Abstract
Gene expression regulation is gated by promoter methylation states modulating transcription factor binding. The known DNA methylation/unmethylation mechanisms are sequence unspecific, but different cells with the same genome have different methylomes. Thus, additional processes bringing specificity to the methylation/unmethylation mechanisms are required. Searching for such processes, we demonstrated that CpG methylation states are influenced by the sequence context surrounding the CpGs. We used such a property to develop a CpG methylation motif discovery algorithm. The newly discovered motifs reveal “methylation/unmethylation factors” that could recruit the “methylation/unmethylation machinery” to the loci specified by the motifs. Our methylation motif discovery algorithm provides a synergistic approach to the differently methylated region algorithms. Since our algorithm searches for commonly methylated regions inside the same sample, it requires only a single sample to operate. The motifs that were found discriminate between hypomethylated and hypermethylated regions. The hypomethylation-associated motifs have a high CG content, their targets appear in conserved regions near transcription start sites, they tend to co-occur within transcription factor binding sites, they are involved in breaking the H3K4me3/H3K27me3 bivalent balance, and they transit the enhancers from repressive H3K27me3 to active H3K27ac during ES cell differentiation. The new methylation motifs characterize the pluripotent state shared between ES and iPS cells. Additionally, we found a collection of motifs associated with the somatic memory inherited by the iPS from the initial fibroblast cells, thus revealing the existence of epigenetic somatic memory on a fine methylation scale.
Genetic network regulation is driven by transcription factors (TFs) binding to gene target promoters gated by promoter methylation. If the TF binding site (TFBS) surroundings are methylated, the TF cannot bind and the gene will not be expressed. Thus, the promoter methylation is an on/off bistable “digital” switch that allows (in the unmethylated state) the TFs to exert a fine-tuned “analogical” regulation. To model and simulate genetic networks, we need to know the TFBSs and the susceptibility of the DNA loci residing inside the promoters to be methylated or unmethylated.
Numerous techniques have been developed to predict TFBSs (Elnitski et al. 2006; Levitsky et al. 2007; von Rohr et al. 2007) and TF binding motifs (TFBMs) (Müller-Molina et al. 2012). However, few studies have attempted to predict DNA methylation patterns. DNA methylation occurs at C5 cytosine positions, mainly in CpG loci. Some research has focused on CpG islands (Ficz et al. 2011) and on predicting their methylation using computational approaches (Das et al. 2006). Genome-wide methylation next-generation sequencing (NGS) has shown that CpG islands are usually unmethylated (Deaton and Bird 2011; Meissner 2011) and methylation alterations in cancer occur neither in promoters, nor in CpG islands, but in sequences up to 2 kb called CpG island shores (Doi et al. 2009; Irizarry et al. 2009). With some exceptions (Bhasin et al. 2005; Bock et al. 2006), research on CpG methylation prediction outside the aforementioned regions is scarce.
Assuming that CpG methylation and CpG sequence context function independently of each other, uniform distribution of methylated CpGs across the different clones in bisulfite lollipop diagrams might be expected. Nevertheless, such diagrams frequently show CpG columns with methylation distributions departing from the expected average (Fig. 1A). We referred to the significantly low- and high-methylated CpGs as methylation-resistant and methylation-prone CpGs, respectively. We hypothesized that such departures are due to the influence of the DNA sequence surrounding the CpG on the recruitment and interaction of methylation/unmethylation agents and their CpG targets. The MethDB database (Grunau et al. 2001) collects methylation information for more than 20,000 CpGs. We observed the same trend in MethDB as in Figure 1A, but MethDB data are insufficient to predict reliable methylation patterns. After all, the human genome has over 28 million CpGs. We benefited from the plethora of data from NGS methylomics surveys compiling an NGS methylomics collection that comprises a high percentage of CpGs with high coverage (Table 1). In this way, we collected enough data to verify our hypothesis that CpG methylation depends on the sequence context and we developed a computational method to discover CpG methylation motifs (CpGMMs).We expect that the DNA methylation differences revealed by the CpGMMs at the CpG level are biologically relevant since they could work as recognition sites for the agents that perform DNA methylation and demethylation. Actually, it has already been found (Mohn and Schübeler 2009; Lienert et al. 2011) that the methylation changes due to single CpG mutations have biological effects.
Figure 1.
DNA methylation patterns are nonuniformly distributed and are influenced by their DNA context. (A) Analysis of the methylation state of nestin (NES) second intron enhancer region. The lollipop diagram in the top panel shows the observed 58.6% global methylation (Han et al. 2009). Such methylation is nonuniformly distributed along CpG “columns.” There are CpG “columns” with higher and lower probability to be methylated. In simulations with a uniform distribution of the methylation states, we can observe lollipop diagrams such as the one shown in the bottom panel, with the same methylation percentage as the one experimentally observed, but with nonpreferential “column” methylation distributions. (B) Heatmaps of the frequencies of the similarity of the methylation between the two DNA strands versus the similarity of the sequence of the two DNA strands for each CpG word. (C) Violin plots of the frequencies of the methylation similarity between the two DNA strands for low (≤0.1) and high (≥0.9) sequence similarity between the two DNA strands. The similarities are calculated genome wide, and their frequencies are represented in log10 scale by gray color bars.
Table 1.
DNA methylome data sets

The capacity for proliferation and pluripotency make embryonic stem (ES) cells promising candidates for regenerative medicine and drug screening applications. However, due to technical and ethical issues, human ES cell experiments are restricted. Cellular reprogramming (Takahashi and Yamanaka 2006) converts unipotent (Okita et al. 2007) or multipotent cells (Kim et al. 2009) into pluripotent cells called induced pluripotent stem (iPS) cells. Although reprogramming is almost a laboratory routine these days, its molecular mechanism is poorly understood and is assumed to be based on the stochastic crosstalk between genetic and epigenetic networks (Artyomov et al. 2010). Concerns remain about the extent to which iPS cells resemble ES cells (Kim et al. 2010, 2011), even though transcriptomics experiments show small differences (Boué et al. 2010). A lot of research has gone into searching the so-called somatic memory (Polo et al. 2010). Such a memory would be the fingerprint of the iPS cell's somatic origin. Transcriptomics characterization of iPS cells reveals very little memory (Kim et al. 2008, 2009). However, on a methylomics level, some fingerprints of such a memory remain (Laurent et al. 2010; Bock et al. 2011; Lister et al. 2011; Ruiz et al. 2012).
To validate our hypothesis that CpG methylation depends on the sequence context surrounding the CpG, we analyzed the correlation between methylation and sequence similarity in the two DNA strands. We used NGS data from human fibroblasts (FBs), and ES and iPS cells from different laboratories (Table 1). Then we searched for sequences with similar methylation, and developed an algorithm for CpGMM discovery that groups such sequences into clusters based on the similarity of both sequence and methylation. For each cell type, we created two types of clusters: one for sequences that are resistant to being methylated, and another for sequences prone to being methylated. We built representative motifs of the cluster members. Such representatives are the CpGMMs, and we tested their capability to discriminate between methylated and unmethylated regions using a scanning approach. To characterize and validate the CpGMMs, we integrated and correlated the CpGMM targets with TFBSs, histone marks, and transcriptomics information. We took advantage of the properties of the CpGMMs to analyze pluripotent cells and to disclose the crosstalk between DNA methylation and TFs in genetic networks. Our method allowed us to obtain CpGMMs specific to the pluripotent state shared by iPS and ES cells and to discover reprogramming somatic memory CpGMMs.
Results
The extreme methylation states are conserved across different cell types and define cell type-specific methylation profiles
To obtain the DNA methylomes specific to the different cell types, we filtered out the cell line noise, producing a “conserved” DNA methylome for each cell type. The filter is intended for preserving the CpGs with low methylation fluctuations across the different cell lines (Supplemental Methods). The scatter plots of the positive DNA strand of the conserved methylomes of each cell type are depicted in Supplemental Figure S1a. High-density methylation regions are revealed in the corner of each panel of Supplemental Figure S1a. These regions show a higher contrast compared with nonconserved CpGs (Supplemental Fig. S1b), where the methylation values appear scattered across the whole methylation range. This conveys that the variability among cell lines of the same cell type happens mainly in the middle range methylation, while the extreme methylation states (hypo- and hypermethylation) are conserved. Thus, the middle range methylation state either has an intrinsic biological variability or is more difficult to be determined by NGS-based methylomics. Therefore, we focused on the stable methylation cases (very low or very high methylation). Additionally, Supplemental Figure S1a (iPS vs. ES cells) shows that iPS and ES have similar methylomes with two common signatures in the corners of the first diagonal—one concentrated at a very low methylation level, and another denser one at a very high methylation level. This feature portrays a common pluripotency methylation signature. Some low-density traces appear in the second diagonal corners, revealing a methylation somatic memory fingerprint. Interestingly, such regions vanish at the transcriptomics level (Supplemental Fig. S1c). The iPS–ES transcriptomics scatter plot shows that although some differently expressed genes appear slightly scattered around the diagonal, well-defined differently behaving regions, as in the corresponding panels in the same column in Supplemental Figure S1, a and b, do not exist. In summary, the somatic memory effect is more pronounced at the methylomics level than at the transcriptomics level.
