Abstract
Genetic alterations of the maternal UBE3A allele result in Angelman syndrome (AS), a neurodevelopmental disorder characterized by severe developmental delay, lack of speech, and difficulty with movement and balance. The combined effects of maternal UBE3A mutation and cell type-specific epigenetic silencing of paternal UBE3A are hypothesized to result in a complete loss of functional UBE3A protein in neurons. However, the allelic specificity of UBE3A expression in neurons and other cell types in the brain has yet to be characterized throughout development, including the early postnatal period when AS phenotypes emerge. Here we define maternal and paternal allele-specific Ube3a protein expression throughout postnatal brain development in the mouse, a species which exhibits orthologous epigenetic silencing of paternal Ube3a in neurons and AS-like behavioral phenotypes subsequent to maternal Ube3a deletion. We find that neurons downregulate paternal Ube3a protein expression as they mature and, with the exception of neurons born from postnatal stem cell niches, do not express detectable paternal Ube3a beyond the first postnatal week. By contrast, neurons express maternal Ube3a throughout postnatal development, during which time localization of the protein becomes increasingly nuclear. Unlike neurons, astrocytes and oligodendrotyes biallelically express Ube3a. Notably, mature oligodendrocytes emerge as the predominant Ube3a-expressing glial cell type in the cortex and white matter tracts during postnatal development. These findings demonstrate the spatiotemporal characteristics of allele-specific Ube3a expression in key brain cell types, thereby improving our understanding of the developmental parameters of paternal Ube3a silencing and the cellular basis of AS.
Keywords: Angelman syndrome, AS model mice, genomic imprinting, epigenetic silencing, E6-AP
INTRODUCTION
In 1965, Harry Angelman first described the neurodevelopmental disorder that is known today as Angelman syndrome (AS) (Bower and Jeavons, 1967). AS is characterized by a constellation of symptoms that emerge during early childhood, including developmental delay, ataxia, lack of speech, paroxysmal laughter, seizures, and frequently comorbid autism (Peters et al., 2004; Williams et al., 2006; Thibert et al., 2013). Large maternal deletions (approximately 6 megabases) of the 15q11-q13 chromosomal region, which encompasses UBE3A, are the most common genetic cause of AS (Williams et al., 2010). UBE3A encodes a HECT (homologous to E6-associated protein carboxyl-terminus) domain E3 ubiquitin ligase (Scheffner et al., 1993; Yamamoto et al., 1997), and is the only gene within the 15q11-q13 region whose expression is unequivocally maternally biased (Leung et al., 2011). This bias is due to neuron-specific epigenetic silencing of the paternal UBE3A allele (Albrecht et al., 1997; Yamasaki, 2003; Landers et al., 2004). Critical UBE3A function may thus be completely lost in neurons when maternal UBE3A is compromised, and this may be the proximal molecular cause of AS. Indeed, deletions or mutations of maternal UBE3A alone are sufficient to result in an AS diagnosis, including those that selectively abrogate the ubiquitin ligase functionality of UBE3A protein (Cooper et al., 2004). Co-occurring haploinsufficiency of biallelically expressed genes in the 15q11-q13 region is thought to enhance the severity of AS phenotypes in large deletion cases (Moncla et al., 1999; Lossie et al., 2001; Gentile et al., 2010).
The hypothesis that maternal UBE3A loss in neurons is sufficient to cause AS, however parsimonious, has yet to be directly tested. In fact, relatively little is known about allelic contributions to UBE3A protein expression in neurons and glia during brain development. Three overarching questions remain to be answered: 1) Is paternal UBE3A completely silenced in neurons and, if so, by what developmental time point? 2) Which glial cells in the brain express UBE3A? 3) What are the allelic contributions to glial UBE3A expression? The mouse is a suitable model species in which to address these questions; the orthologous mouse Ube3a gene resides within a chromosomal region that is syntenic with human 15q11-q13 and is subject to paternal epigenetic silencing in neurons (Nicholls and Knepper, 2001). Moreover, mice lacking a functional copy of maternal Ube3a (AS mice) exhibit AS-like phenotypes (Jiang et al., 1998; Miura et al., 2002). Some AS-relevant phenotypes are more severe in Ube3a double knock-out mice than in AS mice (Jiang et al., 1998; Heck et al., 2008). This suggests that paternal Ube3a expression also contributes to brain development and function, though paternal Ube3a deletion per se does not cause AS-like phenotypes (Jiang et al., 1998; Miura et al., 2002; Heck et al., 2008; Mulherkar and Jana, 2010). It is possible that immature neurons express and require paternal Ube3a expression during early postnatal development (Leung et al., 2009; Sato and Stryker, 2010). Consequently, neuronal function may be maximally impaired in Ube3a double knock-out mice. Glial cells are presumed to biallelically express Ube3a, regardless of their maturational status (Yamasaki, 2003). Glia may also require two functional Ube3a alleles for optimal development and function.
In the present study we took advantage of preexisting mouse models and complementary immunohistochemical approaches to reliably and extensively track allele-specific Ube3a protein expression. As a result, we further resolved the developmental dynamics of maternal and paternal Ube3a protein expression in neurons and glia and, in turn, improved our understanding of how maternal UBE3A loss might contribute to AS pathogenesis.
MATERIALS AND METHODS
Breeding and genotyping mice
Ube3a knock-out mice were generated by the laboratory of A. Beaudet (Jiang et al., 1998) and backcrossed to a congenic C57BL/6J background by Y. Jiang. Angelman syndrome (AS) model mice (Ube3am–/p+) maternally inherit the Ube3a knock-out allele and were typically generated by mating wild-type (Ube3am+/p+) male mice to female mice with paternal inheritance of the Ube3a knock-out allele (Ube3am+/p–). Female Ube3am+/p– mice were mated to male Ube3am+/p– mice to produce litters comprised of Ube3am+/p+, Ube3am+p/–, Ube3am–/p+, and double knock-out (Ube3am–/p–) offspring. Ube3a knock-out mice were genotyped via polymerase chain reaction (PCR). PCR primers used to amplify the wild-type Ube3a allele were: Ube3a Forward 5’-TGAGTTTCCCCTGATGGAAG -3’ and Ube3a Reverse 5’-GGAAATTCACTACTACCTCCCAAA -3’. Primers used to amplify the Ube3a knock-out allele were: Neomycin Forward 5’-AGACAATCGGCTGCTCTGAT-3’ and Neomycin Reverse 5’-ATACTTTCTCGGCAGGAGCA -3’.
Knock-in mice expressing yellow fluorescent protein (YFP) fused to the carboxyl-terminus of Ube3a were generated and generously provided by the laboratory of A. Beaudet (Dindot et al., 2008) and were maintained on a congenic C57BL/6J background. Mice with maternal Ube3a-YFP inheritance (Ube3amYFP/p+) were generated by mating wild-type males to paternal Ube3a-YFP (Ube3am+/pYFP) females. The reciprocal mating scheme was used to generate Ube3am+/pYFP offspring. EGFP Forward 5’-CACATGAAGCAGCACGACTT-‘3 and EGFP Reverse 5’-AGTTCACCTTGATGCCGTTC-‘3 genotyping primers were used to amplify the Ube3a-YFP knock-in allele.
A cross-sectional approach was employed to assess allele-specific expression patterns of endogenous Ube3a and Ube3a-YFP throughout postnatal development. For each genetic cross of interest, brains from at least 3 sets of littermates and from at least two independent litters were analyzed. All research procedures using mice were approved by the Institutional Animal Care and Use Committee at the University of North Carolina, Chapel Hill and conformed to NIH guidelines.
Histology and immunostaining
Postnatal mice were deeply anesthetized with sodium pentobarbital (60 mg/kg i.p.) prior to transcardial perfusion with room temperature phosphate-buffered saline (PBS) immediately followed by room temperature phosphate-buffered 4% paraformaldehyde (pH 7.3). Perfused brains were removed from their skulls and post-fixed overnight at 4°C prior to being cryoprotected via sequential 12-hour incubations in 10%, 20%, and 30% sucrose in PBS (pH 7.5). Cryoprotected brains were frozen on dry ice and cut into 40 μm-thick sections with a sliding microtome (Thermo Scientific, Kalamazoo, MI, USA). Sections were stored in a cryopreservative solution (by volume: 45% PBS, 30% ethylene glycol, 25% glycerol) at −20°C until they were processed for free-floating immunohistochemistry.
For chromogenic staining, sections were rinsed several times in Tris-buffered saline containing 0.3% Triton-X-100 (TBST) prior to antigen retrieval with 10 mM Na Citrate pH 6.0 with 0.05% Triton-X-100 (heated to 75°C for 20 minutes). Additional rinsing in TBST preceded two blocking steps. First, sections were incubated for 5 minutes in H2O2 (0.3% in MeOH) to quench endogenous peroxidases, followed by TBST rinsing. Second, sections were blocked in 4% nonfat milk in TBST (Blotto) for 25 minutes at room temperature. Blocked tissue sections were incubated in primary antibodies diluted in Blotto for 48-72 hours at 4°C. After incubation in primary antibodies, sections were rinsed several times in Blotto before incubation for 2 hours at room temperature with biotinylated donkey anti-mouse secondary antibodies (Jackson Immunoresearch, West Grove, PA, 715-066-150) diluted 1:1,000 in Blotto. Sections were then rinsed in TBST prior to tertiary amplification with the ABC elite avidin-biotin-peroxidase system (Vector Labs, Burlingame, CA, USA). Further rinsing in TBST preceded a 10 minute incubation at room temperature in 3’3’-diaminobenzidine (DAB) chromogenic substrate (0.02% DAB and 0.01% H2O2 in TBST) to visualize immune complexes amplified by avidin-biotin-peroxidase.
For immunofluorescent staining, sections were rinsed several times in PBS before blocking in PBS plus 5% normal goat serum and 0.2% Triton-X-100 (NGST) for 1 hour at room temperature. Blocked tissue sections were incubated in primary antibodies diluted in NGST for 48 hours at 4°C. Sections were then rinsed several times in PBS containing 0.2% Triton-X-100 (PBST) before incubation in secondary antibodies (also diluted in NGST) for 1 hour at room temperature. The following secondary antibodies from Invitrogen (Carlsbad, CA, USA) were used at a 1:500 dilution: goat anti-mouse Alexa 633 (A21052); goat anti-rabbit Alexa 568 (A11011); goat anti-chicken Alexa 488 (A11039); goat anti-rabbit Alexa 633 (A21052); goat anti-mouse IgG1 Alexa 568 (A21124); goat anti-mouse IgG2a Alexa 488 (A21131). In most experiments, 4',6-diamidino-2-phenylindole (DAPI, Invitrogen D1306) was added during the secondary antibody incubation at a concentration of 700 ng/mL for nuclear counterstaining. Brain sections compared within figures were stained within the same experiment, under identical conditions.
