Abstract
Purpose
Genome-scale clinical sequencing is being adopted more broadly in medical practice. The National Institutes of Health developed the Clinical Sequencing Exploratory Research (CSER) program to guide implementation and dissemination of best practices for the integration of sequencing into clinical care. This study describes and compares the state of the art of incorporating whole-exome and whole-genome sequencing results into the electronic health record, including approaches to decision support across the six current CSER sites.
Methods
The CSER Medical Record Working Group collaboratively developed and completed an in-depth survey to assess the communication of genome-scale data into the electronic health record. We summarized commonalities and divergent approaches.
Results
Despite common sequencing platform (Illumina) adoptions, there is a great diversity of approaches to annotation tools and workflow, as well as to report generation. At all sites, reports are human-readable structured documents available as passive decision support in the electronic health record. Active decision support is in early implementation at two sites.
Conclusion
The parallel efforts across CSER sites in the creation of systems for report generation and integration of reports into the electronic health record, as well as the lack of standardized approaches to interfacing with variant databases to create active clinical decision support, create opportunities for cross-site and vendor collaborations.
Keywords: clinical decision support, clinical sequencing, decision support rules, electronic health record, electronic medical record, next-generation sequencing
INTRODUCTION
Background to the study
Clinical sequencing is an area of rapid growth, as laboratories, medical centers, and hospitals adopt this method of comprehensively scanning the human genome for genomic variants associated with clinical disease or pharmacogenomic effects.1 Clinical sequencing impacts multiple facets of clinical care, such as processing of large-scale genomic data; reporting of results, including results to diagnose the presenting condition; integration of results into the medical record; and support of genome-enabled clinical decision making. To guide the development of best practices for the integration of clinical sequencing into clinical care, as well as to research the ethical, legal, and psychosocial implications of delivering broad genomic data into the clinic, the National Human Genome Research Institute, in collaboration with the National Cancer Institute, funded the Clinical Sequencing Exploratory Research (CSER) Program.2 Currently composed of six leading academic medical centers, CSER is working collaboratively to incorporate sequence data into the clinical care of patients and to examine the relevant ethical, legal, and psychosocial issues. Each site is completing a clinical genomic trial (whole-exome sequencing (WES) or whole-genome sequencing) with a unique patient population. The acceleration in uptake of WES outside the research setting is illustrated by the Baylor experience, in which clinical WES for direct patient care was offered beginning in November 2011. In the first 18 months, the laboratory has reported more than 1,000 WES results and currently receives ~200 clinical test requests monthly. The CSER consortium members awarded funding by the National Institutes of Health in 2011 are Baylor College of Medicine (BCM), Brigham and Women's Hospital–Harvard Medical School (BWH), The Children's Hospital of Philadelphia (CHOP), the Dana-Farber Cancer Institute (DFCI), the University of North Carolina (UNC), and the University of Washington (UW).
The CSER Electronic Medical Record Working Group (CSER EMR WG) was created to explore informatics issues related to annotation and prioritization of genomic variants, as well as to the integration of genomic results into the EMR/electronic health record (EHR) and EHR-enabled clinical decision support (CDS). The CSER EMR WG (chaired by P.T.-H.) includes representation from each CSER site and the National Institutes of Health. The initial goal was to systematically describe and compare the current state across the CSER sites regarding how WES and whole-genome sequencing results are incorporated into the EHR, including approaches to decision support. This work is complementary to the special issue article on “Opportunities for Genomic Clinical Decision Support Interventions”3 by the Electronic Medical Records and Genomics Network,4 which describes opportunities for genomic CDS informed by the literature on traditional computerized CDS. Our work takes a bottom-up approach (examining current practices at the CSER sites), whereas the Electronic Medical Records and Genomics article takes top-down approach (describing an ideal state and requirements to achieve this state) to elucidating the future directions for genomic CDS.
Background on the six current CSER sites
The six initial CSER sites5 represent projects looking at the issue of generating and incorporating next-generation sequencing (NGS) data (WES and whole-genome sequencing) across a broad range of clinical settings to study the implementation and impact of genomic medicine. The sites are exploring outcomes including (i) the communication of results to their target populations, (ii) the preferences of their participants with respect to return of incidental findings, (iii) the impact of reporting NGS to participants, (iv) how best to report and store NGS data, (v) the incorporation of novel decision support technologies and solutions, and (vi) the use of NGS as a diagnostic modality. Table 1 describes the patient populations and unique features at each site.
Table 1.
