Abstract
Multi-scale modeling of complex biological systems remains a central challenge in the systems biology community. A method of dynamic knowledge representation known as agent-based modeling enables the study of higher level behavior emerging from discrete events performed by individual components. With the advancement of computer technology, agent-based modeling has emerged as an innovative technique to model the complexities of systems biology. In this work, the authors describe SPARK (Simple Platform for Agent-based Representation of Knowledge), a framework for agent-based modeling specifically designed for systems-level biomedical model development. SPARK is a stand-alone application written in Java. It provides a user-friendly interface, and a simple programming language for developing Agent-Based Models (ABMs). SPARK has the following features specialized for modeling biomedical systems: 1) continuous space that can simulate real physical space; 2) flexible agent size and shape that can represent the relative proportions of various cell types; 3) multiple spaces that can concurrently simulate and visualize multiple scales in biomedical models; 4) a convenient graphical user interface. Existing ABMs of diabetic foot ulcers and acute inflammation were implemented in SPARK. Models of identical complexity were run in both NetLogo and SPARK; the SPARK-based models ran two to three times faster.
Keywords: Agent-Based, Computer Simulation, Framework, Models, SPARK
BACKGROUND
Computational models in systems biology are intended to describe biological phenomena at various scales (Kitano, 2002). However, the ability to transcend multiple scales of biological phenomenon within a single, cohesive computational model remains a significant challenge to the biomedical modeling community. This capacity is particularly important for the goal of translating underlying mechanistic knowledge to the level of clinical relevance. Recently, translational systems biology has been introduced as a means of utilizing dynamic mathematical models and engineering principles to aid in the optimization of clinical practice (Vodovotz, 2008; An, 2008).
Traditional mathematical models in systems biology are built using statistics or differential equations. These models are best suited for circumstances in which the dimensions of the modeled biological problems are few. However, for most biological systems with a high degree of complexity, the models themselves quickly become intractable in terms of both analysis and computation. Agent-based modeling is an alternative technique with which to model complex biological systems. This type of modeling incorporates an object-oriented, rule-based, discrete event method of model construction (An, 2001; An, 2009; Bankes, 2002; Bonabeau, 2002; Grimm, 2005).
Earlier implementations of ABM-building software were geared towards developing models in the social sciences, such as Ascape (Inchiosa, 2002) and Repast (North, 2006), or towards general-purpose discrete-event simulations, such as MASON (Luke, 2003) and NetLogo (Wilensky, 1999). Among these, NetLogo is currently one of the most popular, particularly for nonformally-trained programmers, due to its user-friendly interface and the natural language-like syntax of its Logo-based programming language. These features greatly simplify the programming of ABMs for novice programmers. Many biomedical models have been developed successfully by using NetLogo (Mi, 2007; Li, 2008; An, 2004; Bailey, 2009). However, despite its utility, we believe that the construction of biomedical ABMs would benefit from some capabilities currently not found in NetLogo and similar software. These features include the ability to vary agent size, to employ continuous model space, to organize code into modules that can map to biological processes, as well as offering the potential for parallelization in distributed computer architectures.
These criteria motivated the development of a new agent-based modeling platform – SPARK (Simple Platform for Agent-based Representation of Knowledge). This modeling platform incorporates a number of features currently offered by NetLogo, and offers several features designed to facilitate biological modeling. In SPARK, modelers can build models using a user-friendly language and graphical user interface. In addition, the software allows for agents of various sizes, sophisticated image effects, and facilitates multiscale modeling. We describe these features of SPARK in detail below.
IMPLEMENTATION
SPARK is implemented in the Java programming language. SPARK code can run on all machines with Java Standard Edition runtime environment version 1.5 or 1.6. The SPARK source code is freely available under the MIT license and can be retrieved from the SPARK repository at http://code.google.com/p/spark-abm/. The compiled SPARK packages, along with the tutorials, can be downloaded from the official SPARK website (www.pitt.edu/~cirm/spark).
There are several third-party libraries used in SPARK:
JFreeChart (http://www.jfree.org/jfreechart/)
Java OpenGL (JOGL, https://jogl.dev.java.net/)
JFreeChart is used for creating and visualizing line plots and histograms in real time. JOGL is used for visualization of a simulation. Colt is a library for high performance scientific and technical computing in Java.