Nonshared methylomics regions appear in the second diagonal corners when comparing ES or iPS with FB (FB vs. ES cells and iPS vs. FB) (see Supplemental Fig. S1a). These regions correspond to high methylation in pluripotent cells and low methylation in FBs, thus marking a distinct signature between the FB and pluripotent methylation profiles. We obtained similar results from the analysis of the methylation data of the negative strand.
The methylation variability between the two DNA strands is higher for nonpalindromic sequences
The lollipop diagram in Figure 1A illustrates a typical example (Han et al. 2009) of a promoter with several CpGs showing a trend to be methylation resistant and others to be methylation prone. To analyze to what extent this is a common feature, we benefited from the two DNA strands' methylation information provided by NGS. If the methylation/unmethylation mechanisms can discriminate between CpG occurring in different sequence contexts, these mechanisms will produce a more variable methylation distribution in CpG loci having different sequences in both strands (different sequence context) than in loci with similar sequences in both strands (palindromes) (algorithm description is provided in the Methods section). The results from this analysis are shown in Figure 1, B and C. Since DNA sequences are made of four different nucleotides, the expected similarity between a sequence in a DNA strand and the opposite sequence in the pairing strand is 0.25 (one potential mismatch among four different bases). This is reflected in the heatmaps (Fig. 1B) by the higher density around the 0.25 sequence similarity region. Irrespective of the cell type (FB, iPS, or ES cells), higher sequence similarity corresponds to lower variation in the methylation similarity (Fig. 1C), thus revealing that the methylation/unmethylation machinery recognizes the CpGs appearing in different sequence contexts (one context for each DNA strand) as different. These results demonstrate the context dependency of the CpG methylation. Thus, the level of resolution at which the sequence specificity of the methylation takes place is reduced from the length of CpG islands or CpG island shores to the level of individual CpGs.
The CpGMM discovery algorithm discloses CpGMMs that discriminate between methylation-resistant and methylation-prone loci
We have demonstrated that the CpG methylation state depends on the CpG sequence genomic context. Additionally, similar sequences have similar methylation ratios for the same cell type. This feature is more profound for longer and thus more specific CpG sequences (Supplemental Fig. S2e). To characterize the genomic context that drives the CpG methylation state, we developed a DNA methylation motif search algorithm (for a detailed description of the algorithm, see Methods) (Supplemental Fig. S5). To assess the discrimination capability of the CpGMMs, we used a binding energy scanning paradigm that provides us with a framework to search for the regions where the DNA methylation motifs have the best match. For each motif, the scanning method produces two matching distributions, one for low- and another for high-methylation regions. Their statistically significant dissimilarity is addressed with two checks. Two examples of matching discriminative distributions are depicted in Supplemental Figure S2, one for a methylation-prone (Supplemental Fig. S2c) and another for a methylation-resistant (Supplemental Fig. S2d) CpGMM. By using a bootstrapping method (Efron and Tibshirani 1993), we found that 78% of the CpGMMs are stable (Supplemental Fig. S2g,h). A comprehensive list of annotated motifs with their corresponding scanned distributions, two quality scores, their gene targets, and the associated gene ontologies can be downloaded from Supplemental Material and from our web server at http://computational-biology.mpi-muenster.mpg.de/publications/MethylationMotifs/. These methylation-prone and methylation-resistant CpGMMs can discriminate between methylation-prone and methylation-resistant regions, validating their capacity to distinguish between the two region types.
Pluripotent cells share specific methylation-resistant CpGMMs
The collection of CpGMMs obtained through the CpGMM discovery algorithm is classified according to cell type-specific CpGMMs as described in the Supplemental Methods. Figure 2 depicts the Pearson correlation distributions for the pairwise comparisons of the three cell-type pools. We found (Fig. 2A, upper panel) that a collection of highly correlated, methylation-resistant CpGMMs is shared between the pluripotent cell lines (ES and iPS). Contrarily, fewer shared motifs appear when comparing pluripotent cell lines with FB (Fig. 2A, middle and bottom panels). These results indicate the existence of pluripotency-specific, methylation-resistant CpGMMs. Interestingly, Figure 2B shows equal distributions between all the cell types for the methylation-prone CpGMMs, indicating that there are less candidates for specific pluripotent methylation-prone CpGMMs. We used these correlations (Supplemental Methods) to calculate the number of CpGMM clusters of each cell type and the number of their intersections. The numbers for the merged DNA strands are shown in the Venn diagrams in Figure 2, C and D. The Venn diagrams for the positive and negative strands are shown in Supplemental Figure S3. We found 3999 (Fig. 2C) methylation-resistant and 840 (Fig. 2D) methylation-prone CpGMMs specific to pluripotent cells (the whole collection of CpGMMs is provided on our web server). From such motifs, we found 119 pluripotent methylation-resistant CpGMMs and 13 pluripotent methylation-prone CpGMMs, targeting at least one pluripotent gene from a list of 17 human pluripotency markers (Table 2). A selection of them is listed in Table 3. The collection of pluripotency-specific CpGMMs (provided on our web server) is useful to understand the epigenomic component of the pluripotency network that arises from the crosstalk between methylation-resistant CpGMMs and TFs (see Fig. 7C,D).
Figure 2.
Methylation-resistant CpGMMs are more abundant and are highly correlated among pluripotent populations. Histograms of the Pearson correlation coefficients of pairwise pools of different cell types for methylation-resistant (A) and methylation-prone (B) CpGMMs. Vertical lines mark the low and high correlation boundaries. Venn diagram of the numbers of methylation-resistant (C) and methylation-prone (D) CpGMM clusters in each cell type. ES CpGMM regions are marked in white, iPSs in gray, and fibroblasts in black. The numbers enclosed by the circular segments are the numbers of cell type-specific motifs.
Table 2.
Number of somatic memory and pluripotency-specific CpGMMs with enrichment of at least one corresponding marker

Table 3.
Selection of CpGMMs specific to pluripotent populations

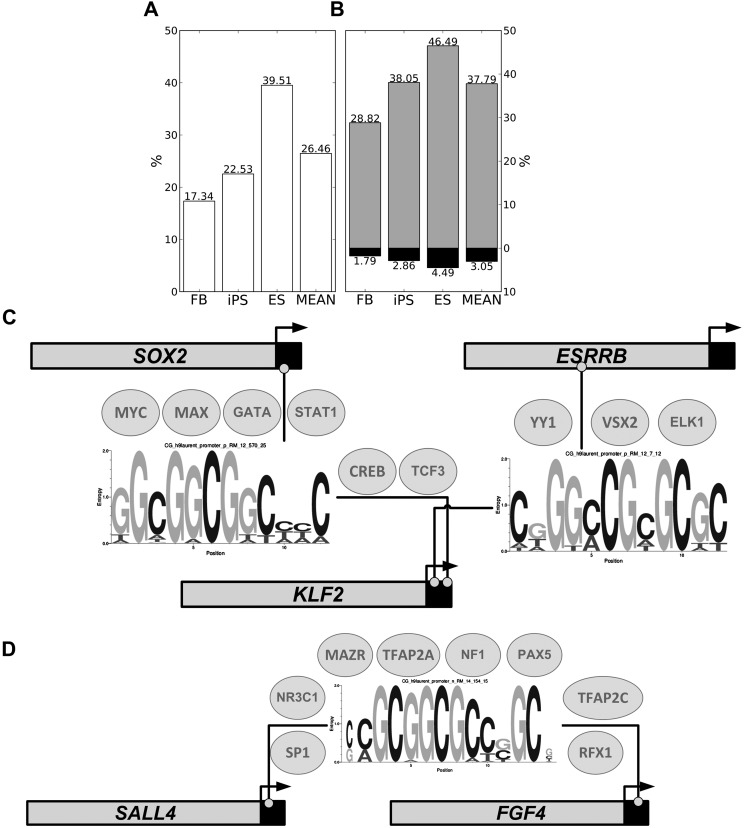
Figure 7.
