Abstract
Gene and poly(A) trappings are high-throughput approaches to capture and interrupt the expression of endogenous genes within a target genome. Although a number of trapping vectors have been developed for investigation of gene functions in cells and vertebrate models, there is still room for the improvement of their efficiency and sensitivity. Recently, two novel trapping vectors mediated by Sleeping Beauty (SB) transposon have been generated by the combination of three functional cassettes that are required for finding endogenous genes, disrupting the expression of trapped genes, and inducing the excision of integrated traps from their original insertion sites and then inserting into another gene. In addition, several other strategies are utilized to improve the activities of two trapping vectors. First, activities of all components were examined in vitro before the generation of two vectors. Second, the inducible promoter from the tilapia Hsp70 gene was used to drive the expression of SB gene, which can mediate the excision of integrated transposons upon induction at 37 °C. Third, the Cre/LoxP system was introduced to delete the SB expression cassette for stabilization of gene interruption and bio-safety. Fourth, three stop codons in different reading frames were introduced downstream of a strong splice acceptor (SA) in the gene trapping vector to effectively terminate the translation of trapped endogenous genes. Fifth, the strong splicing donor (SD) and AU-rich RNA-destabilizing element exhibited no obvious insertion bias and markedly reduced SD read-through events, and the combination of an enhanced SA, a poly(A) signal and a transcript terminator in the poly(A) trapping vector efficiently disrupted the transcription of trapped genes. Thus, these two trapping vectors are alternative and effective tools for large-scale identification and disruption of endogenous genes in vertebrate cells and animals.
Keywords: insertional mutagenesis, transposon, Sleeping Beauty, gene trapping, poly(A) trapping
The rapid advent of high-throughput DNA sequencing technologies has enabled the completion of genome projects for a large number of species including human and other model species. The availability of these genomes has in turn advanced biological researches into the post-genome era, which integrates classical experimental approaches with those of “omics” technologies to elucidate functions of identified genes in various processes such as embryonic development and human diseases. A number of mutagenic methods have long been developed to decipher functions of genes in model animals. For instance, chemical mutation by N-ethyl-N-nitrosourea (ENU) has produced a large number of zebrafish mutants with distinct phenotypes1-3 but identification of mutated genes responsible for these phenotypes by positional cloning is extremely laborious and time consuming.
To address these limitations, retroviral and transposable DNA elements have been used as insertional mutagens,4-7 which has greatly facilitated the identification of mutated genes since the gene loci are tagged by the inserted DNA sequence. However, the mutation rates generated by retroviral or transposon vectors are approximately 7–10 fold lower than that of ENU mutation. Insertional mutagenesis mediated by the pseudotyped retrovirus was found to be very effective for the generation of zebrafish mutants with altered phenotypes in 1 out of 80 to 100 such insertions,4,8 but for some reason the retroviral vectors have a preference for integrating in the 5′-transcriptional regulatory regions of genes9 and tend to create mutations in genes that are expressed during early developmental stages. Apart from the 5′ integration bias, retroviral vectors have intrinsic drawbacks such as limited packaging size, gene expression silencing and ectopic reporter gene expression.
