This work reports a plant transcription factor, RSD, that is required for symbiosome development and symbiotic nitrogen fixation (SNF) in the model legume Medicago truncatula. RSD represses the expression of the secretory pathway gene VAMP721a, which suggests that alteration in this pathway is important for SNF.
Abstract
Transcription factors (TFs) are thought to regulate many aspects of nodule and symbiosis development in legumes, although few TFs have been characterized functionally. Here, we describe REGULATOR OF SYMBIOSOME DIFFERENTIATION (RSD) of Medicago truncatula, a member of the Cysteine-2/Histidine-2 (C2H2) family of plant TFs that is required for normal symbiosome differentiation during nodule development. RSD is expressed in a nodule-specific manner, with maximal transcript levels in the bacterial invasion zone. A tobacco (Nicotiana tabacum) retrotransposon (Tnt1) insertion rsd mutant produced nodules that were unable to fix nitrogen and that contained incompletely differentiated symbiosomes and bacteroids. RSD protein was localized to the nucleus, consistent with a role of the protein in transcriptional regulation. RSD acted as a transcriptional repressor in a heterologous yeast assay. Transcriptome analysis of an rsd mutant identified 11 genes as potential targets of RSD repression. RSD interacted physically with the promoter of one of these genes, VAMP721a, which encodes vesicle-associated membrane protein 721a. Thus, RSD may influence symbiosome development in part by repressing transcription of VAMP721a and modifying vesicle trafficking in nodule cells. This establishes RSD as a TF implicated directly in symbiosome and bacteroid differentiation and a transcriptional regulator of secretory pathway genes in plants.
INTRODUCTION
Symbiotic nitrogen fixation (SNF) by bacteria called rhizobia within legume nodules injects ∼40 million tons of nitrogen into agricultural systems each year (Peoples et al., 2009) and is central to sustainable agriculture. Legume nodule development is initiated by an exchange of chemical signals between plant root cells and soil rhizobia (Murray, 2011; Oldroyd, 2013). In response to plant-secreted flavonoids, rhizobia produce and release nodulation (Nod) factors that are perceived by plant root hair cells, triggering asymmetric growth (curling) of the root hair cells and division of cortical cells (Limpens et al., 2003; Radutoiu et al., 2003). Rhizobia enter and traverse epidermal and underlying cortical cells via infection threads (ITs) and are eventually released, via endocytosis, into the cytoplasm of postmeristematic cells of the nodule primordium. This results in bacteria surrounded by a plant membrane, called the symbiosome membrane (SM), which creates a novel organelle termed the symbiosome (Goodchild and Bergersen, 1966; Robertson and Lyttleton, 1984; Roth and Stacey, 1989). Finally, rhizobia undergo a process of differentiation that involves induction of nitrogen fixation genes and repression of ammonium assimilation genes, making symbiosomes into ammonium-exporting organelles of the plant (Udvardi and Day, 1997; Udvardi and Poole, 2013).
Early molecular events involved in nodule initiation are quite well understood as a result of forward genetic studies of two model legumes, Medicago truncatula and Lotus japonicus (Popp and Ott, 2011; Oldroyd, 2013). SNF mutants can be divided into two major categories: mutants unable to form nodule primordia (Nod−) and mutants that develop nodule-like structures with impaired nitrogen fixation (Nod+/Fix−). Mutations in the early recognition and signaling genes produce the Nod− phenotype (Oldroyd, 2013). Several genes have been implicated in IT growth and bacterial colonization of nodule cells, including Mt-Interacting Protein of DMI3 (IPD3)/Lj-CYCLOPS, Mt-VAPYRIN, the flotillin genes Mt-FLOT2 and Mt-FLOT4, Mt-SYMBIOTIC REMORIN1, a putative long coiled-coil protein (Mt-Rhizobium-directed polar growth), and pectate lyase (Lj-Nodulation pectate lyase) (Arrighi et al., 2008; Yano et al., 2008; Haney and Long, 2010; Lefebvre et al., 2010; Murray et al., 2011; Horváth et al., 2011; Xie et al., 2012). Relatively little is known about plant genes and proteins that act after bacterial endocytosis and are required for symbiosome development. A series of seven Nod+/Fix- mutants of M. truncatula designated as defective in nitrogen fixation (dnf1-7) provide some insight into these later processes (Starker et al., 2006). However, the defective genes in only two of these mutants have been characterized. DNF1 encodes a subunit of the signal peptidase complex, which is required to process and target plant proteins to symbiosomes, and DNF2 is a phosphatidylinositol-specific phospholipase C, the precise role of which remains unknown (Van de Velde et al., 2010; Wang et al., 2010; Bourcy et al., 2013). Reverse-genetic studies of M. truncatula have linked a few transcription factor (TF) genes to nitrogen fixation but not nodulation per se (i.e., mutants in these genes exhibit Nod+/Fix− phenotypes); these genes include alfalfa (Medicago sativa) Kruppel-like Zinc finger Protein2-1, Mt-Heme Activator Protein2.1 (HAP2.1), Mt-EFD, and a basic helix loop helix-TF involved in nodule vasculature development (Frugier et al., 2000; Combier et al., 2006; Vernié et al., 2008; Godiard et al., 2011). Forward genetic studies with L. japonicus have identified roles for the following proteins in the later stages of nodule and/or bacteroid development and differentiation: a sulfate transporter, Symbiotic Sulphate Transporter1; an ankyrin-repeat membrane protein, Ineffective Greenish Nodules1; homocitrate synthase, Failure in ENlargement of infected cells1; a putative iron transporter, SEN1; a syntaxin Syntaxin of plants71; and a citrate transporter, Multidrug And Toxic compound Extrusion1 (Krusell et al., 2005; Kumagai et al., 2007; Hakoyama et al., 2009, 2012a, 2012b; Takanashi et al., 2013).
M. truncatula forms indeterminate nodules that contain a persistent apical meristem (zone I), a bacterial invasion zone (zone II), interzone II-III, a nitrogen-fixing zone (zone III), and a senescence zone that is proximal to the root (zone IV, only present in older nodules; Vasse et al., 1990). In zone II, plant cells undergo successive rounds of endoreduplication, resulting in nuclear DNA content ranging from 2C to 64C (Mergaert et al., 2006). In the distal part of the invasion zone, bacteria are released from ITs and symbiosomes are formed. In this zone, bacteroids resemble their free-living counterparts and are called type I bacteroids (Vasse et al., 1990). Type-I bacteroids undergo several rounds of cell division, followed by endoreduplication, which results in enlarged, type II bacteroids in the proximal region of zone II (Kereszt et al., 2011). In contrast with the 1C/2C DNA content of free-living Sinorhizobium meliloti, the DNA content of bacteroids peaks at 24C (Mergaert et al., 2006). In zone III, endoreduplication of both plant and bacterial cells ceases and type II bacteroids differentiate into type III bacteroids, which exhibit cytoplasmic heterogeneity that increases as type IV nitrogen-fixing bacteroids are formed (Vasse et al., 1990; Cebolla et al., 1999).
Endoplasmic reticulum and Golgi-derived vesicles fuse with the SM continuously during symbiosome maturation (Kereszt et al., 2011). Recently, two vesicle-associated membrane proteins, Mt-VAMP721d (for VESICLE-ASSOCIATED MEMBRANE PROTEIN 721d) and Mt-VAMP721e, were shown to be required for symbiosome formation (Ivanov et al., 2012). Numerous plant proteins are delivered to the symbiosome throughout its development. These include a small family of calmodulin-like proteins, Early Nodulin8, a nodule-specific esterase (Liu et al., 2006; Coque et al., 2008), and nodule-specific Cys-rich (NCR) peptides (Van de Velde et al., 2010). Some of the NCR peptides promote endoreduplication of type I bacteroids (Van de Velde et al., 2010). In the M. truncatula dnf1 mutant impaired in the nodule-specific signal peptidase complex, bacteroid differentiation is halted at the type I stage because the signal sequences of NCR peptides cannot be cleaved and these peptides are unable to reach bacteroids (Van de Velde et al., 2010; Wang et al., 2010). In dnf2 mutants, bacteroids are unable to complete differentiation and are subject to early senescence probably due to NCR peptide toxicity (Bourcy et al., 2013).
Symbiosome development culminates in nitrogen fixation at which stage many bacterial housekeeping genes inside the symbiosomes are repressed, while nitrogen fixation (nif and fix) genes are induced (Terpolilli et al., 2012). Micro-aerobic conditions inside nodules are not only prerequisite for activity of oxygen-labile nitrogenase in bacteroids, but also appear to be necessary for induction of bacterial nif and fix genes: Suppression of plant leghemoglobin gene expression in L. japonicus nodules via RNA interference resulted in higher oxygen concentrations inside nodules, lower bacterial nif and fix gene expression, and absence of nitrogenase activity (Ott et al., 2005, 2009). In M. truncatula, S. meliloti 2011 mutants in nifH, nifA, fixG, fixJ, and fixK give rise to nodules in which bacteroids are fully elongated (type IV), but exhibit early senescence (Maunoury et al., 2010). Similarly, L. japonicus Symbiotic Sulphate Transporter1 nodules, which are defective in transport of sulfate from the plant cell cytoplasm to the bacteroids, show low levels of nitrogen fixation (fix+/− phenotype) and early senescence (Krusell et al., 2005).
Interplay between different TFs regulates nodule development. Four TFs from M. truncatula, NODULATION SIGNALING PATHWAY1 (NSP1), NSP2, ERF REQUIRED FOR NODULATION1, and NODULE INCEPTION, have been found to act at early stages of nodule development in response to Nod factor signaling (Schauser et al., 1999; Oldroyd and Long, 2003; Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Marsh et al., 2007; Middleton et al., 2007). Additionally, Mt-Nuclear transcription factor Y subunit alpha1 (Mt-HAP2-1) has been implicated in nodule meristem formation and nodule development (Combier et al., 2006). Only one TF, EFD, has been connected with symbiosome development, albeit indirectly and via an unknown mechanism (Vernié et al., 2008).
Nodule development and cellular differentiation involve changes in the expression of thousands of plant genes (Colebatch et al., 2004; Benedito et al., 2008). Hundreds of TFs are expressed during nodule development, but the roles of only a few of these have been determined (see above). We have taken a systematic, reverse-genetic approach to elucidate the role of uncharacterized M. truncatula TF genes in SNF. We began by annotating 3692 TF genes in the genome (Young et al., 2011) and targeting several nodule-specific TFs for further study. Here, we present functional characterization of a nodule-specific Cysteine-2/Histidine-2 (C2H2) TF from M. truncatula that we have REGULATOR OF SYMBIOSOME DIFFERENTIATION (RSD).
RESULTS
RSD Encodes a C2H2 TF with Nodule-Specific Expression
Using the M. truncatula Gene Expression Atlas (MtGEA; Benedito et al., 2008), we identified a putative C2H2 TF gene (MtGEA: Mtr.38404.1.S1_at) with nodule-specific expression (see Supplemental Figure 1 online), which we named RSD for reasons explained below. Comparison of cloned cDNA and genomic DNA sequences indicated that the gene contains a single exon, which encodes a putative C2H2 TF of 151 amino acids (see Supplemental Figure 2 online).