Crosstalk between CpGMMs and TFs. Percentages of CpGMM targets of each cell type (A) and methylation-resistant and methylation-prone CpGMM targets of each cell type (B) that co-occupy TFBSs. The methylation-resistant percentages are in gray, the methylation-prone in black, and the merged ones in white. The rightmost bars correspond to the mean across all populations. (C,D) Pluripotent networks arising from the crosstalk between CpGMMs and TFs. The CpGMMs are those whose loci targets have significant enrichment of pluripotent genes. The small circles indicate the positions of the methylation-resistant CpGMM target loci. The ellipses over the CpGMMs enclose the names of the TFs expressed >4.5 FPKM and whose TFBMs resemble (Pearson correlation ≥0.85) the CpGMM. The horizontal bars represent the promoters of the CpGMM targets. The light gray region of the bar represents the gene promoter itself; the black part represents a small portion of the coding region; and the beginning of each horizontal arrow marks the TSSs.
CpGMMs reveal iPS epigenetic somatic memory
The CpGMM discovery algorithm developed here provides a tool to analyze the somatic memory at the methylomics level. For such analysis, we pooled the collection of CpGMMs from all the cell lines into three cell-type categories. We searched for two types of somatic memory. One type is reflected in the persistent somatic CpGMMs that, while present in FBs and iPS, are absent in ES cells. The other type includes the absent somatic CpGMMs that, while absent in both FBs and iPS, show up in ES cells. This memory is reflected by the ES-specific CpGMMs.
The collection of somatic memory CpGMMs due to CpGMMs persistent in iPS and absent in ES cells is provided on our web server. Table 4 presents the somatic memory due to CpGMMs persistent in iPS with enrichment of at least one FB marker. We used 23 FB markers from Yu et al. (2007). We found a clear methylomics fingerprint of the somatic reprogramming memory, with 480 and 608 (Fig. 2C,D) CpGMMs that remain in iPS cells after cellular reprogramming and that are inherited from the initial FB populations, and three and 46 (Fig. 2C,D) CpGMMs characteristic of ES cells that iPS cells have not managed to generate. The number of somatic memory CpGMMs with enrichment of at least one corresponding marker is given in Table 2. There are eight methylation-prone CpGMMs targeting FB markers and somatic memory markers, while 19 methylation-resistant CpGMMs target somatic memory markers.
Table 4.
Selection of somatic memory CpGMMs persistent in iPS cells

The methylation-resistant CpGMMs are CG enriched and their targets occur in highly conserved regions near TSSs
The four nucleotides are differently distributed among the methylation-resistant and methylation-prone CpGMMs (Fig. 3A). The methylation-resistant motifs are specially enriched by guanines and, to a lesser extent, by cytosines, while they are depleted of adenines and thymines. Thus, they are CG enriched, but the methylation-prone motifs have a homogeneous nucleotide composition. Wilcoxon-Mann-Whitney two-sample tests for each of the four nucleotides show that the difference of the distributions between methylation-resistant and methylation-prone CpGMMs is statistically significant (P = 0.0). To understand the biological meaning of the CpGMMs, we looked for common features shared by their targets. First, we checked the loci evolutionary conservation and found that the methylation-resistant CpGMMs loci are considerably more conserved than the methylation-prone CpGMM loci. Indeed, whereas the abundance of the conservation score of the methylation-prone CpGMM targets decreases steadily, the methylation-resistant CpGMM targets have a peak of high abundance around the 1.0 conservation score (Fig. 3B). Under the DNA-sequence conservation-function paradigm, this result points to the functional role of the methylation-resistant CpGMMs. The CpGMM target distributions (Fig. 3C) show that the methylation-resistant CpGMMs targets are mainly located around the TSSs, whereas the methylation-prone CpGMMs targets are depleted around the TSSs, and uniformly distributed in the upstream-extended promoter region. These results pinpoint a potential role of the methylation-resistant CpGMMs in controlling gene transcription.
Figure 3.
General discriminative features between all the methylation-resistant and methylation-prone CpGMMs. Histograms of (A) the 4 nt distributions across the discovered CpGMMs; (B) the conservation of all the CpGMM targets, where the sequence conservation scores were taken from primates phastCons 46-way (see Supplemental Material; Siepel et al. 2005); and (C) the distances of all the CpGMM targets to the TSSs. The methylation-resistant features are in gray; the methylation-prone, in black. Vertical lines mark the position of the median.
Methylation-resistant CpGMM targets inside CpG islands regulate pluripotent cells, whereas differentiated cells are regulated outside CpG islands
A plethora of publications on methylation center their studies around CpG islands (Das et al. 2006; Fang et al. 2006; Bird 2011). To provide an unbiased view, we departed from such constraints, analyzing the DNA methylation state independently of the CpG island localization. We found that, indeed, ∼47% of CpGMM targets occur outside CpG islands (Fig. 4A), thus confirming the importance of the outside CpG island regions. There is a very distinct distribution of CpGMM targets between FBs and pluripotent cells (Fig. 4A). ES cells have a high percentage (72%) of CpGMM targets in CpG islands, indicating that the ES cell's regulation is controlled by methylation switches inside CpG islands. FBs, on the contrary, have 39% of CpGMM targets in CpG islands, and thus are mainly regulated by CpG-poor regions. The CpGMM targets inside CpG islands are dominated by methylation-resistant CpGMMs, again with a very distinct distribution between FBs and pluripotent cells (Fig. 4B). Eighty-six percent of the CpGMM targets in ES cells correspond to methylation-resistant motifs; in FBs the percentage is reduced to 66%, indicating that the methylation-resistant CpGMMs could direct DNA-methylation-mediated repression during lineage specification as observed in different studies (Mohn and Schübeler 2009). Figure 4C shows that the methylation-resistant CpGMM targets occupy higher CG content regions in the CpG islands than the methylation-prone, concording with the high CG content found in the methylation-resistant CpGMMs (Fig. 3A).
Figure 4.
Correlation of methylation-resistant and methylation-prone CpGMM targets with CpG islands for each cell type. Percentage of CpGMM targets of each cell type that lie inside CpG islands (A) and methylation-resistant and methylation-prone CpGMM targets of each cell type that lie inside CpG islands (B). The methylation-resistant percentages are in gray, the methylation-prone in black, and the merged ones in white. The rightmost bars correspond to the mean across all the populations. (C) Histograms of the CG content of the CpGMM targets inside CpG islands for the pool of all samples (ALL), fibroblast (FB), iPS, and ES. The dashed and gray vertical lines mark the positions of the mean of the methylation-resistant and methylation-prone distributions, respectively.
