Abstract
Lipoxygenases, important enzymes in inflammation, can regulate their substrate specificity by allosteric interactions with its own hydroperoxide products. In the current work, addition of both 13-(S) hydroxy-9Z,11E-octadecadienoic acid (13-(S)-HODE) and 13-(S)-hydroperoxy-6Z,9Z,11E-octadecatrienoic acid (13-(S)-HOTrE) to human epithelial 15-lipoxygenase-2 (15-LOX-2) increases the kcat/KM substrate specificity ratio of arachidonic acid (AA) and (γ)-linolenic acid (GLA) by 4-fold. 13-(S)-HODE achieves this change by activating kcat/KM AA but inhibiting kcat/KM GLA, which indicates that the allosteric structural changes at the active site discriminates between the length and unsaturation differences of AA and GLA to achieve opposite kinetics effects. The substrate specificity ratio is further increased, 11-fold total, by increasing pH, suggesting mechanistic differences between the pH and allosteric effects. Interestingly, the loss of the PLAT domain affects substrate specificity, but does not eliminate the allosteric properties of 15-LOX-2, indicating that the allosteric site is located in the catalytic domain. However, the removal of the PLAT domain does change the magnitude of the allosteric effect. These data suggest that the PLAT domain moderates the communication pathway between the allosteric and catalytic sites, thus affecting substrate specificity. These results are discussed in the context of protein dimerization and other structural changes.
Keywords: lipoxygenase, arachidonic acid, pH effect, allosteric effect, gamma linolenic acid, substrate specificity
Introduction
Lipoxygenases (LOX) represent a class of non-heme iron containing enzymes which catalyze the stereo-specific peroxidation of polyunsaturated fatty acids (PUFAs) containing at least one 1,4-cis-pentadiene moiety.(1–4) There are three main types of human LOXs, 5-LOX, 12-LOX and 15-LOX, that are named according to their positional specificity with arachidonic acid (AA).(5) The crystal structures of two mammalian LOXs indicate a single polypeptide chain folding into a two-domain structure.(6–8) The C-terminal catalytic domain consists of primarily α–helices and possesses the catalytic site, with the as-isolated non-heme ferrous ion, which is coordinated by water and endogenous ligands. The smaller N-terminal domain consists of primarily β–sheets and resembles the C2-domain of human lipases, known as PLAT domains (Polycystin-1, Lipoxygenase, α-Toxin),(9) and has been implicated in membrane binding (10, 11) and auto-inactivation.(12, 13)
Humans ingest/synthesize over 7 important fatty acid which are substrates to the 6 human LOX isozymes, as well as to cyclooxygenase (COX) and cytochrome P450.(14) This complex web of interactions translates into an extensive list of possible LOX/COX/P450 products in the body, of which only a fraction are fully understood from a biological perspective. The LOX endogenous products differ in carbon chain length, number/position of unsaturation points, and degree of oxidation, depending on the fatty acid substrate and LOX isozyme. Many of these LOX products are biologically active compounds that not only mediate cellular pathways directly but also act as precursors for potent chemical mediators, such as leukotrienes,(15, 16) resolvins,(17) lipoxins (18–22) and protectins.(17, 18) Regulation of the production of these LOX products involves a variety of cellular mechanisms,(18) with allostery emerging as one of the more intriguing pathways, due to its effect on substrate specificity.(23, 24)
Allostery is the means by which cells communicate the effects of the physical interactions of their machinery, such as proteins, nucleotides and small molecules. Since allostery is fundamentally thermodynamic in nature, the communication across a particular protein can be mediated by both conformational and dynamic fluctuations.(25) Typically, an allosteric effector regulates the activity of an enzyme, as is the case with phosphofructokinase-1 (PFK-1), where ATP inhibits activity and AMP increases activity.(26–28) However, in a few cases, allostery can regulate substrate specificity. The most notable system is that of ribonucleotide reductase (RNR), which produces deoxynucleotides from ribonucleotides.(29–31) RNR must maintain proportional concentrations of the four deoxynucleotides in the cell and utilizes allostery to do this. RNR regulates its enzyme activity for a particular ribonucleotide by binding a deoxynucleotide, such as GDP and dTTP, respectively. The binding of the deoxynucleotide effector molecule to the specificity site alters the structure of a shared loop with the catalytic site, thus altering RNR’s substrate specificity from 2- to 10-fold.(31)
5-LOX was the first LOX isozyme to be shown to possess allostery,(32–35) with calcium and ATP activating its Vmax/KM by over 25-fold.(35) The substrate specificity was also affected by calcium and ATP, with the Vmax/KM ratio of AA/EPA increasing 1.8-fold.(35) With respect to the human 15-LOXs, our laboratory first observed that the kinetic isotope effect of 15-LOX-1 increased with the addition of oleyl sulfate, suggesting a ternary complex and allostery.(36) It was then observed that LOX products changed the substrate specificity for both 15-LOX-1 and 15-LOX-2.(23, 24) For 15-LOX-1, it was shown that 13-(S)-HODE, the product of 15-LOX with linoleic acid (LA), increased the kcat/KM AA/LA substrate specificity ratio.(23) 13-(S)-HODE also affected the kinetics of 15-LOX-2, however the effect was more complicated. 13-(S)-HODE appeared to have a tighter affinity to 15-LOX-2 than 15-LOX-1 and the allosteric effect was pH dependent. It was observed that pH increased the kcat/KM AA/LA substrate specificity ratio of 15-LOX-2, yielding a pH titration curve (pKa of 7.7 ± 0.1), which suggested the possible involvement of a histidine residue.(23, 24) Based on this data, homology modeling indicated that the possible location of the allosteric site could be between the catalytic domain and the PLAT domain, with His627 at the bottom of the cleft to interact with the carboxylic acid of 13-(S)-HPODE.(24). As mentioned above, the PLAT domain of LOX is important for membrane association, since removal of the PLAT domain from various LOX orthologues displayed a reduction in liposome affinity.(12, 13, 37–39) The loss of the PLAT domain also lowered activity for almost all the LOX isozymes investigated, however, the PLAT domain was not implicated in affecting substrate specificity. Given the pH dependency of 15-LOX-2’s substrate specificity and the importance of allostery due to the availability of various LOX substrates in the cellular milieu, the current work investigated the kinetic nature of 15-LOX-2’s allostery and its pH dependence more thoroughly. In addition, the location of the allosteric site was further probed by removal of the PLAT domain and its effects on the catalytic and allosteric response of 15-LOX-2 probed.
Materials and Methods
Materials
All the commercial fatty acids were purchased from Nu Chek Prep, Inc. (MN, USA) and Sigma-Aldrich Chemical Company). The fatty acids were further re-purified using a Higgins HAISIL column (5 μm, 250 X 10 mm) C-18 column. An isocratic elution of 85% A (99.9% methanol and 0.1% acetic acid): 15% B (99.9% water and 0.1% acetic acid) was used to purify all the fatty acids. Post purification, the fatty acids were stored at −80 °C for a maximum of 6 months. Different lipoxygenase products were generated by reacting fatty acids with the appropriate LOX isozyme, such as 13-(S)-HPODE from soybean LOX-1 with LA and 13-(S)-HPOTrE (γ) from 15-LOX-2 with (γ)-linolenic acid (GLA)). Briefly, the protocol involved reacting 50 μM of substrate to completion with the enzyme in 500 mL reaction buffer. A small sample from the big reaction was monitored on the UV Spectrometer till complete turnover. The products were then extracted using dichloromethane, reduced with trimethylphosphite, evaporated to dryness and reconstituted in methanol. The products were HPLC purified using an isocratic elution of 75% A (99.9% methanol and 0.1% acetic acid): 25% B (99.9% water and 0.1% acetic acid). The products were tested for their purity using LC-MS/MS and were found to have >98% purity. All other chemicals were of high quality and used without further purification.