Populations and features of the six current Clinical Sequencing Exploratory Research sites
| Site–Project name | Population | Unique site features |
|---|---|---|
| BCM – BASIC3 | Pediatric patients with newly diagnosed high-risk solid tumors including brain tumors | • Tablet-based platform and graphical display • Return of results to pediatric oncologists and parents • Both germline and tumor sequencing reported for each patient |
| BWH/HMS – MedSeq | Patients with cardiomyopathy and a healthy population | • Whole-genome sequencing rather than whole-exome sequencing • Creating and testing novel ways of integrating next-generation sequencing data into the care of patients • Randomized controlled trial |
| CHOP – PediSeq | Pediatric patients with hearing impairments, mitochondrial disorders, sudden cardiac arrest, and intellectual disability | • Enabling substantial phenotype capture at the point of care to assist with variant decision processes |
| DFCI/Broad – CanSeq | Patients with metastatic lung or colon adenocarcinoma | • Utilizing data to inform treatment decisions • Tumor sequencing |
| UNC – NCGENES | Patients with selected clinical conditions with a likely genetic etiology: familial cancer, neuromuscular disorders, microcephaly, cardiomyopathy, retinitis | • Implementing whole-exome sequencing in traditionally underserved populations throughout North Carolina • Randomized controlled trial |
| UW – NEXT MEDICINE | Patients in whom a hereditary predisposition toward colon cancer and/or polyps is suspected | • Identification of novel CRCP-related genes • Randomized controlled trial |
BCM, Baylor College of Medicine; BWH, Brigham and Women's Hospital–Harvard Medical School; CHOP, The Children's Hospital of Philadelphia; CRCP, colo-rectal cancer polyposis; DFCI, Dana-Farber Cancer Institute; UNC, University of North Carolina; UW, University of Washington.
MATERIALS AND METHODS
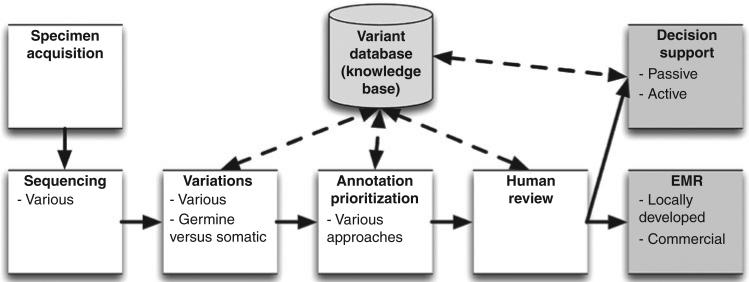
The CSER EMR WG developed a survey to describe and compare across the CSER sites how NGS results are being incorporated into the EHR, including approaches to decision support. Table 2 provides an overview of the survey, and Figure 1 illustrates graphically in shaded form the scope of the survey.
Table 2.
Overview of survey
| Area of survey | Characteristics |
|---|---|
| Context | Germline vs. somatic sequencing of tumors, sequencing platform, sequencing laboratory, approach to CLIA certification, data sources used for annotation and referenced in reports, approach to “binning” of results, and nature of the study population |
| Variant databases – knowledge bases | Which databases are used to curate variants and annotations, how these databases are integrated with clinical workflow, how variants are presented to molecular diagnostic staff for interpretation and sign-out, how databases assist with report writing and/or rule generation, and linkages between variant databases and EHR decision support tools |
| Reporting of results into the EHR | Which EHR(s) are used, if the NGS/EHR process is generalized or specific to the site's CSER, systems currently used for EHR report generation, NGS laboratory information management systems used, interfaces to the EHR for reporting, structure of reports, report generation processes, customization of reports by the audience, routing of results, and reporting tools external to the EHR being utilized |
| Communication of results to providers | Whether consultation regarding NGS results is provided and if so, by whom; length of consultation; and current capacity to scale up |
| Handling of changing variant/annotation information | Content included in the consent process, whether reports are updated subsequent to receiving new information, and if so, how this is achieved |
| Clinical decision support | Type of decision support (passive vs. active), mechanisms to ensure that all providers can view key content, which subset of results are used for decision support, and further characterization of active decision support systems and of how these systems operate |
CLIA, Clinical Laboratory Improvement Amendments; CSER, Clinical Sequencing Exploratory Research; EHR, electronic health record; NGS, next-generation sequencing.
Figure 1. This figure created by the National Institutes of Health Clinical sequencing exploratory Research (CSER) electronic Medical Records Working Group (EMR WG) shows the typical CSER site workflow from specimen acquisition through the reporting of whole-exome or whole-genome results into the EMR.
As described in the text, each of the six CSER sites has its own site-specific workflow and its own variant database/knowledge base. The focus of the CSER EMR WG (and the survey presented in this article) is indicated by the shaded areas of the figure.
Over a 3-month period, the CSER EMR WG members drafted and refined the survey, which was then completed by each of the six sites in early 2013. Each site analyzed and summarized the results for one of the six main sections of the survey, using a qualitative approach. For certain questions, additional clarifying information was sought during the analysis and summarization, which was reviewed by all the sites. The discussion section includes material from CSER EMR WG teleconferences and in-person meetings in which the data from the survey and site-specific approaches to NGS integration into the EHR were presented and discussed.
RESULTS
Context
All six CSER network sites are performing massively parallel sequencing using the Illumina HiSeq platform (San Diego, CA) and analyzing sequence data for germline mutations associated with inherited disease and/or risk of disease. BCM and DFCI are focused on cancer (Table 1) and thus they are also analyzing tumor samples for the presence of somatic alterations.
Five CSER sites are performing NGS locally, and one site is outsourcing NGS followed by on-site variant confirmation by Sanger sequencing. BCM and BWH are performing NGS in a Clinical Laboratory Improvement Amendments (CLIA)–certified laboratory setting. The other sites are performing NGS in a research setting followed by Sanger confirmation of variants in a CLIA–certified laboratory (Table 3). Most sites are planning for a transition of NGS to a CLIA–certified laboratory in the near future.
Table 3.