The methylation-resistant CpGMM targets break the H3K4me3/H3K27me3 bivalent balance and shift the enhancers from repressive H3K27me3 to active H3K27ac during ES cell differentiation
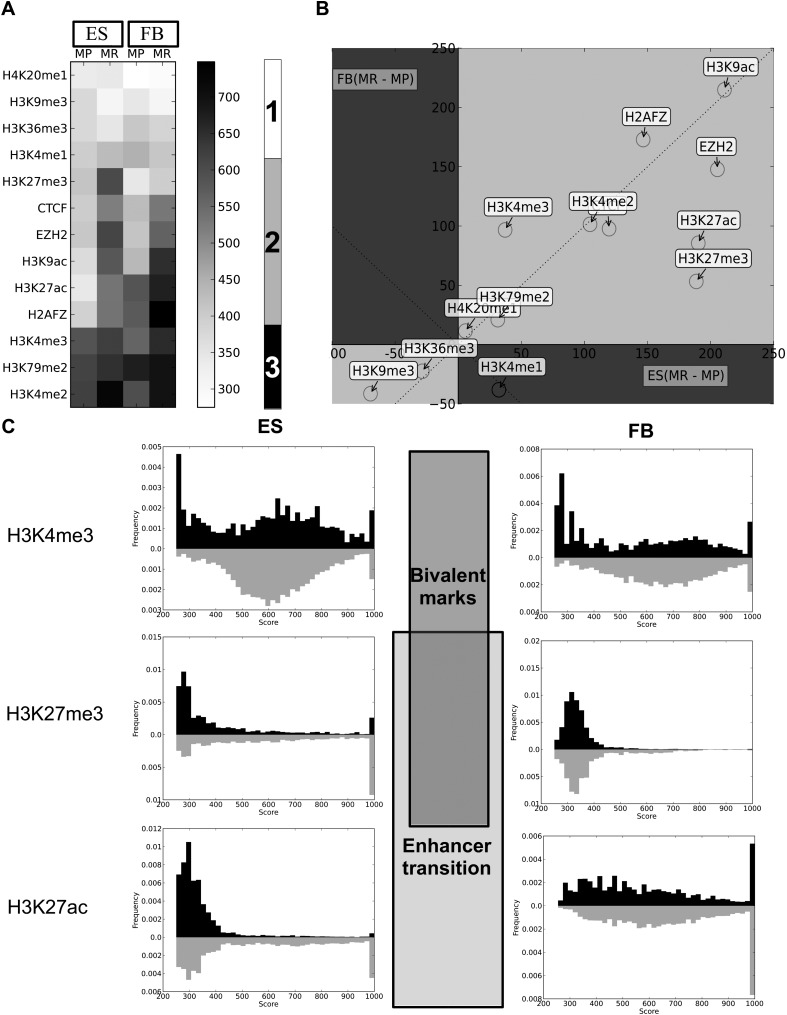
We analyzed the co-occurrence of the ES and FB CpGMM targets with 12 histone marks and CTCF binding sites from the ENCODE Project (The ENCODE Project Consortium 2011). The co-occurrence algorithm description is provided in the Supplemental Methods and in Supplemental Figure S6. We found that in general (Fig. 5A), the cell methylation-prone CpGMMs are less correlated with histone marks than the methylation-resistant CpGMMs in both ES cells and in FBs. This shows that the methylation-resistant CpGMM targets require a more finely tuned regulation than the methylation-prone ones, because once a region is methylated it is not necessary to exert further regulation. We found three types of correlation (Fig. 5A). The first type corresponds to low correlated repressive H3K36me3 and active transcription (H4K20me1, H3K9me3, H3K4me1) marks with CpGMM targets independent of the cell or the CpGMM type. The second type corresponds to marks (H3K27me3, CTCF, EZH2, H3K9ac, H3K27ac, H2AFZ) of higher correlation with methylation-resistant than with methylation-prone CpGMMs. For these marks, interestingly, the transition of enhancers from the inactive to the active state during ES cell differentiation tagged by the change of H3K27 from the repressive H3K27me3 to the active H3K27ac (Creyghton et al. 2010; Rada-Iglesias et al. 2011) is accompanied by the depletion of H3K27me3 and enrichment of H3K27ac methylation-resistant CpGMM targets from ES cell to FBs (Fig. 5C). The last type is formed by the active transcription marks (H3K4me2, H3K79me2, H3K4me3) (Sims and Reinberg 2006; Koch et al. 2007; Steger et al. 2008) that have the highest correlation with CpGMM targets, independently of the cell or the CpGMM type.
Figure 5.
Correlation between ES cell CpGMM loci targets and histone mark signals. (A) Heatmap of the mean signal of the co-occurrences of histone marks with methylation-prone (MP) and methylation-resistant (MR) ES cell CpGMM targets that share loci (calculated as in Supplemental Fig. S6) with ES and fibroblast histone marks. The labeled rectangles mark the three types of correlation patterns. The gray color bar shows the color codification of the mean signal. (B) Scatter plot of the differences of resistant minus prone averages of histone mark signals' co-occurrences with ES cell CpGMM targets in fibroblasts versus ES cells. (C) Histograms of CpGMM targets co-occurring with the H3K4me3, H3K27me3, and H3K27ac histone marks. Only the signals with a score of at least 250 are plotted. The frequencies associated with methylation-resistant and methylation-prone CpGMM targets are in gray and black, respectively.
To clarify the distinctive role of the correlation of CpGMMs with histone marks in pluripotent and somatic cells, for each histone mark signal of FB and ES cells we split the loci between methylation-resistant and methylation-prone CpGMM targets, calculated their co-occurrence (Supplemental Fig. S6), averaged the signals of both methylation types, and depicted the difference of the resistant- minus the prone-associated signals of FB versus ES cells (Fig. 5B). In general, there was a strong correlation of the differences between FB and ES cells in the H3K9ac, H2AFZ, CTCF, H3K4me2, H3K79me2, H4K20me1, H3K36me3, and H3K9me3 marks. The least positively correlated cases are EZH2, H3K27ac, H3K27me3, and H3K4me3. EZH2 is the catalytic subunit of the polycomb repressive complex (PRC2) that trimethylates lysine 27 of histone 3 (H3K27me3), which then recruits PRC1 to modify chromatin in order to enforce gene silencing (Margueron and Reinberg 2011). We observed that the release of polycomb repression during ES cell differentiation is associated with a smaller imbalance of methylation-resistant minus methylation-prone CpGMM targets in FBs. H3K27ac and H3K27me3 are involved in the enhancer activation during ES cell differentiation (Creyghton et al. 2010; Rada-Iglesias et al. 2011), which is also associated with a smaller imbalance of methylation-resistant minus methylation-prone CpGMM targets in FBs (Fig. 5C). Only H3K4me1 has a small negative correlation for quite weak signals. Remarkably, the CpGMM targets exactly reflect the bivalent domain property of H3K4me3 and H3K27me3, which makes the cell linage promoter poised (Bernstein et al. 2006). Figure 5C shows that H3K4me3 acts as a balance, and overlaps equally with methylation-prone and methylation-resistant CpGMM targets, while H3K27me3 unbalances the H3K4me3 effect and overlaps dominantly with the methylation-resistant CpGMM targets to repress the activation of H3K4me3. After differentiation from FBs, this balance is no longer needed. Therefore, the H3K27me3 signal in the FBs methylation-resistant CpGMM targets disappears, and the activation mark H3K4me3 can do its job for expression of the cell-lineage-specific genes.
Crosstalk between CpGMMs and CTCFs reflected in gene expression regulation
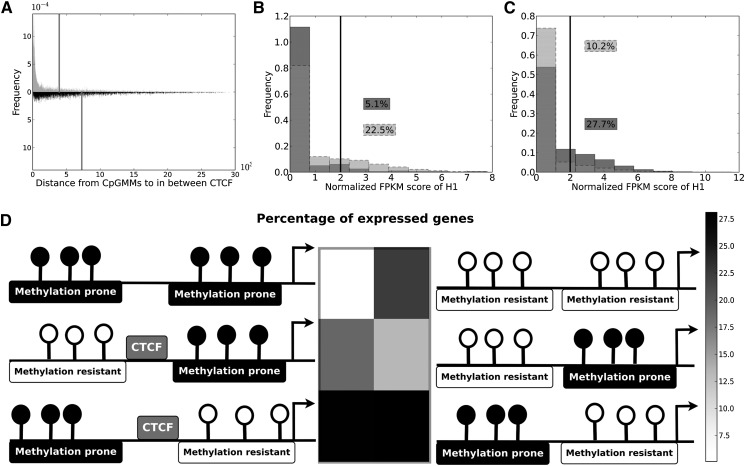
Based on the promoter CpGMM composition, we classified the genes into three groups: the first with only methylation-resistant CpGMMs in their promoters; the second with only methylation-prone CpGMMs and the third with a mixed, bivalent composition of CpGMMs. For the genes with bivalent CpGMM composition, we found many more CTCF binding sites in the regions embraced by methylation-resistant and methylation-prone CpGMMs than expected by chance. In FBs, 66% of such regions have at least one CTCF when the expected value is 5%; this overrepresentation is similar to that in ES cells (64% of the regions between methylation-resistant and methylation-prone CpGMMs have at least one CTCF when the expected value is 4%). These results complement the observation from Figure 5A, showing that CTCF binding sites do not interfere with homogeneous methylation-resistant or methylation-prone CpGMM regions.