Transposon systems have recently been exploited as potential tools for insertional mutagenesis in mouse and zebrafish10-12 due to their less demanding of laboratory and technical conditions when compared with those of psedotyped retrovirus. These systems were also used to develop enhancer-trapping and gene-trapping cassettes and a number of transgenic fish lines with different reporter gene expression patterns were generated.13,14 The transgenic efficiency of Sleeping Beauty (SB)-based gene delivery vector was improved about 6-fold (from 5% to 31%)15 and SB-based insertional mutagenesis screens were introduced in mouse16 and zebrafish17 to elucidate gene functions in all kinds of genetic networks and pathways. Indeed, the SB system was successfully used to identify temporal and spatial expression and functions of genes in developmental pathways7 and has proved to be a highly instrumental tool to induce tumors in experimental animals for the purpose of uncovering the genetic basis of diverse cancers.18,19 In addition, it has been shown that Tol2 transposon-based enhancer trapping renders the comparable mutagenic efficiency with that of retroviral insertions20 and Tol2-based gene trap systems were demonstrated to be useful for the study of developmental processes and gene discovery in zebrafish.14,21,22 Therefore, transposon-mediated insertional mutagenesis offers great potential for loss-of-function screening in model animals. However, tremendous efforts are being made to increase the mutagenic potential of transposon-based gene trapping constructs currently available.23-25
The conventional gene trapping vector contains a promoterless marker/reporter gene flanked by an upstream splice acceptor (SA) and a downstream poly(A) signal. Once inserted into an intron or exon of active genes, the trap cassette is transcribed from the endogenous promoter and the SA intercepts endogenous normal splicing through the generation of a fused transcript between upstream exon and the marker/reporter gene. Since the fusion transcript is prematurely terminated at the foreign poly(A) site, it encodes a truncated and often non-functional version of cellular protein and the marker/reporter.26,27 The key cassette in nearly all gene trapping constructs contains a highly efficient SA and a poly(A) signal. Without such a transcriptional termination cassette, the splicing of endogenous gene transcripts around the trapping vector can readily occur and thus result in an insertion that may not effectively interrupt the gene function at the insertion locus.6,28
Recently, we have developed a novel gene trapping vector pT2/Gene-Trap (Fig. 1).29 The activities of all components in this vector were tested in cultured HeLa cells using an exon trapping vector pSPL3.30 When inserted into an intron of the pSPL3, the SA properly spliced with the upstream exon and efficiently terminated the transcription of downstream exons. In the circumstance of integration into an exon of the modified pSPL3, the trapping cassette directly disrupted the exon and blocked the transcription of downstream exons. The activity of this vector in trapping endogenous genes was further examined in HeLa cells and developing zebrafish embryos. A number of transposon inserts were identified in G418-selected cell colonies and the normal splicing of trapped endogenous genes was totally disrupted by the mutation cassette, which contains a SA signal originated from the carp β-actin gene and a short exon sequence. The short exon contains three stop codons in different reading frames (TGA ATT AGT GA) and can efficiently truncate the translation of trapped gene transcripts. It has been shown that the internal ribosomal entry sequence (IRES) inserted in front of a marker/reporter gene can mediate the independent translation of marker/reporter gene and the IRES-based vectors are able to capture a wide range of genes expressed in a variety of tissues and embryos at different developmental stages.31,32 In our trapping vector, the IRES containing 12 AUGs and an A6 (AAAAAA) bifurcation loop, which performs better in zebrafish than another IRES element containing 10 AUGs and an A7 (AAAAAAA) bifurcation loop.29 In addition, the SA signal from the carp β-actin gene coupled with a poly(A) signal of ocean pout antifreeze gene can severely disrupt the expression of trapped genes by producing a truncated fusion transcript.22 However, conventional gene trapping is not suitable to capture genes that are not or poorly expressed.
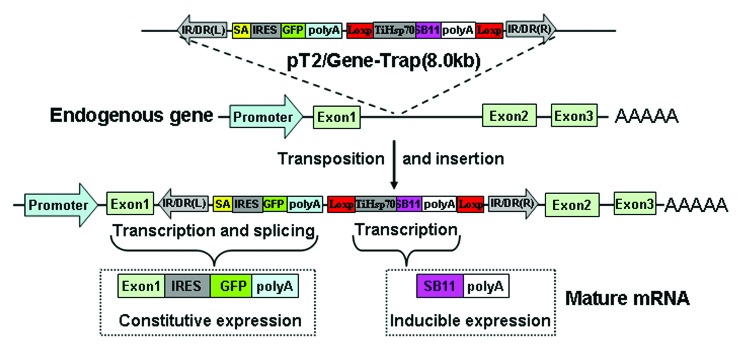
Figure 1. The flowchart of gene trapping with pT2/Gene-Trap. Insertion of the gene-Trap cassette into an intron of transcriptionally active loci can generate a fusion transcript that contains the upstream exon and the reporter marker. The expression of SB11 gene was driven by the heat-inducible promoter TiHsp70. IR/DR(L) and IR/DR(R), left and right inverted repeat/directed repeat of the SB transposon; SA, splice acceptor; IRES, internal ribosome entry site; GFP, GFP reporter gene; poly(A), poly(A) signal; TiHsp70, tilapia Hsp70 promoter; SB11, SB11 transposase gene.