Overview of the SPARK Structure
SPARK is implemented as a client-server application. There are five main components which constitute SPARK (Figure 1), including a specialized SPARK programming language (SPARK-PL) that can be used for model development. The use of SPARK-PL is optional; SPARK models can be written directly in Java as well.
Figure 1.
Main components of SPARK
User models are created with the functionality provided by the SPARK library, which contains definitions of the main SPARK concepts: Agents, Spaces, Data Layers and the Observer.
The simulation engine runs user models. It uses classes from the SPARK library to communicate with user models.
The user interface allows working with SPARK models interactively. Users can load models, set parameters of their choice, run simulations, and visualize results.
The simulation engine is completely independent from the user interface. The user interface (which runs on a client machine) communicates with the simulation engine (which runs on the server) by sending commands to the server and by receiving data from the server. The received data can be saved on a disk, analyzed, or visualized by the client. In general, data sent by the server contains information about the state of a simulation: values of variables, positions of agents in a space, etc.
There are several possible implementations of the SPARK client and server. One option is to implement them inside the same program. In this case, the communication process will be very fast, but the client and server need to share the resources of the same machine. Another option is to implement the client and server as two separate programs, and establish the communication between them using a network interface. In this situation, it is optimal not to send all data after each simulation step. Instead, data can be sent at specific time intervals using data buffers, or data can be collected and sent only after the end of a simulation. The two versions of the implementations of client and server are designed for different purposes of simulations. The first version can be applied for models with relatively short simulation time but requiring quick visualization of output. The latter version is suitable for large scale simulations where the users may want to view the results after thousands of simulation steps. In the standard SPARK distributions, both versions of the client and the server are available.
From a modeler’s point of view, the most important components of SPARK are 1) the SPARK library; 2) SPARK-PL; and 3) the SPARK user interface. Key concepts of the SPARK library, the SPARK programming language, and the user interface are described in the next sections.
KEY CONCEPTS IN THE SPARK LIBRARY
SPARK Model
SPARK models are created by using basic components of the SPARK library. A SPARK model consists of the following elements: definitions of agents and a global model description. A model description includes the initialization process for a model (the “setup” method) and contains the global variables and parameters. It also includes methods which are called before and after each simulation step, and which are primarily used for changing global variables and collecting statistics.
Overview of Main SPARK Model Components
The main components of SPARK models are Space, Data Layers, Agents, Links, and the Observer (Figure 2). As a demonstration of SPARK, we implemented a published NetLogo ABM of skin wound healing in the setting of diabetic foot ulceration (DFU) (Mi, 2007), The DFU ABM incorporates the behavior of several cell types and mediators involved in the pathophysiology of DFU, as well as a variable that represents damaged tissue. The model is intended to represent the dynamics of the interrelated inflammation/wound healing process. The tissue damage variable serves both as a mechanism by which inflammation becomes self-sustaining (through a cycle of inflammation → tissue damage → inflammation), as well as a means of quantifying injury/healing (i.e. an outcome measure).
Figure 2.
UML diagram showing connections between main SPARK model components
Space is analogous to the physical space, and provides a context within which the model evolves. The DFU ABM contains spaces representing tissue and blood.
Data Layers provide a means of tracking multiple variables within the same model. In the DFU model, we use separate data layers to track the amounts of the inflammatory cytokines tumor necrosis factor-α (TNF-α) and transforming growth factor-β1 (TGF-β1) in both space and time.
Agents can move, perform functions, interact with each other, and also interact with the space they occupy. In the DFU ABM, classes of agents correspond to different cell types involved in inflammation and wound healing. Each agent has its own set of behaviors and rules of action. For example, inflammatory cells (macrophages and neutrophils) are chemo-attracted and activated by platelets.
Links can be used to connect two or more agents together with various strengths and distances. Not all biomedical systems can be modeled using a number of independent agents. In some systems, agents are expected to link together. For instance, in normal situation, platelets in the blood can be represented as separate agents in a SPARK model; once these platelets are activated, however, they would change their shape and tightly bind together and also to the injured tissue. In this case, we need a way to build the connection among these agents.