Phylogenetic analysis clustered Mt-RSD with a protein from L. japonicus and two from soybean (Glycine max), which shared between 50 and 62% sequence identity with Mt-RSD (Figures 1A and 1B). Mt-RSD is also related to Arabidopsis SUPERMAN and M. truncatula PALMATE-LIKE PENTAFOLIATA1 (PALM1) proteins (Figure 1B), which regulate different aspects of development (Bowman et al., 1992; Chen et al., 2010). All of the proteins closely related to Mt-RSD share a single C2H2 DNA binding domain near the N terminus and a hexapeptide sequence (DLELRL), known as an EAR domain, at the C terminus (Figure 1A). The EAR domain confers transcriptional repression upon DNA binding proteins (Hiratsu et al., 2003; Ikeda and Ohme-Takagi, 2009).
Figure 1.
Protein Sequence, Conserved Domains, and Phylogenetic Analysis of RSD.
(A) Alignment of Mt-RSD amino acid sequence with that of putative orthologs in L. japonicus and soybean. Conserved amino acids are highlighted. The C2H2 DNA binding domain and EAR domain are marked.
(B) Unrooted phylogenetic unweighted pair group method with arithmetic mean tree of Mt-RSD homologs/orthologs in M. sativa (Ms), Arabidopsis (At), Populus trichocarpa (Pt), G. max (Gm), L. japonicus (Lj), and Oryza sativa (Os). Sequences were identified via BLASTP searches of the National Center for Biotechnology Information database of nonredundant sequences and aligned at the protein level using multiple sequence comparison by log expectation (see Supplemental Data Set 1 online). At-ZAP6, which has a C2H2 DNA binding domain but lacks the EAR domain, was used as an outgroup. Numbers represent the percentage of 1000 bootstrap replications to assess robustness of nodes. The asterisk indicates the nodulation-specific subgroup of proteins. The bar represents estimated amino acid change per sequence position.
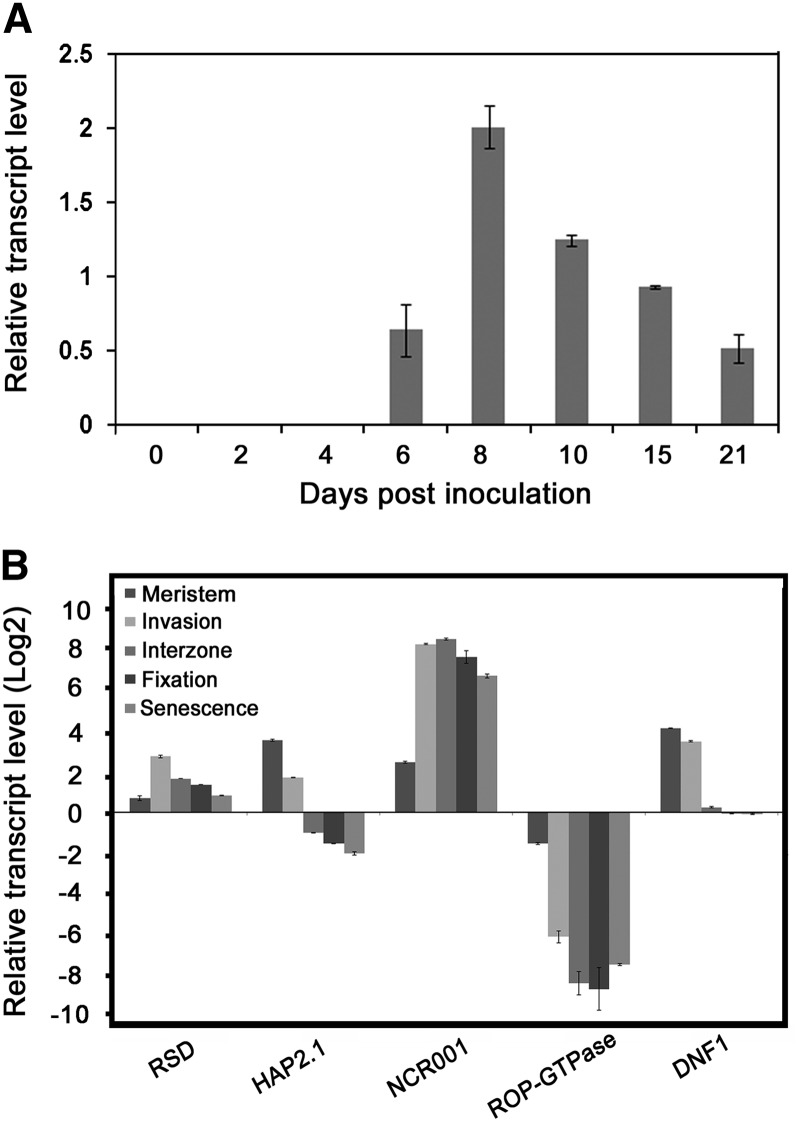
To confirm and extend the RSD gene expression data from MtGEA (see Supplemental Figure 1 online), we measured transcript levels in roots and in nodules of genotypes A17 and R108 at different stages of development or in different nodule zones, using quantitative RT-PCR (qRT-PCR). Preliminary work had established that the onset of nitrogen fixation exhibited similar kinetics in these two genotypes under our experimental conditions (see Supplemental Figure 3 online). RSD transcripts were barely detectable in roots before and 2 and 4 d after inoculation (DAI) with rhizobia in the wild-type R108 genotype (Figure 2A). Transcript levels in developing nodules increased by 6 DAI and peaked at 8 DAI, with a level 4000-fold higher than in roots, before declining steadily at 10, 15, and 21 DAI (Figure 2A). Thus, RSD expression during nodule development was qualitatively similar in the R108 and A17 genotypes. Acetylene reduction assays and qRT-PCR showed that RSD transcript accumulated before the onset of nitrogen fixation (see Supplemental Figure 3 online; Figure 2A). RSD was expressed in all nodule zones at 28 DAI, with maximal levels in the invasion zone in A17 nodules (zone II; Figure 2B). HAP2.1, Nodule-specific Cystine-Rich peptide001, ROP-GTPase, and DNF1 transcript levels were also measured in the five nodule zones to assess our nodule zonation data for RSD. Promoter–β-glucuronidase or in situ hybridization results for expression of these genes in nodules are available in the literature (Mergaert et al., 2003; Combier et al., 2006; Wang et al., 2010; Limpens et al., 2013). Spatially resolved expression data for each of the four marker genes (Figure 2B) matched the published data with the exception of NCR001, transcripts of which were detected previously only in interzone II-III and the nitrogen-fixing zone, using in situ hybridization (Mergaert et al., 2003). Our qRT-PCR data indicated a high level of NCR001 gene expression in the invasion zone in addition to interzone II-III and the nitrogen-fixing zone, which may reflect some cross-contamination between the invasion zone and interzone II-III in our hand-sectioned material. Nonetheless, the higher levels of transcripts for RSD and the marker genes HAP2.1 and DNF1 in the invasion zone compared with in interzone II-III cannot be explained by cross-contamination of cells from the former with the latter zone. To summarize, our data show that induction of RSD expression preceded nitrogen fixation, with maximal expression level in the invasion zone and moderate levels in all other zones of 28-DAI nodules.
Figure 2.
Temporal and Spatial Expression Profiles of Mt-RSD.
(A) Expression profile of RSD during nodule development in M. truncatula genotype R108. qRT-PCR was used to measure RSD transcript levels in the infection-susceptible zone of roots (0 to 4 DAI) and in developing nodules (6 to 21 DAI)
(B) Relative transcript levels of RSD in different zones of nodules of genotype A17 at 28 DAI. HAP2.1, NCR001, ROP-GTPase, and DNF1 relative transcript levels are included for comparison. Mean and sd of three biological replicates are presented in each case.
RSD Is Required for Normal Nodule Development and SNF
To illuminate the function of RSD in nodules, we isolated two independent Tnt1 (for transposable element of Nicotiana tabacum cell type1) insertion rsd mutants of M. truncatula R108 and determined their symbiotic phenotypes. In the mutant line Noble Foundation (NF) 11265, Tnt1 was found in the open reading frame of RSD (rsd-1 allele), whereas in line NF3492, it was in the 3′-untranslated region (UTR), 382 bp after the stop codon (rsd-2 allele; Figure 3A; see Supplemental Figure 2 online). Homozygous rsd mutants of both lines usually developed small, spherical, white nodules that turned brown within 3 weeks of inoculation with S. meliloti strain 2011 (Figure 3B). rsd-1 was unable to fix nitrogen, which resulted in retarded growth relative to the wild type (Figures 3B and 3C). The rsd-2 mutant allele appeared to be weaker than that of rsd-1, as rsd-2 mutants produced occasional light pink nodules and fixed nitrogen, according to the acetylene reduction assay, albeit significantly less than the wild type (Figures 3B and 3C). The leakiness of the rsd-2 allele was likely due to the location of Tnt1 in the 3′-UTR of the gene. Subsequent work on rsd mutants focused on the presumptive complete loss-of-function allele rsd-1, which bears Tnt1 in the RSD coding sequence.
Figure 3.
Symbiotic Phenotypes of rsd Mutants and Complementation by p35S-GFP-MtRSD.
(A) Schematic representation of the Mt-RSD gene with Tnt1 insertion sites (arrowheads) in two independent lines. The black region represents the protein coding region, and UTRs are shown in gray. Bar = 100 bp.
(B) Wild-type R108 (left), rsd-1 (NF11265; middle), and rsd-2 (NF3492; right) at 21 DAI with S. meliloti strain 2011. Close-up views of root nodules of the wild type (left), rsd-1 (middle), and rsd-2 (right) are shown below.
(C) Acetylene reduction activity (ARA) of whole nodulated roots of R108, rsd-1, and rsd-2 at 15 DAI. Data are the mean of 41 plants for R108, 32 plants for rsd-1, and 12 plants for rsd-2. Vertical bars represent se. n.d., not detected.
(D) to (G) Complementation of the rsd-1 (NF11265) mutant with a p35S-GFP-MtRSD construct via Agrobacterium rhizogenes–mediated hairy root transformation. Stereomicroscope images under light ([D] and [F]) and fluorescent illumination ([E] and [G]). Bars = 1 mm.
(D) and (E) Spheroidal nodules on control roots transformed with empty vector at 28 DAI.
(F) and (G) Cigar-shaped nodules on roots transformed with p35S-GFP-MtRSD at 28 DAI. The inset in (F) shows a typical, pink complemented nodule.
Despite the reduced nodule size of rsd-1 mutants (1.2 ± 0.2 mm compared with 2.3 ± 0.3 mm of the wild type at 21 DAI; n = 16 plants; ±sd), nodule number was unaffected at 21 DAI (11.4 ± 4.0 versus 11.2 ± 4.9 of the wild type; n = 22 plants).
Progeny of a heterozygous rsd-1 mutant segregated with an 81:24 ratio of wild-type (Fix+) to mutant (Fix-) phenotypes, fitting the 3:1 ratio expected of a monogenic, recessive mutation (χ2 = 0.257; P > 0.05). Genotyping of Fix− individuals confirmed that all were homozygous for the rsd-1 Tnt1 insertion allele. A similar result was obtained after backcrossing the rsd-1 mutant to wild-type R108, self-pollinating a resultant rsd-1 heterozygote, and analyzing its progeny: 60 Fix+ and 21 Fix− individuals, again fitting the expected 3:1 ratio (χ2 = 0.037; P > 0.05).