Western Blotting
Mice were deeply anesthetized with sodium pentobarbital (60 mg/kg i.p.) prior to decapitation and brain removal. The neocortical hemispheres and hippocampi from each harvested brain were rapidly dissected in ice-cold PBS and then immediately snap-frozen with liquid nitrogen and stored at −80°C. Frozen tissue samples were homogenized in a glass tissue homogenizer (Wheaton, Millville, NJ, USA) with ice-cold RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% triton x-100, 0.1% SDS, 0.5% Na Deoxycholate) supplemented with 2 mM EDTA and a protease inhibitor cocktail (Sigma, Saint Louis, MO, USA). Tissue homogenates were cleared by 16,000 × g centrifugation for 20 minutes at 4°C and protein concentrations of the supernatants were determined using the BCA assay (Thermo Scientific). Protein samples (30 μg per lane) were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked for 1 hour at room temperature in Odyssey blocking buffer (Li-COR, Lincoln, NE, USA) prior to incubation overnight at 4°C with primary antibodies diluted in a 1:1 solution of blocking buffer and Tris-buffered saline containing 0.1% Tween-20 (TBSTween). Membranes were subsequently washed repeatedly with TBSTween prior to incubation for 1 hour at room temperature with secondary antibodies prepared in the same diluent as the primary antibodies. The following secondary antibodies were used: 1:10,000 donkey anti-mouse 800CW (LI-COR, 926-32212); 1:5,000 donkey anti-rabbit Alexa 680 (Invitrogen, A10043). Finally, blots were washed repeatedly in TBSTween followed by TBS alone prior to imaging with the Odyssey imaging system (LI-COR).
Antibody characterization
The list of employed primary antibodies is provided in Table 1. APC antibody weakly stained astrocytes and immature oligodendrocytes and intensely stained mature oligodendrocytes in white matter tracts and in deep layers of the cerebral cortex, consistent with manufacturer's specifications and previous reports (Dimou et al., 2008).
Table of Primary Antibodies used
| Antigen | Immunogen | Manufacturer | Dilution used |
|---|---|---|---|
| APC | recombinant peptide corresponding to a.a.1-226 of APC | Millipore (Billerica, MA), mouse monoclonal, clone CC-1, OP80 | 1:500 |
| Cplx3 | recombinant full-length mouse Cplx3 | Synaptic Systems (Goettingen, DE), rabbit polyclonal, 122-302 | 1:1,000 |
| DARPP-32 | peptide corresponding to a.a. 20-40 of mouse DARPP-32 | Millipore, rabbit polyclonal, AB10518 | 1:250 |
| GAPDH | GAPDH purified from rabbit muscle | Millipore, mouse monoclonal, clone 6C5, MAB374 | 1:5,000 |
| GFAP | GFAP purified from cow spinal cord | Dako (Glostrup, DK), rabbit polyclonal, Z0334 | 1:1,000 |
| GFP | purified recombinant GFP | Aves Labs (Tigard, OR), chicken polyclonal, GFP-1020 | 1:2,500 |
| Ki67 | Prokaryotic recombinant fusion protein corresponding to a 1088 bp Ki67 motif-containing cDNA fragment | Leica (Wetzlar, DE), rabbit polyclonal, Ki67p | 1:2,000 |
| Olig2 | recombinant mouse Olig2 | Millipore, rabbit polyclonal, AB9610 | 1:500 |
| Nestin | Nestin purified from embryonic rat spinal cord | Millipore, mouse monoclonal, clone rat-401, MAB353 | 1:2,000 |
| NeuN | purified nuclei from mouse brain | Millipore, mouse monoclonal, clone A60, MAB377 | 1:500 |
| Ube3a | recombinant full-length human UBE3A | Sigma-Aldrich (St. Louis, MO), mouse monoclonal, clone E6AP-330, E8655 | 1:200 (chromogenic staining) |
| Ube3a | peptide corresponding to a.a. 315-415 (isoforms 1 and 3) or a.a. 336-436 (isoform 2) of mouse Ube3a | Sigma-Aldrich, mouse monoclonal, clone 3E5, SAB1404508 | 1:750 (fluorescent staining) 1:1,000 (Western blotting) |
Note: a.a., amino acids; bp, base pair; APC, adenomatous polyposis coli; Cplx3, Complexin 3; DARPP-32, Dopamine and cAMP-Regulated Phosphoprotein-32 kD; GAPDH, Glyceraldehyde-3-phosphate dehydrogenase; GFAP, Glial Fibrillary Acid Protein; GFP, Green Fluorescent Protein; Olig2, Oligodendrocyte transcription factor 2; NeuN, Neuronal Nuclei; Ube3a, Ubiquitin protein ligase e3a.
Cplx3 antibody does not stain brain tissue from Cplx3–/– mice (Reim et al., 2009). In our hands this antibody specifically stained a subpopulation of subplate neurons in the cerebral cortex, as predicted by a previous study (Hoerder-Suabedissen and Molnar, 2013).
DARPP-32 antibody recognizes a single band of 32 kD on Western blots of rat brain lysate (manufacturer's specifications). We observed this antibody to specifically stain patches of cells in the early postnatal brain that co-localized with tyrosine-hydroxylase-stained axon terminals (data not shown).
GAPDH antibody recognized a single band of approximately 37 kDa in Western blotting experiments, consistent with previous experiments (Jones et al., 2008).
GFAP antibody was previously shown to exhibit a lack of staining in GFAP–/– brain tissue (Hanbury et al., 2003). We observed this antibody to exclusively stain NeuN-negative cells with characteristic astrocytic morphologies throughout the brain.
GFP antibody was previously shown to stain only brain cells in mice that express transgenic GFP (Xu et al., 2006). We observed GFP antibody to produce staining patterns in Ube3a-YFP knockin mice that were nearly identical to endogenous Ube3a expression patterns. We observed no staining in wild type mice using this antibody.
Ki67 antibody has been shown repeatedly to co-localize with short-pulse BrdU-labeled proliferating cells (Mathews et al., 2010; Tseng et al., 2010). We found this antibody to stain sparse populations of Nestin-positive cells in known proliferative niches of the postnatal brain.
Olig2 antibody specifically labels cells of the oligodendrocyte lineage (Tripathi and McTigue, 2008), consistent with our own observations.
Nestin antibody was previously shown to specifically stain neural precursors (Kao et al., 2008). Consistent with this observation, we found that this antibody specifically stains cells in the adult subventricular zone and subgranular zone of the dentate gyrus in postnatal mice in which proliferating, Ki67-positive cells were also prominently localized.
NeuN antibody stains only neuronal perikarya and nuclei (Encinas et al., 2011). We consistently observed this antibody to stain neuronal populations and never observed this antibody to co-localize with glial markers.
Ube3a antibodies used in this study consistently showed drastically reduced immunoreactivity in Ube3am–/p+ brain tissue samples and a complete lack of immunoreactivity in Ube3am–/p– brain tissue samples.
Imaging and figure production
Images of brain sections stained with DAB histochemistry were acquired with a Nikon Eclipse 80i microscope (Nikon, Melville, NY, USA) equipped with Surveyor mosaic imaging software (Objective Imaging Ltd., Cambridge, United Kingdom). Images of brain sections stained using fluorophore-conjugated secondary antibodies were acquired with a Zeiss LSM 710 confocal microscope equipped with ZEN imaging Software (Zeiss, Jena, Germany). Images compared within figures were taken within the same imaging session using identical acquisition parameters. Occasionally, images within figure panels were linearly adjusted for brightness and contrast using Image J software (Schneider et al., 2012). All images to be compared underwent identical manipulations with the exception that the relative brightness of the Ube3a signal for some Ube3am–/p– images had to be enhanced so that low levels of non-specific background staining could be appreciated. This was due to the large dynamic range of the signal and our preference to avoid saturating the Ube3a signal in Ube3am+/p+ samples during imaging. These instances are clearly noted in the figure legends. Figures were prepared using Adobe Illustrator software (Adobe Systems Inc., San Jose, CA, USA).
RESULTS
To begin mapping allele-specific patterns of Ube3a expression in the developing postnatal brain, we utilized an immunohistochemical protocol that was previously shown to specifically stain Ube3a protein in the adult brain (Gustin et al., 2010). We applied this protocol to brain sections from wild-type (Ube3am+/p+) mice across postnatal development and, in general, we observed a broad distribution of staining (Fig. 1A). Upon closer inspection, we saw discrete patterns of Ube3a staining within the deepest laminae of the neocortex and in patches of cells in the striatum during the first postnatal week (Fig. 1C, D, M, N). These patterns were less apparent by the end of the second postnatal week (Fig. 1E, O), as staining in adjacent layers (neocortex) or surrounding cells (striatum) became equivalent. A broad, relatively uniform distribution of Ube3a staining persisted into adulthood (Fig. 1F, G, P, Q).
Figure 1.
Profiling Ube3a expression in the postnatal brain using an established immunostaining protocol. A and B: DAB immunostaining for Ube3a (as per Gustin et al., 2010) in coronal brain sections from wild-type (A, Ube3am+/p+) and AS (B, Ube3am–/p+) mice across postnatal development. Lowmagnification reveals broadly distributed Ube3a staining in the brain at all ages, which is grossly absent in Ube3am–/p+ mice (B). C-G: In Ube3am+/p+ mice, intermediate-magnification (corresponding to boxed regions in panel A) reveals discrete patterns of Ube3a staining in the deepest neocortical layers (arrows, panels C, D) and in patches of striatal cells (double arrows, panels C, D) from P0-P7. Ube3a staining within the neocortex and striatum becomes more evenly distributed by P14 (E). This staining pattern persists into adolescence (F) and throughout adulthood (G). H-L: Intermediate-magnification of boxed regions corresponding to panel B demonstrate lack of specific staining patterns in Ube3am–/p+ mice. M-Q: Highmagnification (boxed regions, panels C-G), provides greater resolution of Ube3a staining patterns in deep neocortical layers (arrows, panels M, N) and in striatal patches (double arrows, panels M, N), which become less salient over development (O-Q). R-V: High-magnification corresponding to boxed regions in H-L. No specific staining is detected in Ube3am–/p+ images, confirming that paternal Ube3a immunoreactivity is not reliably detected in the brain at any postnatal age using this protocol. Ctx, neocortex; Str, striatum; wm, white matter. Scale bar: 2 mm for A and B; 650 μm for C-L; 170 μm for M-V.