To circumvent the limitation, a number of poly(A)-trap vectors have been generated. A basic poly(A)-trap vector contains a reporter/selectable marker gene flanked by an upstream constitutive promoter and a downstream splice donor (SD). Integration of a trap cassette upstream of a functional poly(A) signal within a genome leads to the generation of a stable premRNA and the proper splicing of the SD with a SA downstream of the insertion site and thus gives rise to a fusion transcript that encodes the reporter/selectable marker and an N-terminal truncated version of endogenous protein. Theoretically, poly(A)-trapping is able to capture endogenous genes almost equally regardless of their transcriptional status in a target genome.33-36 Nevertheless, later findings indicate that these basic poly(A)-trap vectors have a bias toward the last introns of trapped genes because of the activation of nonsense-mediated mRNA decay (NMD) mechanism.37 Moreover, most of poly(A)-trap vectors currently available still suffer from other problems such as the background SD read-through events. Therefore, we constructed a novel trap vector pT2/poly(A)-Trap (Fig. 2), which can effectively capture endogenous genes independent of their transcriptional status and inevitably disrupted the transcription of trapped genes in Hela cells and zebrafish.38 Importantly, the poly(A) trap vector contains an AU-rich RNA-destabilizing element that is highly effective for reduction of the background SD read-through events.
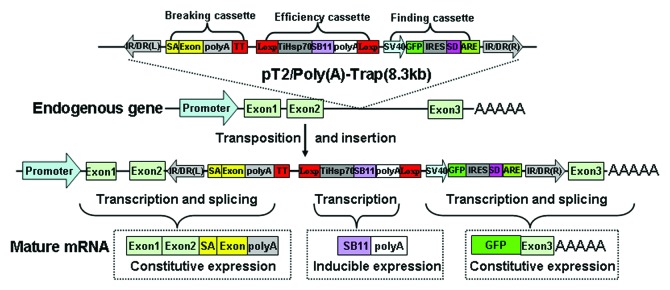
Figure 2. The flowchart of gene trapping with pT2/Poly(A)-Trap. Insertion of the poly(A)-trap cassette into an intron before a functional poly(A) sequence for an endogenous gene leads to the generation of a stable pre-mRNA and the proper splicing between the trap SD with a SA downstream of the insertion site and gives rise to a fusion transcript for GFP expression. The proper expression of trapped gene was disrupted by the formation of another fusion transcript that contains exons upstream of the insert site. The expression of SB11 gene was driven by the heat-inducible promoter TiHsp70. IR/DR(L) and IR/DR(R), left and right inverted/directed repeats of the SB transposon; SA, splice acceptor from carp β-actin gene; Exon, the exon 2 from the carp β-actin gene; SV40 poly(A), SV40 polyadenylation sequence; TT, transcript terminator sequence. TiHSP70, tilapia Hsp70 promotor; SB11, SB11 transposase gene. polyA, a poly(A) signal from carp β-actin gene; SV40, SV40 promoter; GFP, GFP reporter gene; IRES, internal ribosome entry site; SD, splice donor; ARE, AU-riched element.