To confirm that the Tnt1 insertion in the RSD gene caused the defective symbiotic phenotypes, we placed a wild-type copy of the gene under the control of the constitutive cauliflower mosaic virus 35S promoter (35S-MtRSD) and expressed it in transgenic hairy roots of the rsd-1 mutant. Wild-type-like nodules containing fully elongated bacteroids developed on transformed roots of the mutant following inoculation with rhizobia (cf. Figures 3F and 3G with 3D and 3E and Supplemental Figures 4A to 4I online).
In contrast with the growth defect of rsd-1 mutants under symbiotic conditions with low soil mineral nitrogen (Figure 3B; see Supplemental Figures 5A and 5B online), rsd-1 mutants exhibited no significant differences in growth or development of vegetative or reproductive organs when grown in the presence of high soil mineral nitrogen (see Supplemental Figure 5 online).
RSD Affects Symbiosome and Bacteroid Development
No delay in nodule initiation and no defects in IT formation or propagation were observed in rsd-1 mutants (see Supplemental Figures 6A to 6C online). In addition, no significant differences in the numbers of bacterial microcolonies or ITs in the susceptible zone of roots were observed between rsd-1 and wild-type R108 plants (see Supplemental Figure 6C online). Staining of beta-galactosidase (lacZ)-expressing rhizobia revealed a slightly smaller population of bacteria in rsd-1 mutant nodules compared with those of the wild type at 6 DAI and a substantially smaller population by 8 DAI (cf. Figures 4A and 4B with 4D and 4E). By 21 DAI, brown material, possibly polyphenolic compounds involved in plant defense, accumulated proximal to the invasion zone in the rsd-1 mutant, but not in the wild type (cf. Figures 4C and 4F).
Figure 4.
Bacterial Colonization Phenotypes of Nodules from Wild-Type and rsd-1 Mutant Plants.
Wild-type (R108) nodules ([A] to [C]) and rsd-1 mutant nodules ([D] to [F]) at 6 DAI ([A] and [D]), 8 DAI ([B] and [E]), and 21 DAI ([C] and [F]). Bars = 100 μm in (A), (B), (D), and (E) and 200 μm in (C) and (F). lacZ-expressing S. meliloti strain 2011 cells are blue from X-Gal staining.
Despite the retarded growth of developing rsd-1 nodules, no significant defect was apparent at the early stage in zone I (meristem) of the rsd-1 mutant in comparison to the wild type (cf. Figures 4C and 4F with Supplemental Figures 6D and 6E online). However, the meristematic activity ceased rapidly in rsd-1, and nodules were usually spherical instead of cylindrical. Marked differences between the mutant and wild type were observed in the older cell layers of zone II (invasion) and in interzone II-III. Although release of bacteria from ITs and their subsequent division and differentiation into type I bacteroids in the distal part of zone II appeared normal in rsd-1 (Figures 5B and 5F; see Supplemental Figures 6F and 6G online), further differentiation reflected by bacteroid elongation was retarded in the mutant compared with the wild type (Figures 5B to 5D and 5F to 5H; see Supplemental Figures 6F to 6K online). The most striking difference between rsd-1 and wild-type nodules was the presence of plant cells heavily packed with bacteria at the distal end of the invasion zone in the case of the mutant but not the wild type (cf. Figures 5A and 5E with 5B and 5F; cf. Supplemental Figures 6D and 6F with 6E and 6G online).
Figure 5.
Differentiation of Bacteroids Is Impaired in rsd-1 Mutant Nodules.
(A) to (H) Phenotype of R108 ([A] to [D]) and rsd-1 ([E] to [H]) nodules at 12 DAI with S. meliloti strain 1022 containing the mCherry plasmid. Nodules were stained with FM1-43 (green) to visualize membranes ([A], [C], [E], and [G]). The asterisk in (E) marks autofluorescence. Bars = 100 μm in (A) and (E), 10 μm in (B), (C), (F), and (G), and 1 μm in (D) and (H).
(B) and (F) Light micrographs of semithin sections from 12-DAI nodules stained with toluidine blue-O.
(B), (C), (F), and (G) Cells of the invasion zone.
(D) and (H) Transmission electron micrographs of infected cells showing bacteroids in zone II.
(I) to (L) Distribution of ploidy levels of plant and bacteroid cells in nodules at 18 DAI with S. meliloti 1021.
(I) and (J) DNA content of plant cells from wild-type R108 and rsd-1 nodules, respectively. DNA content of individual nuclei was estimated by relative DAPI fluorescence. 2C, 4C, 8C, 16C, 32C, and 64C DNA content are indicated.
(K) and (L) DNA content of DAPI-stained bacteroids measured by flow cytometry.
On the proximal (i.e., root) side of these cells, one or two layers of cells were packed with incompletely elongated bacteroids in the mutant (Figure 5F; see Supplemental Figure 6G online). Furthermore, the characteristic orientation of symbiosomes around the central vacuole of wild-type cells was not observed in rsd-1 mutant cells (cf. Supplemental Figures 6J and 6K online). Overall, invasion zone II was larger in the mutant than in the wild type. Type II bacteroids reach lengths of up to 10 μm (Figures 5C and 5D) in wild-type nodules. However, no bacteroids longer than 5 μm were observed in rsd-1 nodules (Figures 5G and 5H). Moreover, cytoplasmic heterogeneity (i.e., distinct electron-dense and translucent regions imaged by electron microscopy; Vasse et al., 1990) was not observed in bacteroids of mutant nodules. Instead, premature senescence of bacteroids occurred by 12 DAI proximal to zone II of mutant nodules (Figure 5E; see Supplemental Figure 7 online). To distinguish the bacteroid from the SM, we employed rhizobia expressing the mCherry (red) fluorescent protein and the membrane stain, FM1-43 (green). In zone II of rsd-1 nodules, some cells showed mCherry-labeled bacteria within FM1-43–stained SMs, while other cells exhibited FM1-43–stained symbiosomes without mCherry-labeled bacteria, possibly indicating that bacteroid degradation began before disintegration of the SM (cf. Supplemental Figures 6H and 6I online).
To test the viability of bacteria inside rsd-1 nodules, we used a live/dead staining procedure with a mixture of the two fluorescent nucleic acid dyes, SYTO9 and propidium iodide (PI; Haag et al., 2011). Live bacteria with intact cytoplasmic membranes are stained by membrane-permeable SYTO9 (green), while dead bacteria with degraded cytoplasmic membranes are stained by membrane-impermeable PI (red). As shown in Supplemental Figures 7A to 7C online, bacteria in wild-type cells stained green throughout the invasion and fixation zones. By contrast, green-stained bacteria were observed only inside ITs and in a few cells of the invasion zone in rsd-1 nodules at 12 DAI (see Supplemental Figures 7F to 7H online). In the distal part of the invasion zone of rsd-1 nodules, some cells contained both red- (dead) and green-stained (live) bacteria (see Supplemental Figure 7G online). All bacteria except those inside ITs appeared to be dead (stained red) in the proximal part of the invasion zone (see Supplemental Figure 7G online). Transmission electron microscopy showed that degrading symbiosomes, indicated by an enlarged symbiosome space containing multiple bacteria, occurred in plant cells containing intact nuclei (see Supplemental Figures 6K, 7I, and 7J online). Taken together, the microscopy results indicate that bacteria lose viability within the invasion zone of rsd-1 mutant nodules but not as a consequence of plant cell senescence.
Given the microscopy data and the known correlation between cell size and ploidy, we measured the ploidy of both plant and bacteroid cells from rsd mutant and wild-type nodules. There was a slight shift to lower ploidy levels among plant cells harvested from rsd-1 mutants compared with the wild type (Figures 5I and 5J), indicating a reduction in endoreduplication in some of the mutant cells. Similarly, a shift in ploidy distribution to lower levels in bacteroids of the rsd-1 mutant indicated fewer rounds of endoreduplication in the mutant compared with the wild type (Figures 5K and 5L), consistent with the observed reduction in bacteroid size (cf. Figures 5C and 5D with 5G and 5H) and the rapid loss of bacterial viability noted above.
Placing RSD within the Nodule Developmental Program
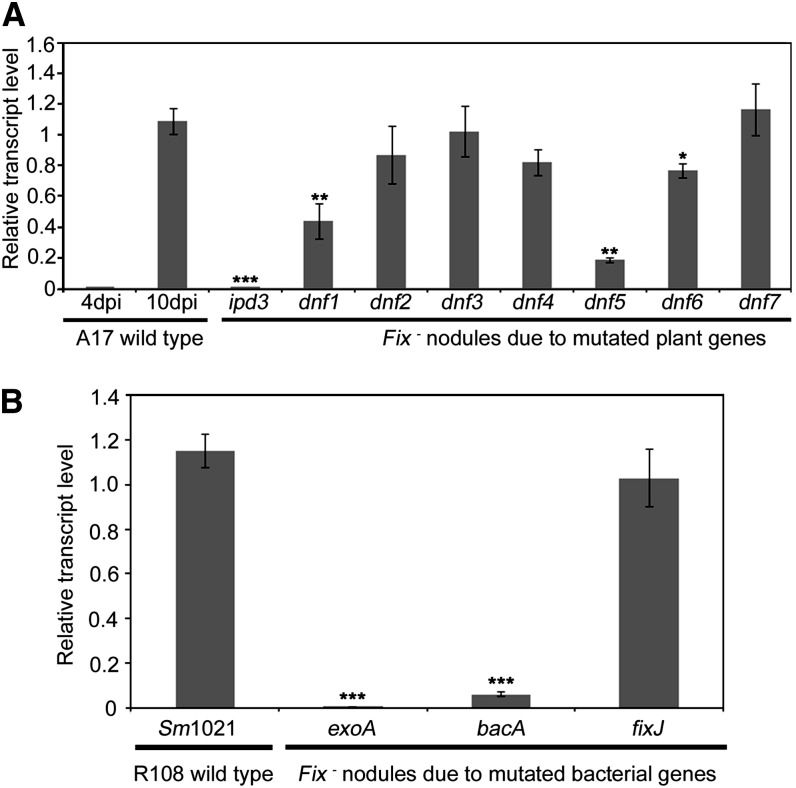
To place RSD function into context relative to other genes that are required for nodule development and/or differentiation, we quantified Mt-RSD transcript levels in various plant mutants. Because RSD gene expression in the wild type commenced around 6 DAI (Figure 2A), after formation of nodule primordia, we excluded Nod− mutants that do not form primordia from this analysis. The mutant with the earliest nodulation defect that we analyzed was ipd3 (Benaben et al., 1995; Horváth et al., 2011). ipd3 mutants make small nodule-like structures in which bacteria are very rarely released from ITs. RSD expression was significantly lower in the ipd3 mutant than in the wild type at 10 DAI (P ≤ 0.001) (Figure 6A). Likewise, RSD expression was significantly lower in dnf1, dnf5 (P ≤ 0.01) , and dnf6 (P ≤ 0.05) mutants at 10 DAI (Figure 6A).
Figure 6.
Expression of RSD in Various Fix− Nodules.
Relative transcript levels of Mt-RSD in different types of Fix− nodules measured at 10 DAI. Asterisks indicate a significant difference in RSD expression with respect to the Fix+ control: *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. Mean and sd of three biological replicates are presented in each case.
(A) Various Fix− plant mutants.
(B) Various Fix− bacterial mutants in wild-type plants.