Overview of Clinical Laboratory Improvement Amendments (CLIA) model, variant annotation, and report generation by CSER site
| BCM | BWH | CHOP | DFCI | UNC | UW | |
|---|---|---|---|---|---|---|
| Clinical Laboratory Improvement Amendments (CLIA) model | CLIA sequencing and validation | CLIA sequencing and validation | Research sequencing followed by CLIA confirmation by Sanger sequencing. Transitioning to CLIA-certified lab sequencing | Research sequencing followed by CLIA confirmation by Sanger sequencing. Transitioning to CLIA-certified lab sequencing | Research sequencing followed by CLIA confirmation by Sanger sequencing or other appropriate method. Likely transitioning to CLIA-certified lab sequencing | Research sequencing followed by CLIA confirmation by Sanger sequencing Transitioning to CLIA-certified lab sequencing but will still validate by Sanger |
| Major data sources used for variant annotation | HGMD, 1000 Genomes, ESP, local data on variant frequencies | HGMD, Entrez, ESP, dbSNP, 1000 Genomes, Condel, PolyPhen2, NNSplice, EBI Variant Effect Predictor, Alamut, ClinVar, locally developed GeneInsight software | HGMD, HGNC, 1000 Genomes, ESP, dbSNP, PubMed, Human Phenotype Ontology, PolyPhen2, SIFT, Alamut, local data on variant frequencies | HGMD, Catalogue of Somatic Mutation in cancer (COSMIC), MSigDB, Cancer Gene Census, ESP database, GET-Evidence, ClinVar, SNPedia, PharmGKB, local CanSeq Actionability Database | HGMD, 1000 Genomes, VarDB, dbSNP, RefSeq, PolyPhen2, SIFT, local data on variant frequencies | HGMD, dbSNP, PharmGKB, PubMed, Genomic Evolutionary Rate Profiling (GERP) scores, PolyPhen2, local UW Exome Variant Server |
| Websites provided in reports | RefSeq for mutation citation, OMIM, PubMed | Reports synthesize data from external sites but do not provide websites | Reports synthesize data from external sites but do not provide websites | OMIM, Clinicaltrials.gov, dbSNP | RefSeq for mutation citation, GeneTests, OMIM, dbSNP, other sites as relevant to variant(s) reported | OMIM, PubMed, Gene Reviews, PharmGKB, other sites as relevant to variant(s) reported |
| Major categories used to classify reportable variants | Focused report: (i) deleterious mutations related to disease phenotype, (ii)VUS related to the disease phenotype, (iii)medically actionable mutations, (iv)autosomal recessive carrier status (opt out), (v)pharmacogenetic; expanded report: includes deleterious mutations in other disease-associated genes and truncating mutations in unannotated genes. Cancer exome report is distinct | (i)Potentially diagnostic results: pathogenic, likely pathogenic, uncertain significance, likely benign, benign; (ii)pharmacogenomic: responsive, resistant, high metabolizer, low metabolizer | (i)Primary diagnosis; (ii)incidental findings binned into four categories: (a)immediately medically actionable, (b)medically actionable childhood-onset disease, (c)medically actionable adult-onset disease, (d)carrier status. All immediately medically actionable variants are reported, whereas other incidental finding categories are optionally reported based on patient preference | Somatic: predictive (responsive or resistant to anticancer agents), prognostic, diagnostic; Germline: cancer-risk, non-cancer risk, pharmacogenomic, carrier status | (i)Primary diagnosis: deleterious mutations and VUS (missense, truncating, splicing) related to disease phenotype; (ii)Incidental findings are reported based on medical action ability and patient preference. All immediately medically actionable variants are reported; randomized patients can choose whether to receive different categories of non-medically actionable incidental findings | (i)Primary diagnosis: potentially diagnostic results related to colorectal cancer/polyposis; (ii)incidental findings restricted to well supported, clinically important and actionable, split into high genetic risk, moderate genetic risk, and pharmacogenomic |
ESP, National Heart, Lung, and Blood Institute GO Exome Sequencing Project exome variant server (evs.gs.washington.edu/EVS); HGMD, Human Gene Mutation Database (http://www.hgmd.cf.ac.uk); HGNC, HUGO Gene Nomenclature Committee (http://www.genenames.org); OMIM, Online Mendelian Inheritance in Man (http://www.omim.org).
BCM, Baylor College of Medicine; BWH, Brigham and Women's Hospital–Harvard Medical School; CHOP, The Children's Hospital of Philadelphia; CLIA, Clinical Laboratory mprovement Amendments; CSER, Clinical Sequencing Exploratory Research; DFCI, Dana-Farber Cancer Institute; UNC, University of North Carolina; UW, University of Washington; VUS, variant of uncertain significance.
Each site has built a local bioinformatics workflow for variant annotation and interpretation (Table 3). All sites have incorporated multiple public data sources for variant annotation as part of their bioinformatics workflow. In addition, each site has built tools to incorporate locally derived NGS data for variant annotation, such as local allele frequencies or tracking of previously ascertained sequence variants. All sites incorporate manual or semi-automated curation of sequence variants by searching the medical literature and/or relevant locus-specific databases to determine the clinical significance of sequence variants before reporting.