The Observer contains information about spaces and all agents in the model and also controls the simulation process.
Simulation Process
A SPARK model consists of agents that have some predefined behavioral rules, and a global description of the model itself: classes of agents in the model, initialization process, global variables. Generally, the simulation proceeds as follows (see Figure 3): first, the model is initialized. Then, the simulation loop begins. A global function, defined in a model class, is called at the beginning of each simulation step. Next, all agents are asked to perform their steps (execute their behavior function). After that, a global function for finalizing a step is called. The simulation ends when a predefined number of simulation steps is achieved, or when some user-defined conditions are met.
Figure 3.
SPARK model and simulation process
Agents
Agents are the central components of a SPARK model; the modeling process consists of creating agents and programming their behavior. Each agent can be characterized by two main features: its class (like inflammatory cell class) and its internal state. The internal state of an agent is the collection of variables and their values, which completely describe the agent. Agents of the same class have identical rules (the same functionality). In keeping with object-oriented specifications, classes of agents form a hierarchical structure, with the base class called, simply, “Agent”. The class is the first thing that is defined for any agent. Many operations in SPARK work directly with “Agent” classes. For example, there are functions that return all agents of a specific class possessing some special properties (e.g., occupying some location in a space). An important feature of an agent class is that all agents of the same class can be scheduled to act at specific time points (see the description of the Observer for more details).
The behavior of each agent is specified in the “step” function defined for any agent. This function is the same for all agents of the same class. The only parameter of the “step” function is the information about time passed from the beginning of a simulation.
Another important function defined for each agent is the function “die”. It kills the agent and removes it from a simulation. The exact behavior of that function depends on the current execution mode and is described later.
Agents can change their own state variables. If an agent wants to change some variables of another agent, it can do so directly, but this is not recommended. If an agent changes variables of another agent, it can create inconsistencies in a parallelized environment. Agents can change states of other agents indirectly. For example, an agent can change some value in a data layer or a global variable and this, in turn, will change the state of other agents. Agents may read the state of another agent, though care should be taken in the case of parallelized simulation, since several agents that read the same variable of another agent during a simulation step can get distinct values depending on the time when the variable was read.
Links
Links are special agents that can connect two other agents, so that each agent knows to which other agents it is connected. Links are agents, so they have the “step” function and they have specific behavioral rules. An important class of links is the “SpaceLink” which can be used to connect SpaceAgents (which are described later) in a space. SpaceLinks have a special function that returns its length, i.e., the distance between two connected agents.
One particular example of a link is a model for a physical spring that connects two agents. In this case, the behavior of a link (spring) is the application of forces to connected agents if the length of the link (spring) has been changed. For instance, in vascular tissues, smooth muscle cells are normally attached to each other, which can be simulated by these physical springs. We can adjust the spring coefficient and the rest length of the spring to fit the desired behavior of smooth muscle cells. When there are forces applied to these smooth muscle cells, these springs may change their lengths and also generate forces against the original force. In this case, we would be able to see the dynamic changes of smooth muscle cells under internal and external forces.
Space
A space is a structure which groups SPARK agents based on their positions. Each SPARK model may contain several spaces with different properties; however, one “base” space needs to be defined as the default space for that model. A space facilitates interactions between agents, and provides additional organization and structure for their interactions. It is not required for an agent to exist in a space; it is possible to define agents that have no spatial properties. Examples of such agents include those that modify values in a data layer at each simulation step, or those that collect some statistics.
Agents that can exist in a space are derived from a SPARK class called “SpaceAgent”. All SpaceAgents can freely move in a space where they are located, and they can change their size and shape. When a new SpaceAgent is created, it is automatically put into the default space of a model. Any SpaceAgent should be located in only one space at one time step, but they can move from one space to another.