We examined the expression of RSD in Fix− nodules elicited by the bacterial mutants exoA, bacA, and fixJ (Leigh et al., 1985; David et al., 1988; Glazebrook et al., 1993; Oke and Long, 1999; Mitra et al., 2004). exoA is an exopolysaccharide EPS I–deficient bacterial mutant that elicits nodule primordium formation but fails to invade nodules (Leigh et al., 1985; Mitra et al., 2004). RSD expression was barely detectable in nodules elicited by the exoA mutant (Figure 6B). Bacterial differentiation A (bacA) mutant bacteria show normal endocytosis but are unable to differentiate (Glazebrook et al., 1993; Oke and Long, 1999). The absence of BacA results in hypersensitivity toward antimicrobial NCR peptides and rapid death of undifferentiated bacteria (Haag et al., 2011). In the Fix− nodules elicited by the bacA mutant, RSD expression was significantly lower than in nodules containing wild-type bacteria after 10 DAI (P ≤ 0.001), although transcript levels were substantially higher in nodules containing the bacA mutant than in those elicited by the exoA mutant (Figure 6B). FixJ is a response regulator, and fixJ mutants elicit wild-type-like nodules that fix nitrogen but undergo early senescence (David et al., 1988; Oke and Long, 1999). RSD expression was normal in these nodules at 10 DAI (Figure 6B).
We also measured the expression of leghemoglobin (Medtr1g011540.1) in all of the above-mentioned Fix− nodules. In dnf1, dnf5 (both plant mutants), and bacA (a bacterial mutant) nodules, expression of RSD was detectable, but expression of leghemoglobin was not (see Supplemental Figure 8 online). Likewise, leghemoglobin gene expression was not induced in the rsd-1 mutant, indicating that expression of RSD precedes that of leghemoglobin (see Supplemental Figure 8 online,).
RSD Is a Nuclear Protein
To gain insight into the possible subcellular location of Mt-RSD protein, we transformed tobacco (Nicotiana tabacum) leaf epidermal cells with DNA encoding a green fluorescent protein (GFP)-MtRSD fusion, which revealed a nuclear location for the protein (see Supplemental Figures 9A and 9B online). Expression of GFP alone in tobacco cells led to GFP fluorescence in both nucleus and cytoplasm (see Supplemental Figures 9C and 9D online).
RSD Acts as a Transcriptional Repressor when Expressed in Yeast
To test whether Mt-RSD, which contains an EAR (for Ethylene-responsive element binding factor-associated Amphiphilic Repression) repressor domain, can repress gene transcription, we used a yeast one-hybrid assay. Mt-RSD or Mt-RSD without the EAR domain–encoding sequence (MtRSDΔEAR) were cloned into vector pGBT9 between the Gal4 DNA binding domain (Gal4-DBD) and the activation domain of herpes simplex virus protein VP16 (Sadowski et al., 1988), generating effector constructs (Figure 7A). Another construct containing six Gal4-DNA binding motifs and the pCYC1 minimal promoter controlling the lacZ gene was used as reporter in transactivation assays (Figure 7A). After coexpressing reporter and effector constructs in yeast, β-galactosidase assays were performed. Mt-RSD repressed the transactivation activity of VP16 by over 90% (comparing pGBT9:MtRSD-VP16 to the control, pGBT9:GFP-VP16; Figure 7A). Removal of the EAR domain of Mt-RSD (pGBT9:MtRSDΔEAR-VP16) led to partial recovery of VP16 activity (Figure 7A). These results indicated that the EAR domain of Mt-RSD can act as a repressor of transcription and that additional amino acids in Mt-RSD may contribute to transcriptional repression.
Figure 7.
Mt-RSD Acts as a Transcriptional Repression.
(A) Effector and reporter constructs for testing Mt-RSD transcriptional activation and repression activity in yeast. Gal4-DBD, Gal4 DNA binding domain; VP16, activation domain of the VP16 protein; GAL4-DB motif, Gal4-DBD binding site. β-Galactosidase assays showing activation of lacZ expression by each construct. Average and standard deviations are calculated from three independent experiments.
(B) Expression profile of Mt-RSD and Mt-VAMP721a. qRT-PCR was used to measure Mt-RSD and Mt-VAMP721a transcript levels at 0, 6, and 8 DAI in M. truncatula R108 and rsd-1 nodules.
(C) ChIP followed by quantitative PCR showed a significant enrichment in Medtr4g022570 (Mt-VAMP721a) promoter DNA detected with primer set 2 (pSET2) from RSD-transformed hairy roots compared with controls transformed with the empty vector. The presumptive binding motif is indicated.
(D) EMSA of the 288-bp promoter region (fragment −1870 to −1581, with respect ATG) as the probe labeled with biotin in the presence (lanes 2 to 4) or absence (lane 1) of purified His-tagged Mt-RSD. Unlabeled promoter fragment (−1870 to −1581) in 25-fold (lane 3) or 50-fold (lane 4) molar excess relative to the biotin-labeled sequence was used as specific competitor.
Transcriptome Perturbations Resulting from Loss of Mt-RSD Function
To identify potential targets of RSD transcriptional repression, we performed transcriptome analysis of mutant and wild-type plants using Medicago Affymetrix GeneChips. Bearing in mind that RSD expression began around 6 DAI and reached a maximal level by 8 DAI (Figure 2A), we sought genes that were expressed at significantly higher levels in nodules of the rsd-1 mutant than in the wild type at both of these time points, using 0 DAI as a point of reference for each genotype (see Supplemental Data Set 1 online). Eleven genes satisfied this criterion (Table 1; see Supplemental Data Set 1 online). To test more broadly for a negative correlation between transcript levels of each of these 11 genes and RSD, we used the MtGEA nodule developmental data and nodule zonation data (see Methods; Table 1). Transcript levels of only one of the genes, VAMP721a, showed a moderate negative correlation (R2 = 0.64) with RSD transcript levels (see Supplemental Figure 10 online). To validate the microarray data, we performed qRT-PCR to quantify RSD and VAMP721a gene expression in wild-type (R108) and rsd-1 roots and nodules at 6 and 8 DAI (Figure 7B). A strong negative correlation between RSD and VAMP721a expression was found (R2 = 0.85). Furthermore, the upregulation ratio of VAMP721a in rsd-1 nodules to that of wild-type R108 nodules at 6 and 8 DAI were 1.5- and 2.13-fold, respectively (Figure 7B), substantiating the microarray data.
Table 1. Regression Analysis for Genes Expressed at Significantly Higher Levels in Nodules of the rsd-1 Mutant Than in the Wild.
| MtGEA Probe Sets | Putative Annotation | MTGI9 | IMAGA3.5v4 Gene ID | Fold Change rsd-1 0 DAI/R108 0 DAI | Fold Change rsd-1 6 DAI/R108 6 DAI | Fold Change rsd-1 8 DAI/R108 8 DAI | Regression (R) | Regression Coefficient (R2) |
|---|---|---|---|---|---|---|---|---|
| Mtr.1810.1.S1_at | Hypothetical protein | BE202654 | Not present | 0.80 | 2.39 | 2.02 | −0.33 | 0.11 |
| Mtr.22685.1.S1_at | Ser hydroxymethyltransferase | IMGA|Medtr7g070020.1 | 1.02 | 3.06 | 3.36 | 0.43 | 0.19 | |
| Mtr.23138.1.S1_s_at | MuDR family transposase protein, partial (61%) | Not present | 1.02 | 48.53 | 49.62 | 0.60 | 0.36 | |
| Mtr.29527.1.S1_at | Hypothetical protein | TC140926 | Not present | 0.63 | 6.30 | 4.48 | −0.23 | 0.05 |
| Mtr.40617.1.S1_at | maiA maleylacetoacetate isomerase | TC121481 | ILLUM|contig_55748_1 | 1.29 | 3.19 | 2.20 | 0.31 | 0.09 |
| Mtr.42809.1.S1_at | Probable peptidyl-tRNA hydrolase 2 | DW016763 | IMGA|Medtr3g109700.1 | 1.26 | 2.89 | 2.72 | 0.39 | 0.15 |
| Mtr.46130.1.S1_at | VAMP721 | TC119955 | IMGA|Medtr4g022570.1 | 1.4 | 2.47 | 2.68 | −0.80 | 0.64 |
| Mtr.48599.1.S1_at | Phosphatidylethanolamine binding protein | TC129531 | IMGA|Medtr7g104460.1 | 0.60 | 2.52 | 2.00 | −0.43 | 0.19 |
| Mtr.48791.1.S1_at | PLAC8 family protein | TC123158 | IMGA|Medtr2g101660.1 | 1.27 | 2.44 | 4.39 | 0.007 | 0.000049 |
| Mtr.583.1.S1_at | Transposase activity | Not present | 1.63 | 2.19 | 2.26 | 0.56 | 0.31 | |
| Mtr.7517.1.S1_at | Protein transport protein Sec24-like At3g07100 | TC128707 | Not present | 1.01 | 4.02 | 3.65 | 0.23 | 0.05 |
Transcript levels of each gene were plotted against the corresponding level for the RSD gene at each time point. Time points used for this analysis are indicated in Supplemental Figure 10 online. Blank cells in column three indicate that gene IDs were not available. MUDR, Mutator (MU) Class II family of transposons.
To identify potentially pleiotropic effects of loss of RSD activity, we also sought genes that were expressed at significantly lower levels in nodules of the rsd-1 mutant than in the wild type at 6 and/or 8 DAI (see Supplemental Data Set 2 online). In nodulated roots of wild-type plants, transcript levels of 1093 plant genes increased significantly (P ≤ 0.05) and substantially (more than twofold increase/decrease in transcript level) by 6 DAI compared with the 0-DAI root control (see Supplemental Figures 11A to 11C and Supplemental Data Set 3 online). Far fewer (479) genes were induced at 6 DAI in nodulated roots of the rsd-1 mutant compared with mutant roots at 0 DAI (see Supplemental Figures 11A to 11C and Supplemental Data Set 3 online). By 8 DAI, 2003 genes were induced in the wild type and 1742 genes were induced in the mutant, relative to their respective 0-DAI controls (see Supplemental Figures 11A to 11C and Supplemental Data Set 3 online). Interestingly, 506 of the genes induced by 6 DAI in wild-type nodules were induced later, only at 8 DAI in rsd-1 mutant nodules (see Supplemental Figures 11A to 11C and Supplemental Data Set 3 online), including many nodule-specific NCR genes (see Supplemental Figure 11D online) and other nodule-specific genes (see Supplemental Data Set 3 online). These microarray results were validated by qRT-PCR analyses of 22 genes, all of which gave qualitatively similar results (see Supplemental Figures 12B to 12E online).
Bacteroid differentiation leading to SNF involves many genes that are induced during nodule development (Oke and Long, 1999; Starker et al., 2006). To determine the stage at which bacteroid differentiation was blocked in the rsd-1 mutant, we measured transcript levels of three bacterial genes: bacA, which is required for S. meliloti survival and differentiation after release from ITs (Haag et al., 2011); nifA, a regulator of nitrogenase gene expression (Starker et al., 2006); and nifH, which encodes a subunit of nitrogenase (Starker et al., 2006). qRT-PCR analysis revealed that bacA transcript levels were similar in bacteroids of rsd and wild-type nodules at 15 DAI (see Supplemental Figure 12A online). By contrast, no expression of the nifA and nifH genes was detected in the mutant, despite high levels of expression of these genes in wild-type nodules at 15 DAI (see Supplemental Figure 12A online). Parallel measurements of plant leghemoglobin gene expression in these nodules revealed high expression levels in the wild type but no expression in rsd-1 mutant nodules at 15 DAI (see Supplemental Figure 12A online).
