 2.
Allele and genotype frequencies and percentage of SNPs of CYP2B6 and CYP2C8 genes in study population
| Gene | Allele | Allele | Genotype frequency (%) | ||||
|---|---|---|---|---|---|---|---|
| SNP | Exon | Frequency | wt/wt | wt/mut | mut/mut | ||
| CYP2B6 | CYP2B6∗2A | c.64C>T | 1 | 41 (5.69) | 325 (90.3) | 28 (7.8) | 7 (1.9) |
| CYP2B6∗6A,∗7A, ∗9A | c.516G>T | 4 | 307 (42.6) | 112 (31.1) | 191 (53.1) | 57 (15.8) | |
| CYP2B6∗4A,∗6A, ∗7A | c.785A>G | 5 | 295 (40.9) | 131 (36.4) | 165 (45.8) | 64 (17.8) | |
| CYP2B6∗3 | c.777C>A | 5 | 0 (0.0) | 360 (100) | 0 (0.0) | 0 (0.0) | |
| CYP2B6∗5A, ∗7A | c.1459C>T | 9 | 3 (0.41) | 357 (99.2) | 3 (0.8) | 0 (0.0) | |
| CYP2C8 | CYP2C8∗2 | c.805A>T | 5 | 115 (15.9) | 246 (68.3) | 111 (30.8) | 3 (0.8) |
| CYP2C8∗3 | c.416G>A | 3 | 35 (4.86) | 327 (90.8) | 31 (8.6) | 2 (0.6) | |
| CYP2C8∗3 | c.1196A>G | 8 | 0 (0.0) | 360 (100) | 0 (0.0) | 0 (0.0) | |
| CYP2C8∗4 | c.792C>G | 5 | 3 (0.41) | 356 (98.9) | 4 (1.1) | 0 (0.0) | |
Abbreviations: Wt = Wild-type, mut = Mutant.
Table 3.
Frequencies and percentage of single nucleotide polymorphisms (SNPs) in CYP2B6 and CYP2C8 genes per gender
| SNP | Allele | Male (n = 522) | Female (n = 198) | P value |
|---|---|---|---|---|
| Frequency (%) | Frequency (%) | |||
| CYP2B6 | ||||
| c.64C>T | C | 496 (95.0) | 186 (93.9) | 0.524 |
| T | 26 (4.98) | 12 (6.0) | ||
| c.516G>T | G | 313 (59.9) | 103 (52.0) | 0.060 |
| T | 209 (40.0) | 95 (47.9) | ||
| c.785A>G | A | 318 (60.9) | 111 (56.0) | 0.207 |
| G | 204 (39.1) | 87 (43.9) | ||
| c.777C>A | C | 522 (100) | 198 (100) | n.d |
| A | 0 (0.0) | 0 (0.0) | ||
| c.1459C>T | C | 517 (99.0) | 196 (98.9) | 0.184∗ |
| T | 5 (0.95) | 2 (1.0) | ||
| CYP2C8 | ||||
| c.805A>T | A | 438 (83.9) | 166 (83.8) | 0.790 |
| T | 84 (16.1) | 32 (16.1) | ||
| c.416G>A | G | 501 (95.9) | 186 (93.9) | 0.356 |
| A | 21 (4.0) | 12 (6.0) | ||
| c.1196A>G | A | 522 (100) | 198 (100) | n.d |
| G | 0 (0.0) | 0 (0.0) | ||
| c.792C>G | C | 520 (99.6) | 196 (98.9) | 0.304∗ |
| G | 2 (0.38) | 2 (1.0) | ||
Abbreviations: n = Number of alleles, n.d = Not done, (∗) = Fisher-exact test.