In parallel, we stained brain sections from littermate AS model mice (Ube3am–/p+ mice). The Ube3a null allele produces neither transcript nor protein (Jiang et al., 1998); consequently, any staining in Ube3am–/p+ mice is likely owed to the presence of protein generated from the paternal Ube3a allele. We did not detect paternal Ube3a staining at any of the postnatal time points we examined (Fig. 1B, H-L, R-V). This was somewhat surprising considering that paternal Ube3a constitutes about 5% of total Ube3a protein as demonstrated by comparative Western analysis of Ube3am+/p+ and Ube3am–/p+ brain lysates (Gustin et al., 2010).
We considered insufficient assay sensitivity to be the most likely explanation for our failure to detect paternal Ube3a staining with this previously published protocol. Therefore, we developed an immunofluorescent Ube3a staining protocol with the goal of detecting low levels of sparsely distributed paternal Ube3a protein. With low magnification, we observed gross patterns of Ube3a staining across development that were nearly identical to what we previously observed; Ube3a was broadly distributed throughout the Ube3am+/p+ brain yet most notable in deep cortical laminae and patches of striatal cells during the first postnatal week (Fig. 2A-C). Staining was grossly absent in age-matched Ube3am–/p+ brain sections (Fig. 2D-F), save for an intriguing pattern of signal in the superficial neocortex of perinatal mice (Fig. 2D). However, with increased magnification we were able to detect paternal Ube3a staining at all postnatal ages in small cells resembling glia (Fig. 2J-L), a staining pattern which was absent in Ube3am–/p– brain sections (Fig. 2G-I, M-O). These results indicated that our immunofluorescent Ube3a staining protocol was sensitive enough to detect very low levels of paternal Ube3a in the brain. We implemented this new protocol in a series of co-immunostaining studies designed to characterize the spatiotemporal dynamics of maternal and paternal Ube3a protein expression during postnatal brain development (see below).
Figure 2.
Improved immunofluorescent detection of Ube3a in the postnatal brain. A-C: Immunofluorescent staining for Ube3a in coronal sections from wild-type (Ube3am+/p+) mice at P0 (A), P7 (B), and P28 (C). Gross patterns of staining during the first postnatal week are nearly identical to those observed with DAB staining, with notable Ube3a immunoreactivity in deep laminae of the neocortex (arrows, A, B) as well as in patches of striatal cells (double arrow, A); staining is more uniformly distributed at P28 (C). D-F: Immunofluorescent staining for paternal Ube3a in matched brain sections from AS mouse (Ube3am–/p+) littermates. Low levels of paternal Ube3a expression are consistently observed during postnatal development in specific structures and cell types throughout the brain, especially in the superficial neocortex at P0 (D). G-I: Gross immnofluorescent staining for Ube3a is not observed in negative control sections from double-knockout (Ube3am–/p–) mice. J-L: High-magnification of the boxed regions in D-F reveals paternal Ube3a staining in presumptive glia (arrowheads). M-O: No specific staining is apparent in corresponding high-magnification images from Ube3am–/p– mice. Ctx, neocortex; Str, striatum. Scale bar: 2 mm for A-I; 120 μm for J-O.
Dynamic patterns of allele-specific Ube3a expression reflect neuronal maturation in the postnatal brain
Cytoplasmic versus nuclear expression of maternal Ube3a shifts during neuronal maturation
Ube3a staining was most salient in deep neocortical laminae and in patches of striatal cells in the early postnatal brain (Figs. 1A, B and 2A, B). These notable patterns were absent in Ube3am–/p+ mice (Figs. 1F, G and 2D, E), indicating that they were solely the product of expression from the maternal Ube3a allele. Patchy maternal Ube3a staining in the striatum was highly reminiscent of the anatomical compartments referred to as striosomes, which can be distinguished from surrounding matrix compartments by predominantly limbic-related afferent inputs (Gerfen, 1989; Eblen and Graybiel, 1995) as well as the expression of distinct neurochemical markers such as the dopamine- and cAMP-regulated phosphoprotein of 32 kilodaltons (DARPP-32) (Foster et al., 1987). We found patches of maternal Ube3a staining that co-localized perfectly with patches of DARPP-32 staining at postnatal day (P) 0 (Fig. 3A-D) up until P7 (Fig. 3E-H) when the pattern became somewhat less salient. The most salient maternal Ube3a staining in the perinatal neocortex was restricted to a thin band of cells immediately adjacent to the subcortical white matter. We correctly assumed that these were residual neurons of the transient subplate layer as evidenced by co-staining with Complexin3 (Cplx3) (Fig. 3I-L), a known marker of surviving subplate neurons in the postnatal cortex (Hoerder-Suabedissen and Molnar, 2013). This co-localization pattern was still apparent at P7 (Fig. 3M-P), although we also detected maternal Ube3a-positive neuronal nuclei throughout the infragranular neocortical layers at this time point.
Figure 3.
Ube3a expression is salient in the striosomes and subplate during early postnatal development in wild-type (Ube3am+/p+) mice. A-H: Images of Ube3a and DARPP-32 immunostaining in P0 (A-D) and P7 (EH) brains. Increased magnification shows that salient Ube3a staining in the P0 striatum is specific to DARPP-32-positive striosomes (arrows, B-D). Striosomal Ube3a staining is less salient by P7 (arrows, FH). I-P: Images of Ube3a and Cplx3 immunoreactivity in the P0 (I-L) and P7 (M-P) brains. In the perinatal (J-L) and early postnatal (N-P) neocortex, Cplx3-positive subplate neurons exhibit intense Ube3a staining with a predominantly nuclear localization. Ctx, neocortex; wm, white matter. Scale bar: 435 μm for A, E, I, and M; 70 μm for B-D, F-H, J-L, and N-P.
Subplate and striosomal neurons are among the earliest-born neurons in the neocortex (Kostovic and Rakic, 1980; Chun and Shatz, 1989; Price et al., 1997) and striatum (Fishell and van der Kooy, 1987; van der Kooy and Fishell, 1987), respectively. Therefore, the precocious salience of maternal Ube3a staining in these neurons may reflect their advanced maturation relative to later-born neurons overlying or surrounding them. To address this possibility, we evaluated changes in nuclear Ube3a staining in the context of laminar gradients of neocortical maturation, since nuclear localization is a key factor contributing to the salience of the maternal Ube3a stain. We found that a prominent nuclear distribution of maternal Ube3a staining was nearly exclusive to subplate neurons at P0 (Fig. 4A). By P7, neurons throughout infragranular neocortex exhibited prominent nuclear Ube3a staining, whereas staining in overlying superficial layers was still largely cytoplasmic (Fig. 4B). Prominent nuclear maternal Ube3a staining appeared latest in neurons of the most superficial layers (Fig. 4C) in accordance with a deep to superficial (“inside-out”) gradient of cortical development (Angevine and Sidman, 1961; Rakic, 1974). Furthermore, we detected a profound cytoplasmic to nuclear shift in the distribution of maternal Ube3a staining within neocortical layers during development (Fig. 4D-L). Collectively, these findings suggested that the subcellular localization of maternal Ube3a becomes increasingly nuclear as neurons mature.
Figure 4.
Developmental changes in Ube3a expression reflect laminar gradients of neocortical maturation. A-C: Low-magnification images of Ube3a immunostaining in the developing neocortex of wildtype (Ube3am+/p+) mice. At P0, subplate neurons most prominently exhibit nuclear Ube3a immunoreactivity (A). By P7, neurons with notable nuclear staining are seen throughout the infragranular layers (B), but are not observed in supragranular layers until later (C). D-L: Within the superficial cortex, the developmental shift in subcellular Ube3a localization is apparent in high-magnification images captured using thin (1.2 μm) optical sectioning. Note the increase in Ube3a staining (D-F) in the nuclear compartment (labeled with DAPI, G-I and J-L) from P0 to P28. Conversely, Ube3a staining in dendritic processes (arrows) progressively declines over the first postnatal month. cp, cortical plate; sp, subplate. Scale bar: 225 μm for A-C; 25 μm for D-L.
Paternal Ube3a expression in neurons is exclusive to immature cells
Only a few paternal Ube3a staining patterns were conspicuous in the brains of Ube3am–/p+ mice. One particularly remarkable pattern was a deep (low) to superficial (high) gradient in the P0 neocortex (Figs. 2D and 5A). Superficial neocortical neurons are later-born and less mature than their underlying counterparts. Accordingly, the staining gradient for NeuN, a mature neuron marker, was reciprocal to the paternal Ube3a staining gradient (Fig. 5B-D). Neurons beyond the superficial extent of the NeuN gradient stained most intensely for paternal Ube3a (Fig. 5E-G). By P7, we could no longer detect paternal Ube3a staining in neurons, and we observed an overall increase in the intensity of the NeuN staining, which was uniformly distributed across layers (Fig. 5H-N). Western blotting of P0 and P7 lysates confirmed the developmental downregulation of paternal Ube3a expression in the Ube3am–/p+ neocortex and hippocampus as neurons mature during the first postnatal week (Fig. 5O).
Figure 5.
Immature neurons of the perinatal cortex express paternal Ube3a. A-N: Images of paternal Ube3a and NeuN immunostaining in the developing neocortex of AS (Ube3am–/p+) mice. Paternal Ube3a is more prominent at P0 (A) than at P0 (H), when a distinct deep (low) to superficial (high) gradient of staining is observed. At P0, the paternal Ube3a laminar gradient opposes the staining gradient for NeuN, a mature neuron marker, as shown in high-magnification images captured using thin (1.2 μm) optical sectioning (BD). In digitally zoomed images (E-G), paternal Ube3a staining is apparent in presumptive immature neurons (double arrows) beyond the superficial extent of NeuN staining and in nearby, weakly NeuN-positive neurons (arrows) slightly deeper in the cortical plate. At P7, only presumptive NeuN-negative glia show paternal Ube3a staining, as shown with high-magnification (I-K; arrow, L-N). O: Western blotting of total cortical and hippocampal lysates from P0 and P7 wild-type and AS mouse littermates. Paternal Ube3a is more abundant in AS lysates at P0 than at P7, corroborating the immunostaining data. cp, cortical plate; wm, white matter. Scale bar: 210 μm for A and H; 35 μm for B-D and I-K; 20 μm for E-G and L-N.