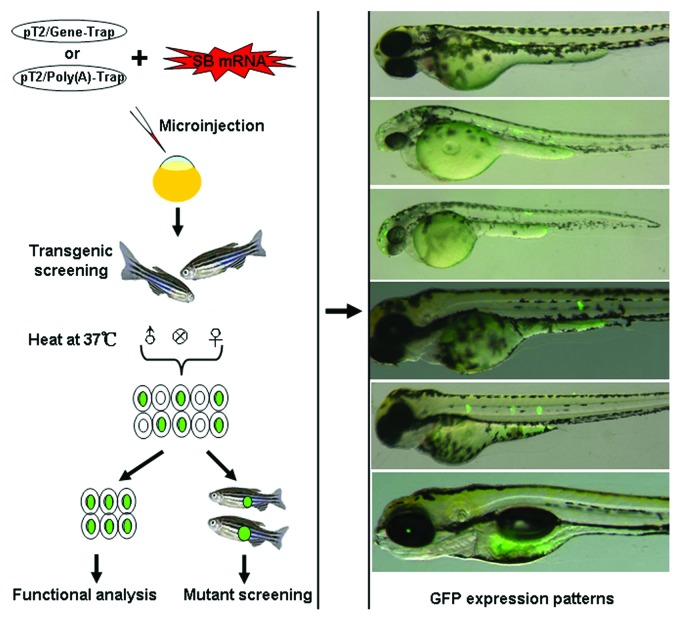
The integration of two gene trapping cassettes into target genomes is meditated by the SB transposon. It is known that exons account for about 1% of the human genome and approximately 24% of the genome is spanned by introns.39 There still exist large fractions of non-coding sequences in the human genome. Given that the copy numbers of SB-mediated inserts are usually less than 10 copies per genome,40,41 the possibility of mutating a gene across a target genome by limited number of transposon inserts remains relatively low. However, a low copy number of inserts allows easier attribution of a mutation to a phenotypic effect and favors the investigation of gene functions. To balance the requirements, an effective cassette was introduced into our gene- and poly(A)-trapping vectors. This cassette controls the expression of SB11 transposes by a Hsp70 promoter from tilapia. The heat-inducible expression of SB transposes leads to the remobilization of transposons inserted in non-coding regions of a target genome and thus improves the efficiency of trapping endogenous genes. Indeed, the trapping cassette can be excised from the original insertion site and new integration patterns were generated after the inducible expression of SB11 transposase.29 Recently, we have obtained 22 stable zebrafish lines from 300 individuals with different patterns of GFP expression by using these two novel trapping vectors (Fig. 3), their trapping rates were about the same as that of the gene trap vector pT2/PAT5.7To avoid the leaky expression of SB transposes gene, the TiHsp7-SB11 cassette was flanked by two Cre/loxP sites for inducible deletion of this cassette by microinjection of capped Cre recombinase mRNA into zebrafish embryos to stabilize the mutants.
Figure 3. The schematic of gene trapping in zebrafish. The transposon-based gene trapping constructs pT2/Gene-Trap or pT2/Poly(A)-Trap and in vitro synthesized SB11 mRNA were co-microinjected into one-cell stage zebrafish embryos to generate transgenic zebrafish lines carrying the gene trapping cassette. Transgenic individuals expressing GFP can be directly used for the analysis of interrupted genes, while other adults showing no GFP expression can be treated with heat-shock during spermatogenesis or oogenesis to obtain offspring with novel insertions and GFP expression. A few of zebrafish individuals expressing GFP in distinct tissues are shown in the right panel.
DNA transposons have a unique ability to move about in the genome. This inherent feature allows these transposable elements to be harnessed as gene vectors for genetic manipulation. In invertebrate models such as C. elegans and Drosophila, transposons have been successfully employed for transgenesis and insertional mutagenesis, but until the reactivation of the Sleeping Beauty transposon in 1997,42 there was no indication of such transposons sufficiently active in vertebrates. The reconstructed SB system is composed of a DNA transposon and a transposase that are both indispensable for transposition. The transposon contains two inverted repeat/direct repeat (IR/DR) elements, in which any DNA sequence such as gene of interest or trapping cassette can be introduced. The transposase was resurrected through the correction of accumulated mutations in extinct transposase sequences found in the genomes of salmonid fish. Typically, binding of two transposase molecules to each IR/DR is required for SB transposition via a “cut-and-paste” mechanism. This system was later improved by further corrections of mutation in IR/DR and SB gene to generate the T2 transposon,43 SB11 transposase44 and hyperactive SB 100X transposase.45 These new versions of the SB system appear to be more active than the original one and are highly effective in vertebrates.45-48
Over the past two decades, the SB transposon system was found to be highly active in human cells, fish, mouse, frog and rat.49 Accordingly, the SB transposon system is successfully used for long-term expression in transgenesis,47,48,50 insertional mutagenesis in animals11 and therapeutic somatic gene vehicle.51 Moreover, SB-mediated integration exhibits less regional preference than retroviruses and is not significantly influenced by transcriptional activity.52,53 Therefore, the SB transposon is widely accepted as one of ideal tools for gene transfer and insertional mutagenesis. Obviously, our SB-based trapping vectors are alternative tools for large-scale mutagenesis in cells and vertebrates and will contribute to the saturation gene mutation across the genomes of model animals including mouse and zebrafish.