Discussion
The present study evaluated the prevalence of the most commonly observed SNPs in the CYP2B6 and CYP2C8 genes in individuals from Mozambique. As these polymorphisms may have an impact on the metabolic clearance of several important drugs, including anti-HIV and antimalarial drugs, some genotypes might be associated with changes in the pharmacokinetics of drugs, modulating the therapeutic responses of drug substrates of CYP2B6 and CYP2C8 enzymes. The presence of SNPs in genes coding for proteins of the cytochrome P450 complex has been described by several studies in different populations, indicating high genetic diversity among different ethnic populations (19,20,24,25,33).
No subjects with variant CYP2B6∗3 and CYP2C8∗3 alleles (Table 2) were identified in the studied population. These results indicated that these variants are very rare in the Mozambican population, as the individuals enrolled in the study were recruited from three different geographic regions of the country. In our study, CYP2B6∗6A (c.516 T, 42.6% and c.785 G, 41.0%) emerged as the most frequent variant in the study population; allele variants c.64 T (5.7%) (CYP2B6∗2A) and c.1459 T (0.4%) (CYP2B6∗5A) were less predominant. The CYP2B6∗6A allele frequency in African populations was reported by (34), who observed a frequency of 32% in malaria subjects, and (17) and (20) reported frequencies of 49% and 42%, respectively. The frequencies reported in these studies are comparable to our study. In our study, only 0.4% of subjects carried the c.1459C>T variant. Data in the literature concerning this variant refer to a frequency of 0.3% in an Asian population (19) and 14% in a Caucasian population (10).
The CYP2B6∗6A variant has been associated with Efavirenz plasma levels, which were significantly elevated and associated with a neuropsychological effect compared to noncarriers (35–37). Additionally, CYP2B6 variant alleles have been found to have an effect on the metabolism and pharmacokinetics of bupropion (an antidepressant drug) (9) and cyclophosphamide (an anticancer and immunosuppressive drug) (38). Although there is limited information regarding the role of CYP2B6 polymorphisms in the metabolism/pharmacokinetics of these drugs, nothing has been reported regarding their effect on antimalarial drugs. The novel and valuable antimalarial drugs, artemisinin and its derivatives, are rapidly metabolised by CYP450 enzymes; peak plasma concentrations occur within 30 min to 2 hours after administration and then decline rapidly (39,40). Whereas the pharmacological significance of drug metabolism polymorphisms on antimalarial drugs can be argued, numerous studies involving malaria patients of different origins have reported large interindividual variations in pharmacokinetic parameters, adverse effects and toxic levels of artemisinin, artesunate and dihydroartemisinin (39,40). This interindividual variability of response appears to include polymorphisms in drug metabolism genes. Although the phenotypic consequences of these polymorphisms are yet to be determined, the relatively high frequency of CYP2B6∗6 observed in our study population may be of interest. Indeed, the identification of genetic variation within genes encoding drug metabolising enzymes and drug targets within different populations is useful due to the drug-drug interactions that result from enzyme inhibition or induction and can improve the quality of healthcare in specific populations.
A comparison of the SNP frequencies by gender was not significantly different; however, the frequencies in female subjects were higher than those in male subjects (Table 3). A similar trend was observed between Caucasian and Asian populations (1,19).
In the present study, we also observed the frequencies of the CYP2C8∗2, ∗3 and ∗4 alleles in the CYP2C8 gene. According to the literature, CYP2C8∗2 occurs at a low frequency (1.6%) and is almost non-existent in Caucasian populations, whereas the CYP2C8∗3 allele (14%) and CYP2C8∗4 allele (7.4%) were more common (25,41). In contrast, the CYP2C8∗2 allele has been observed more frequently in African and African-American populations (18%), whereas the CYP2C8∗3 (2%) and CYP2C8∗4 (0.6%) alleles were rare (25). The CYP2C8∗2 variant allele was observed in 16% of our study population (Table 2). These data are in agreement with previously published data from other populations of African origin, such as Ghana (16.7%) (42), (17.9%) (43), Burkina Faso (11.5%) (29) and Zanzibar (13.9%) (28). The CYP2C8∗3 and CYP2C8∗4 variant alleles were not detected in our study population. These variant alleles have rarely been reported in other African populations (28,42). It has been reported that the SNPs c.416G>A and c.1196A>G usually have close linkage (25, 31). However, in our study, a linkage between the two SNPs was not observed. None of the subjects with the c.416G>A SNP carried the c.119A>G SNP. An isolated presence of the c.416G>A SNP in our samples may be a possible explanation.