We wondered if other populations of immature neurons might preferentially stain for paternal Ube3a. We focused on granule cells in the dentate gyrus and the cerebellum, whose generation and maturation is ongoing in the early postnatal brain (Altman and Das, 1966; Bayer and Altman, 1975). At P7, paternal Ube3a staining was clearly enriched in the dentate gyrus, especially in deeper granule cell layers adjacent to the hilus (Fig. 6A) and in the cerebellum, preferentially deep within the external granule cell layer (Fig. 6H). We further analyzed patterns of paternal Ube3a and NeuN co-localization within these structures. We observed a clear deep (high) to superficial (low or absent) gradient of paternal Ube3a staining across the dentate granule cell layers (Fig. 6B, E), and this gradient was reciprocal to that of NeuN (Fig. 6C, D, F, G). The relationship between paternal Ube3a and NeuN expression was reminiscent of what we observed in the perinatal cortex (Fig. 5A-G). Moreover, the staining pattern was consistent with the established superficial to deep (“outside-in”) gradient of neuronal maturation across layers (Schlessinger et al., 1975; Mathews et al., 2010). Ube3a staining in the early postnatal cerebellum was most salient within the inner margin of the external granule cell layer, wherein newly generated neurons are beginning a radial phase of migration toward their final position in the internal granule cell layer (Fig. 6I, L) (Komuro and Rakic, 1998). These cells were weakly NeuN-positive prior to and during radial migration (Fig. 6J, K, M, N), reflecting their immature status. Taken together, these findings suggested that paternal Ube3a protein expression is a generalizable feature of immature neurons in the developing brain.
Figure 6.
Immature granule cells of the hippocampus and cerebellum express paternal Ube3a. A: Lowmagnification image of paternal Ube3a immunostaining in the dentate gyrus of P7 AS (Ube3am–/p+) mice. Note the relatively intense staining in the inner band of neurons adjacent to the hilus. B-G: Highmagnification images captured using thin (1.2 μm) optical sectioning show that paternal Ube3a staining in the dentate gyrus (panel B) is inversely correlated with staining for the mature neuronal marker NeuN (C and D). Paternal Ube3a is clearly observed in weakly NeuN-positive neurons (arrows) in the inner dentate granule cell layers, whereas it is seemingly absent from strongly NeuN-positive neurons in outer layers (double arrows) as shown in corresponding, digitally zoomed images (E-G). H: Low-magnification image of paternal Ube3a staining in the P7 cerebellum. Note the staining enrichment in this relatively immature structure. I-N: High-magnification shows enriched paternal Ube3a staining deep within the external granule cell layer (panel I), localized to immature neurons that are weakly NeuN-positive (J and K). In digitally zoomed images (L-N), paternal Ube3a/NeuN co-positive neurons are shown beginning (arrows) or continuing (double arrow) their migration past Purkinje cells (asterisks) into the internal granule cell layer. CA1, Cornu Ammonis area 1; H, hilus; IC, inferior colliculus; igl, internal granule layer; ml, molecular layer; egl, external granule layer. Scale bar: 105 μm for A; 275 μm for H; 40 μm for B-D and I-K; 19 μm for E-G and L-N.
Postnatal neural stem cells biallelically express Ube3a
Immature neurons express paternal Ube3a. This led us to question whether neural progenitor cells might also express this protein. We addressed this question by co-immunostaining for Ube3a and two markers of proliferating, self-renewable neural stem cells – Nestin and Ki67. We observed a pattern of paternal Ube3a staining in P28 Ube3am–/p+ mice which resembled the subgranular zone (SGZ) of the dentate gyrus – the site of postnatal hippocampal neurogenesis (Fig. 7B). Paternal Ube3a-stained cells within this region co-stained extensively with Nestin (Fig. 7D, E, G, I) and restrictively with Ki67 (Fig. 7D, F, H, I). Because such staining patterns were absent in Ube3am–/p– mice (Fig. 7C), we were confident in the specificity of paternal Ube3a staining in these neural stem cells. Ube3a staining in the SGZ was much more difficult to appreciate in Ube3am+/p+ mice due to the overwhelming intensity of maternal Ube3a staining in surrounding neurons (Fig. 7A).
Figure 7.
Postnatal hippocampal stem cells biallelically express Ube3a. A-C: Low-magnification images of Ube3a staining in the dentate gyrus of P28 wild-type (A, Ube3am+/p+), AS (B, Ube3am–/p+), and doubleknockout (C, Ube3am–/p–) mice. Paternal Ube3a staining is conspicuous within the inner granule cell layer (B) but absent in Ube3am–/p– mice (C). The signal gain in C was digitally increased so that low levels of nonspecific staining could be appreciated. D-I: High-magnification of the boxed region in B, depicting paternal Ube3a, Nestin, and Ki67 staining. Paternal Ube3a (D) and Nestin (E) colocalize (G). Paternal Ube3a and Ki67 (F) also colocalize (H). Cells that express all three markers (arrow, I) are readily detected. J-L: Low-magnification images of Ube3a-YFP staining in the dentate gyrus of P28 maternal Ube3a-YFP (J, Ube3amYFP/p+), paternal Ube3a-YFP (K, Ube3am+/pYFP), and wild-type (L, Ube3am+/p+) mice. Maternal (J) and paternal (K) Ube3a-YFP staining in the inner granule cell layer is clearly evident, whereas there is no Ube3a-YFP staining in wild-type mice (L). M-R: High-magnification of the boxed region in J, depicting maternal Ube3a-YFP, Nestin, and Ki67 staining. S-X: High-magnification of the boxed region in K, depicting paternal Ube3a-YFP, Nestin, and Ki67 staining. Maternal (arrow, P-R) and paternal (arrow, V-X) Ube3a-YFPexpressing cells are found to co-express Nestin and Ki67, indicating that Ube3a is biallelically expressed by proliferating stem cells in the postnatal dentate gyrus. CA1, Cornu Ammonis area 1; DG, dentate
To determine if SGZ neural stem cells biallelically express Ube3a, we performed similar immunostaining experiments in Ube3a-YFP fusion protein knock-in mice, which facilitate the tracking of allele-specific Ube3a protein expression (Dindot et al., 2008). By immunostaining with GFP antibodies, which also recognize YFP, we were able to detect an SGZ-like pattern of maternal Ube3a-YFP expression in the dentate gyrus of maternal Ube3a-YFP (Ube3amYFP/p+) mice (Fig. 7J). Notably, this pattern was not obscured as it was with Ube3a staining in Ube3am+/p+ mice (Fig. 7A), perhaps owing to relatively decreased expression or stability of the maternal Ube3a-YFP protein in neurons. Paternal Ube3a-YFP staining in Ube3am+/pYFP mice also yielded an SGZ-like pattern (Fig. 7K), which was strikingly similar to the paternal Ube3a staining pattern observed in the Ube3am–/p+ dentate gyrus (Fig. 7B). Importantly, we did not detect Ube3a-YFP staining in the dentate gyrus of Ube3am+/p+ mice (Fig. 7L), confirming the specificity of the staining patterns that we observed. Both maternal (Fig. 7M-R) and paternal (Fig. 7S-X) Ube3a-YFP-stained cells within the SGZ proved to be Nestin and Ki67 co-positive, fully supporting that SGZ neural stem cells biallelically express Ube3a. From these same co-immunostaining experiments, we also determined that neural stem cells of the postnatal subventricular zone express both maternal and paternal Ube3a-YFP (data not shown). Therefore, neural progenitors within major postnatal neural stem cell niches appear to biallelically express Ube3a.
Oligodendrocytes and astrocytes biallelically express Ube3a
Olig2-expressing glia biallelically express Ube3a during neocortical gliogenesis
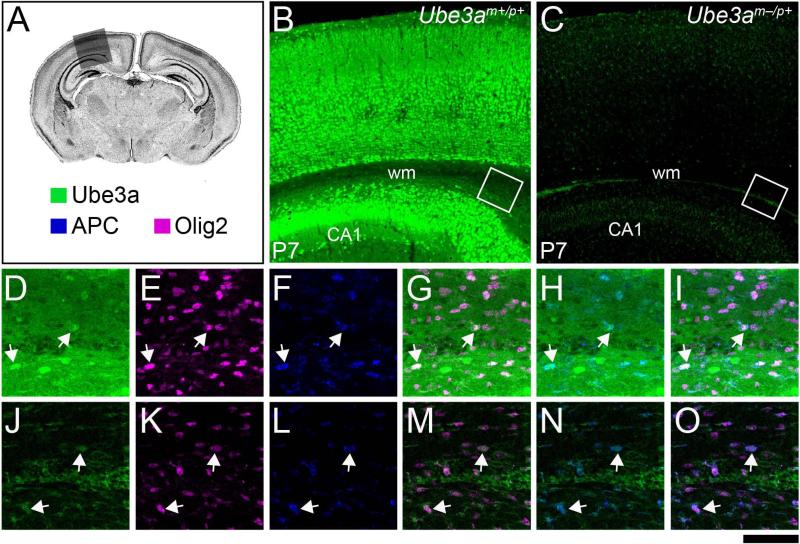
We observed paternal Ube3a staining of glia throughout the neocortex and underlying white matter of Ube3am–/p+ mice at P7 (Figs. 5I-N and 8C), the peak of neocortical gliogenesis (Bandeira et al., 2009). We also detected Ube3a-stained glial profiles among stained axons in the white matter of Ube3am+/p+ mice (Fig. 8B, D), though glial staining in the overlying grey matter was obscured by maternal Ube3a staining in neurons. The vast majority of Ube3a-stained glia in Ube3am+/p+ (Fig. 8D, E, G, I) and Ube3am–/p+ mice (Fig. 8J, K, M, O) co-stained for the oligodendrocyte lineage marker Olig2. Quantitative analyses (Fig. 9) supported our observations; nearly 90% of paternal Ube3a-stained glia in both the grey and white matter proved to be Olig2 co-positive (Fig. 9A-G). Most Ube3a/Olig2 co-positive glia also stained for adenomatous polyposis coli (APC). A subset of these cells, mostly located in or near the white matter, stained very intensely for APC, thus revealing them to be mature, myelinating oligodendrocytes (Figs. 8D, F, H, I and J, L, N, O; 9A, D, G) (Bhat et al., 1996). We observed similar patterns of Olig2/APC co-localization with maternal (Fig. 10B, E-J) and paternal (Fig. 10C, K-P) Ube3a-YFP, although maternal Ube3a-YFP-stained glia were more difficult to distinguish against a backdrop of stained axons. We never observed glial Ube3a-YFP staining in negative control brain sections from Ube3am+/p+ mice (Fig. 10D). Collectively, these data suggested that glia of the Olig2-expressing lineage, including myelinating oligodendrocytes, biallelically express Ube3a during gliogenesis.