Citation: Song G, Cui Z. Novel strategies for gene trapping and insertional mutagenesis mediated by Sleeping Beauty transposon. Mobile Genetic Elements 2013; 3:e26499; 10.4161/mge.26499
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/26499
References
- 1.Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 2.Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 3.de Bruijn E, Cuppen E, Feitsma H. Highly Efficient ENU Mutagenesis in Zebrafish. Methods Mol Biol. 2009;546:3–12. doi: 10.1007/978-1-60327-977-2_1. [DOI] [PubMed] [Google Scholar]
- 4.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–7. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–40. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- 6.Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–44. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Sivasubbu S, Balciunas D, Davidson AE, Pickart MA, Hermanson SB, Wangensteen KJ, Wolbrink DC, Ekker SC. Gene-breaking transposon mutagenesis reveals an essential role for histone H2afza in zebrafish larval development. Mech Dev. 2006;123:513–29. doi: 10.1016/j.mod.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Allende ML, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10:3141–55. doi: 10.1101/gad.10.24.3141. [DOI] [PubMed] [Google Scholar]
- 9.Wang D, Jao LE, Zheng N, Dolan K, Ivey J, Zonies S, Wu X, Wu K, Yang H, Meng Q, et al. Efficient genome-wide mutagenesis of zebrafish genes by retroviral insertions. Proc Natl Acad Sci U S A. 2007;104:12428–33. doi: 10.1073/pnas.0705502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding Y, Liu W, Deng Y, Jomok B, Yang J, Huang W, Clark KJ, Zhong TP, Lin X, Ekker SC, et al. Trapping cardiac recessive mutants via expression-based insertional mutagenesis screening. Circ Res. 2013;112:606–17. doi: 10.1161/CIRCRESAHA.112.300603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grabundzija I, Izsvák Z, Ivics Z. Insertional engineering of chromosomes with Sleeping Beauty transposition: an overview. Methods Mol Biol. 2011;738:69–85. doi: 10.1007/978-1-61779-099-7_5. [DOI] [PubMed] [Google Scholar]
- 12.Ivics Z, Li MA, Mátés L, Boeke JD, Nagy A, Bradley A, Izsvák Z. Transposon-mediated genome manipulation in vertebrates. Nat Methods. 2009;6:415–22. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawakami K, Abe G, Asada T, Asakawa K, Fukuda R, Ito A, Lal P, Mouri N, Muto A, Suster ML, et al. zTrap: zebrafish gene trap and enhancer trap database. BMC Dev Biol. 2010;10:105. doi: 10.1186/1471-213X-10-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trinh A, Fraser SE. Enhancer and gene traps for molecular imaging and genetic analysis in zebrafish. Dev Growth Differ. 2013;55:434–45. doi: 10.1111/dgd.12055. [DOI] [PubMed] [Google Scholar]
- 15.Davidson AE, Balciunas D, Mohn D, Shaffer J, Hermanson S, Sivasubbu S, Cliff MP, Hackett PB, Ekker SC. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev Biol. 2003;263:191–202. doi: 10.1016/j.ydbio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Keng VW, Villanueva A, Chiang DY, Dupuy AJ, Ryan BJ, Matise I, Silverstein KA, Sarver A, Starr TK, Akagi K, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27:264–74. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGrail M, Hatler JM, Kuang X, Liao HK, Nannapaneni K, Watt KE, Uhl JD, Largaespada DA, Vollbrecht E, Scheetz TE, et al. Somatic mutagenesis with a Sleeping Beauty transposon system leads to solid tumor formation in zebrafish. PLoS One. 2011;6:e18826. doi: 10.1371/journal.pone.0018826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Donnell KA, Keng VW, York B, Reineke EL, Seo D, Fan D, Silverstein KA, Schrum CT, Xie WR, Mularoni L, et al. A Sleeping Beauty mutagenesis screen reveals a tumor suppressor role for Ncoa2/Src-2 in liver cancer. Proc Natl Acad Sci U S A. 2012;109:E1377–86. doi: 10.1073/pnas.1115433109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.March HN, Rust AG, Wright NA, ten Hoeve J, de Ridder J, Eldridge M, van der Weyden L, Berns A, Gadiot J, Uren A, et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat Genet. 