The lack of a significant analysis of the SNPs observed was the main limitation of the present study. Previous studies have shown that CYP2B6 polymorphisms affect both the regulation of gene expression and bioavailability of the enzyme (44,45). Lang and colleagues (10) have reported an association between the presence of the c.1459C>T variant with eight-fold lower CYP2B6 catalytic activity, which had a considerable impact on the enzyme activity of CYP2B6. The CYP2C8∗2 and ∗3 polymorphisms are associated with a decrease in the enzymatic degradation activity. The CYP2C8∗2 variant allele resulted in a 15% reduction in the metabolism of the anticancer agent paclitaxel in vitro compared to the wild-type allele (25), whereas the CYP2C8∗3 variant allele resulted in a 50% reduction in paclitaxel metabolism (31). This reduction led to a poor metaboliser phenotype, which could potentially cause drug toxicity (25). Further studies are warranted to understand the influence of CYP2B6 and CYP2C8 polymorphisms on antimalarial drugs and the significant impact of other functional polymorphisms in genes such as CYP2C9, UGT1A9 and UGT2B7 on the metabolism of antimalarials.
Conclusion
In conclusion, the current study has identified allelic variants of CYP2B6 and CYP2C8 in the Mozambican population. The frequencies obtained are comparable to data previously reported in other populations of African origin. This study provides a framework for future studies concerning the role of polymorphic variants of the CYP2B6 and CYP2C8 genes in the genesis of various diseases or the design of future pharmacogenetic and pharmacokinetic studies of CYP2B6 and CYP2C8 substrates in the Mozambican population.
Acknowledgments
We thanks the blood donor volunteers and the staffing service of the “Banco de Sangue” of Hospital Central de Maputo, Hospital Central de Nampula, Hospital Central da Beira, Hospital Provincial de Pemba, Hospital Geral José Macamo, Hospital Geral de Mavalane and Hospital Rural de Chókwè, for help on specimens collection.
Footnotes
Conflict of interest
None.
Funds
This work was supported by the Ministry of Health Mozambique (MISAU) and by the “Laboratório de Biologia Molecular Aplicado a Micobactérias” (IOC- FIOCRUZ) and CNPq Scholarship number: 190080/2008-6, Brasil.
Authors’ contributions
Conception and design: PA, RET, ARS
Analysis and interpretation of the data: PA, ARS Drafting of the article: PA, PNS, ARS
Critical revision of the article for the important intellectual content and obtaining of funding: RET, PNS, ARS
Final approval of the article: ARS
Provision of study materials or patient: MQL
Statistical expertise: PA
Collection and assembly of data: PA, MQL
References
- 1.Lamba V, Lamba J, Yasuda K, Strom S, Davila J, Hancock ML, et al. Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Ther. 2003;307(3):906–922. doi: 10.1124/jpet.103.054866. doi: 10.1124/jpet.103.054866 . [DOI] [PubMed] [Google Scholar]
- 2.Grace JM, Aguilar AJ, Trotman KM, Peggins JO, Brewer TG. Metabolism of beta-arteether to dihydroqinghaosu by human liver microsomes and recombinant cytochrome P450. Drug Metab Dispos. 1998;26(4):313–317. [PubMed] [Google Scholar]