Figure 8.
Olig2-expressing glia express paternal Ube3a during gliogenesis. A: DAPI-stained coronal section from a P7 wild-type mouse. The shaded box indicates the region of interest and the color key indicates the identity of immunostaining targets. B and C: Low-magnification images of Ube3a immunostaining in wildtype (B, Ube3am+/p+) and AS (C, Ube3am–/p+) mice in the region of interest. The paternal Ube3a staining that remains in AS mice is uniformly distributed in small cells reminiscent of glia (C). D-I: Highmagnification of the boxed region in B, depicting Ube3a, Olig2, and APC staining in the subcortical white matter. Against a backdrop of Ube3a-stained axons, small Ube3a-stained glia are seen (D), nearly all of which are co-stained with Olig2 (E and G). A subset of these glia is heavily co-stained with APC (arrows, F, H, and I). J-O: High-magnification of the boxed region in C, depicting paternal Ube3a, Olig2, and APC staining in the subcortical white matter. Paternal Ube3a-stained glia are particularly salient as surrounding axonal staining is lost. Nearly all glia co-express Olig2 (K and M) but only a subset strongly co-express APC (arrows, L, N, and O). CA1, Cornu Ammonis area 1; wm, white matter. Scale bar: 275 μm for B and C; 60 μm for D-O.
Figure 9.
Quantification of paternal Ube3a co-localization with glial markers. A and H: Images of P7 (A) and P28 (H) Ube3am–/p+ somatosensory cortex stained for paternal Ube3a and the glial markers Olig2 and APC. The boxed regions indicate the areas sampled for co-localization analysis. Dashed lines indicate white matter boundaries. B, C, E, and F and I, J, L, and M: Paternal Ube3a co-localization with glial markers in digitally zoomed images of P7 (B, C, E, and F) and P28 (I, J, L, and M) somatosensory grey matter. Arrows indicate Ube3a co-localization with Olig2 in the absence of APC; double arrows indicate co-localization of Ube3a and Olig2 with weak APC staining (APC Lo); arrowheads indicate colocalization of Ube3a and Olig2 with APC staining that is still visible after thresholding (APC Hi). Thresholding for APC Hi helped to select for the intense APC labeling characteristic of mature oligodendrocytes. D, G, K, and N: Proportionality of distinct paternal Ube3a-expressing glial subpopulations in the grey matter (D and K) and white matter (G and N) of somatosensory cortex at P7 (D and G) and P28 (K and N). Values represent the mean percentage of total paternal Ube3a-stained cells that co-stained with the indicated combination of immunofluorescent markers. O: Pie chart color key. N = 4 animals per developmental time point. Optical section thickness = 1.4 μm. Scale bar: 280 μm for A and H; 60 μm for B, C, E, and F and I, J, L, and M.
Figure 10.
Olig2-positive glia biallelically express Ube3a-YFP during gliogenesis. A: DAPI-stained coronal section from a P7 wild-type mouse. The shaded box indicates the region of interest and the color key indicates the identity of immunostaining targets. B-D: Low-magnification images of Ube3a-YFP immunostaining in maternal Ube3a-YFP (B, Ube3amYFP/p+), paternal Ube3a-YFP (C, Ube3am+/pYFP), and wildtype (D, Ube3am+/p+) mice. Relative to maternal Ube3a-YFP staining (B), paternal Ube3a-YFP staining (C) is sparse and evenly distributed in small cells. No specific YFP staining is observed in wild-type mice (D). E-J: High-magnification of the boxed region in B, depicting maternal Ube3a-YFP, Olig2, and APC staining in the subcortical white matter. Stained glial cells are difficult to notice amid axons that are also maternal Ube3a-YFP-positive (E). Nearly all maternal Ube3a-YFP-stained glia are Olig2-stained (F and H) and a minor subset also co-stain with APC (arrows, G, I, and J). K-P: High-magnification of the boxed region in C, depicting paternal Ube3a-YFP, Olig2, and APC staining in the subcortical white matter. There is no apparent paternal Ube3a-YFP staining in axons, so stained glia are readily detected (K). Most paternal Ube3a-YFP stained glia also express Olig2 (L and N) and a few also express APC (arrows, M, O, and P). CA1, Cornu Ammonis area 1; wm, white matter. Scale bar: 340 μm for B-D; 60 μm for E-P.
Glial Ube3a expression becomes increasingly restricted to mature oligodendrocytes
A majority of astrocytes express Olig2 during peak gliogenesis (Cai et al., 2007), indicating that Olig2 expression is not exclusive to the oligodendrocyte lineage until later in development. To determine if early postnatal populations of paternal Ube3a-expressing glia comprise astrocytes in addition to oligodendrocytes, we co-stained for the astrocyte marker glial fibrillary acid protein (GFAP). A subset of paternal Ube3a-stained glia co-stained for GFAP in both the neocortical (Fig. 11A-D) and hippocampal (Fig. 11E-H) grey matter of P7 Ube3am–/p+ mice. Paternal Ube3a-stained glia also co-stained for GFAP in the hippocampi of P28 mice (Fig. 11I-L). This suggested that subpopulations of astrocytes express Ube3a throughout postnatal development.
Figure 11.
Co-expression of paternal Ube3a and GFAP. A-D: Paternal Ube3a (B), NeuN (C), and GFAP (D) staining in the P7 Ube3am–/p+ somatosensory cortex. E-H: Paternal Ube3a (F), NeuN (G), and GFAP (H) staining in the P7 Ube3am–/p+ hippocampus. I-L: Paternal Ube3a (J), NeuN (K), and GFAP (L) staining in the P28 Ube3am–/p+ hippocampus. Arrowheads indicate representative glia that are clearly co-stained with both paternal Ube3a and GFAP. pyr, pyramidal layer of hippocampus; rad, stratum radiatum of hippocampus. Optical section thickness = 1.4 μm. Scale bar: 45 μm for all panels.
By P28, astrocytes in the neocortical grey matter no longer express GFAP. However, they continue to express low levels of APC and are Olig2-negative. We determined that astrocytes defined by this staining profile constitute approximately 30% and 20% of all paternal Ube3a-expressing glia in the neocortical grey matter (Fig. 9H-K) and white matter (Fig. 9H, N), respectively. By contrast, maternal (Fig. 12B, E, F, H, J) and paternal (Fig. 12C, K, L, N, P) Ube3a/Olig2 co-staining was much more prevalent. Over 60% of paternal Ube3a-stained glia in the grey matter (Fig. 9H, I, M, K) and nearly 80% of paternal Ube3a-stained glia in the white matter (Fig. 9H, N) also stained for Olig2, which is a dedicated oligodendrocyte marker by P28. Many maternal (Fig. 12E, G, I, J) and paternal (Fig. 12K, M, O, P) Ube3a-stained oligodendrocytes proved to be mature, myelinating cells as evidenced by intense APC co-staining. In fact, mature oligodendroctyes accounted for almost 40% of all paternal Ube3a-stained glia in the grey matter (Fig. 9H, I, L, M, K) and over 50% of such cells in the white matter (Fig. 9H, N). This was a marked proportional increase from what we observed at P7 (Fig. 9D, G), indicating that Ube3a expression in glia becomes increasingly restricted to mature oligodendrocytes over the first postnatal month. Finally, we observed extensive Olig2/APC co-localization with both maternal (Fig. 13B, E-J) and paternal (Fig. 13C, K-P) Ube3a-YFP at this later postnatal period. From these observations we drew two main conclusions. First, astrocytes and oligodendrocytes are the two major glial cell types that biallelically express Ube3a. Second, presumptive myelinating oligodendrocytes emerge as the predominant Ube3a-expressing glial cell type during postnatal development.
Figure 12.
Mature oligodendrocytes express paternal Ube3a. A: DAPI-stained coronal section from a P28 wild-type mouse. The shaded box indicates the region of interest and the color key indicates the identity of immunostaining targets. B-D: Low-magnification images of Ube3a immunostaining in wild-type (B, Ube3am+/p+), AS (C, Ube3am–/p+), and double-knockout (D, Ube3am–/p–) mice in the region of interest. The majority of staining observed in wild-type mice (B) is lost in AS mice (C), but glial profiles that are concentrated in the white matter persist. These patterns are absent in double-knockout mice (D). E-J: High-magnification of the boxed region in B, depicting Ube3a, Olig2, and APC staining in the callosum. Glial cells and surrounding axons are stained for Ube3a (E). Most Ube3a-stained glia in this myelinated tract costain for Olig2 (F and H) and APC (arrows, G, I, and J). K-P: High-magnification of the boxed region in C, depicting paternal Ube3a, Olig2, and APC staining in the callosum. Paternal Ube3a staining in glia is apparent in the absence of axonal maternal Ube3a staining (K). Paternal Ube3a-stained glia in the callosum are frequently co-stained with both Olig2 (L and N) and the mature oligodendrocyte marker APC (arrows, M, O, and P). cc, corpus callosum; dhc, dorsal hippocampal commissure. Scale bar: 340 μm for B-D; 60 μm for E-P.
Figure 13.
Mature oligodendrocytes biallelically express Ube3a-YFP. A: DAPI-stained coronal section from a P28 wild-type mouse. The shaded box indicates the region of interest and the color key indicates the identity of immunostaining targets. B-D: Low-magnification images of Ube3a-YFP immunostaining in maternal Ube3a-YFP (B, Ube3amYFP/p+), paternal Ube3a-YFP (C, Ube3am+/pYFP), and wild-type (D, Ube3am+/p+) mice. Maternal Ube3a-YFP staining is intense and distributed broadly in neurons (B), whereas paternal Ube3a-YFP staining is concentrated in callosal glia (C). These patterns are absent in wild-type mice (D). EJ: High-magnification of the boxed region in B, depicting maternal Ube3a-YFP, Olig2, and APC staining in the callosum. Maternal Ube3a-YFP is present in glia (E) which are largely Olig2 (F and H) and APC co-positive (arrows, G, I, and J). K-P: High-magnification of the boxed region in C, depicting paternal Ube3a-YFP, Olig2, and APC staining in the callosum. Paternal Ube3a-YFP staining is present in glia but not axons in the callosum (K). Paternal Ube3a-YFP-stained glia are nearly always co-stained with Olig2 (L and N) and frequently co-stained with the mature oligodendrocyte marker APC (arrows, M, O, and P). cc, corpus callosum; dhc, dorsal hippocampal commissure. Scale bar: 340 μm for B-D; 60 μm for E-P.