Overexpression and purification of 15-LOX-2 and 15-LOX-2NoPLAT
Purification of wild-type 15-LOX-2 (15-LOX-2) and 15-LOX-2 PLAT domain truncation mutant (15-LOX-2NoPLAT) was carried out using a construct encoding a fusion protein consisting of an N-terminal His8-tag fused to maltose binding protein (MBP). The parent plasmid used in these studies, pVP68K, is available from the Protein Structure Initiative Materials Repository, http://www.psimr.asu.edu. The 15-LOX-2 fusion and the 15-LOX-2NoPLAT fusion was expressed using the pVP68K-102188 and pVP68K-Ndelta188 plasmids respectively in Escherichia coli (E. coli) BL21 (DE3), where the first 118 amino acids were removed for the truncated enzyme, 15-LOX-2NoPLAT. For expression of the proteins, the host cells were grown to 0.6 OD at 37° C, induced by dropping the temperature to 20° C and grown overnight (16 h). The cells were harvested in 2 L fractions at a velocity of 5,000 g, then snap frozen in liquid nitrogen. The cell pellets were re-suspended in buffer A (25 mM Hepes, pH 8, 150 mM NaCl), and lysed using a Power Laboratory Press. The cellular lysate was centrifuged at 40,000 g for 25 min, and the supernatant was loaded onto an NTA-Ni affinity column. The column was washed with 15 mM imidazole in buffer A, followed by elution with 250 mM imidazole in buffer A (no NaCl). For 15-LOX-2, the fractions were collected, pooled together and then dialyzed in 25 mM Hepes, pH 7.5, containing 150 mM NaCl, followed by overnight treatment with His6-TEV protease at 4° C. For 15-LOX-2NoPLAT, the pooled fractions were dialyzed for 1 h against 25 mM Hepes, pH 7.5. It was then removed from the dialysis bag and cleaved with His6-TEV protease for 1.5 h, followed by second dialysis for 1 h in 25 mM Hepes, pH 6.5, containing 150 mM NaCl. The use of TEV protease was as previously described.(40) The proteolyzed 15-LOX-2 sample was applied to an NTA-Co2+ column, and eluted in buffer A containing 15 mM imidazole. However, the proteolyzed 15-LOX-2NoPLAT protein required a different procedure, which entailed an NTA-Ni2+ column and then an amylose column to bind residual MBP-tagged proteins. Both of these procedures were found to yield protein with greater than 90% purity. It should be noted that after TEV cleavage, an un-natural serine is left on the N-terminus of both 15-LOX-2 proteins. The resulting non-tagged 15-LOX-2 and 15-LOX-2NoPLAT were concentrated by ultrafiltration (30 kDa molecular mass cutoff), combined with glycerol to 20% (v/v) and then snap frozen under liquid nitrogen. Overexpression and purification of soybean LOX-1 followed a protocol outlined previously.(41) The enzyme purity for all the isozymes was evaluated by SDS-PAGE. Iron content of the LOX enzymes was determined with a Thermo Element XR inductively coupled plasma mass spectrometer (ICP-MS), using Scandium (EDTA) or Cobalt (EDTA) as internal standards. Iron concentrations were compared to standard iron solutions. All the kinetic data was normalized to the iron content. The protein concentration was determined using the Bradford Assay, with Bovine Serum Albumin (BSA) as a protein standard. Briefly, Bradford protein dye reagent was diluted 1:5 using deionized water. Different solutions of BSA in deionized water were prepared, ranging from 0 mg/mL to 1 mg/mL (linear range of the assay for BSA). Diluted Bradford reagent and BSA stocks were mixed in 50:1 ratio, vortex and incubated for 5 minutes. Similarly, protein samples with diluted Bradford reagent were made in duplicate. All the samples were then spun down and the absorbance was recorded at 595 nm on Perkin Elmer Lambda 40 instrument. The concentration of the proteins was extrapolated from the standard curve of BSA.
Effect of pH using the competitive substrate capture method
The competitive substrate capture method experiments were performed on the AA-LA substrate pair at pH 7.5 and pH 8.5 with 15-LOX-2 (from the SF9 and E. coli expression system) and 15-LOX-2NoPLAT using the previously described protocol.(23) Briefly, the AA-LA mixture was prepared with a molar ratio of 1:1. The reaction was initiated by adding (all normalized to the Fe content) ~20 nM 15-LOX-2 (E. coli), ~50nM 15-LOX-2 (SF9) and ~90 nM 15-LOX-2NoPLAT to a 30 mL reaction cuvette (25 mM Hepes, pH 7.5 (or pH 8.5)) at room temperature, with a total substrate concentration of 1 μM. The reaction was quenched at 5% substrate turnover using 1% glacial acetic acid. The reaction mixture was then extracted with DCM and evaporated to dryness under vacuum. The dried sample was reconstituted in 100 μL methanol, centrifuged and stored at −20° C. The substrate capture ratio (kcat/KM) of AA and LA was determined from the peak areas and normalized based on the ratio of the substrates present in the initial mixture.
Effect of pH on the steady-state substrate specificity kinetics
Lipoxygenase activity was assayed spectro-photometrically (Perkin Elmer lambda 40) by monitoring an increase in absorbance at 234 nm due to formation of a conjugated diene product (ε = 25 000 M−1 cm−1). The reaction was carried out in 25 mM Hepes buffer (pH 7.5 or pH 8.5), constant ionic strength of 200 mM, room temperature, a final reaction volume of 2 mL and substrate concentrations ranging from 1 to 20 μM. Assays were initiated by adding 45–55 nM 15-LOX-2 and 100–200 nM 15-LOX-2NoPLAT (concentration normalized to iron content) and were constantly stirred using a magnetic stir bar. Assays for the different substrates were performed on the same day to allow a direct comparison and minimize error between AA and GLA data. The substrate concentrations were quantitatively determined by allowing the enzymatic reaction to go to completion. Initial rates were recorded at each substrate concentration and fitted to the Michaelis-Menten equation using KaleidaGraph (Synergy) to determine kcat and KM values.
Effect of LOX products on steady-state substrate specificity kinetics
GLA steady-state kinetics experiments were performed in the presence of two different lipoxygenase products, 13-(S)-HODE and 13-(S)-HOTrE(γ) at pH 7.5 and pH 8.5 with product concentrations ranging from 0, 5, 15 and 30 μM. For the AA kinetics, initially the effect of both the products was tested with and without 15 μM 13-(S)-HODE and 13-(S)-HOTrE (γ) at both pH values. Further, the effect of 13-(S)-HODE on AA kinetics was investigated in detail at lower concentrations of 13-(S)-HODE ranging from 0, 1, 3 and 5 μM at pH 7.5. Enzymatic assays were conducted using the same conditions as mentioned above (25 mM Hepes, pH 7.5 or pH 8.5, constant ionic strength, 200 mM, and at room temperature). The enzyme was incubated with product first (15 sec), to facilitate their interaction, followed by an initiation of the reaction by adding the substrate. The plots of KM and kcat/KM versus product concentration were fit to the hyperbolic inhibition equations (vide infra).
CD half-transition temperature measurements
Protein unfolding was monitored by CD measurements at 220 nm using an AVIV CD Spectrometer model 62DS in a 0.1 cm quartz cuvette. Both 15-LOX-2 and 15-LOX-2NoPLAT samples consisted of 5 μM enzyme, 200 μL, 25 mM Hepes, 150 mM NaCl and pH 7.5. Measurements were made between 10–80° C, at intervals of 5° C, except close to T50, where the intervals were decreased to 2° C. Each temperature recording was after a 10 min equilibration period.
Results and Discussion
Overexpression and purification of 15-LOX-2 and 15-LOX-2NoPLAT
Methods developed for structural genomics studies of eukaryotic proteins were applied to the expression and purification of 15-LOX-2 and 15-LOX-2NoPLAT.(42) After 16 hours of low-temperature growth (20° C) and no IPTG induction, approximately 40 mgs of the His8-MBP-fusion protein per 2 liter of medium was isolated, at approximately 80% purity after the first IMAC column. After cleavage of the His8-MBP tag with TEV protease,(40) and subtractive IMAC, approximately 75% of the 15-LOX-2 and 15-LOX-2NoPLAT were recovered, giving a final yield of approximately 30 mgs of pure protein per 2 liter of culture medium. The final proteins isolated from E. coli were 90% pure as judged by SDS-PAGE, which was comparable to the purity previously achieved using the SF9 expression system.(43) However, ICP-MS data indicated that 15-LOX-2 and 15-LOX-2NoPLAT had iron content ranging from 25–45%, which is greater than our previous SF-9 15-LOX-2 preparations.(23, 24, 44) The steady-state kinetics of this E. coli preparation of 15-LOX-2 was also compared to the SF9 preparation and found to be significantly more active per active site iron (Table 1) (23). With AA as a substrate, the kcat was 2-fold greater and the kcat/KM was 4-fold greater than the SF9 preparation of 15-LOX-2.(23, 24) Therefore, the unique benefits of this expression/purification method were its high yield in E. coli, its increased metal content, and its increased activity.