All sites report different categories of information (Table 3) based on whether a variant is related to the disease phenotype, a medically actionable incidental finding, or other incidental but otherwise deemed reportable finding (e.g., carrier status, pharmacogenetic information). Each site has developed its own strategy, described in detail elsewhere,6 for how to classify and report incidental information. Several sites incorporate patient choice to receive certain types of incidental genomic information with opt-in or opt-out categories. All sites provide an indication-specific, focused report for potentially diagnostic information and also report incidental findings not related to the disease phenotype as additional information or in a separate report. In addition to relevant literature references, most of the CSER sites provide links in their reports to websites that provide additional information regarding variant classification and interpretation, such as OMIM,7 PubMed,8 RefSeq,9 dbSNP,10 GeneTests,11,12 GeneReviews,13–15 Clinicaltrials.gov,16 and PharmGKB,17 as relevant. As a result of the heterogeneity apparent in Tables 1 and 3, each site has developed its own custom annotation workflow. Currently, these bioinformatics tools are not being shared, and commercial tools are not being used.
Variant databases/knowledge bases
The capabilities and characteristics of site-specific, accumulated variant databases/knowledge bases (VDBKBs) have implications for what structured and computable data can be sent to the EHR. External and internal VDBKBs are a critical component of NGS diagnostic processes. The goal of these processes is to determine whether any of the tens of thousands to millions of variants identified by NGS are clinically important (e.g., associated with the indication for testing, a medically actionable incidental finding, or having a significant pharmacogenomic association). The need for these assessments and the creation of local VDBKBs stems from the fact that the NGS platforms currently available do not include tools for annotation of the variants they report nor are they integrated with VDBKBs.
The sites reported the use of different external variant and gene databases (Table 3). External variant databases that catalog reported associations between specific variants and known clinical phenotypes serve as a first-pass filter for annotation. However, thoroughly evaluating NGS data often requires verifying reported associations and searching for other potentially important variants that have not yet been associated with clinical phenotypes. This task is particularly challenging because most annotation features available for filtering provide suboptimal sensitivity and/or specificity. External databases with information on variant frequencies or gene-level annotations are particularly useful in this context.
In addition to utilizing these external databases, each CSER site has developed a custom local VDBKB to manage its internal variant information and support its reporting process (Table 4). These systems record the variants identified in each patient and most CSER sites are also recording their internal variant assessments. Investigating the clinical implications of variants can be a time-consuming process; therefore, it is useful to store variant classifications so that they can be leveraged if a variant is identified again in another patient. BWH uses VDBKB annotation software (GeneInsight, Boston, MA)6 that is commercially available. The other sites have developed their own systems.
Table 4.
Overview of variant database, report generation, EHR, reporting, and decision support by CSER site
| BCM | BWH | CHOP | DFCI | UNC | UW | |
|---|---|---|---|---|---|---|
| VDBKB | Custom local | Custom local (available commercially GeneInsight) | Custom local | Custom local | Custom local | Custom local |
| Report generation | Custom semi-automated | Custom semi-automated | Manual | Custom semi-automated | Custom semi-automated | Custom semi-automated |
| Report types | (a)Focused exome report, (b)expanded reporta, (c)tumor report | (a)Indication report, (b)general genome report | One report | (a)EHR report, (b)tumor board report | (a)Indication report, (b)incidental findings report, (c)Research report summarizing WES analysis parameters | (a)Indication report, (b)incidental finding report |
| LIMS | Custom local | GeneInsight Lab plus custom local linked to PowerPath | ThermoFisher Nautilus | CoPath | Geneus | |
| Currently operational EHR | Epic | Custom EHR | Epic | Custom EHR | WebCis custom EHR | Epic – outpatient, Sorian – one inpatient site, Cerner – two inpatient sites and oncology |
| Partner sites/EHRs | N/A | N/A | N/A | N/A | Vidant Health/East Carolina University and potentially others. Variety of EHRs | Group Health. Epic EHR |
| Passive decision support | ||||||
| Machine-readable structured reporting | No | Yes, machine readable (XML, coded variants) | No | Yes, machine readable (coded variants – hg19 and HCNG) | No | Yes, machine readable (coded gene-linked results) |
| Active decision support | No | Yes (custom) | No | No | No | Yes (leveraging Cerner) |
| Other decision support | iPad application with clickable links | No | No | No | Clickable links | Clickable links |
| Disclosure of new genomic informationb | Yes | Yes | Yes | No | Yes | No |
BCM, Baylor College of Medicine; BWH, Brigham and Women's Hospital–Harvard Medical School; CHOP, The Children's Hospital of Philadelphia; CSER, Clinical Sequencing Exploratory Research; DFCI, Dana-Farber Cancer Institute; EHR, electronic health record; HCNG, HUGO Gene Nomenclature Committee; LIMS, laboratory information management system; N/A, not applicable; UNC, University of North Carolina; UW, University of Washington; VDBKB, variant database/knowledge base; WES, whole-exome sequencing.
For the Baylor CSER project only the focused exome report is provided. Outside physicians can request the expanded report after receiving the focused report.
“Disclosure of New Genomic Information”: Each CSER site has thus considered the challenge of reinterpreting genomic events and what, if any, actions are required when new information is available for a patient's data.