DISCUSSION
We extensively mapped allele-specific Ube3a protein expression in the developing postnatal brain. Our development of an immunofluorescent Ube3a staining protocol sensitive enough to detect low levels of paternal Ube3a protein in Ube3am–/p+ mice was critical to the success of this venture. Nevertheless, we were conscientious of the possibility that transcription from the paternal Ube3a locus or stability of the resultant transcript or protein might be abnormal in an AS-like developmental context. Therefore, we undertook a complementary approach, staining for paternal Ube3a-YFP in Ube3am+/pYFP mice that express an intact copy of maternal Ube3a. We observed a high degree of correspondence between paternal Ube3a and paternal Ube3a-YFP staining patterns, which bolstered our confidence in their veracity. Reciprocal Ube3a-YFP staining experiments in Ube3amYFP/p+ mice, performed in parallel, allowed us to isolate maternal Ube3a contributions to the Ube3a protein pool. A limitation of using such parallel approaches is that paternal and maternal Ube3a cannot be distinguished from one another within the same animal. Consequently, we were unable to address the possibility of more subtle maternal expression bias within brain cells that biallelically express Ube3a (e.g., neural progenitors of the SGZ and oligodendrocytes) as has been seen in peripheral tissues (Herzing et al., 2002; Gustin et al., 2010). We keep these experimental advantages and limitations in mind as we discuss our findings in broader contexts.
Developmental changes in Ube3a subcellular localization: causes and consequences
We observed a striking cytoplasmic to nuclear shift in Ube3a subcellular localization in neurons over postnatal development. Potential causes of this shift may include changes in the expression of Ube3a protein isoforms and/or interacting proteins that affect Ube3a trafficking. Differential amino-terminal splicing produces 3 isoforms of human UBE3A (Yamamoto et al., 1997). Proteomic analysis of UBE3A-interacting proteins in heterologous systems has revealed a limited number of isoform-specific UBE3A protein partners that could reflect (or perhaps even dictate) unique patterns of subcellular distribution (Martinez-Noel et al., 2012). Whether such interactions occur in neurons, change over development, or affect UBE3A localization is not yet known.
Mice also express three known Ube3a isoforms. Isoform II encodes the entire Ube3a open reading frame, whereas I and III encode isoforms lacking the first 21 amino-terminal amino acids. Isoform I may also lack 87 carboxy-terminal amino acids that are critical for E3 ligase function (Greer et al., 2010; Miao et al., 2013). Miao et al. (2013) overexpressed each of these isoforms individually in neurons lacking endogenous Ube3a expression due to short hairpin RNA knock-down. In their study, overexpressed isoforms I and II localized preferentially to the cytoplasm, while overexpressed isoform III localized to the nucleus. Interestingly, only isoform II overexpression resulted in a statistically significant rescue of dendritic morphogenesis in these experiments, suggesting that cytoplasmically localized Ube3a plays a preferential role in this process. Curiously, Miao et al. (2013) also found that isoform III mRNA was progressively down-regulated over development. This finding is seemingly at odds with our observation that neurons preferentially localize Ube3a to the nucleus as they mature. The antibody that we used to immunofluorescently stain for Ube3a was generated using an immunogen sequence shared by all three isoforms (Table 1). Isoform-specific Ube3a antibodies and Ube3a knock-in mice, when available, should help to clarify putative relationships between Ube3a isoform expression, protein localization, and function.
Paternal Ube3a expression in immature versus mature neurons
Relaxed paternal Ube3a imprinting in developing neurons has been invoked as a hypothesis to explain the absence (Sato and Stryker, 2010) or differential expression (Heck et al., 2008) of certain phenotypes in Ube3am–/p+ and Ube3am–/p– mice. However, due in part to insufficient paternal Ube3a expression mapping, this hypothesis has lacked much empirical basis. Here, we produced the first paternal Ube3a expression map of notable spatiotemporal resolution. As a result, we found that paternal Ube3a expression is a defining feature of immature neurons in the postnatal brain, while more mature neurons expressing high levels of NeuN were devoid of paternal Ube3a expression. These findings support the hypothesis that paternal Ube3a silencing is established during the process of neuronal maturation.
Epigenetic silencing of paternal Ube3a in neurons requires expression in cis of a Ube3a antisense sequence (Ube3a-ATS) located at the 3’ end of a large noncoding RNA transcript (lncRNA) (Huang et al., 2012; Meng et al., 2012). Ube3a-ATS is hypothesized to interfere with the transcription of paternal Ube3a from the opposite DNA strand (Lalande and Calciano, 2007; Leung et al., 2011). The lncRNA is transcribed from a paternal promoter, which becomes highly active due to allele-specific hypomethylation of a bipartite imprinting center that controls the expression of imprinted genes within the region (Sutcliffe et al., 1994). In immature neurons, transcription of the lncRNA all the way through the Ube3a-ATS region may be inhibited by chromatin condensation. However, as a neuron matures, chromatin downstream of the Snrpn locus is hypothesized to relax, enabling transcription of the lncRNA completely through the Ube3a-ATS region and, in turn, silencing of paternal Ube3a (Leung et al., 2011). Indeed, in the adult mouse brain there are paternal Ube3a-ATS transcripts that correspond to Ube3a promoter regions upstream of Ube3a transcription start site, encouraging speculation that they are required to fully silence paternal Ube3a (Numata et al., 2011), Elegant fluorescence in situ hybridization experiments targeting the Snrpn-Ube3a genomic region have revealed that decondensaton of this paternal genomic region occurs in neocortical neurons sometime during the first two postnatal weeks (Leung et al., 2009). This finding is generally consistent with our observation that neocortical neurons cease to express detectable levels of paternal Ube3a by P7.
Consideration of allele-specific Ube3a expression in the context of AS pathogenesis
The quest to understand the molecular and cellular bases of AS naturally began with a focus on neurons; maternal UBE3A mutations are sufficient to cause AS (Kishino et al., 1997; Matsuura et al., 1997; Sutcliffe et al., 1997), and neurons are the only cell type known to have an extreme maternal UBE3A expression bias. We now know, nearly 15 years after publication of the first Ube3am–/p+ mouse models (Jiang et al., 1998; Miura et al., 2002), that maternal Ube3a deletion adversely affects the function of diverse neuronal populations (Kaphzan et al., 2011; Mabb et al., 2011; Riday et al., 2012; Wallace et al., 2012; Egawa et al., 2012; Condon et al., 2013; Miao et al., 2013). This is generally consistent with the broad distribution of maternal Ube3a in the adult brain (Gustin et al., 2010), but valuable insight is lost whenever phenotypes that emerge developmentally are interpreted in the context of adult expression patterns. Neurons within the thalamus, thalamic reticular nucleus, and cortex of adult mice all express maternal Ube3a, which has led to the hypothesis that sleep disturbances, generalized seizures, and electroencephalographic abnormalities in both Ube3am–/p+ mice (Jiang et al., 1998; Colas et al., 2005) and AS patients (Miano et al., 2004; Pelc et al., 2008; Conant et al., 2009; Thibert et al., 2013) are due to thalamocortical circuit disruption (Gustin et al., 2010). This hypothesis did not initially account for subplate neurons, which are key mediators of thalamocortical circuit development (Kanold and Luhmann, 2010), likely because they only express maternal Ube3a saliently during the early postnatal period.
Paternal Ube3a silencing renders neurons especially vulnerable to maternal Ube3a mutation. Whether maternal Ube3a protein loss leads to haploinsufficiency in cell populations that biallelically express Ube3a is an open question. We discovered that biallelic Ube3a expression in the postnatal brain is largely restricted to two distinct cell populations of particular interest to AS pathogenesis: postnatal neural stem cells and oligodendrocytes. Ube3a haploinsufficiency in postnatal neural stem cells of the hippocampal SGZ could possibly affect cognition (Sahay et al., 2011) and mood (Santarelli et al., 2003) – both of which are altered in individuals with AS (Williams et al., 2006; Thibert et al., 2013). Although functional roles for maternal Ube3a in SGZ neural stem cells remain to be fully elucidated, one study in Ube3am–/p+ mice suggests that neurogenesis per se is not affected (Mardirossian et al., 2009). Rather, maternal Ube3a deficiency moderately limits the potential for postmitotic SGZ-born neural precursors to mature into NeuN-positive neurons (Mardirossian et al., 2009). Because paternal Ube3a appears to become silenced during neuronal maturation, maternal and paternal Ube3a may not be functionally equivalent with regard to this postmitotic phenotypic outcome. Embryonic neural stem cells of the dorsal pallium are also likely to biallelically express Ube3a protein (Yamasaki, 2003), but, similar to SGZ stem cells, may not require maternal Ube3a for neurogenesis as neocortical lamination and size appear to be normal in early postnatal Ube3am–/p+ mice (M.C. Judson and B.D. Philpot, unpublished observations).
White matter abnormalities that have been described in AS patients (Harting et al., 2009; Peters et al., 2011; Tiwari et al., 2012) could potentially be caused by Ube3a haploinsufficiency in oligodendrocytes. White matter abnormalities in AS are widespread, consistent with our observation that Olig2-expressing cells throughout the brain express Ube3a. While the nature of these white matter abnormalities is not yet clear, they may be due in part to a developmental delay in myelination (Harting et al., 2009; Castro-Gago et al., 2010). Interestingly, we observed that mature, presumably myelinating, oligodendrocytes emerge as the predominant Ube3a-expressing glial cell type during the first postnatal month, coincident with the active myelination of major forebrain axon tracts (Vincze et al., 2008). However, it is not yet known if maternal Ube3a protein loss in any way compromises the myelinating capacity of oligodendrocytes. These and other potential cell type-specific consequences of maternal Ube3a loss will ultimately have to be parsed with the aid of conditional Ube3a knock-out mouse models. We expect that the knowledge gained in this study will help to focus experiments in these forthcoming model systems.
OTHER ACKNOWLEDGEMENTS
We thank Jaya Miriyala for valuable technical assistance. We also thank Vladimir Gukassyan and the UNC Neuroscience Center microscopy core facility.