Thus, loss of Mt-RSD function resulted in delayed or no induction of many nodule-specific plant genes as well as bacterial nif genes, some of which are crucial for bacteroid differentiation and SNF. However, given the likely role of RSD as a transcriptional repressor, we considered these effects pleiotropic and focused our attention on the Mt-VAMP721a gene as a putative target of Mt-RSD transcriptional repression.
RSD Binds to the Promoter of VAMP721a
To test the hypothesis that VAMP721a is a direct target of RSD repression, we performed a chromatin immunoprecipitation (ChIP)-PCR assay. First, we introduced a p35S-RSD-GFP construct into M. truncatula hairy roots. We then performed ChIP using an anti-GFP antibody, with three independently transformed hairy root systems, followed by quantitative PCR using several primer pairs designed to bind within and outside the VAMP721a promoter region. Quantitative PCR analysis with primer set 2, which amplified the region between −1645 and −1579 upstream (5′) of the ATG start codon revealed massive and significant enrichment of DNA compared with two other segments in the promoter region and one in exon 5 of the VAMP721a gene (Figure 7C). The sequence ACAGTGTC, and especially the AGT in its core, is known to bind C2H2 zinc-finger proteins from petunia (Petunia hybrida) and Arabidopsis thaliana (Takatsuji and Matsumoto, 1996; Dathan et al., 2002; Ren et al., 2007). We found three instances of the sequence AAAGTGTC between positions −1641 and −2139 relative to the translation start codon of VAMP721a (Figure 7C). To confirm direct binding of RSD to the VAMP721a promoter, we performed an electrophoretic mobility shift assay (EMSA) using a 288-bp fragment (−1870 to −1581: two AAAGTGTC-boxes are present in this region) as the DNA probe and purified His6-MtRSD protein (Figure 7D). RSD protein formed a single distinct complex with this 288-bp fragment, as evidenced by a single retarded band in the EMSA. Competition assays with 25- and 50-fold molar excess of nonlabeled DNA probe indicated specific binding of RSD to the VAMP721a promoter fragment (Figure 7D).
DISCUSSION
The data presented here reveal an important role of RSD in symbiosome development and nitrogen fixation in M. truncatula. To recapitulate, RSD was expressed in a nodule-specific manner, with massive transcript accumulation in nodules by 6 DAI, maximal transcript levels in the invasion zone (II) and high levels in the interzone (II-III) of mature nodules (28 DAI), the zones in which symbiosomes arise and differentiate prior to SNF in zone III (Figure 2). RSD was barely expressed in nodules of plants in which primordia formed, but no bacteria were taken up into symbiosomes, such as those formed by the bacterial exoA mutant and the plant idp3 mutant. On the other hand, Mt-RSD was expressed in nodules in which bacterial endocytosis was normal, but bacteria did not fully differentiate into nitrogen-fixing bacteroids, such as in nodules of the plant dnf1-7 mutant (Figure 6A) and nodules of wild-type plants elicited by bacterial bacA and fixJ mutants (Figure 6B). Furthermore, bacteroids failed to differentiate normally in rsd-1 mutant nodules as indicated by microscopy (Figures 4 and 5; see Supplemental Figure 6 online) and reduced endoreduplication (Figures 5I to 5L). Reduced bacterial enlargement in rsd-1 nodules was observed first in invasion zone II, coincident with the peak of RSD expression in wild-type nodules. Live-dead staining showed that rhizobia lost viability within the invasion zone of rsd-1 nodules (see Supplemental Figure 7 online). Nodules of rsd-1 mutants exhibited early nodule senescence, which is typical of symbioses formed by rhizobia mutants that fail to differentiate and/or cannot fix nitrogen (Starker et al., 2006; Van de Velde et al., 2006).
Mt-RSD is a member of the C2H2 family of plant TFs and contains not only the signature C2H2 domain, but also a conserved EAR domain (Figure 1A), which is known to confer transcriptional repressor activity on some TFs (Bowman et al., 1992; Takatsuji, 1999; Chen et al., 2010). Consistent with this, we found that the EAR domain contributed to transcriptional repression by Mt-RSD of a reporter gene in a yeast transactivation assay (Figures 7A and 7B). Removal of the EAR domain partially alleviated the repressive effect of Mt-RSD. Transcriptome analyses (Supplemental Figure 11 and Supplemental Data Sets 1 to 3 online; Table 1) pointed to at least one potential target of RSD repression in M. truncatula, namely, the VAMP721a gene. Subsequently, we found that RSD binds to the promoter of VAMP721a, confirming the latter as a direct target of RSD repression (Figures 7C and 7D).
VAMP721a belongs to the R-SNARE family, members of which are generally located on the membranes of trafficking vesicles (V-SNAREs) and are required for vesicle fusion with specific target membranes via interaction with target t/Q-SNAREs (Lipka et al., 2007). Recently, it was found that two related v/R-SNAREs in M. truncatula, VAMP721d and VAMP721e, are required to form the SM during nodule development, via an exocytotic pathway (Ivanov et al., 2012). Reduction of VAMP721d and VAMP721e levels via RNAi in M. truncatula led to failure in both release of rhizobia from ITs and subsequent symbiosome proliferation, due to reduced cell wall dissolution around the IT at the site of bacterial entry into plant cells. The authors proposed that the cargo delivered by vesicles bearing VAMP721d and VAMP721e are important for the formation of a cell wall–free interface at the IT prior to symbiosome formation. VAMP721a, VAMP721d, and VAMP721e are strongly expressed in roots, but VAMP721a is repressed dramatically during nodule development, whereas VAMP721d and VAMP721e are not (Ivanov et al., 2012; Table 1, Figure 7B; see Supplemental Figure 9 online). Recently, it was found that the expression patterns of VAMP721a and RSD are mutually exclusive (Limpens et al., 2013). VAMP721a transcript was found in uninfected cells of the fixation zone, but not in the infected cells in which RSD was expressed specifically (Limpens et al., 2013). This indicates that repression of VAMP721a by RSD in infected cells may be crucial for normal symbiosome development.
One hypothesis to explain the symbiotic defect of the rsd-1 mutant is that VAMP721a directs vesicle traffic to an alternative target membrane(s), possibly the plasma membrane, at the expense of vesicle flow to the IT and SMs (Figure 8). Substantially reduced vesicle flow to these symbiotic membranes could compromise bacterial entry and/or symbiosome development, as observed in VAMP721d and VAMP721e RNA interference plants (Ivanov et al., 2012). Alternatively, VAMP721a-bearing vesicles, containing cargo different from that of VAMP721d- and VAMP721e vesicles, might fuse with symbiosomes in the rsd-1 mutant, delivering compounds that interfere with bacteroid differentiation and symbiosome function (Figure 8). Germane to this idea are the roles in plant defense of the closest homologs of Mt-VAMP721a in Arabidopsis, namely, At-VAMP721/722. These proteins are part of the secretory apparatus that targets vesicles containing cell wall–reinforcing material and other defense compounds to the plasma membrane at sites of fungal attack, which is an important component of non-host resistance in plants (Kwon et al., 2008). If Mt-VAMP721a plays an analogous role in plant defense in M. truncatula, it may be important that it is repressed during symbiosis with rhizobia to avoid delivery of defense compounds to the IT and/or developing symbiosomes.
Figure 8.
Hypothetical Mode of Action of Mt-RSD during Nodule Development.
IT delivery and endocytosis of rhizobia (green) and biogenesis of symbiosomes in a single plant cell are depicted. Mt-VAMP721d and Mt-VAMP721e are required to target and fuse specialized vesicles, presumably containing cell wall–degrading enzymes, to the IT membrane, which leads to dissolution of the IT cell wall and bacterial endocytosis (Ivanov et al., 2012). These VAMPs may continue to deliver membrane and cargo to the symbiosomes as they proliferate. The endoplasmic reticulum and Golgi are involved in the biogenesis of these and other types of secretory vesicles. Mt-RSD blocks the production of Mt-VAMP721a (via transcriptional repression), eliminating an alternative secretory pathway that may compete with the pathway(s) required for symbiosome biogenesis or alternatively may act in plant defense. Mt-RSD is crucial for symbiosome development, possibly because of its direct effect on Mt-VAMP721a expression and vesicle trafficking or its indirect effects on other genes, such as the NCR genes, which encode proteins that are delivered to symbiosomes and affect bacteroid differentiation.
At least two phases of plant transcriptional activity accompany differentiation and symbiosome formation during nodule development (Maunoury et al., 2010). Massive induction of secretory and endocytic pathway genes takes place during the first phase of gene activation (Maunoury et al., 2010). Mt-RSD is activated during this phase (Maunoury et al., 2010; Moreau et al., 2011). Loss of RSD activity in the rsd-1 mutant interferes with the induction of many genes that are normally induced during phases one and two, including many of the NCR genes. Although NCR genes are not targets of RSD repression, delayed/no expression of these genes in the rsd-1 mutant (see Supplemental Figure 11D and Supplemental Data Set 3 online) may contribute to the aberrant differentiation of bacteroids observed in rsd-1 nodules, given their known involvement in this process (Van de Velde et al., 2010).
Phylogenetic analysis of Mt-RSD and homologous sequences from other plant species identified one putative ortholog in L. japonicus and two in soybean (Figure 1), all of which are expressed in a nodule-specific manner (L. japonicus, Probset chr1.CM0147.88_at is nodule specific according to the L. japonicus Gene Expression Atlas [Høgslund et al., 2009; Verdier et al., 2013]; soybean, Glyma07g16290 and Glyma18g40360, are nodule specific according to SoyBase [Severin et al., 2010]). It also has been shown the putative ortholog of RSD in L. japonicus is expressed specifically in infected cells of nodules (Kouchi and Hata, 1995; Kumagai and Kouchi, 2003). Therefore, it is likely that the role of these proteins has been conserved among legumes during evolution and is not tied to terminal differentiation of bacteroids in M. truncatula.
In summary, we identified a member of the C2H2 family of TFs, Mt-RSD, as being crucial for symbiosome development and SNF in M. truncatula. This opens a new chapter in symbiosis research by identifying a plant TF involved in this process. We have shown that one of the direct targets of Mt-RSD is Mt-VAMP721a, and ectopic expression of this secretory pathway gene in the rsd-1 mutant could conceivably explain the symbiosome development defect of the mutant. On the other hand, we cannot exclude the possibility that other direct targets of RSD or indirect effects of the rsd-1 mutation are responsible for the complex mutant phenotype. Future work on VAMP721a and any other direct targets of RSD should help to clarify this.
Finally, our demonstration that Mt-RSD is a transcriptional regulator of plant secretory pathway genes provides an entry point into elucidating the genetic regulation of processes that are crucial for many aspects of plant development and response to the environment, including cytokinesis, embryogenesis, gravitropism, plant–microbe interactions, abscisic acid signaling, and abiotic stress responses (Lipka et al., 2007). It will be interesting to determine if other secretory pathway genes contain C2H2 TF binding domains in their promoters and whether transcript levels of such genes correlate negatively or positively with those of C2H2 TFs with or without EAR domains, respectively. Identification, characterization, and utilization of transcriptional regulators of plant secretory pathways will help us to understand better these important processes and to manipulate them to improve plant performance.