Table 1.
Comparison of the kinetic parameters of 15-LOX-2 with various substrates at pH 7.5 and 8.5.a
| kcat/KM (μM−1s−1) | kcat (s−1) | |||
|---|---|---|---|---|
| pH 7.5 | pH 8.5 | pH 7.5 | pH 8.5 | |
| GLA | 0.64±0.02 | 0.37±0.03 | 1.8±0.03 | 1.1±0.02 |
| AA | 0.40±0.02 | 0.76±0.06 | 1.5±0.03 | 2.0±0.04 |
| DGLA | 0.26±0.04 | 0.49±0.08 | 1.5±0.08 | 1.8±0.1 |
| EPA | 0.72±0.03 | 1.4±0.07 | 2.1±0.02 | 2.8±0.04 |
| pH 7.5 | pH 8.5 | pH 7.5 | pH 8.5 | |
| AA/GLA | 0.63±0.04 | 2.1±0.03 | 0.83±0.02 | 1.8±0.05 |
| AA/DGLA | 1.5±0.2 | 1.6±0.3 | 0.99±0.06 | 1.1±0.07 |
| AA/EPA | 0.56±0.04 | 0.54±0.05 | 0.71±0.02 | 0.72±0.02 |
Enzymatic assays were performed in 25 mM Hepes at 22° C.
Substrate specificity determined by competitive substrate capture for 15-LOX-2
Our lab had previously reported a novel and simple method to determine the substrate specificity ratio of lipoxygenases.(23) Although kcat and KM cannot be determined for individual substrates via this method, it is still a very efficient method to compare the kcat/KM ratios of different substrate pairs directly. To confirm that the newly E. coli purified 15-LOX-2 behaves similarly to that of the previously published SF9 purified 15-LOX-2, the substrate specificity ratio for AA/LA at pH 7.5 and 8.5 was determined using the competitive substrate capture method. The kcat/KM AA/LA ratio for E. coli 15-LOX-2 was found to be 2.3 ± 0.4 at pH 7.5 and 4.9 ± 0.5 at pH 8.5, which compares well with the kcat/KM AA/LA ratio values for SF9 15-LOX-2.(24) The results indicate that E. coli 15-LOX-2 shares similar substrate specificity at two pH conditions to that of SF9 15-LOX-2 and supports them being similar enzymes.
Effect of pH on the steady-state substrate specificity kinetics of 15-LOX-2
To further investigate the pH effect on 15-LOX-2, steady-state kinetics were performed at different pH values on AA and GLA (Figure 1). While GLA is not found in the body at concentrations as high as LA, it was chosen for this investigation because it is a 20-fold better substrate than LA, which minimized error and allowed for more detailed allosteric kinetic analysis (vide infra). The kinetic data indicated that as pH increased, GLA became a poorer substrate, with both kcat GLA and kcat/KM GLA decreasing with increasing pH (Table 1). However, for AA both kcat AA and kcat/KM AA increased with increasing pH. This pH effect resulted in a 3.4-fold increase in the kcat/KM AA/GLA ratio (0.63±0.04 at pH 7.5 to 2.1±0.03 at pH 8.5) and a 2.2-fold increase in the kcat AA/GLA ratio (0.83±0.02 at pH 7.5 to 1.8±0.05 at pH 8.5). The other C20 substrates, DGLA and EPA, also became better substrates with increasing pH, with EPA having an almost 2-fold greater kcat/KM than AA and an almost 3-fold greater kcat/KM than DGLA, correlating with their differences in unsaturation. It is unlikely that this difference in kinetic rates is due to formation of micelles since the CMC of AA under these conditions is 43±3 μM as determined by isothermal titration calorimetry (MicroCal VP-ITC),(45) well above the concentrations used in these kinetic experiments. This showed that EPA and DGLA demonstrated similar pH activation to that of AA (Table 1) and indicated a distinct catalytic mechanism for both substrate capture (kcat/KM) and product release (kcat) between GLA and the other three C20 fatty acid substrates. We are currently investigating the enzymatic mechanism of GLA with solvent isotope and viscosity effects in order to determine if its rate limiting steps are comparable to those of AA.(46)
Figure 1.

Structures of AA, LA, 13-(S)-HODE, GLA and 13-(S)-HOTrE(γ).
Effect of LOX products on steady-state substrate specificity kinetics of 15-LOX-2
The allosteric effect of 13-(S)-HODE was previously demonstrated to affect the AA/LA ratio by competitive substrate capture methods,(23, 24) but attempts to investigate the allosteric effect further proved difficult, due to the high error we encountered with LA as the substrate. With the discovery that GLA was a facile substrate and with the more active E. coli 15-LOX-2 preparation, the allosteric effects of 13-(S)-HODE and 13-(S)-HOTrE(γ), were investigated with steady-state kinetics on both AA and GLA, in order to more fully understand their allosteric properties (Table 2). It was observed that 13-(S)-HODE increased the kcat/KM AA (0.40 ± 0.02 μM−1s−1 to 0.66 ± 0.07 μM−1s−1), but decreased the kcat/KM GLA (0.64 ± 0.02 μM−1s−1 to 0.29 ± 0.02 μM−1s−1). 13-(S)-HOTrE(γ) exerted a similar effect by increasing the kcat/KM AA and decreasing the kcat/KM GLA, albeit to a lesser extent (Table 2). As a result, the addition of 13-(S)-HODE and 13-(S)-HOTrE(γ) elicited a 3.7-fold and 2.2-fold increase in the kcat/KM AA/GLA ratio, respectively. These increases in ratio are of comparable direction and magnitude to the 3.3-fold pH effect on kcat/KM AA/GLA, discussed above (Table 1). Interestingly, the kcat AA/GLA ratio decreased slightly with the addition of 13-(S)-HODE (1.3-fold) and 13-(S)-HOTrE(γ) (1.1-fold), which was opposite to the pH effect, where the kcat AA/GLA ratio increased by 2.2-fold (Table 1). These data represent clear similarities and distinctions between the allosteric and pH effects with respect to kcat/KM (substrate capture) and kcat (product release).
Table 2.
Comparison of Kinetic parameters of 15-LOX-2 with AA and GLA, with and without different C18 products (pH 7.5).a
| kcat/KM (μM−1s−1) | kcat (s−1) | |||||
|---|---|---|---|---|---|---|
| AA | GLA | AA/GLA | AA | GLA | AA/GLA | |
| No Product | 0.40±0.02 | 0.64±0.02 | 0.63±0.04 | 1.5±0.03 | 1.8±0.03 | 0.83±0.02 |
| 13-(S)-HODE | 0.66±0.07 | 0.29±0.02 | 2.3±0.3 | 1.3±0.02 | 2.0±0.06 | 0.65±0.02 |
| 13-(S)-HOTrE(γ) | 0.45±0.03 | 0.32±0.02 | 1.4±0.01 | 1.4±0.05 | 1.9±0.04 | 0.73±0.01 |
Enzymatic assays were performed in 25 mM Hepes, pH 7.5, at 22° C with and without 15 μM 13-(S)-HODE and 15 μM 13-(S)-HOTrE(γ).