Grant sponsors: National Institutes of Health (NIH) 5F32NS077686 (to MCJ) and National Institutes of Mental Health (NIMH) 5R01MH093372, Angelman Syndrome Foundation, and Simons Foundation Autism Research Initiative (to BDP). The microscopy core was funded by grants from the National Institute of Neurological Disorders and Stroke (NINDS) P30NS045892 and the National Institute of Child Health and Human Development (NICHD) P30HD03110.
ROLE OF AUTHORS
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: MCJ, JOS-P, JH and BDP Acquisition of data: MCJ, JH, and JOS-P. Analysis and interpretation of data: MCJ, JOS-P, WAD, JH, and BDP. Drafting of the manuscript: MCJ. Critical revision of the manuscript for important intellectual content: MCJ, JOS-P, WAD, JH, and BDP. Statistical analysis: MCJ. Obtained funding: MCJ and BDP. Administrative, technical, and material support: JH. Study supervision: BDP.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
LITERATURE CITED
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nature genetics. 1997;17(1):75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- Altman J, Das GD. Autoradiographic and histological studies of postnatal neurogenesis. I. A longitudinal investigation of the kinetics, migration and transformation of cells incorporating tritiated thymidine in neonate rats, with special reference to postnatal neurogenesis in some brain regions. The Journal of comparative neurology. 1966;126(3):337–389. doi: 10.1002/cne.901260302. [DOI] [PubMed] [Google Scholar]
- Angevine JB, Jr., Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- Bandeira F, Lent R, Herculano-Houzel S. Changing numbers of neuronal and nonneuronal cells underlie postnatal brain growth in the rat. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(33):14108–14113. doi: 10.1073/pnas.0804650106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer SA, Altman J. The effects of X-irradiation on the postnatally-forming granule cell populations in the olfactory bulb, hippocampus, and cerebellum of the rat. Experimental neurology. 1975;48(1):167–174. doi: 10.1016/0014-4886(75)90231-9. [DOI] [PubMed] [Google Scholar]
- Bhat RV, Axt KJ, Fosnaugh JS, Smith KJ, Johnson KA, Hill DE, Kinzler KW, Baraban JM. Expression of the APC tumor suppressor protein in oligodendroglia. Glia. 1996;17(2):169–174. doi: 10.1002/(SICI)1098-1136(199606)17:2<169::AID-GLIA8>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bower BD, Jeavons PM. The “happy puppet” syndrome. Archives of disease in childhood. 1967;42(223):298–302. doi: 10.1136/adc.42.223.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, Parada LF, Lu QR. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134(10):1887–1899. doi: 10.1242/dev.02847. [DOI] [PubMed] [Google Scholar]
- Castro-Gago M, Gomez-Lado C, Eiris-Punal J, Rodriguez-Mugico VM. Abnormal myelination in Angelman syndrome. European journal of paediatric neurology : EJPN : official journal of the European Paediatric Neurology Society. 2010;14(3):292. doi: 10.1016/j.ejpn.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Chun JJ, Shatz CJ. The earliest-generated neurons of the cat cerebral cortex: characterization by MAP2 and neurotransmitter immunohistochemistry during fetal life. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1989;9(5):1648–1667. doi: 10.1523/JNEUROSCI.09-05-01648.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas D, Wagstaff J, Fort P, Salvert D, Sarda N. Sleep disturbances in Ube3a maternal-deficient mice modeling Angelman syndrome. Neurobiology of disease. 2005;20(2):471–478. doi: 10.1016/j.nbd.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Conant KD, Thibert RL, Thiele EA. Epilepsy and the sleep-wake patterns found in Angelman syndrome. Epilepsia. 2009;50(11):2497–2500. doi: 10.1111/j.1528-1167.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- Condon KH, Ho J, Robinson CG, Hanus C, Ehlers MD. The Angelman syndrome protein Ube3a/E6AP is required for Golgi acidification and surface protein sialylation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33(9):3799–3814. doi: 10.1523/JNEUROSCI.1930-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper EM, Hudson AW, Amos J, Wagstaff J, Howley PM. Biochemical analysis of Angelman syndrome-associated mutations in the E3 ubiquitin ligase E6-associated protein. The Journal of biological chemistry. 2004;279(39):41208–41217. doi: 10.1074/jbc.M401302200. [DOI] [PubMed] [Google Scholar]
- Dimou L, Simon C, Kirchhoff F, Takebayashi H, Gotz M. Progeny of Olig2-expressing progenitors in the gray and white matter of the adult mouse cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28(41):10434–10442. doi: 10.1523/JNEUROSCI.2831-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Human molecular genetics. 2008;17(1):111–118. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- Eblen F, Graybiel AM. Highly restricted origin of prefrontal cortical inputs to striosomes in the macaque monkey. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1995;15(9):5999–6013. doi: 10.1523/JNEUROSCI.15-09-05999.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa K, Kitagawa K, Inoue K, Takayama M, Takayama C, Saitoh S, Kishino T, Kitagawa M, Fukuda A. Decreased tonic inhibition in cerebellar granule cells causes motor dysfunction in a mouse model of Angelman syndrome. Science translational medicine. 2012;4(163):163ra157. doi: 10.1126/scitranslmed.3004655. [DOI] [PubMed] [Google Scholar]
- Encinas JM, Hamani C, Lozano AM, Enikolopov G. Neurogenic hippocampal targets of deep brain stimulation. The Journal of comparative neurology. 2011;519(1):6–20. doi: 10.1002/cne.22503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G, van der Kooy D. Pattern formation in the striatum: developmental changes in the distribution of striatonigral neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1987;7(7):1969–1978. doi: 10.1523/JNEUROSCI.07-07-01969.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster GA, Schultzberg M, Hokfelt T, Goldstein M, Hemmings HC, Jr., Ouimet CC, Walaas SI, Greengard P. Development of a dopamine- and cyclic adenosine 3′:5′-monophosphate-regulated phosphoprotein (DARPP-32) in the prenatal rat central nervous system, and its relationship to the arrival of presumptive dopaminergic innervation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1987;7(7):1994–2018. doi: 10.1523/JNEUROSCI.07-07-01994.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile JK, Tan WH, Horowitz LT, Bacino CA, Skinner SA, Barbieri-Welge R, Bauer-Carlin A, Beaudet AL, Bichell TJ, Lee HS, Sahoo T, Waisbren SE, Bird LM, Peters SU. A neurodevelopmental survey of Angelman syndrome with genotype-phenotype correlations. Journal of developmental and behavioral pediatrics : JDBP. 2010;31(7):592–601. doi: 10.1097/DBP.0b013e3181ee408e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR. The neostriatal mosaic: striatal patch-matrix organization is related to cortical lamination. Science. 1989;246(4928):385–388. doi: 10.1126/science.2799392. [DOI] [PubMed] [Google Scholar]
- Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, Ploegh HL, Chowdhury S, Worley PF, Steen J, Greenberg ME. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140(5):704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin RM, Bichell TJ, Bubser M, Daily J, Filonova I, Mrelashvili D, Deutch AY, Colbran RJ, Weeber EJ, Haas KF. Tissue-specific variation of Ube3a protein expression in rodents and in a mouse model of Angelman syndrome. Neurobiology of disease. 2010;39(3):283–291. doi: 10.1016/j.nbd.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanbury R, Ling ZD, Wuu J, Kordower JH. GFAP knockout mice have increased levels of GDNF that protect striatal neurons from metabolic and excitotoxic insults. The Journal of comparative neurology. 2003;461(3):307–316. doi: 10.1002/cne.10667. [DOI] [PubMed] [Google Scholar]
- Harting I, Seitz A, Rating D, Sartor K, Zschocke J, Janssen B, Ebinger F, Wolf NI. Abnormal myelination in Angelman syndrome. European journal of paediatric neurology : EJPN : official journal of the European Paediatric Neurology Society. 2009;13(3):271–276. doi: 10.1016/j.ejpn.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Heck DH, Zhao Y, Roy S, LeDoux MS, Reiter LT. Analysis of cerebellar function in Ube3a-deficient mice reveals novel genotype-specific behaviors. Human molecular genetics. 2008;17(14):2181–2189. doi: 10.1093/hmg/ddn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzing LB, Cook EH, Jr., Ledbetter DH. Allele-specific expression analysis by RNA-FISH demonstrates preferential maternal expression of UBE3A and imprint maintenance within 15q11- q13 duplications. Human molecular genetics. 2002;11(15):1707–1718. doi: 10.1093/hmg/11.15.1707. [DOI] [PubMed] [Google Scholar]
- Hoerder-Suabedissen A, Molnar Z. Molecular diversity of early-born subplate neurons. Cerebral cortex. 2013;23(6):1473–1483. doi: 10.1093/cercor/bhs137. [DOI] [PubMed] [Google Scholar]
- Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, Sciaky N, Dutton JW, Jr., Lee HM, Chen X, Jin J, Bridges AS, Zylka MJ, Roth BL, Philpot BD. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481(7380):185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21(4):799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Jones LG, Prins J, Park S, Walton JP, Luebke AE, Lurie DI. Lead exposure during development results in increased neurofilament phosphorylation, neuritic beading, and temporal processing deficits within the murine auditory brainstem. The Journal of comparative neurology. 2008;506(6):1003–1017. doi: 10.1002/cne.21563. [DOI] [PubMed] [Google Scholar]
- Kanold PO, Luhmann HJ. The subplate and early cortical circuits. Annual review of neuroscience. 2010;33:23–48. doi: 10.1146/annurev-neuro-060909-153244. [DOI] [PubMed] [Google Scholar]
- Kao HT, Li P, Chao HM, Janoschka S, Pham K, Feng J, McEwen BS, Greengard P, Pieribone VA, Porton B. Early involvement of synapsin III in neural progenitor cell development in the adult hippocampus. The Journal of comparative neurology. 2008;507(6):1860–1870. doi: 10.1002/cne.21643. [DOI] [PubMed] [Google Scholar]
- Kaphzan H, Buffington SA, Jung JI, Rasband MN, Klann E. Alterations in intrinsic membrane properties and the axon initial segment in a mouse model of Angelman syndrome. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(48):17637–17648. doi: 10.1523/JNEUROSCI.4162-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nature genetics. 1997;15(1):70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Komuro H, Rakic P. Distinct modes of neuronal migration in different domains of developing cerebellar cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18(4):1478–1490. doi: 10.1523/JNEUROSCI.18-04-01478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostovic I, Rakic P. Cytology and time of origin of interstitial neurons in the white matter in infant and adult human and monkey telencephalon. Journal of neurocytology. 1980;9(2):219–242. doi: 10.1007/BF01205159. [DOI] [PubMed] [Google Scholar]