2011;43:1202–9. doi: 10.1038/ng.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagayoshi S, Hayashi E, Abe G, Osato N, Asakawa K, Urasaki A, Horikawa K, Ikeo K, Takeda H, Kawakami K. Insertional mutagenesis by the Tol2 transposon-mediated enhancer trap approach generated mutations in two developmental genes: tcf7 and synembryn-like. Development. 2008;135:159–69. doi: 10.1242/dev.009050. [DOI] [PubMed] [Google Scholar]
- 21.Abe G, Suster ML, Kawakami K. Tol2-mediated transgenesis, gene trapping, enhancer trapping, and the Gal4-UAS system. Methods Cell Biol. 2011;104:23–49. doi: 10.1016/B978-0-12-374814-0.00002-1. [DOI] [PubMed] [Google Scholar]
- 22.Clark KJ, Balciunas D, Pogoda HM, Ding Y, Westcot SE, Bedell VM, Greenwood TM, Urban MD, Skuster KJ, Petzold AM, et al. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat Methods. 2011;8:506–15. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni TT, Lu J, Zhu M, Maddison LA, Boyd KL, Huskey L, Ju B, Hesselson D, Zhong TP, Page-McCaw PS, et al. Conditional control of gene function by an invertible gene trap in zebrafish. Proc Natl Acad Sci U S A. 2012;109:15389–94. doi: 10.1073/pnas.1206131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maddison LA, Lu J, Chen W. Generating conditional mutations in zebrafish using gene-trap mutagenesis. Methods Cell Biol. 2011;104:1–22. doi: 10.1016/B978-0-12-374814-0.00001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trinh A, Hochgreb T, Graham M, Wu D, Ruf-Zamojski F, Jayasena CS, Saxena A, Hawk R, Gonzalez-Serricchio A, Dixson A, et al. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes Dev. 2011;25:2306–20. doi: 10.1101/gad.174037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–23. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 27.Skarnes WC, Auerbach BA, Joyner AL. A gene trap approach in mouse embryonic stem cells: the lacZ reported is activated by splicing, reflects endogenous gene expression, and is mutagenic in mice. Genes Dev. 1992;6:903–18. doi: 10.1101/gad.6.6.903. [DOI] [PubMed] [Google Scholar]
- 28.Stanford WL, Cohn JB, Cordes SP. Gene-trap mutagenesis: past, present and beyond. Nat Rev Genet. 2001;2:756–68. doi: 10.1038/35093548. [DOI] [PubMed] [Google Scholar]
- 29.Song G, Li Q, Long Y, Gu Q, Hackett PB, Cui Z. Effective gene trapping mediated by Sleeping Beauty transposon. PLoS One. 2012;7:e44123. doi: 10.1371/journal.pone.0044123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang XQ, Luk JM, Leung PP, Wong BW, Stanbridge EJ, Fan ST. Alternative mRNA splicing of liver intestine-cadherin in hepatocellular carcinoma. Clin Cancer Res. 2005;11:483–9. [PubMed] [Google Scholar]
- 31.Bonaldo P, Chowdhury K, Stoykova A, Torres M, Gruss P. Efficient gene trap screening for novel developmental genes using IRES beta geo vector and in vitro preselection. Exp Cell Res. 1998;244:125–36. doi: 10.1006/excr.1998.4208. [DOI] [PubMed] [Google Scholar]
- 32.Zhu P, Narita Y, Bundschuh ST, Fajardo O, Schärer YP, Chattopadhyaya B, Bouldoires EA, Stepien AE, Deisseroth K, Arber S, et al. Optogenetic Dissection of Neuronal Circuits in Zebrafish using Viral Gene Transfer and the Tet System. Front Neural Circuits. 2009;3:21. doi: 10.3389/neuro.04.021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niwa H, Araki K, Kimura S, Taniguchi S, Wakasugi S, Yamamura K. An efficient gene-trap method using poly A trap vectors and characterization of gene-trap events. J Biochem. 1993;113:343–9. doi: 10.1093/oxfordjournals.jbchem.a124049. [DOI] [PubMed] [Google Scholar]