Technically, SpaceAgents are not directly contained inside a space. Instead, each Space-Agent contains a variable of type “SpaceNode”, which associates an agent and a space. In other words, SpaceAgents are attached to SpaceNodes, and SpaceNodes are entities that exist in a space. A SpaceNode contains all information about “physical” appearance of an agent in a space, such as its coordinates, size and shape. An agent can be thought of as a brain that controls a body (SpaceNode). This approach allows changing SpaceNodes for an agent dynamically. For instance, all SpaceNodes have a fixed shape, and if an agent wants to change its shape, then it is enough to create a new SpaceNode that has a desired shape and attach an agent to this new node. There are circular SpaceNodes, square SpaceNodes; this is done for efficient collision detection algorithms.
Each space has some topological structure. It is not required to have a Euclidean coordinate system in a space, but in the current version of SPARK only Euclidean spaces are available. There are two main types of spaces in SPARK: a continuous space and a grid (discrete) space. Each type can be either two-dimensional or three-dimensional. The basic topologies of these spaces are rectangular, cylindrical, or toroidal. A continuous space has the usual Euclidean coordinate system. In a grid (discrete) space, some operations have special meaning. A grid space is divided into cells - squares in a two-dimensional space, cubes in a three-dimensional space - and all agents in the same cell are considered to be at the same location on the grid.
There are several operations available in a space itself. One can get all agents at a specific location, get all agents of a given class at a specific location, or all agents that overlap with another specified agent type. Using space allows for more efficient operation than iteration over all agents. Other operations in space include getting space size and space topology.
Data Layers
Data layers are used to define a scalar field in a space. A data layer associates a numerical value with each point of a space, making it easy to work with these values and to perform operations on them. Data layers can be implemented in different ways. The simplest way is to define a grid structure in the bounded space. Each cell of the grid keeps some numerical value, and all points in the space inside this cell are associated with this value. Computations involving grids can be performed quite efficiently since each grid stores numeric values in a rectangular table.
The data layers can be used to represent the location specific values which can have smaller scale than agents. For example, if agents represent cells and a space represents a tissue then data layers represent chemical compounds in the tissue. In SPARK, data layers can be of different scale. Some data layers can be coarse, other can be fine. Each data layer is associated with one space for which the data layer is defined. Each space contains a complete list of all data layers inside it. SpaceAgents can read and change values of any data layer at any position of a space. In particular, it is common to work with data layers at the location of an agent itself. There are special operations defined for data layers which allow reading and modifying values at the location of a specific agent. Reading and changing values at a specific point of a data layer are local operations for a data layer. The most important global operations defined for data layers are diffusion and evaporation. There are two implementations of a diffusion operation in SPARK. One is a simple diffusion (similar to NetLogo), which diffuses a value of a data layer cell to all adjacent neighbors. Another version of a diffusion operation is an improved version of a simple diffusion that allows diffusion to all neighbors at a specific distance.
Observer
The Observer has two main functions: it is the main container for all other SPARK components and it schedules and executes actions of other components. Agents and spaces are contained directly in the Observer.
Basic functions of the Observer allow retrieval of information about created agents (for example, a number of agents of a given class) and sets of agents. When a new agent is created it is automatically added to the Observer. When an agent dies, it is removed from the Observer. The Observer also can be used to get information about spaces defined in a model; data layers are stored in spaces, so to get information about a data layer, it is required to get space first, and then retrieve a data layer.
There are two main modes in which the Observer executes actions of agents. The first mode is called the serial mode. In this mode, agents carry out their steps one after another and all actions of agents have an immediate effect. For example, if an agent hatches another agent, then the hatched agent is immediately added to the set of existing agents and all other agents are able to see it. The second operation mode is called the concurrent mode. In this mode, many actions of agents are postponed until the end of a simulation step, and executed only after all agents have made their steps. For instance, if the Observer runs in the concurrent mode, then a newly created agent will not be added to the set of agents immediately. It will be added only after all agents finish their steps. The main idea of the concurrent mode is to make the order of agents’ actions irrelevant. This mode is particularly useful for implementation of a parallelized version of SPARK because it prevents synchronization problems. In many cases no changes are required for a serial model to be able to run in the concurrent mode.