- Lalande M, Calciano MA. Molecular epigenetics of Angelman syndrome. Cellular and molecular life sciences : CMLS. 2007;64(7-8):947–960. doi: 10.1007/s00018-007-6460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers M, Bancescu DL, Le Meur E, Rougeulle C, Glatt-Deeley H, Brannan C, Muscatelli F, Lalande M. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic acids research. 2004;32(11):3480–3492. doi: 10.1093/nar/gkh670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KN, Chamberlain SJ, Lalande M, LaSalle JM. Neuronal chromatin dynamics of imprinting in development and disease. Journal of cellular biochemistry. 2011;112(2):365–373. doi: 10.1002/jcb.22958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KN, Vallero RO, DuBose AJ, Resnick JL, LaSalle JM. Imprinting regulates mammalian snoRNA-encoding chromatin decondensation and neuronal nucleolar size. Human molecular genetics. 2009;18(22):4227–4238. doi: 10.1093/hmg/ddp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossie AC, Whitney MM, Amidon D, Dong HJ, Chen P, Theriaque D, Hutson A, Nicholls RD, Zori RT, Williams CA, Driscoll DJ. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. Journal of medical genetics. 2001;38(12):834–845. doi: 10.1136/jmg.38.12.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabb AM, Judson MC, Zylka MJ, Philpot BD. Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes. Trends in neurosciences. 2011;34(6):293–303. doi: 10.1016/j.tins.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardirossian S, Rampon C, Salvert D, Fort P, Sarda N. Impaired hippocampal plasticity and altered neurogenesis in adult Ube3a maternal deficient mouse model for Angelman syndrome. Experimental neurology. 2009;220(2):341–348. doi: 10.1016/j.expneurol.2009.08.035. [DOI] [PubMed] [Google Scholar]
- Martinez-Noel G, Galligan JT, Sowa ME, Arndt V, Overton TM, Harper JW, Howley PM. Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Molecular and cellular biology. 2012;32(15):3095–3106. doi: 10.1128/MCB.00201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews EA, Morgenstern NA, Piatti VC, Zhao C, Jessberger S, Schinder AF, Gage FH. A distinctive layering pattern of mouse dentate granule cells is generated by developmental and adult neurogenesis. The Journal of comparative neurology. 2010;518(22):4479–4490. doi: 10.1002/cne.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nature genetics. 1997;15(1):74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- Meng L, Person RE, Beaudet AL. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Human molecular genetics. 2012;21(13):3001–3012. doi: 10.1093/hmg/dds130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano S, Bruni O, Leuzzi V, Elia M, Verrillo E, Ferri R. Sleep polygraphy in Angelman syndrome. Clinical neurophysiology : official journal of the International Federation of Clinical Neurophysiology. 2004;115(4):938–945. doi: 10.1016/j.clinph.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Miao S, Chen R, Ye J, Tan GH, Li S, Zhang J, Jiang YH, Xiong ZQ. The Angelman syndrome protein Ube3a is required for polarized dendrite morphogenesis in pyramidal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33(1):327–333. doi: 10.1523/JNEUROSCI.2509-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Kishino T, Li E, Webber H, Dikkes P, Holmes GL, Wagstaff J. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiology of disease. 2002;9(2):149–159. doi: 10.1006/nbdi.2001.0463. [DOI] [PubMed] [Google Scholar]
- Moncla A, Malzac P, Voelckel MA, Auquier P, Girardot L, Mattei MG, Philip N, Mattei JF, Lalande M, Livet MO. Phenotype-genotype correlation in 20 deletion and 20 non-deletion Angelman syndrome patients. European journal of human genetics : EJHG. 1999;7(2):131–139. doi: 10.1038/sj.ejhg.5200258. [DOI] [PubMed] [Google Scholar]
- Mulherkar SA, Jana NR. Loss of dopaminergic neurons and resulting behavioural deficits in mouse model of Angelman syndrome. Neurobiology of disease. 2010;40(3):586–592. doi: 10.1016/j.nbd.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annual review of genomics and human genetics. 2001;2:153–175. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- Numata K, Kohama C, Abe K, Kiyosawa H. Highly parallel SNP genotyping reveals high-resolution landscape of mono-allelic Ube3a expression associated with locus-wide antisense transcription. Nucleic acids research. 2011;39(7):2649–2657. doi: 10.1093/nar/gkq1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelc K, Boyd SG, Cheron G, Dan B. Epilepsy in Angelman syndrome. Seizure : the journal of the British Epilepsy Association. 2008;17(3):211–217. doi: 10.1016/j.seizure.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Peters SU, Beaudet AL, Madduri N, Bacino CA. Autism in Angelman syndrome: implications for autism research. Clinical genetics. 2004;66(6):530–536. doi: 10.1111/j.1399-0004.2004.00362.x. [DOI] [PubMed] [Google Scholar]
- Peters SU, Kaufmann WE, Bacino CA, Anderson AW, Adapa P, Chu Z, Yallampalli R, Traipe E, Hunter JV, Wilde EA. Alterations in white matter pathways in Angelman syndrome. Developmental medicine and child neurology. 2011;53(4):361–367. doi: 10.1111/j.1469-8749.2010.03838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DJ, Aslam S, Tasker L, Gillies K. Fates of the earliest generated cells in the developing murine neocortex. The Journal of comparative neurology. 1997;377(3):414–422. [PubMed] [Google Scholar]
- Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183(4123):425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- Reim K, Regus-Leidig H, Ammermuller J, El-Kordi A, Radyushkin K, Ehrenreich H, Brandstatter JH, Brose N. Aberrant function and structure of retinal ribbon synapses in the absence of complexin 3 and complexin 4. Journal of cell science. 2009;122(Pt 9):1352–1361. doi: 10.1242/jcs.045401. [DOI] [PubMed] [Google Scholar]
- Riday TT, Dankoski EC, Krouse MC, Fish EW, Walsh PL, Han JE, Hodge CW, Wightman RM, Philpot BD, Malanga CJ. Pathway-specific dopaminergic deficits in a mouse model of Angelman syndrome. The Journal of clinical investigation. 2012;122(12):4544–4554. doi: 10.1172/JCI61888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahay A, Scobie KN, Hill AS, O'Carroll CM, Kheirbek MA, Burghardt NS, Fenton AA, Dranovsky A, Hen R. Increasing adult hippocampal neurogenesis is sufficient to improve pattern separation. Nature. 2011;472(7344):466–470. doi: 10.1038/nature09817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S, Weisstaub N, Lee J, Duman R, Arancio O, Belzung C, Hen R. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301(5634):805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- Sato M, Stryker MP. Genomic imprinting of experience-dependent cortical plasticity by the ubiquitin ligase gene Ube3a. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(12):5611–5616. doi: 10.1073/pnas.1001281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75(3):495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Schlessinger AR, Cowan WM, Gottlieb DI. An autoradiographic study of the time of origin and the pattern of granule cell migration in the dentate gyrus of the rat. The Journal of comparative neurology. 1975;159(2):149–175. doi: 10.1002/cne.901590202. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Jiang YH, Galijaard RJ, Matsuura T, Fang P, Kubota T, Christian SL, Bressler J, Cattanach B, Ledbetter DH, Beaudet AL. The E6-Ap ubiquitin-protein ligase (UBE3A) gene is localized within a narrowed Angelman syndrome critical region. Genome research. 1997;7(4):368–377. doi: 10.1101/gr.7.4.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Nakao M, Christian S, Orstavik KH, Tommerup N, Ledbetter DH, Beaudet AL. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nature genetics. 1994;8(1):52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- Thibert RL, Larson AM, Hsieh DT, Raby AR, Thiele EA. Neurologic manifestations of angelman syndrome. Pediatric neurology. 2013;48(4):271–279. doi: 10.1016/j.pediatrneurol.2012.09.015. [DOI] [PubMed] [Google Scholar]
- Tiwari VN, Jeong JW, Wilson BJ, Behen ME, Chugani HT, Sundaram SK. Relationship between aberrant brain connectivity and clinical features in Angelman Syndrome: a new method using tract based spatial statistics of DTI color-coded orientation maps. NeuroImage. 2012;59(1):349–355. doi: 10.1016/j.neuroimage.2011.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi RB, McTigue DM. Chronically increased ciliary neurotrophic factor and fibroblast growth factor-2 expression after spinal contusion in rats. The Journal of comparative neurology. 2008;510(2):129–144. doi: 10.1002/cne.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Gruzdeva N, Li A, Chuang JZ, Sung CH. Identification of the Tctex-1 regulatory element that directs expression to neural stem/progenitor cells in developing and adult brain. The Journal of comparative neurology. 2010;518(16):3327–3342. doi: 10.1002/cne.22402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kooy D, Fishell G. Neuronal birthdate underlies the development of striatal compartments. Brain research. 1987;401(1):155–161. doi: 10.1016/0006-8993(87)91176-0. [DOI] [PubMed] [Google Scholar]
- Vincze A, Mazlo M, Seress L, Komoly S, Abraham H. A correlative light and electron microscopic study of postnatal myelination in the murine corpus callosum. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience. 2008;26(6):575–584. doi: 10.1016/j.ijdevneu.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Wallace ML, Burette AC, Weinberg RJ, Philpot BD. Maternal loss of Ube3a produces an excitatory/inhibitory imbalance through neuron type-specific synaptic defects. Neuron. 2012;74(5):793–800. doi: 10.1016/j.neuron.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CA, Beaudet AL, Clayton-Smith J, Knoll JH, Kyllerman M, Laan LA, Magenis RE, Moncla A, Schinzel AA, Summers JA, Wagstaff J. Angelman syndrome 2005: updated consensus for diagnostic criteria. American journal of medical genetics Part A. 2006;140(5):413–418. doi: 10.1002/ajmg.a.31074. [DOI] [PubMed] [Google Scholar]
- Williams CA, Driscoll DJ, Dagli AI. Clinical and genetic aspects of Angelman syndrome. Genetics in medicine : official journal of the American College of Medical Genetics. 2010;12(7):385–395. doi: 10.1097/GIM.0b013e3181def138. [DOI] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. The Journal of comparative neurology. 2006;499(1):144–160. doi: 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Huibregtse JM, Howley PM. The human E6-AP gene (UBE3A) encodes three potential protein isoforms generated by differential splicing. Genomics. 1997;41(2):263–266. doi: 10.1006/geno.1997.4617. [DOI] [PubMed] [Google Scholar]
- Yamasaki K. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Human molecular genetics. 2003;12(8):837–847. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]