- 3.Svensson US, Ashton M. Identification of the human cytochrome P450 enzymes involved in the in vitro metabolism of artemisinin. Br J Clin Pharmacol. 1999;48(4):528–535. doi: 10.1046/j.1365-2125.1999.00044.x. doi: 10.1046/j.1365-2125.1999.00044.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Björkman A, Andersson TB, Ridderström M, Masimirembwa CM. Amodiaquine clearance and its metabolism to N-desethylamodiaquine is mediated by CYP2C8: a new high affinity and turnover enzymespecific probe substrate. J Pharmacol Exp Ther. 2002;300(2):399–407. doi: 10.1124/jpet.300.2.399. [DOI] [PubMed] [Google Scholar]
- 5.Li XQ, Bjorkman A, Andersson TB, Gustafsson LL, Masimirembwa CM. Identification of human cytochrome P (450)s that metabolise anti-parasitic drugs and predictions of in vivo drug hepatic clearance from in vitro data. Eur J Clin Pharmacol. 2003;59(5–6):429–442. doi: 10.1007/s00228-003-0636-9. doi: 10.1007/s00228-003-0636-9 . [DOI] [PubMed] [Google Scholar]
- 6.Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther. 2003;306(1):287–300. doi: 10.1124/jpet.103.049601. [DOI] [PubMed] [Google Scholar]
- 7.Miksys S, Lerman C, Shields PG, Mash DC, Tyndale RF. Smoking, alcoholism and genetic polymorphisms alter CYP2B6 levels in human brain. Neuropharmacology. 2003;45(1):122–132. doi: 10.1016/s0028-3908(03)00136-9. [DOI] [PubMed] [Google Scholar]
- 8.Ekins S, Vandenbranden M, Ring BJ, Gillespie JS, Yang TJ, Gelboin HV, et al. Further characterization of the expression in liver and catalytic activity of CYP2B6. J Pharmacol Exp Ther. 1998;286(3):1253–1259. [PubMed] [Google Scholar]
- 9.Hesse LM, He P, Krishnaswamy S, Hao Q, Hogan K, von Moltke LL, et al. Pharmacogenetic determinants of interindividual variability in bupropion hydroxylation by cytochrome P450 2B6 in human liver microsomes. Pharmacogenetics. 2004;14(4):225–238. doi: 10.1097/00008571-200404000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Lang T, Klein K, Fischer J, Nüssler AK, Neuhaus P, Hofmann U, et al. Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics. 2001;11(5):399–415. doi: 10.1097/00008571-200107000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Ingelman-Sundberg M. Human drug metabolising cytochrome P450 enzymes: properties and polymorphisms. Naunyn-Schmiedeberg’s Arch Pharmacol. 2004;369(1):89–104. doi: 10.1007/s00210-003-0819-z. doi: 10.1007/s00210-003-0819-z . [DOI] [PubMed] [Google Scholar]
- 12.Santisteban I, Povey S, Shephard EA, Phillips IR. The major phenobarbital-inducible cytochrome P-450 gene subfamily (P450IIB) mapped to the long arm of human chromosome 19. Ann Hum Genet. 1988;52(2):129–135. doi: 10.1111/j.1469-1809.1988.tb01088.x. [DOI] [PubMed] [Google Scholar]
- 13.Rotger M, Tegude H, Colombo S, Cavassini M, Furrer H, Décosterd L, et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther. 2007;81(4):557–566. doi: 10.1038/sj.clpt.6100072. doi: 10.1038/sj.clpt.6100072 . [DOI] [PubMed] [Google Scholar]