The Observer also schedules agent actions. The scheduler works at the level of agent classes, so agents of the same class are scheduled in the same way. The basic operation of the scheduler is to specify at which simulation steps agents of a particular class will act. For example, if some agents can act at each simulation step, while others only each third step, a convenient way to schedule actions is by using the notion of simulation time. In this case each simulation step has a time value associated with it; therefore each simulation step represents the smallest unit of time for the simulation. For each agent class, it is possible to define the time interval between actions of agents of that class. For instance, there could be agents who act after two time units and agents who act after one and half time units (fractional time is also possible). Then, a simulation will proceed as follows: nobody will act during the first simulation step, agents of the second type (one and half time units) will act during the second simulation step, agents of the first type will act during the third simulation step, and after them agents of the second type will also act (three time units passed), etc.
Besides time, agents are assigned priorities. If there are several agents acting at the same time point, priority tells the Observer which agent will act first. If no priority is specified explicitly, or if two agents have the same priority, then lexicographic order on agent class names determines the order of agents’ actions.
THE SPARK PROGRAMMING LANGUAGE (SPARK-PL)
The SPARK programming language (SPARK-PL) was developed to simplify the process of model creation by researchers not experienced in computer programming. Its syntax is derived primarily from the Logo programming language (the inspiration for NetLogo language) and the Java programming language. All models written in SPARK-PL are translated into Java source code first, and then a Java compiler is used to produce the byte code that can be executed by the SPARK simulation engine. Because of this two-step process, it is convenient to extend SPARK-PL with native Java constructions. As a result, SPARK-PL harnesses the full power of the Java language for ABM development. The main features of this language include object-oriented constructions, a static type system with type inference, close ties with SPARK, as well as the concise and expressive syntax of the Logo language.
SPARK-PL includes several object-oriented concepts such as classes and inheritance similar to mainstream general purpose programming languages (such as C++, Java, C#). These hierarchical properties allow for greater ease of organization and scheduling code blocks, which facilitates the development of complex, large-scale models. The source code of each model in SPARK-PL can be separated into several files, and can also be easily organized in a hierarchical format.
SPARK-PL static type system eliminates many programmers’ errors concerning an incorrect use of variables of incompatible type. Type inference feature of the SPARK Language makes it possible to implement models in a way similar to Logo-like languages, such as Netlogo, in which dynamic type system is used. With type inference, it is not required to provide an explicit type for every new variable. The translator is capable of finding the correct type of many variables automatically by looking at the expressions. Another advantage of static type system is that it leads to efficient and fast code, because it is not required to check type consistency in runtime.
Table 1 provides a side-by-side comparison between SPARK-PL and NetLogo language by implementing one simple model using both languages. It is clear to see that the SPARK-PL code has roughly the same length as the NetLogo code and that the syntax in SPARK-PL is both object-oriented and intuitive.
Table 1.
A comparison of SPARK-PL (left) with NetLogo (right)
| agent Agent: SpaceAgent | turtles-own [ life ] |
| var life: number | to setup |
| to create | ca |
| radius = 0.3 | crt 1000 |
| set-random-position | [ |
| life = 20 + random 100 | setxy random-xcor random-ycor |
| color = red | set life (random 100) + 20 |
| end | set color red |
| ] | |
| to step [tick] | end |
| if life < 10 and random 100 < 10 | to go |
| [ | ask turtles |
| ask hatch-one Agent 1 | [ |
| [ | if life < 10 and random 100 < 10 |
| if myself.color == red | [ |
| [ | let c color |
| color = green | hatch 1 |
| ] | [ |
| ] | if c = red |
| ] | [ |
| life -= 1 | set color green |
| if life < 0 [ die ] | set life (random 100) + 20 |
| end | ] |
| ] | |
| model Model | ] |
| space StandardSpace −20 20 −20 20 | set life life - 1 |
| true true | if life < 0 [ die ] |
| ] | |
| to setup | |
| var agentsNumber = 1000 | tick |
| create Agent agentsNumber | end |
| end |
Further information about SPARK-PL can be found in the documentation section of the SPARK website www.pitt.edu/~cirm/spark/documentation.html
RUNNING A SPARK MODEL
Figure 4 depicts the user interface of the DFU ABM, which is used for running simulations. This interface is comprised of a number of windows, which can be customized by modelers according to the specific information they want to visualize during simulations. The main window [A], labeled “Main Frame”, is a visual representation of a running simulation. To monitor the changes of the variable recorded in each data layer, additional windows can be activated for visualization [B]. The figure shows the simulated concentrations of TNF-α and TGF-β1, respectively, in the DFU ABM. The visualization parameters of any window can be modified to better display selected data layers or agents at any time, even while the simulation is running [G]. Graphical appearance of each data layer can also be changed individually [F].