- 34.Zambrowicz BP, Friedrich GA, Buxton EC, Lilleberg SL, Person C, Sands AT. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature. 1998;392:608–11. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 35.Salminen M, Meyer BI, Gruss P. Efficient poly A trap approach allows the capture of genes specifically active in differentiated embryonic stem cells and in mouse embryos. Dev Dyn. 1998;212:326–33. doi: 10.1002/(SICI)1097-0177(199806)212:2<326::AID-AJA17>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 36.Horie K, Yusa K, Yae K, Odajima J, Fischer SEJ, Keng VW, Hayakawa T, Mizuno S, Kondoh G, Ijiri T, et al. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol Cell Biol. 2003;23:9189–207. doi: 10.1128/MCB.23.24.9189-9207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker KE, Parker R. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol. 2004;16:293–9. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Song G, Li Q, Long Y, Hackett PB, Cui Z. Effective expression-independent gene trapping and mutagenesis mediated by Sleeping Beauty transposon. J Genet Genomics. 2012;39:503–20. doi: 10.1016/j.jgg.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1305–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 40.Clark KJ, Geurts AM, Bell JB, Hackett PB. Transposon vectors for gene-trap insertional mutagenesis in vertebrates. Genesis. 2004;39:225–33. doi: 10.1002/gene.20049. [DOI] [PubMed] [Google Scholar]
- 41.Kolacsek O, Krízsik V, Schamberger A, Erdei Z, Apáti A, Várady G, Mátés L, Izsvák Z, Ivics Z, Sarkadi B, et al. Reliable transgene-independent method for determining Sleeping Beauty transposon copy numbers. Mob DNA. 2011;2:5. doi: 10.1186/1759-8753-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–10. doi: 10.1016/S0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 43.Cui Z, Geurts AM, Liu G, Kaufman CD, Hackett PB. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol. 2002;318:1221–35. doi: 10.1016/S0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 44.Geurts AM, Yang Y, Clark KJ, Liu GY, Cui ZB, Dupuy AJ, Bell JB, Largaespada DA, Hackett PB. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–17. doi: 10.1016/S1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 45.Mátés L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–61. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 46.Izsvák Z, Chuah MK, Vandendriessche T, Ivics Z. Efficient stable gene transfer into human cells by the Sleeping Beauty transposon vectors. Methods. 2009;49:287–97. doi: 10.1016/j.ymeth.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 47.Newman M, Lardelli M. A hyperactive sleeping beauty transposase enhances transgenesis in zebrafish embryos. BMC Res Notes. 2010;3:282. doi: 10.1186/1756-0500-3-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mátés L. Rodent transgenesis mediated by a novel hyperactive Sleeping Beauty transposon system. Methods Mol Biol. 2011;738:87–99. doi: 10.1007/978-1-61779-099-7_6. [DOI] [PubMed] [Google Scholar]
- 49.Ivics Z, Izsvák Z. The expanding universe of transposon technologies for gene and cell engineering. Mob DNA. 2010;1:25. doi: 10.1186/1759-8753-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlson DF, Geurts AM, Garbe JR, Park CW, Rangel-Filho A, O’Grady SM, Jacob HJ, Steer CJ, Largaespada DA, Fahrenkrug SC. Efficient mammalian germline transgenesis by cis-enhanced Sleeping Beauty transposition. Transgenic Res. 2011;20:29–45. doi: 10.1007/s11248-010-9386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ammar I, Izsvák Z, Ivics Z. The Sleeping Beauty transposon toolbox. Methods Mol Biol. 2012;859:229–40. doi: 10.1007/978-1-61779-603-6_13. [DOI] [PubMed] [Google Scholar]
- 52.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–94. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]