To check whether the CTCF binding sites insulate methylation-resistant and methylation-prone CpGMM regions, we measured the distance from the methylation-resistant and methylation-prone CpGMM regions to the CTCF binding sites. We found that the methylation-resistant CpGMMs loci appear very close to CTCF sites (Fig. 6A), whereas the methylation-prone CpGMM loci are more dispersed. To check whether such CTCF–CpGMM configurations have a regulatory counterpart, we analyzed their correlation with gene expression. For the nonmixed promoter case, we found that genes with only methylation-resistant CpGMMs promoters are expressed more than those with only methylation-prone CpGMMs (Fig. 6B; Supplemental Fig. S4b), thus validating the putative functional regulatory role of the two motif types. The genes with mixed CpGMM patterns have a higher expression than the total (Fig. 6C; Supplemental Fig. S4c), indicating that the expression of mixed promoter genes could be driven by the methylation-resistant CpGMMs. To analyze whether the CTCF loci influence the way the CpGMMs drive gene expression, we calculated the transcriptomics distributions of the genes with methylation-resistant CpGMMs close to TSSs. We split the distributions into containing and not containing CTCF binding sites between methylation-resistant and methylation-prone CpGMMs. And we repeated the same CTCF splitting analysis for only methylation-prone (Supplemental Fig. S4e,g) and only methylation-resistant (Supplemental Fig. S4d,f) CpGMMs close to TSSs in ES cells and FBs, respectively. We summarized the analysis, estimating the percentage of genes expressed in each case and depicting the results in the heatmap with the promoter structure models for ES cells (Fig. 6D) and FBs (Supplemental Fig. S4h).
Figure 6.
Discriminative features of mixed promoters containing bivalent and monovalent CpGMMs in ES cells. (A) Histograms of the distances of methylation-resistant CpGMM targets (gray), and methylation-prone CpGMM targets (black), to the in-between CTCF for promoters simultaneously containing methylation-resistant and methylation-prone CpGMMs. The vertical lines show mean values of the distances to CTCF of methylation-resistant and methylation-prone CpGMM loci. (B) Histogram of the expression of genes with only methylation-resistant (light gray) and methylation-prone (gray) CpGMMs. (C) Histogram of the expression of all genes (light gray) and genes with promoters simultaneously containing methylation-resistant and methylation-prone CpGMMs (gray). The black vertical lines show the gene expression threshold. The numbers inside boxes are the percentages of expressed genes. (D) Heatmap of the percentage of expressed genes with mixed and unmixed structures of methylation-resistant and methylation-prone CpGMMs 1kb upstream of the TSS. The corresponding genomic structure with positions relative to the TSS (marked with an arrow) of the methylation-resistant and methylation-prone CpGMMs and CTCF binding is represented beside each heatmap cell.
The promoters with methylation-resistant CpGMMs close to TSSs have a higher expression than those with methylation-prone CpGMMs close to TSSs. This effect is even stronger in the ES cells (Fig. 6D), which express more genes than FBs (Supplemental Fig. S4h). These results also show that for the genes with methylation-resistant CpGMM loci near the TSS, expression is independent of neighboring CTCF binding sites, thus revealing that when a gene has methylation-resistant CpGMMs near the TSS, there is a small chance for the CTCF to exert its insulation function. Interestingly, for promoters with methylation-prone CpGMMs near TSSs, the existence of close CTCFs significantly increases gene expression. In this case, the CTCFs exert their insulator role, not allowing potential repressors to bind to the methylation-resistant CpGMMs. On the one side, these results validate the putative role of the CpGMMs identified by our method. The promoters with methylation-resistant CpGMMs have a higher expression than those with methylation-prone CpGMMs. On the other side, they disclose the crosstalk between CpGMMs and CTCFs in the gene regulation process, showing that the CTCFs near methylation-prone CpGMMs close to TSS modulate the repressor effect of the methylation-prone CpGMMs.
The methylation-resistant CpGMM targets have a higher trend than the methylation-prone to co-occur within TFBSs
We hypothesized that the CpGMMs can be used by some DNA sequence-specific binding proteins to recruit the “methylation/unmethylation machinery” to specific loci. The TFs are high-potential recruiting candidates because of their DNA sequence recognition capability. To compare CpGMMs and TFBMs, we designed a technique based on detecting co-occurrences between the targets of the two types of motifs (Supplemental Methods). We found that 38% of the methylation-resistant CpGMMs co-occur with TFBSs, whereas only 3% of methylation-prone CpGMMs do (Fig. 7B). This enrichment of methylation-resistant CpGMMs concurs with the findings that TFs binding to CpG-rich promoters tend to keep them unmethylated (Lienert et al. 2011). A selection of methylation-resistant CpGMMs co-occurring with TFBSs, with the corresponding TFs is given in Table 5. We found the SP1 TFBM to be associated with the TFBSs that co-occur with methylation-resistant CpGMMs targets. It is already known that SP1 is essential for protecting CpG islands from de novo DNA methylation (Brandeis et al. 1994; Macleod et al. 1994; Bird 2011). The pluripotency level corresponds to a larger number of CpGMM targets that co-occur within TFBSs (Fig. 7A). The CpGMMs targets within TFBSs correspond mainly with methylation-resistant CpGMMs (Fig. 7B). These results, together with the conservation of the methylation-resistant CpGMM targets (Fig. 3B) and their enrichment around the TSSs (Fig. 3C) indicate that the methylation-resistant CpGMMs interact with gene transcription regulation.
Table 5.
Selection of methylation-resistant CpGMMs that co-occur with TFBSs

A pluripotent network arises from the crosstalk between methylation-resistant CpGMMs and TFs
To understand the reprogramming mechanism, we searched for CpGMM targets using the Berg–von Hippel method (see Methods) (Berg and Von Hippel 1987) in the promoters of pluripotent markers. We found numerous cases of methylation-resistant CpGMMs that simultaneously target the pluripotent marker promoters with a statistically significant enrichment (P < 0.05). Since the methylation-resistant CpGMM loci have a high correlation of co-occupancy with TFBS (Fig. 7B), we searched for the TFs that share loci with methylation-resistant CpGMMs, filtering out the lowly expressed TFs in ES cells—those with fragments per kilobase of exon per million fragments mapped (FPKM) are <4.5. Thus we found two networks: In one (Fig. 7C), formed by ESRRB, KLF2, and SOX2, KLF2 plays a central role, being simultaneously targeted by two methylation-resistant CpGMMs. On the other side, YY1, targeting ESRRB may direct histone deacetylases and histone acetyltransferases to the promoter in order to activate or repress them (Coull et al. 2000; He and Margolis 2002). The other network (Fig. 7D) is formed by SALL4 and FGF4. These networks are important to elucidate the upstream effectors of the pluripotent genes that control their expression and to understand the reprogramming mechanisms. The methylation-resistant CpGMMs of these networks have the high CG-content composition already revealed in the nucleotide composition analyses (Fig. 3A).
Discussion
This work was prompted by the observation that some CpGs in methylation lollipop diagrams tend to be methylation-resistant or methylation-prone. This phenomenon is not static. Depending on the cell type and the environmental conditions, the methylation state of any CpG can be driven to total unmethylation or full methylation. Thus, the methylation-prone and methylation-resistant CpGs are instantaneous “photographs” of a probabilistic process, in which each CpG, depending on its sequence context, has a variable susceptibility to be methylated or unmethylated. Mammalian DNA methylation is driven by DNA methyltransferases (DNMTs) that perform de novo methylation (DNMT3A and DNMT3B) and maintain it (DNMT1). DNA demethylation occurs through a passive or through poorly understood active mechanisms (Wu and Zhang 2010), where 5-hydroxymethylcytosine could act as a temporary state (Guo et al. 2011). These mechanisms are DNA sequence unspecific. However, DNA methylation is a cell type-specific process, as it regulates gene expression of the different cell lineages. It is paradoxical to reconcile a DNA sequence-unspecific mechanism with cell type-specific events. If the cell types of an individual were isogenic, the sequence-unspecific mechanism would be unable to discriminate among them, and all the cells would have the same methylome, which is false. Our finding of cell type-specific CpGMMs (Fig. 2) sets the ground for such a mechanism. Thus, we hypothesize that additional mechanisms exist that provide specificity to the enzymes modifying the DNA methylation state. Such mechanisms could be based on DNA sequence-specific binders interacting with methylation/unmethylation enzymes and recruiting them to specific DNA loci. This would be similar to the gene-transcription mechanism, in which DNA sequence-specific TFs recruit the transcription machinery to the promoter locus of the genes to be transcribed. Thus, “methylation/unmethylation factors” (MUFs) might exist which are not necessarily different from the TFs.