- 14.Hofmann MH, Blievernicht JK, Klein K, Saussele T, Schaeffeler E, Schwab M, et al. Aberrant splicing caused by single nucleotide polymorphism c.516G>T [Q172H], a marker of CYP2B6∗6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther. 2008;325(1):284–292. doi: 10.1124/jpet.107.133306. doi: 10.1124/jpet.107.133306 . [DOI] [PubMed] [Google Scholar]
- 15.Zukunft J, Lang T, Richter T, Hirsch-Ernst KI, Nussler AK, Klein K, et al. A natural CYP2B6 TATA box polymorphism (-82T--> C) leading to enhanced transcription and relocation of the transcriptional start site. Mol Pharmacol. 2005;67(5):1772–1782. doi: 10.1124/mol.104.008086. doi: 10.1124/mol.104.008086 . [DOI] [PubMed] [Google Scholar]
- 16.Tsuchiya K, Gatanaga H, Tachikawa N, Teruya K, Kikuchi Y, Yoshino M, et al. Homozygous CYP2B6 ∗6 (Q172H and K262R) correlates with high plasma efavirenz concentrations in HIV-1 patients treated with standard efavirenz-containing regimens. Biochem Biophys Res Commun. 2004;319(4):1322–1326. doi: 10.1016/j.bbrc.2004.05.116. doi: 10.1016/j.bbrc.2004.05.116 . [DOI] [PubMed] [Google Scholar]
- 17.Nyakutira C, Röshammar D, Chigutsa E, Chonzi P, Ashton M, Nhachi C, et al. High prevalence of the CYP2B6 516G-->T(∗6) variant and effect on the population pharmacokinetics of efavirenz in HIV/AIDS outpatients in Zimbabwe. Eur J Clin Pharmacol. 2008;64(4) doi: 10.1007/s00228-007-0412-3. doi: 10.1007/s00228-007-0412-3 . [DOI] [PubMed] [Google Scholar]
- 18.Cabrera Figueroa S, Iglesias Gómez A, Sánchez Martín A, de la Paz Valverde Merino M, Domínguez-Gil Hurlé A, Cordero Sánchez M. Long-Term Efficacy and Safety of Efavirenz Dose Reduction to 200 mg Once Daily in a Caucasian Patient with HIV. Clin Drug Investig. 2010;30(6):405–411. doi: 10.1007/BF03256910. doi: 10.2165/11535320-000000000-00000 . [DOI] [PubMed] [Google Scholar]
- 19.Guan S, Huang M, Chan E, Chen X, Duan W, Zhou S. Genetic polymorphisms of cytochrome P450 2B6 gene in Han Chinese. Eur J Pharm Sci. 2006;29(1):14–21. doi: 10.1016/j.ejps.2006.04.004. doi: 10.1016/j.ejps.2006.04.004 . [DOI] [PubMed] [Google Scholar]
- 20.Mehlotra RK, Ziats MN, Bockarie MJ, Zimmerman PA. Prevalence of CYP2B6 alleles in malaria-endemic populations of West Africa and Papua New Guinea. Eur J Clin Pharmacol. 2006;62(4):267–275. doi: 10.1007/s00228-005-0092-9. doi: 10.1007/s00228-005-0092-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Totah RA, Rettie AE. Cytochrome P450 2C8: substrates, inhibitors, pharmacogenetics, and clinical relevance. Clin Pharmacol Ther. 2005;77(5):341–352. doi: 10.1016/j.clpt.2004.12.267. [DOI] [PubMed] [Google Scholar]
- 22.Klose TS, Blaisdell JA, Goldstein JA. Gene structure of CYP2C8 and extrahepatic distribution of the human CYP2Cs. J Biochem Mol Toxicol. 1999;13(6):289–295. doi: 10.1002/(sici)1099-0461(1999)13:6<289::aid-jbt1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 23.Delozier TC, Kissling GE, Coulter SJ, Dai D, Foley JF, Bradbury JA, et al. Detection of human CYP2C8, CYP2C9, and CYP2J2 in cardiovascular tissues. Drug Metab Dispos. 