Figure 4.
SPARK user interface
Some variables in a model are adjustable parameters: their values may be changed via a convenient slider bar [D]. Modelers may also plot values of specified model variables in chart windows [C].
Output data from each simulation can be saved manually by clicking the “save” button in the dataset window [E], or automatically by employing a batch mode. In the latter case, the modeler specifies the number of steps in each run, the number of runs, and the name of output data files. Each simulation will run the specified number of steps and the results obtained in each simulation will be saved in files with given file names. This feature is very useful during the stage of parameter calibration in model development.
RESULTS AND DISCUSSION
Features of SPARK
The features of SPARK reflect the goal of developing a platform specialized to the needs of systems biology ABMs, while being lightweight, extensible, and computationally efficient. These characteristics are listed below:
SPARK-PL is a simple and convenient programming language. SPARK-PL is a Logo-like language with additional features and syntax similar to modern objected oriented languages such as Java.
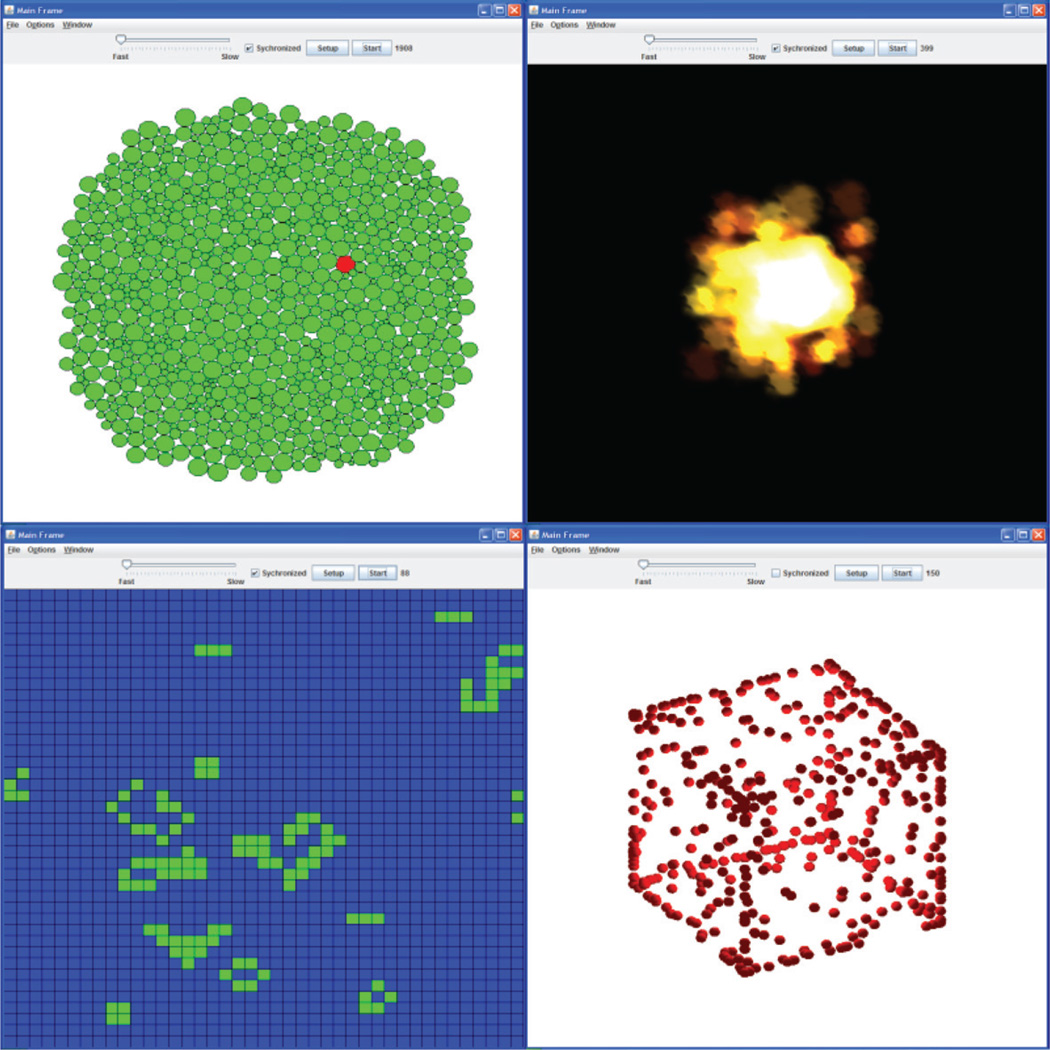
SPARK allows multiple types of spaces. SPARK ABMs can be implemented using various simulation spaces, such as discrete space (Figure 5, Panel C), continuous two-dimensional space, and continuous three-dimensional space (Figure 5, Panel D) SPARK also provides a convenient method by which modelers can create and visualize multiple spaces for the same simulation. This feature allows for the synthesis of agent behavior across multiple scales of space.