We found a significant percentage of methylation-resistant CpGMMs co-binding with TFs (Fig. 7A,B), and many similar to TFBMs (web server) (Table 2), such as the SP1 motif. Thus, some TFs could play a dual role, recruiting the transcription machinery when acting as TFs, and recruiting the methylation/unmethylation machinery when acting as MUFs. Such behavior is, for example, observed in cancer cells (Croce et al. 2002; Carbone et al. 2006) or in the repression capability of DAXX via DNA methyltransferase recruitment (Puto and Reed 2008). CpGMMs could be signals recognized by specific MUFs that in turn recruit the methylation/unmethylation machinery near the CpG loci. Different cell types can express specific MUFs inducing diverse methylation states to the same CpG loci. Thus, the CpG context provides specificity to discriminate between the different CpGs of the same cell type (Fig. 1B), and MUFs expressed by each cell type provide specificity to discriminate between the same CpG words appearing in different cellular contexts.
We found that the methylation-resistant CpGMMs are in crosstalk with TFs in gene expression regulation (Fig. 7). Such crosstalk could be explained by at least two mechanisms. One was proposed by Schübeler's group (Lienert et al. 2011), where the TFs binding to DNA regions protect them from being methylated. Another mechanism might be that the methylation-resistant CpGMMs signal the TFs to recruit DNA sequence-specific unmethylation machinery. Both mechanisms might not be exclusive and might apply cooperatively. For example, the reprogramming process needs a DNA methylation sequence-specific mechanism that recognizes fibroblast DNA specific sequences methylated in the promoters of pluripotent genes such as POU5F1 (OCT4) or NANOG (Takahashi and Yamanaka 2006). This mechanism recruits the unmethylation machinery to unmethylate them in the reprogrammed iPS cells. Thus, the unmethylation recruitment mechanism seems more suitable to explain the unmethylation of specific promoter regions of pluripotent genes during the reprogramming than the methylation protection. Subsequently, it is possible that some TFs bind to the promoters of the pluripotent genes in iPS cells and keep them unmethylated.
In general, the different cell line CpGMMs are reproducible for each cell type; however, methylation-resistant CpGMMs are more reproducible in pluripotent cells, and methylation-prone ones are more reproducible in FBs (Supplemental Fig. S2f). This could indicate that pluripotency requires a tighter maintenance of unmethylation of specific regions.
The results produced by our algorithms are complementary to those obtained with DMR search methods. DMR methods analyze the same locus in different samples, searching for differentially methylated loci (Zhang et al. 2011), and require at least two samples. Our method searches for loci that in different regions have similar methylation, thus providing CpGMMs without the necessity of control samples. The controls are the different methylation states of different loci of the same sample.
DMR search approaches are useful to characterize methylomes produced under different experimental conditions, but understanding the methylation mechanism from DMRs is complicated. DMR search approaches need to set in advance the region length, and usually operate with iso-length regions across the genome. These features hinder them from detecting methylomics signatures at a single CpG level. Inversely, the CpGMM discovery method is a region length-adaptive system that adjusts its results depending on the degree of similarity among different genomic regions. The CpGMM discovery method performs a complementary methylomics analysis that in synergy with DMRs can help us better understand the methylation/unmethylation mechanism.
The algorithms developed and the CpGMMs found here are useful tools to investigate the sequence specificity of the DNA methylation/unmethylation process and for searching potential MUFs. They open up the door for understanding the methylation readout mechanism. Once potential CpGMMs are disclosed, the proteins that have a binding specificity for such motifs remain to be found. The search for DNA methylation aberration patterns in cancer cells is another potential application of the method.
Pluripotent methylomics profiles (Supplemental Fig. S1a,b) show higher variability than pluripotent transcriptomics profiles (Supplemental Fig. S1c). The methylomics variability is reflected in the different number of motifs found in each data set (Fig. 2C,D). Moreover, it indicates that methylomics profiles are more sensible to detected variations between reprogrammed cells than transcriptomics profiles. Thus, methylomics could be more suitable than transcriptomics methods for understanding the cellular reprogramming mechanism and disclosing the somatic memory.
One of the aims of our study is to find insights into the reprogramming mechanism. Therefore, we search for CpGMMs that could be associated to the methylation/unmethylation mechanisms. We used the CpGMM discovery method for exploring the somatic memory. We found a clear methylomics fingerprint of the somatic memory associated with a collection of methylation-resistant (Fig. 2C) and methylation-prone (Fig. 2D) CpGMMs. Interestingly, the somatic memory found by Ruiz et al. (2012) based on differentially methylated CpG sites (DMSs) identified a reprogramming-specific epigenetic signature composed of nine aberrantly methylated genes. Though our approach, which is based on CpGMMs, scrutinizes the somatic memory from a different perspective, our results are in line with theirs. Three (FAM19A5, DPP6, and PTPRT) out of nine genes co-occur with four somatic memory CpGMMs. Thus, we speculate that demethylation is strongly remembered in contrast to de novo methylation. These results differ from those obtained with the DMR search (Bock et al. 2011) which detected very few somatic memory regions. One of the reasons for such a discrepancy is that the DMR-based method cannot disclose such small hot spots due to noise filtering. Our technique filters out the noise by averaging similar methylation signals emerging in multiple regions with a similar sequence dispersed across the whole genome, and not by averaging through windowing the methylation signal over a specific area. Our technique takes a step forward in relating the somatic memory with the way that some specific DNA sequences are recognized to become more or less methylated during reprogramming.
Methods
Data collection, mapping, and annotation
The general features of the DNA methylomes used for different cell lines of FB, iPS, and ES are described in Table 1. The raw data (fastq files) of each methylome were downloaded from the European Nucleotide Archive (ENA) (http://www.ebi.ac.uk/ena/home) and processed with the procedure described in detail in the Supplemental Methods.
Generation of “conserved” DNA methylome for each cell type
The methylomes of different cell lines from the same cell type are grouped. Then all the CpGs are aligned based on the cytosine genomic positions. Thus, we define a CpG site as conserved when the methylation ratios across all the cell lines are low fluctuating. The methylation ratios fluctuation significance analysis is described in Supplemental Methods.
Analysis of sequence and methylation similarity between the two DNA strands
We define the methylation dissimilarity MetDis(CpGi) of the methylation in each CpG locus i (i marks the cytosine position) of the two DNA strands as the absolute difference between the methylation ratios of the cytosines at the positive MR(CpG+) and negative MR(CpG−) strands:
Since the methylation ratios are determined in the range [0 1], we can define the methylation similarity MetSim as
We define the sequence similarity SeqSim(CpGiw) of length w sequences at a locus i, as the match between the sequences of length w in the positive CpGiw+ and negative CpGiw− strands centered in the CpG of each loci i, normalized by the sequence length w,
 |
where CpGiw+(j) and CpGiw−(j) are the nucleotides at position j of the CpG sequences at the genomic position i of the positive and negative strands, respectively. The sequence match is the Hamming distance between each nucleotide in the positive strand, and the paired nucleotide in the negative strand. This similarity is a measurement of the degree of palindromy of the sequence. We applied Equations 1–3 for 12 ≤ w ≤ 52, with a step of 2 (1 nt in both directions outward of the central CpG, whose length is also accounted for in the sequence length) and depicted the methylation similarity versus the sequence similarity using heatmaps that represent the dot density on a gray color scale.
Discriminative CpGMM discovery algorithm
The algorithm is based on the compilation of CpG word dictionaries: one collected from low-methylated, and another from high-methylated regions. Each CpG word can have a different length w. To avoid small words inside big ones, we designed a fusion procedure of CpG words. After fusion, the words are clustered with a hierarchical algorithm. All the sequences inside a cluster are defined as a cluster prototype, whose matrix of nucleotide frequencies defines a CpGMM.
The motifs with the capability to discriminate between low- and high-methylated regions are selected with a scanning method based on a binding energy analogy. We applied the following pipeline to both DNA strands. In order to avoid cumbersome notations, we describe the procedure for the positive strand. The method workflow is depicted in Supplemental Figure S5.