2007;35(4):682–688. doi: 10.1124/dmd.106.012823. doi: 10.1124/dmd.106.012823 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daily EB, Aquilante CL. Cytochrome P450 2C8 pharmacogenetics: a review of clinical studies. Pharmacogenomics. 2009;10(9):1489–1510. doi: 10.2217/pgs.09.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai D, Zeldin DC, Blaisdell JA, Chanas B, Coulter SJ, Ghanayem BI, et al. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics. 2001;11(7):597–607. doi: 10.1097/00008571-200110000-00006. [DOI] [PubMed] [Google Scholar]
- 26.Dreisbach AW, Japa S, Sigel A, Parenti MB, Hess AE, Srinouanprachanh SL, et al. The Prevalence of CYP2C8, 2C9, 2J2, and soluble epoxide hydrolase polymorphisms in African Americans with hypertension. Am J Hypertens. 2005;18(10):1276–1281. doi: 10.1016/j.amjhyper.2005.04.019. doi: 10.1016/j.amjhyper. 2005.04.019 . [DOI] [PubMed] [Google Scholar]
- 27.King LM, Gainer JV, David GL, Dai D, Goldstein JA, Brown NJ, et al. Single nucleotide polymorphisms in the CYP2J2 and CYP2C8 genes and the risk of hypertension. Pharmacogenet Genomics. 2005;15(1):7–13. doi: 10.1097/01213011-200501000-00002. [DOI] [PubMed] [Google Scholar]
- 28.Cavaco I, Piedade R, Gil JP, Ribeiro V. CYP2C8 polymorphism frequencies among malaria patients in Zanzibar. Eur J Clin Pharmacol. 2005;61(1):15–18. doi: 10.1007/s00228-004-0871-8. doi: 10.1007/s00228-004-0871-8 . [DOI] [PubMed] [Google Scholar]
- 29.Parikh S, Ouedraogo J, Goldstein JA, Rosenthal PJ, Kroetz DL. Amodiaquine metabolism is impaired by common polymorphisms in CYP2C8: implications for malaria treatment in Africa. Clin Pharmacol Ther. 2007;82(2):197–203. doi: 10.1038/sj.clpt.6100122. doi: 10.1038/sj.clpt.6100122 . [DOI] [PubMed] [Google Scholar]
- 30.Cavaco I, Piedade R, Gil JP, Ribeiro V. CYP2C8 polymorphism among the Portuguese. Clin Chem Lab Med. 2006;44(2):168–170. doi: 10.1515/CCLM.2006.030. [DOI] [PubMed] [Google Scholar]
- 31.Bahadur N, Leathart JBS, Mutch E, Steimel-Crespi D, Dunn SA, Gilissen R, et al. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6alpha-hydroxylase activity in human liver microsomes. Biochem Pharmacol. 2002;64(11):1579–1589. doi: 10.1016/s0006-2952(02)01354-0. [DOI] [PubMed] [Google Scholar]
- 32.Nakajima M, Fujiki Y, Noda K, Ohtsuka H, Ohkuni H, Kyo S, et al. Genetic polymorphisms of CYP2C8 in Japanese population. Drug Metab Dispos. 2003;31(6):687–690. doi: 10.1124/dmd.31.6.687. [DOI] [PubMed] [Google Scholar]
- 33.Hiratsuka M, Takekuma Y, Endo N, Narahara K, Hamdy SI, Kishikawa Y, et al. Allele and genotype frequencies of CYP2B6 and CYP3A5 in the Japanese population. Eur J Clin Pharmacol. 2002;58(6):417–421. doi: 10.1007/s00228-002-0499-5. doi: 10.1007/s00228-002-0499-5 . [DOI] [PubMed] [Google Scholar]
- 34.Ferreira PE, Veiga MI, Cavaco I, Martins JP, Andersson B, Mushin S, et al. olymorphism of antimalaria drug metabolizing, nuclear receptor, and drug transport genes among malaria patients in Zanzibar, East Africa. Ther Drug Monit. 2008;30(1):10–15. doi: 10.1097/FTD.0b013e31815e93c6. doi: 10.1097/FTD.0b013e31815e93c6 . [DOI] [PubMed] [Google Scholar]