METHODS
Plant Growth and Bacterial Strains
Medicago truncatula ecotype R108 and homozygous rsd mutants (rsd-1 and rsd-2) were used. Seeds were scarified for 10 min in H2SO4 followed by sterilization for 3 min with 30% (v/v) commercial bleach containing a few drops of Tween 20. Surface-sterilized seeds were placed on inverted plates containing damp, sterile filter paper in the dark for 3 d at 4°C and 1 d at 20°C. Germinated seedlings were transferred to plastic cones containing a 2:1 ratio of turface:vermiculite. Plants were cultivated under a 16-h/8-h light/dark regime with 200 μE m−2 s−1 light irradiance at 21°C and 40% relative humidity. After 7 d of growth, plants were inoculated with 50 mL of Sinorhizobium meliloti (strains 1021 and 2011 with pXLGD4 plasmid [Boivin et al., 1990] or 1022 with mCherry-expressing plasmid). S. meliloti was grown overnight in yeast mannitol broth liquid medium, with shaking at 250 rpm, to OD600 ≈ 1.0, pelleted by centrifugation, and resuspended in half-strength Broughton and Dilworth (Broughton and Dilworth, 1971) with 0.5 mM KNO3 at OD600 ≈ 0.02. For root transformation, ARqua1 Agrobacterium rhizogenes was used, while for Nicotiana benthamiana transient expression, Agrobacterium tumefaciens strain LBA4404 was used.
Gene Identification and Phylogenetic Analysis
The Mt-RSD gene was identified by its nodule-specific expression profile (probe set ID Mtr.38404.1.S1_at), using the MtGEA Web server (Benedito et al., 2008) available at http://mtgea.noble.org/v2/ (He et al., 2009). Alignment of the deduced amino acid sequences of Mt-RSD and other proteins of this family (see Supplemental Data Set 4 online) was performed using multiple sequence comparison by log expectation in the Geneious software suite (Biomatters). The phylogenetic tree was built using an unweighted pair group method with arithmetic mean with 1000 bootstrap replicates.
Insertional Mutant Screening, Genotyping, and Backcrossing
Generation of the M. truncatula Tnt1 insertional mutant population and growth of R1 seeds were described previously (Tadege et al., 2008). Reverse genetic screening for Tnt1 retrotransposon insertions in Mt-RSD was performed using a nested PCR approach (Cheng et al., 2011). Homozygous rsd-1 and rsd-2 plants were further isolated from R1 seeds by using the primers mentioned in Supplemental Data Set 5 online. Backcrossing of mutants with the R108 accession was performed as described (Taylor et al., 2011).
Complementation of the rsd Mutant Phenotype
The open reading frame of Mt-RSD was amplified from cDNA synthesized with total RNA extracted from mature nodules of the ecotype R108 using the Trizol RNA extraction method (Life Technologies) and Superscript III reverse transcriptase (Life Technologies). PCR products were cloned into Gateway destination vector pK7WGF2 (Karimi et al., 2002). This generated GFP-MtRSD fusion protein. The empty vector expressing only the GFP gene was used as a control for hairy root transformation. pK7WGF2 vectors harboring GFP-MtRSD or GFP sequences were transformed into A. rhizogenes strain ARqua 1. Agrobacteria were infected on M. truncatula (R108) seedlings to generate composite plants (Boisson-Dernier et al., 2001). Transformed hairy roots were screened under UV light.
Nodulation Assays and Sample Collection
For nodulation time-course assays, susceptible zones were harvested for 0- to 4-DAI samples. From 6 DAI onwards, only portions of the roots carrying visible nodules were excised and immediately frozen under liquid nitrogen. For the mutants defective in nitrogen fixation (dnf; Mitra and Long, 2004; Starker et al., 2006) and TE7 mutants (Benaben et al., 1995) and for bacterial mutants (bacA, fixJ and exoA; Maunoury et al., 2010) samples were collected at 10 DAI. To measure Mt-RSD expression by qRT-PCR along the nodule zones, 28-DAI wild-type nodules (A17) were dissected into the meristematic zone, invasion zone, interzone II-III, nitrogen fixation, and senescence zone. Hand-dissection was performed on unfixed nodules, under a dissecting microscope, using leghemoglobin coloration and overall nodule morphology to discriminate between different nodule zones. For nodule zonation data, nodule zones were isolated for each zone by three independent biological replicates. qRT-PCR was performed on each of these biological replicates. Samples were collected at 15 DAI for bacterial gene expression quantification.
RNA Extraction, Microarray, and qRT-PCR Analyses
Total RNA was extracted using TRIZOL reagent (Life Technologies; Chomczunski and Mackey, 1995), followed by genomic DNA removal by DNase 1 (Ambion), and additional column purification with RNeasy MinElute CleanUp kit (Qiagen). Three independent biological replicates were included and hybridized to the Affymetrix GeneChip (Medicago Genome Array-Affymetrix). Array hybridization and scanning was done according to the manufacturer’s recommendations (Affymetrix), and scanning of arrays was performed as described previously (Benedito et al., 2008). Microarray data were submitted to MIAMExpress with accession number E-MEXP-3723. Raw data were normalized by robust multichip averaging, as described earlier (Irizarry et al., 2003). Presence and absence calls for probe sets were obtained using the dCHIP algorithm (Li and Wong, 2001). Differentially expressed genes in mutant and overexpressing lines were identified using the associative analysis (Dozmorov and Centola, 2003). Type I error rate was reduced using a Bonferroni corrected P value (threshold 0.05). To identify differentially regulated genes, we used a Bonferroni corrected P value threshold of 5% and at least a twofold difference.
For qRT-PCR, reverse transcription was performed as described previously (Kakar et al., 2008). Transcript levels were normalized using the geometric average of three housekeeping genes, Ubiquitin-Conjugating enzyme E2 (TC106312), Polypyrimidine Tract-Binding protein (TC111751) Ubiquitin (TC102473), whose transcript levels were very stable across all the samples analyzed. All primer sequences used in this analysis are given in Supplemental Data Set 5 online. For qRT-PCR analysis three technical replicates were used.
Phenotypic Analysis, Histochemical Staining, and Confocal and Transmission Electron Microscopy
Images of whole-mount nodulated roots were captured using an Olympus SZX12 stereomicroscope (Olympus) equipped with a Nikon DXM1200C digital camera (Nikon Instruments). Histochemical staining with X-Gal (5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside) was performed as described previously (Boivin et al., 1990). Detached nodules were embedded in 5% low-melting-point agarose and sliced into 50-µm-thick sections with a 1000 Plus Vibratome (The Vibratome Co, Technical Product International). For confocal microscopy, sample preparation was done according to Haynes et al. (2004). Nodules were sliced using a vibratome. Forty-micrometer sections were prepared and stained with SYTO13, Calcofluor (Life Technologies), and/or FM1-43 (Life Technologies). Images were acquired with a Leica TCS SP2 AOBS confocal laser scanning microscope. For transmission electron microscopy, nodules were fixed with 3% glutaraldehyde (v/v) and 4% paraformaldehyde (v/v) in cacodylate buffer, postfixed in 1% osmium tetroxide, dehydrated in a graded ethanol series, and embedded in LR White resin (London Resin Company). Serial 0.1-µm sections were cut with a diamond knife on a Leica EM UC7 ultramicrotome (Leica Mikrosysteme). Ultrathin sections were then put onto carbon-coated copper grids and stained with 2% uranyl acetate followed by staining with Sato’s lead. Specimens were observed under a Zeiss 10A transmission electron microscope operated at 80 kV. Images captured onto negative films were scanned and digitalized. All digital micrographs were processed using Adobe Photoshop CS4.
In Situ Live/Dead Staining of Rhizobia
Live/dead staining was performed according to Haag et al. (2011). Briefly, wild-type R108 and rsd-1 were grown and inoculated with S. meliloti strain Sm1021. Nodules were harvested 12 DAI and embedded in 6% (w/v) agarose. Nodule sections of 100-µm thickness were prepared with a 1000 Plus Vibratome (The Vibratome Co, Technical Product International) and incubated for 20 min in live/dead staining solution (1:1000 dilution of 1.67 mM SYTO9 and 18.3 mM PI [Molecular Probes; L7007] in 50 mM Tris, pH 7.0, buffer). Sections were removed from staining solution and mounted in deionized water for microscopy. Images were acquired with a Leica TCS SP2 AOBS confocal laser scanning microscope.
Acetylene Reduction Assay
Acetylene reduction assays were performed as described previously (Oke and Long, 1999). Plants were grown on a mixture of vermiculite and turface, watered with B&D solution containing 0.5 mM nitrate, and inoculated with rhizobia Sm1021. At 15 DAI, entire root systems were place into sealed test tubes for the assays.
Flow Cytometry
M. truncatula R108 and rsd-1 mutant plants were cultivated and inoculated with S. meliloti in aeroponic factories. Measurements of the DNA content of plant nuclei and bacteroids were done according to Cebolla et al. (1999) and Mergaert et al. (2006), respectively. Briefly, freshly collected nodules at 18 DAI were sliced in 0.5 mL of Galbraith buffer containing 1% Triton X-100, nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (5 μg/mL), and 5000 nuclei were analyzed per measurement. In the case of bacteria, the total bacteroid population was analyzed. DNA content of nuclei and bacteria was measured with a MoFlo Astrios flow cytometer from Beckman-Coulter driven by Summit 6.1 software.
Transient Expression in N. benthamiana
Suspensions of Agrobacterium strain LBA4404 carrying the p35S-GFP-MtRSD construct or the empty vector (pK7WGF2) were infiltrated into leaves of N. benthamiana, as previously described (Goodin et al., 2002).
Transcriptional Repression in Yeast
The vector pGBT9 (Clontech Laboratories) was used to construct effector plasmids for transcriptional repression activity assays. The reporter yeast strain carrying six Gal4-UASs upstream of the lacZ reporter and the pGBT-9 vector in which Gal4-UAS is fused to the herpes simplex virus VP16-activation domain were described previously (Shen et al., 2012). Mt-RSD was cloned into pGBT9 as a fusion protein with Gal4-UAS or Gal4-UAS plus VP16 transactivation domain. The effector plasmid was transformed into the yeast reporter strain according to the yeast protocols handbook (Clontech). β-Galactosidase assays were performed as described in the Matchmaker one-hybrid system user manual (Clontech).