Combined pH and product effects on the steady-state substrate specificity kinetics of 15-LOX-2
The above steady-state kinetic data demonstrated that both pH (Table 1) and the two LOX products at pH 7.5 (Table 2) affected the kinetic parameters of GLA and AA. The ability of 13-(S)-HODE and 13-(S)-HOTrE(γ) to affect the kinetics was then investigated at pH 8.5, in order to determine if the pH and product effects were additive. As seen in Table 3, addition of 13-(S)-HODE and 13-(S)-HOTrE(γ) at pH 8.5 demonstrated the same trends in substrate specificity as at pH 7.5, with kcat/KM AA/GLA increasing and kcat AA/GLA remaining unchanged. However, the magnitude of the AA/GLA substrate specificity ratios increased significantly, indicating that the pH and product effects were partially additive. For example, the kcat/KM AA/GLA substrate specificity ratio changed from 0.63±0.04 at pH 7.5 with no product present (Table 2), to a ratio of 6.7±0.8 at pH 8.5 with 15 μM 13-(S)-HODE present (Table 3). This is an 11-fold increase in the substrate specificity ratio when both effects are taken into account and suggests the mechanisms of the pH and allosteric effects are similar but distinct.
Table 3.
Comparison of kinetic parameters of 15-LOX-2 with AA and GLA as substrates and different C18 products added (pH 8.5).a
| kcat/KM (μM−1s−1) | kcat (s−1) | |||||
|---|---|---|---|---|---|---|
| AA | GLA | AA/GLA | AA | GLA | AA/GLA | |
| No Product | 0.76±0.06 | 0.37±0.03 | 2.1±0.2 | 2.0±0.04 | 1.1±0.02 | 1.8±0.05 |
| 13-(S)-HODE | 1.4±0.1 | 0.21±0.02 | 6.7±0.8 | 1.9±0.03 | 1.1±0.02 | 1.7±0.04 |
| 13-(S)-HOTrE(γ) | 0.87±0.09 | 0.22±0.02 | 4.0±0.5 | 1.8±0.04 | 1.1±0.03 | 1.6±0.06 |
Enzymatic assays were performed in 25 mM Hepes, pH 8.5, at 22° C with and without 15 μM 13-(S)-HODE and 15 μM 13-(S)-HOTrE(γ).
Hyperbolic inhibition of 15-LOX-2 by LOX products
The overall trends for the 13-(S)-HODE and 13-(S)-HOTrE(γ) effects were comparable, however, their magnitudes were different (Table 2 and 3). This data suggested that the additional double bond at C6 for 13-(S)-HOTrE(γ) (Figure 1), did register functional allosteric differences. This exquisite selectivity of the allosteric site between 13-(S)-HODE and 13-(S)-HOTrE(γ) is consistent with our previous data which demonstrated that 12-(S)-HETE and 15-(S)-HETE do not register any allosteric effect with 15-LOX-2.(23) Given this functional allosteric difference between 13-(S)-HODE and 13-(S)-HOTrE(γ), both products were titrated against 15-LOX-2, at pH 7.5 and 8.5 with GLA as the substrate. From the data, it was observed that 15-LOX-2 exhibited a hyperbolic response to increasing amounts of 13-(S)-HOTrE(γ) with an increase in KM(app) for GLA from 2.8 μM to ~9.8 μM (Figure 2) and a decrease in kcat/KM (app) from 0.64 μM −1s−1 to ~0.28 μM −1s−1 (Figure 3). The saturation behavior of KM(app) and kcat/KM is indicative of hyperbolic inhibition (i.e. partial inhibition), as seen previously for soybean 15-LOX-1 and the inhibitor, oleyl sulfate.(36) This data indicates the presence of an allosteric binding site that affects the catalysis by changing the microscopic rate constants of 15-LOX-2, as described in Scheme 1. From Scheme 1, equations 1–4 allow for the determination of Ki, the strength of binding, α, the change in KM, β, the change in kcat and β/α, the change in kcat/KM.
Figure 2.
Effect of 13(S)-HOTrE (γ) on KM (app) of 15-LOX-2 with GLA. (pH 7.5, 25 mM Hepes, 1–20 μM GLA at each product point) The data is fit to equation 2 (Scheme 1), where KM = 2.8 μM. α and Ki were determined to be 3.5 ± 0.2 and 4.9 ± 0.7 respectively.
Figure 3.
Effect of 13(S)-HOTrE (γ) on kcat/KM (app) of 15-LOX-2 with GLA. (pH 7.5, 25 mM Hepes, 1–20 μM GLA at each product point) The data is fit to equation 3 (Scheme 1), where KM = 2.8 μM, kcat = 0.65 s−1, α = 3.5 and Ki = 4.9 μM. β was determined to be 1.2 ± 0.02.
Scheme 1.

| (1) |
| (2) |
| (3) |
| (4) |
A plot of KM(app) with the addition of 13-(S)-HOTrE(γ) at pH 7.5 (Figure 2), when fitted with equation 2, yielded an α of 3.5 ± 0.2 and a Ki of 4.9 ± 0.7 μM. The values of α and Ki were then utilized in equation 3 and fit to the kcat/KM data (Figure 3), which yielded a β of 1.2 ± 0.02. The value of β was also determined from the kcat data with equation 4, with the above values of α and Ki applied (data not shown), which yielded a β of 1.2 ± 0.05 and matched well with the β value from the kcat/KM plot. These values indicate that the kinetics show mixed hyperbolic allostery, α > 1 (K-type inhibition) and β > 1 (V-type activation),(47) with the majority of kinetic change being seen in the value of KM (α = 3.5 ± 0.2) and only a slight effect on the kcat value (β = 1.2 ± 0.02). This comparison is best observed in the β/α value, the allosteric effect on kcat/KM, which is less than 1 (β/α = 0.34) and indicates kcat/KM allosteric inhibition. The hyperbolic data suggest the formation of a catalytically active ternary complex (I•E•S) between 15-LOX-2 and 13-(S)-HOTrE(γ) and are consistent with our previous finding of an allosteric site in 15-LOX-2.(24) Similarly, graphing of steady-state kinetic data of 15-LOX-2 with 13-(S)-HOTrE(γ) at pH 8.5 gave an α of 2.0 ± 0.1 and a Ki of 6.1 ± 1 from the KM plot (Supplemental Data, Figure S1) and a β of 0.9 ± 0.04 from the kcat/KM (Supplemental Data, Figure S2). From this data it was observed that while the α and β values have decreased with pH, the Ki value for 13-(S)-HOTrE(γ) showed little pH dependence. Therefore, the pH change of the AA/GLA substrate specificity ratio was not due to a change in the affinity of 13-(S)-HOTrE(γ) at higher pH, but more likely a pH dependent structural change, which affected the allosteric communication between the catalytic and allosteric sites (α and β), but not 13-(S)-HOTrE(γ) binding.