SPARK allows varied agent sizes and shapes. Agents can be created with sizes proportional to their real-world physical sizes (Figure 5, Panel A). This feature allows for the reproduction of the differential relative sizes of biological objects, and may enhance the fidelity of representation of a particular system.
SPARK allows data layers at multiple resolutions. SPARK facilitates the implementation of multiple levels of granularity, as some grids of data layers can be very fine and others can be quite coarse depending on the needs of the particular model. It is computationally efficient to use coarse grids, while fine grids may capture subtle but potentially significant differences.
SPARK utilizes advanced graphics. The OpenGL library is a standard library for generating 2D and 3D graphics. SPARK applies the OpenGL library to create many realistic graphical effects (Figure 5, Panel B). For instance, modelers may use a real picture or cartoon of a cell to represent the cell agent in the model. This feature may appeal to those wanting to use ABM’s for educational purposes.
Figure 5.
Examples of SPARK models
SPARK Performance Test
For most biomedical ABMs, simulations consist of the computation associated with the execution of agent rules and operations within the data layer. SPARK was evaluated with the following actions: 1) simple cell movement and proliferation; 2) chemical diffusion; and 3) a full biomedical ABM of an Innate Immune Response model initially implemented by An (2004) in NetLogo (An 2004). For comparison purposes, the same actions were run in NetLogo. As shown in Table 2, for the selected tasks, SPARK simulations generally require approximately half the time that it takes NetLogo to complete the same task. The comparison becomes even more favorable when the simulations are run without visualizations, a setting in which SPARK can run up to three times as fast as NetLogo. The tasks that were chosen for testing spanned a range of model complexity, demonstrating that SPARK can execute basic computational tasks with a high degree of efficiency even in complex tasks.
Table 2.
SPARK performance test
| Model 1 | Time with visualization (second) | Time without visualization (second) |
| SPARK | 6.7 | 2.9 |
| NetLogo | 14.5 | 5.3 |
| Model 2 | Time with visualization (second) | Time without visualization (second) |
| SPARK | 17.2 | 3.7 |
| NetLogo | 25 | 10.5 |
| Model 3 | Time with visualization (second) | Time without visualization (second) |
| SPARK | 43 | 23.5 |
| NetLogo | 66 | 51 |
CONCLUSION
We have created a framework, SPARK, for the development of biomedical ABMs. The goal of this software is to provide an efficient, flexible, and powerful tool for researchers in systems biology. We are currently utilizing the SPARK framework to develop multi-scale complex inflammation models in diverse settings such as cancer, viral infection, and spinal cord injury. We are also working to improve the interface and develop a parallelized version of SPARK that can automatically parallelize user-developed models to take advantage of fast-growing computational power.