ChIP
ChIP was performed using GFP-MtRSD–overexpressing hairy roots. The tissues were pulverized in liquid nitrogen. Powdered tissue was taken in cross-linking buffer (0.4 M Suc, 50 mM HEPES-KOH, pH 7.5, 5 mM β-ME, and PI cocktail) containing 1% formaldehyde. Cross-linking was done for 10 min at room temperature and was quenched by the addition of Gly to a final concentration of 125 mM. Following centrifugation of the cross-linked material at 4000g for 20 min, the pellet was resuspended in extraction buffer 2 (0.25 M Suc, 10 mM Tris-Cl, pH 8, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, and Complete protease inhibitor tablets [Roche; PI tablet]). This was followed by another centrifugation at 14,000g for 20 min. The pellet was resuspended in extraction buffer 3 (1.7 M Suc, 10 mM Tris-Cl, pH 8, 2 mM MgCl2, 0.15% Triton X-100, 5 mM β-ME, and proteinase inhibitor cocktail) and centrifuged at 20,000g for 40 min. The resulting pellet was resuspended in sonication buffer (10 mM Tris-aminomethane-hydrochloride, pH 8, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, and PI tablet). This was followed by sonication (10 × 10-s pulses with 1-min interval between each pulse) to shear the chromatin. Cell debris was removed by another centrifugation at 20,000g for 40 min. To be used as the input sample, 100 μL of this clear supernatant containing chromatin was preserved for subsequent analysis. ChIP was performed with rabbit anti-GFP polyclonal antibody (Abcam) bound to Dynabead Protein-G magnetic beads (Life Technologies) for 8 h at 4°C. The beads were washed twice with high-salt wash buffer (500 mM NaCl, 0.2% SDS, 0.5% Triton X-100, 20 mM Tris-HCl, pH 8, 2 mM EDTA, and PI tablet), followed by RIPA buffer (50 mM HEPES-KOH, pH 7.5, 500 mM LiCl, 1 mM EDTA, 1% Nonidet P‑40 or Igepal CA-630, 0.7% Na-deoxycholate, and PI tablet) and Tris-buffered saline. Chromatin was eluted in elution buffer (50 mM Tris-HCl, pH 8, 10 mM EDTA, and 1% SDS) and de-cross-linked (including the inputs) at 65°C for 16 h. Cellular RNA and protein were then removed by RNase (Sigma-Aldrich) and proteinase-K (Applied Biosystems) treatment, respectively. DNA was then precipitated using 2.5 volumes of 100% ethanol and finally dissolved in 50 μL water. Target DNA enrichment for each sample was determined by real-time PCR (PCR primers are given in Supplemental Data Set 5 online) using the equation 2(Ct of Input-Ct of ChIP) (Ct stands for cycle threshold). RSD-dependent DNA enrichment was calculated as the ratio of DNA enrichment in GFP-MtRSD–expressing roots to that in empty vector transformed roots.
EMSA
His6-tagged Mt-RSD was expressed in Escherichia coli BL21 codon plus cells using pDEST17 vector and purified with the QIAexpressionist kit, following the manufacturer’s instructions (Qiagen). EMSA was performed with a Light Shift Chemiluminescent EMSA kit, following the manufacturer’s instructions (Pierce). Briefly, we used ∼100 ng of purified recombinant protein and 20 fmol of biotin-labeled DNA fragment in a 30-μL reaction including 25 mM HEPES, pH 7.5, 50 mM KCl, 6.25 mM MgCl2, 0.13% Nonidet P‑40, 5% glycerol, and 500 ng double-stranded poly(dI/dC). After incubation, the mixture was loaded on a 6% polyacrylamide gel (Life Technologies) and run in 0.5% Tris Borate Ethylene diamine tetra-acetate at 4°C (120 V for 4 h). Biotin-labeled DNA fragment was generated using biotin-labeled forward primer. Primer sequences are listed in Supplemental Data Set 5 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At-ZAP6 (NP_176873), At-SUPERMAN (NP_188954), Os-C2H2 (BAD31832), Gm-PALM1 (ADI60289), Gm-PALM2 (ADI60290), Ms-PALM1 (ADI60287), Mt-PALM1 (XP_003611481), At-TAC1 (NP_187540), At-C2H2-Znf (NP_199167), Pt-C2H2-Znf (XP_002323519), Gm-RSD1 (NP_001235396), Gm-RSD2 (NP_001235081), and Mt-RSD (R108 RSD sequence was generated during this study and submitted to GenBank with accession no. JX307863). For L. japonicus, the protein ID is according to the L. japonicus genome database (Lj-RSD; LjGDB|chr1.CM0147.870). Microarray data generated in this study were submitted to MIAMExpress with accession number E-MEXP-3723.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RSD Gene Expression and Acetylation Reduction in Wild-Type M. truncatula Ecotypes.
Supplemental Figure 2. RSD cDNA and Deduced Amino Acid Sequence from M. truncatula R108.
Supplemental Figure 3. Nitrogenase Acetylene Reduction Activity during Nodule Development of the Two M. truncatula Wild-Type Genotypes A17 and R108.
Supplemental Figure 4. Confocal Microscopy of RSD-Complemented rsd-1 Nodules.
Supplemental Figure 5. Effect of Full Nitrogen and Sinorhizobium meliloti on Wild-Type R108 and the rsd-1 Mutant.
Supplemental Figure 6. Rhizobial Infection Phenotype of rsd-1.
Supplemental Figure 7. Rhizobia in rsd-1 Nodules Undergo Rapid Death.
Supplemental Figure 8. Expression of leghemoglobin in Various Fix− Mutants.
Supplemental Figure 9. Localization of Mt-RSD in Tobacco Epidermal Cells.
Supplemental Figure 10. RSD Regulates VAMP721a (Medtr4g022570: IMGAG3.5) Expression.
Supplemental Figure 11. Nodulation-Induced and -Repressed Genes in the rsd-1 Mutant and Wild-Type Plants.
Supplemental Figure 12. Expression of Plant and Bacterial Genes in rsd-1 Mutant and Wild-Type Nodules.
Supplemental Table 1.http://www.plantcell.org/cgi/content/full/tpc.113.114017/DC1 List of Primers.
Supplemental Data Set 1. List of Genes Expressed at Significantly Higher Levels in Nodules of the rsd-1 Mutant Than in the Wild Type at Both 6 and 8 DAI.
Supplemental Data Set 2. List of Genes Expressed at Significantly Lower Levels in Nodules of the rsd-1 Mutant Than in the Wild Type at 6 and/or 8 DAI.
Supplemental Data Set 3. List of Genes Expressed during Wild-Type Nodule Development, but Delayed or Not Expressed in rsd-1 Nodules.
Supplemental Data Set 4. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 1.
Supplemental Data Set 5. List of Primers Used in This Study.
Acknowledgments
This work was supported by the National Science Foundation Plant Genome Research Program (Grant DBI-0703285), The Samuel Roberts Noble Foundation, and National Science Foundation Major Research Instrumentation Program (NSF DBI-0722635). We thank Phil Poole for providing the mCherry rhizobia strain, Shulan Zhang and Conner Boatright for assistance with the nodulation assay, Colleen Elles, Janie Gallaway, Frank Coker, and Vicki Barrett for technical assistance in maintaining the M. truncatula Tnt1 lines, and Yuhong Tang and Tui Ray for assistance with Affymetrix chip analysis. We also thank Pascal Ratet, Kirankumar S. Mysore, and Million Tadege for the construction of the Tnt1 mutant population, Jiangqi Wen and Xiaofei Cheng for the screening of this population for the Mt-RSD gene, Mark Taylor for backcrossing the mutant, Hui Shen for the yeast strain and constructs, Chenggang Liu for advice on yeast experiments, and Rahul Bahulikar for help with the acetylene reduction assay.
AUTHOR CONTRIBUTIONS
S.S., E.K., K.B., V.A.B., and M.K.U. designed the research. S.S., I.T.-J., K.B., C.I.P., and A.K. performed research. J.N. performed transmission electron microscopy. S.S. and I.T.-J. analyzed data. S.S. and M.K.U. wrote the article.
Glossary
- SNF
symbiotic nitrogen fixation
- IT
infection thread
- SM
symbiosome membrane
- NCR
nodule-specific Cys-rich
- C2H2
Cysteine-2/Histidine-2
- MtGEA
M. truncatula Gene Expression Atlas
- qRT-PCR
quantitative RT-PCR
- DAI
days after inoculation
- UTR
untranslated region
- PI
propidium iodide
- GFP
green fluorescent protein
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- DAPI
4′,6-diamidino-2-phenylindole
- β-ME
mercaptoethanol
References
- Arrighi J.F., Godfroy O., de Billy F., Saurat O., Jauneau A., Gough C. (2008). The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc. Natl. Acad. Sci. USA 105: 9817–9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaben V., Duc G., Lefebvre V., Huguet T. (1995). TE7, an inefficient symbiotic mutant of Medicago truncatula Gaertn. cv Jemalong. Plant Physiol. 107: 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito V.A., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Boivin C., Camut S., Malpica C.A., Truchet G., Rosenberg C. (1990). Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2: 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourcy M., Brocard L., Pislariu C.I., Cosson V., Mergaert P., Tadege M., Mysore K.S., Udvardi M., Gourion B., Ratet P. (2013). Medicago truncatula DNF2 is a PI-PLC-XD- containing protein required for bacteroid persistence and prevention of nodule early senescence and defense-like reactions. New Phytol. 197: 1250–1261 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Sakai H., Jack T., Weigel D., Mayer U., Meyerowitz E.M. (1992). SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114: 599–615 [DOI] [PubMed] [Google Scholar]
- Broughton W.J., Dilworth M.J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla A., Vinardell J.M., Kiss E., Oláh B., Roudier F., Kondorosi A., Kondorosi E. (1999). The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 18: 4476–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., et al. (2010). Control of dissected leaf morphology by a Cys(2)His(2) zinc finger transcription factor in the model legume Medicago truncatula. Proc. Natl. Acad. Sci. USA 107: 10754–10759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Wen J., Tadege M., Ratet P., Mysore K.S. (2011). Reverse genetics in Medicago truncatula using Tnt1 insertion mutants. Methods Mol. Biol. 678: 179–190 [DOI] [PubMed] [Google Scholar]
- Chomczunski P., Mackey K. (1995). Modification of the TRI TM Reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques 19: 942–945 [PubMed] [Google Scholar]
- Colebatch G., Desbrosses G., Ott T., Krusell L., Montanari O., Kloska S., Kopka J., Udvardi M.K. (2004). Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J. 39: 487–512 [DOI] [PubMed] [Google Scholar]
- Combier J.P., Frugier F., de Billy F., Boualem A., El-Yahyaoui F., Moreau S., Vernié T., Ott T., Gamas P., Crespi M., Niebel A. (2006). MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20: 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque L., Neogi P., Pislariu C., Wilson K.A., Catalano C., Avadhani M., Sherrier D.J., Dickstein R. (2008). Transcription of ENOD8 in Medicago truncatula nodules directs ENOD8 esterase to developing and mature symbiosomes. Mol. Plant Microbe Interact. 21: 404–410 [DOI] [PubMed] [Google Scholar]
- Dathan N., Zaccaro L., Esposito S., Isernia C., Omichinski J.G., Riccio A., Pedone C., Di Blasio B., Fattorusso R., Pedone P.V. (2002). The Arabidopsis SUPERMAN protein is able to specifically bind DNA through its single Cys2-His2 zinc finger motif. Nucleic Acids Res. 30: 4945–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M., Daveran M.L., Batut J., Dedieu A., Domergue O., Ghai J., Hertig C., Boistard P., Kahn D. (1988). Cascade regulation of nif gene expression in Rhizobium meliloti. Cell 54: 671–683 [DOI] [PubMed] [Google Scholar]
- Dozmorov I., Centola M. (2003). An associative analysis of gene expression array data. Bioinformatics 19: 204–211 [DOI] [PubMed] [Google Scholar]
- Frugier F., Poirier S., Satiat-Jeunemaître B., Kondorosi A., Crespi M. (2000). A Krüppel-like zinc finger protein is involved in nitrogen-fixing root nodule organogenesis. Genes Dev. 14: 475–482 [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Ichige A., Walker G.C. (1993). A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev. 7: 1485–1497 [DOI] [PubMed] [Google Scholar]
- Godiard L., Lepage A., Moreau S., Laporte D., Verdenaud M., Timmers T., Gamas P. (2011). MtbHLH1, a bHLH transcription factor involved in Medicago truncatula nodule vascular patterning and nodule to plant metabolic exchanges. New Phytol. 191: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild D.J., Bergersen F.J. (1966). Electron microscopy of the infection and subsequent development of soybean nodule cells. J. Bacteriol. 92: 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin M.M., Dietzgen R.G., Schichnes D., Ruzin S., Jackson A.O. (2002). pGD vectors: Versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31: 375–383 [DOI] [PubMed] [Google Scholar]