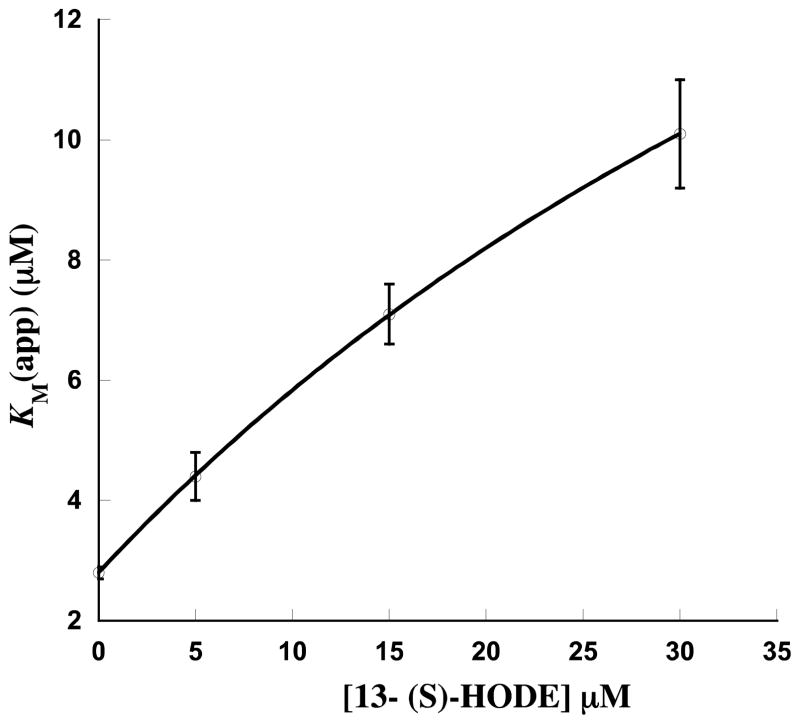
Fitting the 13-(S)-HODE data in a similar manner, showed an α value of 7.3 ± 0.06 (Ktype inhibition) and a Ki value of 9.8 ± 0.2 μM from the KM (app) plot (Figure 4). The α value was double that of the α value for 13-(S)-HOTrE(γ), while the Ki value was comparable to that of 13-(S)-HOTrE(γ), indicative of a stronger allosteric effect on KM for 13-(S)-HODE. This is supported by the lower β/α value of 0.22 for 13-(S)-HODE, as compared to that of 13-(S)-HOTrE(γ) (β/α = 0.34). The β value was determined to be 1.6 ± 0.1(Figure 5), which is V-type activation, similar to that seen for 13-(S)-HOTrE(γ) and highlights a key similarity between the two LOX products. The 13-(S)-HODE data at pH 8.5 could not be fit using hyperbolic inhibition (Scheme 1), nor a simple non-competitive model, therefore, the change of kcat/KM (app) was plotted versus 13-(S)-HODE concentration and fit with a saturation curve (Supplemental Data, Figure S3). Using this method, the KD at pH 8.5 increased to 53 ± 25 μM, considerably greater than that observed at pH 7.5 (8.6 ± 0.7 μM), indicating a lower binding affinity with increasing pH. This lower affinity could account for the smaller change in kcat/KM GLA with 13-(S)-HODE at pH 8.5 than at 7.5 and not necessarily a change in α and β, although we can not determine this directly. It should be noted that this alternative fitting method yielded a KD of 8.6 ± 0.7 μM at pH 7.5 for 13-(S)-HODE, which was comparable to the Ki of 9.8 ± 0.2 μM found by the hyperbolic inhibition fit and validated this fitting method. Interestingly, the fact that 13-(S)-HODE binding was affected by pH but the binding of 13-(S)-HOTrE(γ) was not, suggests that the structural differences between the two LOX products (i.e. degree of unsaturation) changes their allosteric binding modes.
Figure 4.
Effect of 13(S)-HODE on KM (app) of 15-LOX-2 with GLA. (pH 7.5, 25mM Hepes, 1–20 μM GLA at each product point) The data is fit to equation 2 (Scheme 1), where KM = 2.8 μM. α and Ki were determined to be 7.3 ± 0.06 and 9.8 ± 0.2, respectively.
Figure 5.
Effect of 13(S)-HODE on kcat/KM (app) of 15-LOX-2 with GLA. (pH 7.5, 25mM Hepes, 1–20 μM GLA at each product point) The data is fit to equation 3 (Scheme 1), where KM = 2.8 μM, kcat =0.65 s−1, α = 7.3 and Ki = 9.8 μM. β was determined to be 1.6 ± 0.1.
As described above, the addition of 15 μM 13-(S)-HODE and 13-(S)-HOTrE (γ) exhibited the opposite effects depending on the substrate, with the AA kinetics being activated, while the GLA kinetics were inhibited (Table 2). In order to investigate this effect further, titration kinetics with 13-(S)-HODE, the more potent activator, were determined with AA at pH 7.5. It was observed that 13-(S)-HODE affected AA kinetics with an α value of 0.2 ± 0.01 (Ktype activation), a β value of 0.76 ± 0.01 (V-type inhibition), a β/α of 3.8 (kcat/KM allosteric activation) and a Ki value of 1.2 ± 0.03 μM (Figure 6 and 7). The β/α with 13-(S)-HODE/AA is opposite to that seen with 13-(S)-HODE/GLA, which showed kcat/KM allosteric inhibition (β/α= 0.22). In addition, the β value for 13-(S)-HODE/AA indicated V-type inhibition, which was opposite to the β value for 13-(S)-HODE/GLA (V-type activation, β = 1.6 ± 0.1). This difference is significant since 13-(S)-HODE both activates and inhibits, depending on the length and unsaturation of the substrate and suggests subtle structural changes in the active site. It should be noted that we previously reported a slight inhibition of both kcat and kcat/KM at the single concentration of 10 μM 13-(S)-HPODE and AA.(23) We were not able to reproduce this result and considering that the previous data utilized only one product concentration and one pH condition, we consider the current data more reliable for interpretation. In addition, 13-(S)-HODE manifested different thresholds of response, with the allosteric effect on AA catalysis occurring at a lower concentration than the allosteric effect on GLA catalysis (Ki = 1.2 ± 0.03 μM and 9.8 ± 0.02 μM, respectively). These differences in Ki values are difficult to rationalize with the current one site allosteric model. The data may indicate two distinct allosteric sites with distinct Ki values or two distinct 15-LOX-2 species, which bind 13-(S)-HODE with different affinities, such as E•AA and E•GLA. Further studies are required to differentiate these possible explanations. Finally, the Ki for 13-(S)-HODE binding with AA as substrate is considerably greater than was estimated by our competitive substrate capture method with both AA and LA being present.(24) Considering that the Ki for 13-HODE varies with AA and GLA, it could be possible that the presence of LA also affects the Ki value of 13-(S)-HODE, which we are currently investigating further.
Figure 6.
Effect of 13(S)-HODE on KM (app) of 15-LOX-2 with AA
(pH 7.5, 25 mM Hepes, 1–20 μM AA at each product point) The data is fit to equation 2 (Scheme 1), where KM = 3.5 μM. α and Ki were determined to be 0.2 ± 0.01 and 1.2 ± 0.03, respectively.
Figure 7.
Effect of 13(S)-HODE on kcat/KM (app) of 15-LOX-2 with AA
(pH 7.5, 25 mM Hepes, 1–20 μM AA at each product point) The data is fit to equation 3 (Scheme 1), where KM = 3.5 μM, kcat =1.4 s−1, α = 0.2 and Ki = 1.2 μM. β was determined to be 0.76 ± 0.01.
Summary of 15-LOX-2 data
In summary, the wild-type 15-LOX-2 kinetic results showed that GLA and AA had different responses to pH, indicating that the molecular mechanisms for GLA and AA were distinct. Addition of 13-(S)-HODE and 13-(S)-HOTrE(γ), mirrored the kcat/KM pH effect, with an increase in kcat/KM AA/GLA, however, the pH and allosteric effects were additive, with the combined effect being 11-fold, suggesting both mechanistic similarities and differences between the pH and allosteric effects. The two allosteric effectors, 13-(S)-HODE and 13-(S)-HOTrE(γ), displayed comparable hyperbolic inhibition kinetics, however, the Ki of 13-(S)-HODE was pH dependent while that of 13-(S)-HOTrE(γ) was not, indicating a difference in their binding modes and supporting the presence of a specific binding site, since it is unlikely their structural differences would affect non-specific binding. Finally, it was observed that 13-(S)-HODE activated kcat/KM AA but inhibited kcat/KM GLA. This difference is significant and indicates that the allosteric structural changes differentiate the length and unsaturation of AA and GLA. Recently, it has been postulated that addition of 13-(S)-HODE induces dimer formation for rabbit 15-LOX-1 (i.e. 12/15-LOX),(48, 49) however, the above data suggests a more complicated scenario. Considering the opposite effects of 13-(S)-HODE on GLA and AA catalysis, it is unlikely that only dimer formation could account for these two opposing effects (vide infra).
Characterization of the PLAT domain deletion mutant (15-LOX-2NoPLAT)
Thermal stability measurements of 15-LOX-2NoPLAT
Homology modeling of 15-LOX-2 previously suggested that its allosteric site could be located in a cleft between the PLAT and catalytic domains.(24) This hypothesis was investigated by removing the PLAT domain and generating 15-LOX-2NoPLAT. The thermal stability of 15-LOX-2NoPLAT was measured and as seen in Figure 8, both 15-LOX-2 and 15-LOX-2NoPLAT underwent a loss in secondary structure with a two-state unfolding transition and a T50 between 40–45° C for both proteins. These data indicated that the thermal stability of the truncated protein to be approximately the same as the wild-type protein.