Reporting of results into the EHR
The systems used for report generation (before integration of the report into the EHR) are unique to each site (Table 4). Reporting is semi-automated (manual at CHOP), involving a combination of expertise in genetics, genomics, bioinformatics, and laboratory medicine/pathology using custom systems for report generation, including the site's local VDBKB. Four sites send the same report(s) to all providers/patients. CHOP has separate reports for each group. DFCI and BCM have a tumor board–specific report. BCM has a more in-depth clinical report for external providers. The UW report has the first section designed for nongeneticists and later sections aimed at experts in molecular variants. Each of the sites uses a different laboratory information management system (LIMS) (Table 4) that is either locally developed or involves custom adaptations to a commercial platform.
The destination systems for reports are both commercial and custom-developed EHRs (Table 4). Four sites use a single EHR and do not have partner sites, whereas UNC and UW have partner sites with separate EHRs. UW has a heterogeneous EHR environment (Table 4), using Cerner,18 Sorian,19 and Epic20 EHRs. BWH, DFCI, and UNC are using custom-developed EHRs to report NGS results while making plans to report into the commercial EHRs in the future.
Despite the heterogeneity in workflows, VDBKBs, bioinformatics tools, LIMS, and EHRs (Tables 3 and 4), the common end result at all sites is a PDF human-readable structured document designed to be sent to the EHR (analogous to a pathology or other text report). Several CSER sites also report to outside labs that are not part of the CSER project, using their normal reporting process. Some sites also provide structured reporting in machine-readable format. Due to a lack of standards in report content, structure, coding, generation, and LIMS (Table 4), current EHRs are not able to process these structured reports because their format is unique to each site. For a subset of actionable indication and incidental findings, UW uses structured laboratory data (molecular testing results pathway) in the SunQuest clinical laboratory system for active decision support.
Communication of results to providers
All sites feel that it is important to have highly trained personnel in medical/molecular genetics available to the ordering physician. Each laboratory has identified genetic counselors, molecular geneticists, or medical geneticists to communicate with the ordering physician. In addition, the majority of sites make the medical director of the lab available to ordering physicians as needed to explain more complex results.
Sites estimated that conversations to explain the implications of results, discuss interpretation of uncertain variants, and answer questions typically take 15–30 min. The ability to effectively explain the results to ordering physicians was felt to be an important challenge in scaling up the process. The BCM laboratory, currently the largest clinical WES CSER testing site, has already had to increase the number of staff who perform this specific function.
Handling of changing variant/annotation information
As new genomic discoveries are made, genomic findings may be reclassified over time, and other medical knowledge relevant to an NGS analysis or test interpretation may be gained. Therefore, each CSER site has considered the challenge of reinterpreting genomic events and what, if any, actions are required when new information is available for a patient's genome report. The results of this survey demonstrate the wide spectrum of study policies across the sites for the reanalysis of genomic data and the disclosure and reporting mechanisms for new information. The diversity of approaches across the CSER sites highlights the challenges of integrating new genomic information into clinical care given the rapid pace of scientific discovery.
CSER sites have segregated into one of two overarching options when it comes to disclosure of new genomic information that may impact a patient's clinical management: (i) strict nondisclosure of new information (DFCI, UW) or (ii) disclosure of new information if logistically feasible (Table 4). The BWH site has an automated method for disclosure using GeneInsight. Automated e-mail alerts are sent to clinicians when a variant is reclassified in a manner that generates a “high” alert, and clinicians receive “medium-” and “low- priority” alerts in batch form or on a weekly basis.21 A “high” or “medium” alert reflects a possible change in treatment (e.g., uncertain significance to pathogenic or pathogenic to nonpathogenic category), whereas a low-level alert specifies less substantial reclassifications (e.g., benign to likely benign). Other sites have opted for semi-annual reviews (UNC) or periodically as determined by the diagnostic lab (BCM, CHOP).
Regarding incidental findings that do not directly relate to the diagnostic question under evaluation but that could impact decision making in a broader clinical context, one site (UNC) has opted to issue amended reports that include new data. However, all other sites that would issue new information plan to do so only for variants that fall within the original indication for testing where this original assessment may include a “general genome report” that assesses highly penetrant conditions independent of a prior probability of disease.
CDS
Given the growing number of reportable and actionable genes, the ideal approach to NGS CDS in the EHR would include a combination of active (e.g., alerts inside the EHR triggered by context) and passive CDS (e.g., reports requiring providers to seek out and review reports).22
Because all sites use PDF documents for passive CDS (Table 4), we explored how the sites ensure clinicians are made aware of the reports. Two organizations (DFCI and UW) used features built in to the EHR to deliver sequence results (with notification) to a wide range of clinical stake-holders, whereas two locations (BWH and BCM) sent e-mails (no protected health information) outside the EHR to staff at the clinic where the order was placed and provided verbal notification for more complex cases. At BWH, the e-mails provide deep links that enable clinicians to authenticate and access relevant information or navigate to the information through a system integrated with the EHR under a patient genetic summary table. A medical geneticist at a fourth location (UNC) provided results verbally to study subjects and solicited informed consent for results to be entered into the EHR. In addition to the built-in notification features of the EHR, UW also directly contacted the primary provider for a subset of the participants.