Compilation of CpG word dictionaries
As shown in the Supplemental Information file, the minimal wmin = 12 and maximal wmax = 44 CpG word lengths were based on the results depicted in Supplemental Figure S2a. For all the genomic CpGs, and for lengths wmin ≤ w ≤ wmax, with a step of 2, we collected the sequence centered in each CpG (the central CpG is included in the w length). This procedure generates a CpG-centered word genomic dictionary harboring all sequences of length w centered in each CpG. The repeated sequences are grouped into unique ones. We call their repetition numbers FCpGw frequency of the sequence of length w. Hence, we generate a set of unique CpGw sequences of different lengths w. We assume that similar sequences have similar methylation ratios MRCpGw. Therefore, we assigned to each unique sequence the average of the methylation ratios of the CpGs from the same unique group. We denote this step as a unique sequence search step. A final scanning analysis selects the CpGMMs that discriminate between methylation-prone and methylation-resistant regions. Next, to reduce the noise we implemented a filter step. The unique sequences are filtered by their frequencies. Only unique patterns with frequencies of three or more are retained based on the calculation shown in the Supplemental Information file. To check whether the assumption that similar sequences have similar methylation ratios MRCpGw, for each CpG word of length w, we discretized the CpG methylation ratios into three categories according to the method described by Stadler et al. (2011): 0.0 ≤ MRCpGw < 0.1 for unmethylated sites (UMSs), 0.1 ≤ MRCpGw < 0.5 for low-methylated sites (LMSs), and 0.5 ≤ MRCpGw ≤ 1.0 for high-methylated sites (HMSs). Then, for each CpG word, we count its membership percentage to one of the three categories. If the percentage is >90%, we assign a 1 to that word, and a 0, if it is <90%. Finally calculate the percentage of ones in relation to the total number of CpG words.
Fusion of CpG word dictionaries
During the dictionary compilations, the sequences are extended in both directions outward from the central CpG. Therefore, some sequences of length w could appear inside sequences of length w + 2. Hence, we designed a sequence fusion method that avoids shorter submotifs centered inside longer ones. This method is implemented in an iterative way, starting from the shortest length wmin. Thus, for each length w, if a sequence CpGw is included in the center of a sequence CpGw+2 of length w + 2, the shorter sequence is fused inside the longer one. The methylation ratio of the new sequence is updated as the weight averaged methylated ratios of the fused sequences:
 |
Before the sequence fusion step, each unique sequence CpGw+2 has an associated scalar frequency FCpGw+2 that imputes the same frequency to all sequence nucleotides. After the fusion, to keep track of the individual frequency position in the fused sequence, the scalar frequency is converted into a frequency vector FCpGw+2 of length w + 2, which stores for each nucleotide position j its respective frequency. Thus, for the central common positions in the sequence CpGw+2, the vectorial frequencies are FCpGw+2(j) = FCpGw+2(j) + FCpGw(j − 1). The peripheral positions preserve the original frequencies of the longer sequence. The scalar frequency of the new sequence is updated as the sum of the scalar frequencies of the fused sequences FupdateCpGw+2 = FCpGw + FCpGw+2. After fusion, the shorter sequence CpGw is eliminated from the dictionary. The fusion method iterates till reaching the longest sequence length wmax, fusing whenever possible shorter sequences centered in the central CpG into the longer sequences. An example of the distribution of the number of CpG words before and after fusion with respect to their length for the highly methylated CpGs of the negative strand is depicted in Supplemental Figure S2a.
Motif discovery through hierarchical clustering
After the fusion step, the information associated with each CpG word i of length w of each dictionary is integrated into a position occurrence matrix POMiw of dimension (4 × w) that collects in every column j the frequency FCpGw(j), and stores it in the row indexed by the nucleotide in the position j of the sequence CpGiw(j). Initially, the POMiw column has only a non-null value. For each length w, all the POMs are grouped with a hierarchical clustering algorithm, using the cosine metric calculated with Equation 4 and the complete linkage method.
 |
Before calculating the distances, the (4 × 3 w) bidimensional matrices are vectorized into 4w length vectors. The cut-off parameter for the cluster distance was set to 0.75. This parameter is learned from the ADS-iPSC promoter methylome data set, using the silhouettes algorithm (Crooks et al. 2004), in-house implemented in Python. We performed 1001 cutoffs of the hierarchical clustering from 0 ≤ cutoff ≤ 1, with a step of 0.001. As a final cutoff, we chose the one (0.75) that maximizes the average silhouette width. For each cluster c, all the sequences are merged into a new averaged POMcw that represents the motif cluster.
Selection of the motifs with discrimination capability based on a binding energy scanning method
With the given potential discriminative methylation motif sets, one for methylation prone and the other for methylation resistant, we searched for motifs that specifically discriminate between high- and low-methylated CpGs. For this purpose, we took advantage of the analogy of the TF-DNA binding energy, using the Berg–von Hippel method (Berg and Von Hippel 1987). Based on this analogy, we treat the methylation motif as TFBM, and consider that a methylation motif has a good match with a genomic region center in a CpG, if the “virtual” binding energy estimated by the Berg–von Hippel method is high. To perform such an energy calculation, first we normalize the POMs, creating the so-called position weight matrices (PWMs):
 |
Thus, all the motifs PWMw are scanned as in a typical TFBS search (Sarkar et al. 2008) against each CpG sequence of the methylation-prone and methylation-resistant sets, using the binding energy equation of the Berg–von Hippel method,
where β = 0.00001. The addition of β is necessary to avoid division by zero. The specific value of β was chosen after empirical study to maximize the score dynamic range. CpGw is the CpG word of length w centered in the genomic CpG locus. For better computational performance, we used a different but equivalent implementation of Equation 5. Higher matching score corresponds to more specific similarity of the motif with the target sequence. We split the matching scores for each motif into two distributions, one for high and another for low methylation regions. Next, we checked whether the motif can discriminate between the two distributions using the Kolmogorov–Smirnov test (KStest) for two samples (with the stats package of scipy) with a significance level α = 0.00001. The motifs passing this test are retained and subjected to a second filter with a double objective. On one hand, the filter estimates the minimal matching score (threshold of the right tail) that has to have a potential target DNA sequence to be “bound” by the motif. The thresholds Tr of the right tails are computed with the equation
where θ is the threshold of the right tail of the matching score distribution (methylation-prone distribution, if the underlying motif is a potential methylation-prone CpGMM), μ is the matching score distribution mean, σ is the matching score distribution standard deviation, and λ is set to two, based on an empirical study. On the other hand, considering as true targets the sequences that pass the filter (6), to strengthen the discriminating capabilities of the motif, we select only those with a false discovery rate (FDR) ≤ 0.05,
where FalsePositive is the number of scores ≥θ in the methylation-resistant distribution if the underlying motif is a potential methylation-prone CpGMM, TruePositive is the number of scores ≥θ in the methylation-prone distribution if the underlying motif is a potential methylation-prone CpGMM. We use the right tail (high matching score) of the two distributions. The selected motifs are represented as motif logos, using WebLogo 3.0 (Crooks et al. 2004). All the found motifs were annotated with the gene ontology of their corresponding targets using the R-bioconductor package GOstat (Falcon and Gentleman 2007). To assess the stability of the motif discovery, we performed a bootstrapping with 100 time samplings with replacement over the ADS-iPSC methylomics data. We used the percentage of CpGMMs that are recovered in at least half of the bootstrapping samples as an estimation of the CpGMM stability.
Search of cell type-specific and somatic memory CpGMMs
For all the methylation motifs in each pair of cell types (ES/FB/iPS), we computed the Pearson correlation:
 |
where PWMi and PWMj are PWMs of the cell type i and j, respectively. When the two PWMs have different lengths, e.g., if length(PWMi) > length(PWMj), we substitute, in the Pearson correlation (8), the longest matrix PWMi by the overlapped central block of the PWMi matrix with the same length as the shorter matrix PWMj (additional details are described in the Supplemental Methods).
Nucleotide enrichment analysis of CpGMMs
The four types of nucleotides for each methylation-resistant and methylation-prone CpGMM are counted and normalized with the motif length. The Wilcoxon-Mann-Whitney test is applied for each pair distribution of methylation-prone/-resistant CpGMMs of the same nucleotide to find the significantly different enrichments with a significance level α = 0.01.
Analysis of conservation and colocalization of CpGMM targets with genetic loci
For the CpGMM conservation analysis, all targets of methylation-resistant and methylation-prone CpGMMs are mapped to the phastCons46way conservation track in primates (Siepel et al. 2005). From that file we get the conservation score for each nucleotide position. For the analysis of colocalization of CpGMM targets near TSSs, the distances from all targets of methylation-resistant and methylation-prone CpGMMs to the corresponding TSS are computed. All target genes and TSSs annotation are taken from the UCSC Genome Browser RefSeq hg19. For the analysis of colocalization of CpGMM targets with CpG islands, we downloaded the conserved CpG islands' annotation from the UCSC Genome Browser. We counted for each cell type, the number of targets of methylation-resistant and methylation-prone CpGMMs occurring inside or outside CpG islands. For the analysis of colocalization of CpGMM targets with TFBSs, we downloaded the conserved TFBS from the UCSC Genome Browser. We developed the algorithm for searching TFs that shares binding sites with the CpGMMs described in the Supplemental Methods. For the colocalization of CpGMM targets with histone marks, we downloaded the 12 histone marks' broad peak signals from the ENCODE Project (The ENCODE Project Consortium 2011) for ES cells (H1) and FBs (NHLF), and we developed the CpGMM target histone marks colocalization algorithm described in the Supplemental Methods.