Figure 8.
CD half-transition temperature (T50) measurement for 15-LOX-2 (Open circles) and 15-LOX-2NoPLAT (Open squares). The experiment was performed in 25 mM Hepes buffer (150 mM NaCl, pH 7.5) between 10–80° C.
Characterization of steady-state kinetics of 15-LOX-2NoPLAT with AA and GLA
Steady-state kinetics were conducted with 15-LOX-2NoPLAT at pH 7.5, using both AA and GLA to investigate the effect of removing the PLAT domain (Table 4). A loss in activity was observed for 15-LOX-2NoPLAT with a 50% decrease in kcat AA (0.75 ± 0.03 s−1) and was consistent with previous results where an 84% decrease in kcat AA for rabbit 15-LOX-1NoPLAT was observed.(13) The kcat/KM value of 15-LOX-2NoPLAT also decreased approximately 50% (0.21 ± 0.02 μM−1 s−1), which was also consistent with the previous 73% decrease in kcat/KM AA for rabbit 15-LOX-1NoPLAT.(13) In comparing the substrate specificity of 15-LOX-2NoPLAT with AA and GLA, it was observed that the kcat AA/GLA ratio did not change much relative to 15-LOX-2, 0.76 ± 0.06 and 0.83 ± 0.03, respectively. However, the kcat/KM AA/GLA ratio did change, with a ratio of 1.3 ± 0.2 for 15-LOX-2NoPLAT and a ratio of 0.62 ± 0.04 for 15-LOX-2. This data indicated that the PLAT domain affected substrate specificity and warranted further investigation.
Table 4.
Comparison of kinetic parameters of 15-LOX-2 and 15-LOX-2NoPLAT, with AA and GLA as substrates (pH 7.5). a
| kcat/KM (μM−1s−1) | kcat (s−1) | |||||
|---|---|---|---|---|---|---|
| AA | GLA | AA/GLA | AA | GLA | AA/GLA | |
| 15-LOX-2 | 0.40±0.02 | 0.64±0.02 | 0.62±0.04 | 1.5±0.03 | 1.8±0.03 | 0.83±0.03 |
| 15-LOX-2NoPLAT | 0.21±0.02 | 0.16±0.02 | 1.3±0.2 | 0.75±0.03 | 0.99±0.04 | 0.76±0.06 |
Enzymatic assays were performed in 25mM Hepes, pH 7.5, at 22° C
Substrate specificity pH profile of 15-LOX-2NoPLAT
The competitive substrate capture method was employed to explore whether the PLAT domain was responsible for the pH effect on substrate specificity. It was determined that the kcat/KM AA/GLA ratio for 15-LOX-2NoPLAT was found to be 2.5 ± 0.2 at pH 7.5 and 3.6 ± 0.3 at pH 8.5. These values were comparable to that found in the current work with wild-type 15-LOX-2 (vide supra) and indicated that the PLAT domain had a minimal contribution to the pH effect.
Effect of 13-(S)-HODE on GLA and AA steady-state kinetics of 15-LOX-2NoPLAT
Wild-type 15-LOX-2 demonstrated a hyperbolic allosteric effect, with 13-(S)-HODE and GLA (pH 7.5) (Table 5). For 15-LOX-2NoPLAT, increasing amounts of 13-(S)-HODE also evoked a hyperbolic allosteric effect with GLA, revealing an α value of 4.9 ± 2, a β value of 1.6 ± 0.2, a β/α value of 0.33, and a Ki value of 6.1 ± 2 μM (Table 5 and Supplemental Data, Figures S4 and S5). The fact that 15-LOX-2NoPLAT had a similar allosteric response to that of 15-LOX-2 indicates that removal of the PLAT domain does not eliminate allostery and that the location of the allosteric site is in the catalytic domain.
Table 5.
Comparison of hyperbolic fit parameters of 15-LOX-2 and 15-LOX-2NoPLAT with 13-(S)-HODE and AA or GLA as substrates at pH 7.5. a
| 15-LOX-2 | 15-LOX-2NoPLAT | |||||||
|---|---|---|---|---|---|---|---|---|
| Conditions | Ki | α | β | β/α | Ki | α | β | β/α |
| GLA + 13(S)-HODE | 9.8±0.2 | 7.3±0.06 | 1.6±0.1 | 0.22 | 6.1±2 | 4.9±2 | 1.6±0.2 | 0.33 |
| AA + 13(S)-HODE | 1.2±0.03 | 0.2±0.01 | 0.76±0.01 | 3.8 | 2.0±0.6 | 2.3±0.4 | 1.1±0.03 | 0.48 |
Enzymatic assays were performed in 25 mM Hepes (pH 7.5), at 22° C with and without products.
The addition of 13-(S)-HODE to 15-LOX-2NoPLAT with AA as substrate, however, evoked dramatically different kinetic behavior to that of 15-LOX-2. The KM(app) fit revealed an α value of 2.3 ± 0.4, a β value of 1.1 ± 0.03, a β/α value of 0.48 and a Ki value of 2.0 ± 0.6 μM (Table 5 and Supplemental Data, Figures S6 and S7). The fact that the β/α is now 0.48, which is the opposite effect seen for 15-LOX-2 (β/α = 4), represents a significant change in the allosteric effect with removal of the PLAT domain. It should be noted that 13-(S)-HODE also manifests different Ki values for AA and GLA catalysis with 15-LOX-2NoPLAT, similar to that seen for 15-LOX-2. These data support the hypothesis postulated above, that there either are two distinct allosteric sites or two distinct allosteric binding species, such as E•AA and E•GLA. Both of these hypotheses are being investigated further.
Summary of 15-LOX-2NoPLAT data
In summary, removal of the PLAT domain does not affect the overall protein stability and does not affect its kinetic pH dependence, indicating the source of the pH effect is in the catalytic domain and not at the interface of the two domains with H627, as was postulated before.(46) Alignments of the actives sites of 15-LOX-2 and 15-LOX-1(7) do not indicate a potential histidine in the 15-LOX-2 active site, as the pH dependence suggests,(46) however it does not exclude the possibility of a buried histidine emerging into the active site upon catalytic protein motions. However, the removal of the PLAT domain did manifest a 2-fold increase in kcat/KM AA/GLA, indicating that the PLAT domain does influence substrate specificity. Interestingly, 15-LOX-2NoPLAT retained its allosteric properties, indicating that the allosteric site is also located in the catalytic domain, like the pH effect, and not in the cleft between the two domains. Nonetheless, removal of the PLAT domain does affect the degree of allostery (i.e. the sign and magnitude of α and β), suggesting that the PLAT domain does moderate the communication pathway between the allosteric and catalytic sites. Therefore, the PLAT domain not only has a role in membrane affinity,(13) but it also has a role in allostery and substrate specificity. Nonetheless, it is still unclear what specific biophysical changes occur upon allosteric effector binding. As mentioned above, Kuhn and coworkers proposed that dimerization was induced with the addition of 13-(S)-HODE to rabbit 15-LOX-1 and that the postulated dimerization interface was opposite to the PLAT domain, which is consistent with our current results. However, the SAXS data could not determine if dimerization was the only structural change.(48, 49) It could be that the allosteric effect is responsible for not only dimerization, but also for specific structural changes in the active site, as seen in RNR,(29) or for protein dynamic changes, as seen for calbindin D9k.(50) A possible explanation could be similar to that seen for COX-2. COX-2 is a homodimer, but binding of an inhibitor to only one of the monomers establishes a conformational heterodimer, whose active monomer has altered kinetic behavior.(51) 15-LOX-2 could also manifest a similar behavior where allosteric effectors not only induce dimerization, but also bind to only one of the subunits, establishing a functional heterodimer. Continued efforts are underway to refine our understanding of the allosteric properties of 15-LOX-2 in terms of the location of the allosteric site and its molecular mechanism. Synthetic allosteric effectors are also being investigated in the hopes of developing the allosteric site as a possible therapeutic target for LOX regulation by changing the proportions of LOX products produced, as opposed to inhibition of the entire enzymatic activity. If achieved, this could be an important advance, since some LOX products are beneficial while others are detrimental to human health.(17)
Supplementary Material
Acknowledgments
The authors thank Dr. Russell L. Wrobel and Mr. Karl W. Nichols (both from the Center for Eukaryotic Structural Genomics) for producing the clone used in these studies and for discussions on the preparation of lipoxygenase fusion proteins, respectively.