Active decision support rules were functional at two organizations, BWH and UW (Table 4). BWH developed custom decision support external to, but integrated with, its custom (locally built) EHR. This system, along with medication data from the EHR, provides pharmacogenomics decision support and sends notifications when variants are reclassified. UW is using native features in its commercial EHR (Cerner) to implement alerts as part of pilot efforts for selected variants for patients enrolled in the CSER study by building on prior UW pilot work.23
Other CDS approaches are also used. Two organizations (UNC, UW) enhance their reports with clickable links to supporting materials as a variant of passive decision support. BCM has put significant effort into developing an iPad application providing the test report in a more dynamic platform. The tablet application allows control of the content and direct educational links (glossary, OMIM, PubMed) independent of the current limitations of the EHR at the study site as well as the variety of EHRs used by hospitals outside the study receiving test results as a simple PDF.
DISCUSSION
Across the sites there is a common sequencing approach (Illumina HiSeq) and a common end point (PDF text providing passive CDS). Despite this commonality, there is great diversity of workflow, bioinformatics tools, and approaches starting from sequence data and ending with report generation, which presents both opportunities and challenges for the community.
A key reason for this diversity is that each site has its own approach to annotating variants (Table 3), with its own informatics tools and overall workflow. This heterogeneity is independent of NGS platform and results from the manual annotation currently required of NGS data (Figure 1) and different decisions around this annotation process at each site (Table 3). Another cause of diversity in workflow and tools is that each site has its own internal VDBKB to capture site-specific assessments of variants (Table 4). Unfortunately, VDBKB content cannot yet be shared across sites due to lack of standardization. A consequence of this heterogeneity is that the same variant in a sequence may not be annotated or reported on in the same way across the sites. There are a number of National Institutes of Health–supported initiatives, including ClinVar24 and a proposed resource for the identification and dissemination of consensus information on genetic variants relevant for clinical care (described in RFA-HG-12-016, ref. 25) to address the current duplication and heterogeneity of internal VDBKBs.
Passive CDS in the EHR is implemented at all sites as human-readable structured documents. The ability to easily generate a PDF report independent of the workflow leading to the report, and the ability of virtually any commercial or custom EHR system to accept a PDF report, suggests this is likely to be a common first step to genomic decision support. However, this approach has the known risks of passive decision support,22 which are substantially exacerbated by the complexity of NGS.
Active CDS thus will be necessary as NGS-based testing is increasingly adopted, and as the number of genes and variants deemed actionable/reportable for a given patient rises.26 Providers cannot be expected to read and remember all variants for a given patient via a passive CDS. Active CDS triggered by context (e.g., drug–gene interactions at the time of electronic order entry) will be critical to effectively scaling up EHR-based CDS of NGS results. EHRs, particularly those of commercial systems, include decision support engines that can be adapted for active genomic CDS subject to the availability of trigger conditions.27 Institutions have published literature about using CDS engines for pharmacogenomics decision support using different types of genomic data. For example, the University of Utah used limited single-nucleotide polymorphism and allele data to pilot pharmacogenomic decision support for CYP2C9.28 St Jude Hospital (Memphis, TN) used data from the DMET Plus array (Affymetrix, Santa Clara, CA 1,936 genomic variants in 225 genes) and the Cerner EHR to implement a set of pharmacogenomics decision support rules for 29 CYP2D6 alleles and 9 TMPT alleles.29 Two CSER sites are extending this work to NGS (Table 4). UW is building on its proof-of-concept work23 and extending the standard single-gene molecular testing result mechanism to put multiple actionable pharmacogenomics variants into the EHR in a computable representation. These data can then be used by the UW commercial EHR for active CDS. This approach is consistent with the recommendation made by Masys et al.,30 which involves putting only the actionable variants into the EHR as discrete data rather than all the NGS data. This approach is also consistent with the model presented by the Electronic Medical Records and Genomics consortium.31 Representing actionable variants via a clinical LIMS single-gene molecular testing result reporting data structure could generalize to any LIMS and any EMR; however, it is difficult to scale up because a new “test” would need to be created for each gene. BWH is using an external tool (GeneInsight) integrated into the EHR through a single sign-on mechanism that passes patient context. GeneInsight sends structured data into EHRs to support CDS rules that rely on these data (e.g., supplying data to the EHR needed to provide a pharmacogenomic alert that appears at the time of electronic order entry). Both CSER active CDS approaches have the potential to be implemented at other sites.
A challenge to scaling up active CDS is that currently computable rules cannot be automatically derived or created from the VDBKB. Although not explicitly asked in the survey, four of the six sites stated that they felt more sophisticated (and standardized) biomedical informatics tools for interpretation of sequence variants are needed in order to effectively scale up NGS diagnostics. The emerging discipline of translational bioinformatics32 includes as its focus these types of translational tools. The change of variant/annotation information over time magnifies the challenge of maintaining these rules. These challenges are independent of the NGS method and the approach to delivering the results into the EHR and are inherent to the nature of NGS. The need to re-annotate is especially an issue for the sites disclosing new genomic information (Table 4).