- Haag A.F., et al. (2011). Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol. 9: e1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakoyama T., et al. (2009). Host plant genome overcomes the lack of a bacterial gene for symbiotic nitrogen fixation. Nature 462: 514–517 [DOI] [PubMed] [Google Scholar]
- Hakoyama T., et al. (2012b). The integral membrane protein SEN1 is required for symbiotic nitrogen fixation in Lotus japonicus nodules. Plant Cell Physiol. 53: 225–236 [DOI] [PubMed] [Google Scholar]
- Hakoyama T., et al. (2012a). The SNARE protein SYP71 expressed in vascular tissues is involved in symbiotic nitrogen fixation in Lotus japonicus nodules. Plant Physiol. 160: 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney C.H., Long S.R. (2010). Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc. Natl. Acad. Sci. USA 107: 478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes J.G., Czymmek K.J., Carlson C.A., Veereshlingam H., Dickstein R., Sherrier J.D. (2004). Rapid analysis of legume root nodule development using confocal microscopy. New Phytol. 163: 661–668 [DOI] [PubMed] [Google Scholar]
- He J., Benedito V.A., Wang M., Murray J.D., Zhao P.X., Tang Y., Udvardi M.K. (2009). The Medicago truncatula gene expression atlas web server. BMC Bioinformatics 10: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann A.B., Lombardo F., Miwa H., Perry J.A., Bunnewell S., Parniske M., Wang T.L., Downie J.A. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142: 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Høgslund N., et al. (2009). Dissection of symbiosis and organ development by integrated transcriptome analysis of lotus japonicus mutant and wild-type plants. PLoS ONE 4: e6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth B., et al. (2011). Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol. Plant Microbe Interact. 24: 1345–1358 [DOI] [PubMed] [Google Scholar]
- Ikeda M., Ohme-Takagi M. (2009). A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50: 970–975 [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Ivanov S., Fedorova E.E., Limpens E., De Mita S., Genre A., Bonfante P., Bisseling T. (2012). Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc. Natl. Acad. Sci. USA 109: 8316–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakar K., et al. (2008). A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kereszt A., Mergaert P., Kondorosi E. (2011). Bacteroid development in legume nodules: Evolution of mutual benefit or of sacrificial victims? Mol. Plant Microbe Interact. 24: 1300–1309 [DOI] [PubMed] [Google Scholar]
- Kouchi H., Hata S. (1995). GmN56, a novel nodule-specific cDNA from soybean root nodules encodes a protein homologous to isopropylmalate synthase and homocitrate synthase. Mol. Plant Microbe Interact. 8: 172–176 [DOI] [PubMed] [Google Scholar]
- Krusell L., et al. (2005). The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17: 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai H., Hakoyama T., Umehara Y., Sato S., Kaneko T., Tabata S., Kouchi H. (2007). A novel ankyrin-repeat membrane protein, IGN1, is required for persistence of nitrogen-fixing symbiosis in root nodules of Lotus japonicus. Plant Physiol. 143: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai H., Kouchi H. (2003). Gene silencing by expression of hairpin RNA in Lotus japonicus roots and root nodules. Mol. Plant Microbe Interact. 16: 663–668 [DOI] [PubMed] [Google Scholar]
- Kwon C., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840 [DOI] [PubMed] [Google Scholar]
- Lefebvre B., et al. (2010). A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. USA 107: 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J.A., Signer E.R., Walker G.C. (1985). Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 82: 6231–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wong W.H. (2001). Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Limpens E., Moling S., Hooiveld G., Pereira P.A., Bisseling T., Becker J.D., Küster H. (2013). Cell- and tissue-specific transcriptome analyses of Medicago truncatula root nodules. PLoS ONE 8: e64377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V., Kwon C., Panstruga R. (2007). SNARE-ware: The role of SNARE-domain proteins in plant biology. Annu. Rev. Cell Dev. Biol. 23: 147–174 [DOI] [PubMed] [Google Scholar]
- Liu J., Miller S.S., Graham M., Bucciarelli B., Catalano C.M., Sherrier D.J., Samac D.A., Ivashuta S., Fedorova M., Matsumoto P., Gantt J.S., Vance C.P. (2006). Recruitment of novel calcium-binding proteins for root nodule symbiosis in Medicago truncatula. Plant Physiol. 141: 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J.F., Rakocevic A., Mitra R.M., Brocard L., Sun J., Eschstruth A., Long S.R., Schultze M., Ratet P., Oldroyd G.E. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunoury N., et al. (2010). Differentiation of symbiotic cells and endosymbionts in Medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS ONE 5: e9519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P., Nikovics K., Kelemen Z., Maunoury N., Vaubert D., Kondorosi A., Kondorosi E. (2003). A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 132: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P., Uchiumi T., Alunni B., Evanno G., Cheron A., Catrice O., Mausset A.E., Barloy-Hubler F., Galibert F., Kondorosi A., Kondorosi E. (2006). Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. USA 103: 5230–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton P.H., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R.M., Long S.R. (2004). Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiol. 134: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R.M., Shaw S.L., Long S.R. (2004). Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 101: 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S., Verdenaud M., Ott T., Letort S., de Billy F., Niebel A., Gouzy J., de Carvalho-Niebel F., Gamas P. (2011). Transcription reprogramming during root nodule development in Medicago truncatula. PLoS ONE 6: e16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.D. (2011). Invasion by invitation: Rhizobial infection in legumes. Mol. Plant Microbe Interact. 24: 631–639 [DOI] [PubMed] [Google Scholar]
- Murray J.D., et al. (2011). Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65: 244–252 [DOI] [PubMed] [Google Scholar]
- Oke V., Long S.R. (1999). Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32: 837–849 [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E. (2013). Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Long S.R. (2003). Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol. 131: 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott T., Sullivan J., James E.K., Flemetakis E., Günther C., Gibon Y., Ronson C., Udvardi M. (2009). Absence of symbiotic leghemoglobins alters bacteroid and plant cell differentiation during development of Lotus japonicus root nodules. Mol. Plant Microbe Interact. 22: 800–808 [DOI] [PubMed] [Google Scholar]
- Ott T., van Dongen J.T., Günther C., Krusell L., Desbrosses G., Vigeolas H., Bock V., Czechowski T., Geigenberger P., Udvardi M.K. (2005). Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 15: 531–535 [DOI] [PubMed] [Google Scholar]
- Peoples M.B., et al. (2009). The contributions of nitrogen-fixing crop legumes to the productivity of agricultural systems. Symbiosis 48: 1–17 [Google Scholar]
- Popp C., Ott T. (2011). Regulation of signal transduction and bacterial infection during root nodule symbiosis. Curr. Opin. Plant Biol. 14: 458–467 [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Ren S., Mandadi K.K., Boedeker A.L., Rathore K.S., McKnight T.D. (2007). Regulation of telomerase in Arabidopsis by BT2, an apparent target of TELOMERASE ACTIVATOR1. Plant Cell 19: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J.G., Lyttleton P. (1984). Division of peribacteroid membranes in root nodules of white clover. J. Cell Sci. 69: 147–157 [DOI] [PubMed] [Google Scholar]
- Roth L.E., Stacey G. (1989). Bacterium release into host cells of nitrogen-fixing soybean nodules: The symbiosome membrane comes from three sources. Eur. J. Cell Biol. 49: 13–23 [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. (1998). GAL4-VP16 is an unusually potent transcriptional activator. Nature 335: 563–564 [DOI] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Severin A.J., et al. (2010). RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., He X., Poovaiah C.R., Wuddineh W.A., Ma J., Mann D.G., Wang H., Jackson L., Tang Y., Stewart C.N., Jr, Chen F., Dixon R.A. (2012). Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 193: 121–136 [DOI] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Starker C.G., Parra-Colmenares A.L., Smith L., Mitra R.M., Long S.R. (2006). Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol. 140: 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., Cayrel A., Endre G., Zhao P.X., Chabaud M., Ratet P., Mysore K.S. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Takanashi K., Yokosho K., Saeki K., Sugiyama A., Sato S., Tabata S., Ma J.F., Yazaki K. (2013). LjMATE1: A citrate transporter responsible for iron supply to the nodule infection zone of Lotus japonicus. Plant Cell Physiol. 54: 585–594 [DOI] [PubMed] [Google Scholar]
- Takatsuji H. (1999). Zinc-finger proteins: The classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Takatsuji H., Matsumoto T. (1996). Target-sequence recognition by separate-type Cys2/His2 zinc finger proteins in plants. J. Biol. Chem. 271: 23368–23373 [DOI] [PubMed] [Google Scholar]
- Taylor, M., Blaylock, L., Nakashima, J., McAbee, D., Ford, J., Harrison, M., and Udvardi, M. (2011). Medicago truncatula hybridization: Supplemental videos. In Medicago truncatula Handbook Version July 2011 U. Mathesius, E.P. Journet, and L.W. Sumner, eds (Ardmore, Oklahoma: The Samuel Roberts Noble Foundation), pp. 1–5. http://www.noble.org/MedicagoHandbook/
- Terpolilli J.J., Hood G.A., Poole P.S. (2012). What determines the efficiency of N(2)-fixing Rhizobium-legume symbioses? Adv. Microb. Physiol. 60: 325–389 [DOI] [PubMed] [Google Scholar]
- Udvardi M., Poole P.S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64: 781–805 [DOI] [PubMed] [Google Scholar]
- Udvardi M.K., Day D.A. (1997). Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 493–523 [DOI] [PubMed] [Google Scholar]
- Van de Velde W., et al. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327: 1122–1126 [DOI] [PubMed] [Google Scholar]
- Van de Velde W., Guerra J.C., De Keyser A., De Rycke R., Rombauts S., Maunoury N., Mergaert P., Kondorosi E., Holsters M., Goormachtig S. (2006). Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol. 141: 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse J., de Billy F., Camut S., Truchet G. (1990). Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 172: 4295–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J., Torres-Jerez I., Wang M., Andriankaja A., Allen S.N., He J., Tang Y., Murray J.D., Udvardi M.K. (2013). Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. Plant J. 74: 351–362 [DOI] [PubMed] [Google Scholar]
- Vernié T., Moreau S., de Billy F., Plet J., Combier J.P., Rogers C., Oldroyd G., Frugier F., Niebel A., Gamas P. (2008). EFD is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell 20: 2696–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Griffitts J., Starker C., Fedorova E., Limpens E., Ivanov S., Bisseling T., Long S. (2010). A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science 327: 1126–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Murray J.D., Kim J., Heckmann A.B., Edwards A., Oldroyd G.E., Downie J.A. (2012). Legume pectate lyase required for root infection by rhizobia. Proc. Natl. Acad. Sci. USA 109: 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N.D., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]