Correlation analysis between CpGMM targets, CTCF, and gene expression
The transcriptomics RNA-seq data and the CTCF binding data of ES cells (H1) and FBs (NHLF) are downloaded from the ENCODE Project (The ENCODE Project Consortium 2011). We focused on the extended promoter regions as defined in the Data collection, mapping, and annotation section, and developed an algorithm to calculate the correlation between CpGMM targets, CTCF, and gene expression (described in the Supplemental Methods).
Data access
Comprehensive lists of annotated motifs with their corresponding scanned distributions can be downloaded from Supplemental Material and from our web server at http://computational-biology.mpi-muenster.mpg.de/publications/MethylationMotifs/. These lists include the motif found for each of all the cell lines analyzed, the pluripotent-specific motifs, the motifs that reveal the somatic memory, and the motifs shared by TFs.
Acknowledgments
We thank Daniela Gerovska and David Obridge for proofreading the manuscript, Ryan Lister for fruitful clarifications about his methylomics data sets, and Kenjiro Adachi, Karin Hübner, and all the other members of the Department of Cell and Developmental Biology at the Max Planck Institute for Molecular Biomedicine for fruitful discussions and comments during the preparation of this manuscript.
Author contributions: P.-L.L. implemented and executed the computational tools; P.-L.L. and M.J.A.B. analyzed and interpreted the results; H.R.S. provided critical discussion of experimental plans; M.J.A.B. designed the project and wrote the manuscript; and all authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.155960.113.
References
- Artyomov MN, Meissner A, Chakraborty AK 2010. A model for genetic and epigenetic regulatory networks identifies rare pathways for transcription factor induced pluripotency. PLoS Comput Biol 6: e1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg OG, Von Hippel PH 1987. Selection of DNA binding sites by regulatory proteins: Statistical-mechanical theory and application to operators and promoters. J Mol Biol 193: 723–743 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bhasin M, Zhang H, Reinherz EL, Reche PA 2005. Prediction of methylated CpGs in DNA sequences using a support vector machine. FEBS Lett 579: 4302–4308 [DOI] [PubMed] [Google Scholar]
- Bird A 2011. Putting the DNA back into DNA methylation. Nat Genet 43: 1050–1051 [DOI] [PubMed] [Google Scholar]
- Bock C, Paulsen M, Tierling S, Mikeska T, Lengauer T, Walter J 2006. CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure. PLoS Genet 2: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. 2011. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144: 439–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boué S, Paramonov I, Barrero MJ, Izpisúa Belmonte JC 2010. Analysis of human and mouse reprogramming of somatic cells to induced pluripotent stem cells. What is in the plate? PLoS ONE 5: e12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H 1994. SP1 elements protect a CpG island from de novo methylation. Nature 371: 435–438 [DOI] [PubMed] [Google Scholar]
- Carbone R, Botrugno OA, Ronzoni S, Insinga A, Di Croce L, Pelicci PG, Minucci S 2006. Recruitment of the histone methyltransferase SUV39H1 and its role in the oncogenic properties of the leukemia-associated PML-retinoic acid receptor fusion protein. Mol Cell Biol 26: 1288–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen P-Y, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. 2010. Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, Davie JR, Shi Y, Hansen U, Margolis DM 2000. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol 74: 6790–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce LD, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Coco FL, Kouzarides T, Nervi C, et al. 2002. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295: 1079–1082 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Dimitrova N, Xuan Z, Rollins RA, Haghighi F, Edwards JR, Ju J, Bestor TH, Zhang MQ 2006. Computational prediction of methylation status in human genomic sequences. Proc Natl Acad Sci 103: 10713–10716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A 2011. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A, Park I-H, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. 2009. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 41: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Tibshirani R 1993. An introduction to the bootstrap. Chapman &Hall/CRC, Boca Raton, FL [Google Scholar]
- Elnitski L, Jin VX, Farnham PJ, Jones SJM 2006. Locating mammalian transcription factor binding sites: A survey of computational and experimental techniques. Genome Res 16: 1455–1464 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2011. A User's Guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol 9: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S, Gentleman R 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258 [DOI] [PubMed] [Google Scholar]
- Fang F, Fan S, Zhang X, Zhang MQ 2006. Predicting methylation status of CpG islands in the human brain. Bioinformatics 22: 2204–2209 [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W 2011. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473: 398–402 [DOI] [PubMed] [Google Scholar]
- Grunau C, Renault E, Rosenthal A, Roizes G 2001. MethDB: A public database for DNA methylation data. Nucleic Acids Res 29: 270–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming G, Song H 2011. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han DW, Do JT, Araúzo-Bravo MJ, Lee SH, Meissner A, Lee HT, Jaenisch R, Schöler HR 2009. Epigenetic hierarchy governing Nestin expression. Stem Cells 27: 1088–1097 [DOI] [PubMed] [Google Scholar]
- He G, Margolis DM 2002. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol Cell Biol 22: 2965–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Araúzo-Bravo MJ, Ruau D, Han DW, Zenke M, et al. 2008. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454: 646–650 [DOI] [PubMed] [Google Scholar]
- Kim JB, Greber B, Araúzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Schöler HR 2009. Direct reprogramming of human neural stem cells by OCT4. Nature 461: 649–653 [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LIR, et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature 467: 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, Hongguang H, Loh Y-H, Aryee MJ, Lensch MW, et al. 2011. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol 29: 1117–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaöz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, et al. 2007. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res 17: 691–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Sung KWK, Rigoutsos I, Loring J, et al. 2010. Dynamic changes in the human methylome during differentiation. Genome Res 20: 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitsky VG, Ignatieva EV, Ananko EA, Turnaev II, Merkulova TI, Kolchanov NA, Hodgman TC 2007. Effective transcription factor binding site prediction using a combination of optimization, a genetic algorithm and discriminant analysis to capture distant interactions. BMC Bioinformatics 8: 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D 2011. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet 43: 1091–1097 [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, et al. 2011. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471: 68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod D, Charlton J, Mullins J, Bird AP 1994. SP1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev 8: 2282–2292 [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A 2011. Guiding DNA methylation. Cell Stem Cell 9: 388–390 [DOI] [PubMed] [Google Scholar]
- Mohn F, Schübeler D 2009. Genetics and epigenetics: Stability and plasticity during cellular differentiation. Trends Genet 25: 129–136 [DOI] [PubMed] [Google Scholar]
- Müller-Molina AJ, Schöler HR, Araúzo-Bravo MJ 2012. Comprehensive human transcription factor binding site map for combinatory binding motifs discovery. PLoS ONE 7: e49086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S 2007. Generation of germline-competent induced pluripotent stem cells. Nature 448: 313–317 [DOI] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. 2010. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol 28: 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puto LA, Reed JC 2008. Daxx represses RelB target promoters via DNA methyltransferase recruitment and DNA hypermethylation. Genes Dev 22: 998–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S, Diep D, Gore A, Panopoulos AD, Montserrat N, Plongthongkum N, Kumar S, Fung H-L, Giorgetti A, Bilic J, et al. 2012. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc Natl Acad Sci 109: 16196–16201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Siddiquee KAZ, Araúzo-Bravo MJ, Oba T, Shimizu K 2008. Effect of cra gene knockout together with edd and iclR genes knockout on the metabolism in Escherichia coli. Arch Microbiol 190: 559–571 [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, Reinberg D 2006. Histone H3 Lys 4 methylation: Caught in a bind? Genes Dev 20: 2779–2786 [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495 [DOI] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim J-E, Chen J, Lazar MA, et al. 2008. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 28: 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- von Rohr P, Friberg MT, Kadarmideen HN 2007. Prediction of transcription factor binding sites using genetical genomics methods. J Bioinform Comput Biol 5: 773–793 [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y 2010. Active DNA demethylation: Many roads lead to Rome. Nat Rev Mol Cell Biol 11: 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu H, Lv J, Xiao X, Zhu J, Liu X, Su J, Li X, Wu Q, Wang F, et al. 2011. QDMR: A quantitative method for identification of differentially methylated regions by entropy. Nucleic Acids Res 39: e58. [DOI] [PMC free article] [PubMed] [Google Scholar]