A final barrier to putting actionable variant NGS data into the EHR is poor adherence to existing standards to represent genomic variants, such as the guideline proposed by the Human Genome Variation Society33 (in contrast to adopted standardized coding systems such as the International Classification of Diseases, 10th revision).37 This challenge is magnified by the lack of any VDBKB collective that would feed actionable variant results into the EHR. Mark Hoffman (of the Cerner EHR company) has over the past 8 years begun to put into the Cerner system an open source ontology along these lines,34 working toward a vision of a genome-enabled EHR35 to enable a more generalizable and scalable approach to personalized medicine.36 Ultimately, adoption of such standardized ontologic approaches will be key to enabling sharing of active genomic decision support rules across organizations and EHR vendors.
CDS alone is not sufficient and needs to be augmented by a mechanism to provide human-to-human communication. All sites feel it is important to provide the ordering physician access to highly trained personnel in medical/molecular genetics. This need is independent of the EHR environment and reflects broader perspectives in the genetics community. Given the complexity and nuances of interpreting genomic test results, as well as the possible involvement of different medical stakeholders with widely varied perspectives and genomic knowledge, there is a need for better leveraging of standard EHR mechanisms for person-to-person consultation (e.g., provider-to-provider messaging embedded in commercial EHRs such as inboxes and electronic consult workflows).
The issues above and the heterogeneity in approaches to variant identification, annotation, prioritization, VDBKBs, and reporting tools (Tables 3 and 4) has led each CSER site to build bioinformatics tools and workflows for the four intermediate steps between sequence data and decision support outlined here (Figure 1). This represents significant past and ongoing investment in bioinformatics infrastructure within and across the sites. Based on the current study and the work of the CSER EMR WG, it appears that these tools are too closely linked to local variations in approaches (Tables 3 and 4 ) to be generalizable across the sites, even if they are open source. A number of early-phase companies are attempting to fill this gap between the sequence data and the EHR. BWH uses the GeneInsight and Alamut applications in their workflow. CHOP uses Cartagenia and Alamut. The CSER EMR WG is thus exploring opportunities for cross-site collaboration across the steps in Figure 1 as part of the recently formed CSER Coordinating Center.
We conclude that future directions to maximize the ability to scale up NGS for clinical use include (i) more cross-site collaboration in creation, curation, and integration of VDBKBs; (ii) ensuring these knowledge bases are able to generate both human-readable and computable reports (and standardizing the vocabulary or ontology used to code the computable reports); (iii) development of standards for automating the translation of information in VDBKBs into active decision support rules; (iv) development of best practices for integrating biomedical informatics into clinical and communication workflows; and (v) collaboration with vendors on adapting their active CDS (both EHR and related genetic system vendors as well as the emerging third-party NGS decision support vendors).
ACKNOWLEDGMENTS
All the authors contributed equally to the writing of the this article. The authors thank the members of the Clinical Sequencing Exploratory Research (CSER) consortium, in particular all additional members of the informatics teams at each of the six current CSER sites. The following sources of funding supported parts of the work described in this article: National Institutes of Health extramural projects U01HG006507, U01HG00637, U01HG006500, U01HG006492, U01HG006487, U01HG006485, U01HG006546, KG100355, RC1LM010526, UL1RR02574, UL1TR000423, U01HL098188, and 275200800001C-2-0-1; the Susan G. Komen foundation; the Washington State Life Sciences Discovery Fund; the Northwest Institute for Genetic Medicine; and the Dana-Farber Cancer Institute Leadership Council. L.A.H. is a member of the National Human Genome Research Institute CSER staff team, responsible for scientific program management of the CSER program.
Footnotes
DISCLOSURE
S.J.A. and M.L.: Partners HealthCare licensed the GeneInsight technology in November 2012 to a company in which it acquired 100% equity ownership. This transaction was reviewed by the Partners Committee on conflicts of interest in light of Partners’ acquisition of this financial interest, and the Committee, consistent with Partners policy, required that notice of Partners financial interest in the technology be provided to journals and in publications and presentations on the technology. S.J.A. and M.L. are staff and employee. The other authors declare no conflict of interest.
REfERENCES
- 1.Green ED, Guyer MS. National Human Genome Research Institute. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 2.National Human Genome Research Institute Clinical Sequencing Exploratory Research (U01) 2013 http://grants.nih.gov/grants/guide/rfa-files/RFA-HG-10-017.html.
- 3.Overby C, Kohane I, Williams M, et al. Opportunities for genomic clinical decision support interventions. Genet Med. 2013;15:817–823. doi: 10.1038/gim.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarty CA, Chisholm RL, Chute CG, et al. eMERGE Team The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Human Genome Research Institute. Clinical Sequencing Exploratory Research (CSER) 2013 http://www.genome.gov/27546194.
- 6.Berg J, Amendola L, Eng C, et al. Processes and preliminary outputs for identification of actionable genes as incidental findings in genomic sequence data in the Clinical Sequencing Exploratory Research consortium. doi: 10.1038/gim.2013.133. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Online Mendelian Inheritance in Man, OMIM. 2013 http://omim.org. [PubMed]
- 8.PubMed. 2013 http://www.pubmed.org.
- 9.RefSeq: NCBI Reference Sequence Database. 2013 http://www.ncbi.nlm.nih.gov/refseq/
- 10.dbSNP Short Sequence Variation. 2013 http://www.ncbi.nlm.nih.gov/projects/SNP/
- 11.GeneTests Laboratory Directory. 2013 http://www.genetests.org.