Abbreviations
- LOX
lipoxygenase
- 15-LOX-2
human epithelial 15-lipoxygenase-2
- 15-LOX-2NoPLAT
human epithelial 15-lipoxygenase-2 PLAT domain removed
- 15-LOX-1
human reticulocyte 15-lipoxygenase-1
- rabbit 15-LOX
rabbit 15-lipoxygenase
- 12-LOX
human platelet 12-lipoxygenase
- soybean LOX-1
soybean lipoxygenase 1
- 5-LOX
human 5-lipoxygenase
- COX
cyclooxygenase
- P450
cytochrome P450
- TEV protease
tobacco etch virus protease
- BSA
bovine serum albumin
- MBP
maltose binding protein
- NTA
nitrilotriacetic acid
- IMAC
immobilized metal affinity chromatography
- T50
half-transition temperature
- DCM
dichloromethane
- AA
arachidonic acid
- LA
linoleic acid
- 13-(S)-HPODE
13-(S)-hydroperoxy-9Z,11E-octadecadienoic acid
- 13-(S)-HODE
13-(S)-hydroxy-9Z,11E-octadecadienoic acid
- GLA
(γ)-linolenic acid
- 13-(S)-HPOTrE(γ)
13-(S)-hydroperoxy-6Z,9Z,11E-octadecatrienoic acid
- 13-(S)-HOTrE(γ)
13-(S)-hydroxy-6Z,9Z,11E-octadecatrienoic acid
- 9-(S)-HOTrE(γ)
DGLA, dihomo (γ) linolenic acid
- EPA
eicosapentaenoic acid
- kcat
the rate constant for product release
- kcat/KM
the rate constant for fatty acid capture
- KD
equilibrium constant for the dissociation of a ligand from the protein site
- kcat/KM[O2]
the rate constant for O2 capture
Footnotes
Supporting Information Available. Additional kinetic graphics are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ivanov I, Heydeck D, Hofheinz K, Roffeis J, O’Donnell VB, Kuhn H, Walther M. Molecular enzymology of lipoxygenases. Archives of biochemistry and biophysics. 2010;503:161–174. doi: 10.1016/j.abb.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 2.Brash AR. Lipoxygenases: Occurrence, Functions, Catalysis and Acquisition of Substrate. J Biol Chem. 1999;274:23679–23682. doi: 10.1074/jbc.274.34.23679. [DOI] [PubMed] [Google Scholar]
- 3.Noguchi N, Yamashita H, Hamahara J, Nakamura A, Kuhn H, Niki E. The specificity of lipoxygenase-catalyzed lipid peroxidation and the effects of radical-scavenging antioxidants. Biol Chem. 2002;383:619–626. doi: 10.1515/BC.2002.064. [DOI] [PubMed] [Google Scholar]
- 4.Schneider C, Pratt DA, Porter NA, Brash AR. Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem Biol. 2007;14:473–488. doi: 10.1016/j.chembiol.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto S. Mammalian lipoxygenases: molecular structures and functions. Biochim Biophys Acta. 1992;1128:117–131. doi: 10.1016/0005-2760(92)90297-9. [DOI] [PubMed] [Google Scholar]
- 6.Gilbert NC, Bartlett SG, Waight MT, Neau DB, Boeglin WE, Brash AR, Newcomer ME. The structure of human 5-lipoxygenase. Science. 2011;331:217–219. doi: 10.1126/science.1197203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi J, Chon JK, Kim S, Shin W. Conformational flexibility in mammalian 15S-lipoxygenase: Reinterpretation of the crystallographic data. Proteins. 2008;70:1023–1032. doi: 10.1002/prot.21590. [DOI] [PubMed] [Google Scholar]
- 8.Gillmor SA, Villasenor A, Fletterick R, Sigal E, Browner M. The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity. Nature Struct Biol. 1997;4:1003–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 9.Chahinian H, Sias B, Carriere F. The C-terminal domain of pancreatic lipase: functional and structural analogies with c2 domains. Curr Protein Pept Sci. 2000;1:91–103. doi: 10.2174/1389203003381487. [DOI] [PubMed] [Google Scholar]
- 10.May C, Hohne M, Gnau P, Schwennesen K, Kindl H. The N-terminal beta-barrel structure of lipid body lipoxygenase mediates its binding to liposomes and lipid bodies. European Journal of Biochemistry. 2000;267:1100–1109. doi: 10.1046/j.1432-1327.2000.01105.x. [DOI] [PubMed] [Google Scholar]
- 11.Tatulian SA, Steczko J, Minor W. Uncovering a calcium-regulated membrane-binding mechanism for soybean lipoxygenase-1. Biochemistry. 1998;37:15481–15490. doi: 10.1021/bi981062t. [DOI] [PubMed] [Google Scholar]
- 12.Walther M, Hofheinz K, Vogel R, Roffeis J, Kühn H. The N-terminal β-barrel domain of mammalian lipoxygenases including mouse 5-lipoxygenase is not essential for catalytic activity and membrane binding but exhibits regulatory functions. Archives of Biochemistry and Biophysics. 2011;516:1–9. doi: 10.1016/j.abb.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Walther M, Anton M, Wiedmann M, Fletterick R, Kuhn H. The Nterminal domain of the reticulocyte-type 15-lipoxygenase is not essential for enzymatic activity but contains determinants for membrane binding. J Biol Chem. 2002;277:27360–27366. doi: 10.1074/jbc.M203234200. [DOI] [PubMed] [Google Scholar]
- 14.Rouzer CA, Marnett LJ. Endocannabinoid oxygenation by cyclooxygenases, lipoxygenases, and cytochromes P450: cross-talk between the eicosanoid and endocannabinoid signaling pathways. Chemical reviews. 2011;111:5899–5921. doi: 10.1021/cr2002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samuelsson B. Leukotrienes: Mediators of Immediate Hypersensitivity Reactions and Inflammation. Science. 1983;220:568–575. doi: 10.1126/science.6301011. [DOI] [PubMed] [Google Scholar]
- 16.Jampilek J, Dolezal M, Opletalova V, Hartl J. 5-Lipoxygenase, leukotrienes biosynthesis and potential antileukotrienic agents. Curr Med Chem. 2006;13:117–129. doi: 10.2174/092986706775197935. [DOI] [PubMed] [Google Scholar]
- 17.Serhan C, Petasis N. Resolvins and Protectins in Inflammation Resolution. Chemical reviews. 2011;111:5922–5943. doi: 10.1021/cr100396c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol. 2008;8:349–361. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn H, Brash AR, Wiesner R, Alder L. Lipoxygenase catalyzed oxygenation of hydroxy fatty acids to lipoxins. Adv Exp Med Biol. 1988;229:39–49. doi: 10.1007/978-1-4757-0937-7_4. [DOI] [PubMed] [Google Scholar]
- 20.Samuelsson B, Dahlen SE, Lindgren JA, Rouzer CA, Serhan CN. Leukotrienes and Lipoxins: Structures, Biosynthesis, and Biological Effects. Science. 1987;237:1171–1176. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- 21.Kuhn H, Wiesner R, Alder L, Fitzsimmons BJ, Rokach J, Brash AR. Formation of lipoxin B by the pure reticulocyte lipoxygenase via sequential oxygenation of the substrate. Eur J Biochem. 1987;169:593–601. doi: 10.1111/j.1432-1033.1987.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 22.Kuhn H, Wiesner R, Alder L, Schewe T, Stender H. Formation of lipoxin B by the pure reticulocyte lipoxygenase. FEBS Lett. 1986;208:248–252. doi: 10.1016/0014-5793(86)81026-2. [DOI] [PubMed] [Google Scholar]