- 35.Kwara A, Lartey M, Sagoe KW, Kenu E, Court MH. CYP2B6, CYP2A6 and UGT2B7 genetic polymorphisms are predictors of efavirenz middose concentration in HIV-infected patients. AIDS. 2009;23(16):2101–2106. doi: 10.1097/QAD.0b013e3283319908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haas DW, Ribaudo HJ, Kim RB, Tierney C, Wilkinson GR, Gulick RM, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS. 2004;18(18):2391–2400. [PubMed] [Google Scholar]
- 37.Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics. 2005;15(1):1–5. doi: 10.1097/01213011-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 38.Xie H, Yasar U, Lundgren S, Griskevicius L, Terelius Y, Hassan M, et al. Role of polymorphic human CYP2B6 in cyclophosphamide bioactivation. Pharmacogenomics J. 2003;3(1):53–61. doi: 10.1038/sj.tpj.6500157. doi: 10.1038/sj.tpj.6500157 . [DOI] [PubMed] [Google Scholar]
- 39.Krishna S, Planche T, Agbenyega T, Woodrow C, Agranoff D, Bedu-Addo G, et al. Bioavailability and preliminary clinical efficacy of intrarectal artesunate in Ghanaian children with moderate malaria. Antimicrob Agents Chemother. 2001;45(2):509–516. doi: 10.1128/AAC.45.2.509-516.2001. doi: 10.1128/AAC.45.2.509-516.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karunajeewa HA, Reeder J, Lorry K, Dabod E, Hamzah J, Page-Sharp M, et al. Artesunate suppositories versus intramuscular artemether for treatment of severe malaria in children in Papua New Guinea. Antimicrob Agents Chemother. 2006;50(3):968–974. doi: 10.1128/AAC.50.3.968-974.2006. doi: 10.1128/AAC.50.3.968-974.2006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weise A, Grundler S, Zaumsegel D, Klotzek M, Gröndahl B, Forst T, et al. Development and evaluation of a rapid and reliable method for cytochrome P450 2C8 genotyping. Clin Lab. 2004;50(3–4):141–148. [PubMed] [Google Scholar]
- 42.Röwer S, Bienzle U, Weise A, Lambertz U, Forst T, Otchwemah RN, et al. Short communication: High prevalence of the cytochrome P450 2C8∗2 mutation in Northern Ghana. Trop Med Int Health. 2005;10(12):1271–1273. doi: 10.1111/j.1365-3156.2005.01525.x. doi: 10.1111/j.1365-3156.2005.01525.x . [DOI] [PubMed] [Google Scholar]
- 43.Adjei GO, Kristensen K, Goka BQ, Hoegberg LCG, Alifrangis M, Rodrigues OP, et al. Effect of Concomitant Artesunate Administration and Cytochrome P4502C8 Polymorphisms on the Pharmacokinetics of Amodiaquine in Ghanaian Children with Uncomplicated Malaria. Antimicrob Agents Chemother. 2008;52(12):4400–4406. doi: 10.1128/AAC.00673-07. doi: 10.1128/AAC.00673-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ariyoshi N, Miyazaki M, Toide K, Sawamura Yi, Kamataki T. A single nucleotide polymorphism of CYP2b6 found in Japanese enhances catalytic activity by auto activation. Biochem Biophys Res Commun. 2001;281(5):1256–1260. doi: 10.1006/bbrc.2001.4524. doi: 10.1006/bbrc.2001.4524 . [DOI] [PubMed] [Google Scholar]
- 45.Jinno H, Tanaka-Kagawa T, Ohno A, Makino Y, Matsushima E, Hanioka N, et al. Functional characterization of cytochrome P450 2B6 allelic variants. Drug Metab Dispos. 2003;31(4):398–403. doi: 10.1124/dmd.31.4.398. [DOI] [PubMed] [Google Scholar]