- 12.Tarczy-Hornoch P, Covington ML, Edwards J, Shannon P, Fuller S, Pagon RA. Creation and maintenance of Helix, a Web based database of medical genetics laboratories, to serve the needs of the genetics community. Proc AMIA Symp. 1998:341–345. [PMC free article] [PubMed] [Google Scholar]
- 13.GeneReviews. 2013 http://www.ncbi.nlm.nih.gov/sites/GeneTests/review.
- 14.Tarczy-Hornoch P, Shannon P, Baskin P, Espeseth M, Pagon RA. GeneClinics: a hybrid text/data electronic publishing model using XML applied to clinical genetic testing. J Am Med Inform Assoc. 2000;7:267–276. doi: 10.1136/jamia.2000.0070267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagon RA, Tarczy-Hornoch P, Baskin PK, et al. GeneTests-GeneClinics: genetic testing information for a growing audience. Hum Mutat. 2002;19:501–509. doi: 10.1002/humu.10069. [DOI] [PubMed] [Google Scholar]
- 16.ClinicalTrials.gov. 2013 http://www.clinicaltrials.gov.
- 17.PharmGKB: The Pharmacogenomics Knowledgebase. 2013 http://www.pharmgkb.org.
- 18.Cerner Corporation 2013 http://www.cerner.com.
- 19.Sorian HIS, Siemens Corporation 2013 http://www.siemenssoarian.com.
- 20.Epic Corporation 2013 http://www.epic.com.
- 21.Aronson S, Clark E, Varugheese M, et al. Communicating new knowledge on previously reported genetic variants. Genet Med. 2012;14(8):713–719. doi: 10.1038/gim.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobach D, Sanders G, Bright T, et al. Enabling health care decisionmaking through clinical decision support and knowledge management. In: Quality AfHRa, editor. Evidence Report/Technology Assessments. Agency for Healthcare Research and Quality (US) Rockville, MD: 2012. [PMC free article] [PubMed] [Google Scholar]
- 23.Overby CL, Tarczy-Hornoch P, Kalet IJ, et al. Developing a Prototype System for Integrating Pharmacogenomics Findings into Clinical Practice. J Pers Med. 2012;2:241–256. doi: 10.3390/jpm2040241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Center for Biotechnology Information ClinVar aggregates information about sequence variation and its relationship to human health. 2013 http://www.ncbi.nlm.nih.gov/clinvar/
- 25.National Human Genome Research Institute. Clinically Relevant Genetic Variants Resource: A Unified Approach for Identifying Genetic Variants for Clinical Use (U01) 2013 http://grants.nih.gov/grants/guide/rfa-files/rfa-hg-12-016.html.
- 26.Cassa CA, Savage SK, Taylor PL, Green RC, McGuire AL, Mandl KD. Disclosing pathogenic genetic variants to research participants: quantifying an emerging ethical responsibility. Genome Res. 2012;22:421–428. doi: 10.1101/gr.127845.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overby CL, Tarczy-Hornoch P, Hoath JI, Kalet IJ, Veenstra DL. Feasibility of incorporating genomic knowledge into electronic medical records for pharmacogenomic clinical decision support. BMC Bioinformatics. 2010;11(suppl 9):S10. doi: 10.1186/1471-2105-11-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deshmukh VG, Hoffman MA, Arnoldi C, Bray BE, Mitchell JA. Efficiency of CYP2C9 genetic test representation for automated pharmacogenetic decision support. Methods Inf Med. 2009;48:282–290. doi: 10.3414/ME0570. [DOI] [PubMed] [Google Scholar]
- 29.Hicks JK, Crews KR, Hoffman JM, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin Pharmacol Ther. 2012;92:563–566. doi: 10.1038/clpt.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masys DR, Jarvik GP, Abernethy NF, et al. Technical desiderata for the integration of genomic data into Electronic Health Records. J Biomed Inform. 2012;45:419–422. doi: 10.1016/j.jbi.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starren J, Williams MS, Bottinger EP. Crossing the omic chasm: a time for omic ancillary systems. JAMA. 2013;309:1237–1238. doi: 10.1001/jama.2013.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar IN, Butte AJ, Lussier YA, Tarczy-Hornoch P, Ohno-Machado L. Translational bioinformatics: linking knowledge across biological and clinical realms. J Am Med Inform Assoc. 2011;18:354–357. doi: 10.1136/amiajnl-2011-000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Human Genome Variation Society Recommendations for the description of sequence variants. 2013 http://www.hgvs.org/mutnomen/recs/html.
- 34.Hoffman M, Arnoldi C, Chuang I. The clinical bioinformatics ontology: a curated semantic network utilizing RefSeq information. Pac Symp Biocomput. 2005:139–150. doi: 10.1142/9789812702456_0014. [DOI] [PubMed] [Google Scholar]
- 35.Hoffman MA. The genome-enabled electronic medical record. J Biomed Inform. 2007;40:44–46. doi: 10.1016/j.jbi.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Hoffman MA, Williams MS. Electronic medical records and personalized medicine. Hum Genet. 2011;130:33–39. doi: 10.1007/s00439-011-0992-y. [DOI] [PubMed] [Google Scholar]
- 37.World Health Organization. International Classification of Diseases. (10th ed.) 2010 http://apps.who.int/classifications/icd10/browse/2010/en.