ACKNOWLEDGMENT
AS acknowledges support from NIDRR H133E070024. GA acknowledges support from NIDRR H133E070024, NSF 0830-370-V601, IBM Shared University Research Award and NIH P50-GM-53789. YV and QM acknowledge support from NIDRR H133E070024, NIH R01-DC-008290, IBM Shared University Research Award, NIH P50-GM-53789, and a grant from the Commonwealth of Pennsylvania. YV also acknowledges support from NIH R33-HL-089082 and NIH R01-HL080926. LZ and QM acknowledge support from NSF IIS-0938393.
Contributor Information
Alexey Solovyev, University of Pittsburgh, USA.
Maxim Mikheev, University of Pittsburgh, USA.
Leming Zhou, University of Pittsburgh, USA.
Joyeeta Dutta-Moscato, University of Pittsburgh, USA.
Cordelia Ziraldo, University of Pittsburgh, USA.
Gary An, University of Chicago and University of Pittsburgh, USA.
Yoram Vodovotz, University of Pittsburgh, USA.
Qi Mi, University of Pittsburgh, USA.
REFERENCES
- An G. Agent-based computer simulation and sirs: building a bridge between basic science and clinical trials. Shock (Augusta, Ga.) 2001;16:266–273. doi: 10.1097/00024382-200116040-00006. [DOI] [PubMed] [Google Scholar]
- An G. Innate Immune Response. NetLogo User Community Models; 2004. [Google Scholar]
- An G. In-silico experiments of existing and hypothetical cytokine-directed clinical trials using agent based modeling. Critical Care Medicine. 2004;32(10):2050–2060. doi: 10.1097/01.ccm.0000139707.13729.7d. [DOI] [PubMed] [Google Scholar]
- An G, Faeder J, Vodovotz Y. Translational systems biology: introduction of an engineering approach to the pathophysiology of the burn patient. Journal of Burn Care & Research; Official Publication of the American Burn Association. 2008;29:277–285. doi: 10.1097/BCR.0b013e31816677c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Mi Q, Dutta-Moscato J, Vodovotz Y. Agent-based Models in Translational Systems Biology. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2009;1(2):159–171. doi: 10.1002/wsbm.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AM, Lawrence MB, Shang H, Katz AJ, Peirce SM. Agent-based model of therapeutic adipose-derived stromal cell trafficking during ischemia predicts ability to roll on P-selectin. PLoS Computational Biology. 2009;5:e1000294. doi: 10.1371/journal.pcbi.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankes SC. Agent-based modeling: a revolution? Proceedings of the Natl Acad Sci USA. 2002:7199–7200. doi: 10.1073/pnas.072081299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonabeau E. Agent-based modeling: methods and techniques for simulating human systems. Proceedings of the Natl Acad Sci. 2002:7280–7287. doi: 10.1073/pnas.082080899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm V, Revilla E, Berger U, Jeltsch F, Mooij WM, Railsback SF. Pattern-oriented modeling of agent-based complex systems: lessons from ecology. Science. 2005;310:987–991. doi: 10.1126/science.1116681. [DOI] [PubMed] [Google Scholar]
- Inchiosa ME, Parker MT. Overcoming design and development challenges in agent-based modeling using ASCAPE. Proceeding of the Natl Acad Sci. 2002:7304–7308. doi: 10.1073/pnas.082081199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li NY, Verdolini K, Clermont G, Mi Q, Rubinstein EN, Hebda PA, Vodovotz Y. A patient-specific in silico model of inflammation and healing tested in acute vocal fold injury. PLoS ONE. 2008;3:e2789. doi: 10.1371/journal.pone.0002789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke S, Balan GC, Panait L, Claudio CR, Paus S. MASON: A Java Multi-Agent. 2003 [Google Scholar]
- Mi Q, Riviere B, Clermont G, Steed DL, Vodovotz Y. Agent-based model of inflammation and wound healing: insights into diabetic foot ulcer pathology and the role of transforming growth factor-beta1. Wound Repair and Regeneration. 2007;15:671–682. doi: 10.1111/j.1524-475X.2007.00271.x. [DOI] [PubMed] [Google Scholar]
- Vodovotz Y, Csete M, Bartels J, Chang S, An G. Translational systems biology of inflammation. PLoS Computational Biology. 2008;4:e1000014. doi: 10.1371/journal.pcbi.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]