- 23.Wecksler AT, Kenyon V, Deschamps JD, Holman TR. Substrate specificity changes for human reticulocyte and epithelial 15-lipoxygenases reveal allosteric product regulation. Biochemistry. 2008;47:7364–7375. doi: 10.1021/bi800550n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wecksler AT, Kenyon V, Garcia NK, Deschamps JD, van der Donk WA, Holman TR. Kinetic and structural investigations of the allosteric site in human epithelial 15-lipoxygenase-2. Biochemistry. 2009;48:8721–8730. doi: 10.1021/bi9009242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai CJ, del Sol A, Nussinov R. Allostery: absence of a change in shape does not imply that allostery is not at play. Journal of molecular biology. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sola-Penna M, Da Silva D, Coelho WS, Marinho-Carvalho MM, Zancan P. Regulation of mammalian muscle type 6-phosphofructo-1-kinase and its implication for the control of the metabolism. IUBMB Life. 2010;62:791–796. doi: 10.1002/iub.393. [DOI] [PubMed] [Google Scholar]
- 27.Sols A. Multimodulation of enzyme activity. Curr Top Cell Regul. 1981;19:77–101. doi: 10.1016/b978-0-12-152819-5.50020-8. [DOI] [PubMed] [Google Scholar]
- 28.Strater N, Marek S, Kuettner EB, Kloos M, Keim A, Bruser A, Kirchberger J, Schoneberg T. Molecular architecture and structural basis of allosteric regulation of eukaryotic phosphofructokinases. FASEB J. 2011;25:89–98. doi: 10.1096/fj.10-163865. [DOI] [PubMed] [Google Scholar]
- 29.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 30.Kolberg M, Strand KR, Graff P, Andersson KK. Structure, function, and mechanism of ribonucleotide reductases. Biochim Biophys Acta. 2004;1699:1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Reichard P. Ribonucleotide reductases: the evolution of allosteric regulation. Archives of biochemistry and biophysics. 2002;397:149–155. doi: 10.1006/abbi.2001.2637. [DOI] [PubMed] [Google Scholar]
- 32.Ochi K, Yoshimoto T, Yamamoto S, Taniguchi K, Miyamoto T. Arachidonate 5-lipoxygenase of guinea pig peritoneal polymorphonuclear leukocytes. Activation by adenosine 5′-triphosphate. J Biol Chem. 1983;258:5754–5758. [PubMed] [Google Scholar]
- 33.Furukawa M, Yoshimoto T, Ochi K, Yamamoto S. Studies on arachidonate 5-lipoxygenase of rat basophilic leukemia cells. Biochimica et biophysica acta. 1984;795:458–465. [PubMed] [Google Scholar]
- 34.Rouzer CA, Samuelsson B. On the nature of the 5-lipoxygenase reaction in human leukocytes: enzyme purification and requirement for multiple stimulatory factors. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:6040–6044. doi: 10.1073/pnas.82.18.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aharony D, Stein RL. Kinetic mechanism of guinea pig neutrophil 5-lipoxygenase. The Journal of biological chemistry. 1986;261:11512–11519. [PubMed] [Google Scholar]
- 36.Mogul R, Johansen E, Holman TR. Oleyl sulfate reveals allosteric inhibition of Soybean Lipoxygenase-1 and Human 15-Lipoxygenase. Biochemistry. 2000;39:4801–4807. doi: 10.1021/bi992805t. [DOI] [PubMed] [Google Scholar]
- 37.Aleem AM, Jankun J, Dignam JD, Walther M, Kuhn H, Svergun DI, Skrzypczak-Jankun E. Human platelet 12-lipoxygenase, new findings about its activity, membrane binding and low-resolution structure. Journal of molecular biology. 2008;376:193–209. doi: 10.1016/j.jmb.2007.11.086. [DOI] [PubMed] [Google Scholar]
- 38.Dainese E, Angelucci CB, Sabatucci A, De Filippis V, Mei G, Maccarrone M. A novel role for iron in modulating the activity and membrane-binding ability of a trimmed soybean lipoxygenase-1. The FASEB Journal. 2010;24:1725–1736. doi: 10.1096/fj.09-141390. [DOI] [PubMed] [Google Scholar]
- 39.Di Venere A, Salucci ML, van Zadelhoff G, Veldink G, Mei G, Rosato N, Finazzi-Agro A, Maccarrone M. Structure-to-function relationship of mini-lipoxygenase, a 60-kDa fragment of soybean lipoxygenase-1 with lower stability but higher enzymatic activity. J Biol Chem. 2003;278:18281–18288. doi: 10.1074/jbc.M212122200. [DOI] [PubMed] [Google Scholar]
- 40.Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holman TR, Zhou J, Solomon EI. Spectroscopic and functional characterization of a ligand coordination mutant of soybean lipoxygenase-1: first coordination sphere analogue of human 15-lipoxygenase. Journal of the American Chemical Society. 1998;120:12564–12572. [Google Scholar]
- 42.Jeon WB, Aceti DJ, Bingman CA, Vojtik FC, Olson AC, Ellefson JM, McCombs JE, Sreenath HK, Blommel PG, Seder KD, Burns BT, Geetha HV, Harms AC, Sabat G, Sussman MR, Fox BG, Phillips GN., Jr High-throughput purification and quality assurance of Arabidopsis thaliana proteins for eukaryotic structural genomics. Journal of structural and functional genomics. 2005;6:143–147. doi: 10.1007/s10969-005-1908-7. [DOI] [PubMed] [Google Scholar]
- 43.Deschamps JD, Kenyon VA, Holman TR. Baicalein is a potent in vitro inhibitor against both reticulocyte 15-human and platelet 12-human lipoxygenases. Bioorg Med Chem. 2006;14:4295–4301. doi: 10.1016/j.bmc.2006.01.057. [DOI] [PubMed] [Google Scholar]
- 44.Wecksler AT, Jacquot C, van der Donk WA, Holman TR. Mechanistic Investigations of Human Reticulocyte 15-and Platelet 12-Lipoxygenases with Arachidonic Acid. Biochemistry. 2009;48:6259–6267. doi: 10.1021/bi802332j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McAuley WJ, Jones DS, Kett VL. Characterisation of the interaction of lactate dehydrogenase with Tween-20 using isothermal titration calorimetry, interfacial rheometry and surface tension measurements. Journal of pharmaceutical sciences. 2009;98:2659–2669. doi: 10.1002/jps.21640. [DOI] [PubMed] [Google Scholar]
- 46.Wecksler AT, Deschamps JD, Kenyon V, Garcia NK, van der Donk WA, Holman TR. Kinetic and structural investigations of the allosteric site in human epithelial 15-lipoxygenase-2. Biochemistry Manuscript in revisions. 2009 doi: 10.1021/bi9009242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinhart GD. Quantitative analysis and interpretation of allosteric behavior. Methods Enzymol. 2004;380:187–203. doi: 10.1016/S0076-6879(04)80009-0. [DOI] [PubMed] [Google Scholar]
- 48.Shang W, Ivanov I, Svergun DI, Borbulevych OY, Aleem AM, Stehling S, Jankun J, Kuhn H, Skrzypczak-Jankun E. Probing dimerization and structural flexibility of mammalian lipoxygenases by small-angle X-ray scattering. Journal of molecular biology. 2011;409:654–668. doi: 10.1016/j.jmb.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 49.Ivanov I, Shang W, Toledo L, Masgrau L, Svergun DI, Stehling S, Góez H, Di Venere A, Mei G, Lluch JM, Skrzypczak-Jankun E, Gonzáez-Lafont À, Kühn H. Ligand-induced formation of transient dimers of mammalian 12/15-lipoxygenase: A key to allosteric behavior of this class of enzymes? Proteins: Structure, Function, and Bioinformatics. 2012;80:703–712. doi: 10.1002/prot.23227. [DOI] [PubMed] [Google Scholar]
- 50.Maler L, Blankenship J, Rance M, Chazin WJ. Site-site communication in the EF-hand Ca2+-binding protein calbindin D9k. Nature structural biology. 2000;7:245–250. doi: 10.1038/73369. [DOI] [PubMed] [Google Scholar]
- 51.Yuan C, Rieke CJ, Rimon G, Wingerd BA, Smith WL. Partnering between monomers of cyclooxygenase-2 homodimers. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6142–6147. doi: 10.1073/pnas.0601805103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.