Abstract
Several studies with animal models have demonstrated that bioequivalence of generic products of antibiotics like vancomycin, as currently defined, do not guarantee therapeutic equivalence. However, the amounts and characteristics of impurities and degradation products in these formulations do not violate the requirements of the U.S. Pharmacopeia (USP). Here, we provide experimental data with three generic products of meropenem that help in understanding how these apparently insignificant chemical differences affect the in vivo efficacy. Meropenem generics were compared with the innovator in vitro by microbiological assay, susceptibility testing, and liquid chromatography/mass spectrometry (LC/MS) analysis and in vivo with the neutropenic guinea pig soleus infection model (Pseudomonas aeruginosa) and the neutropenic mouse thigh (P. aeruginosa), brain (P. aeruginosa), and lung (Klebisella pneumoniae) infection models, adding the dihydropeptidase I (DHP-I) inhibitor cilastatin in different proportions to the carbapenem. We found that the concentration and potency of the active pharmaceutical ingredient, in vitro susceptibility testing, and mouse pharmacokinetics were identical for all products; however, two generics differed significantly from the innovator in the guinea pig and mouse models, while the third generic was therapeutically equivalent under all conditions. Trisodium adducts in a bioequivalent generic made it more susceptible to DHP-I hydrolysis and less stable at room temperature, explaining its therapeutic nonequivalence. We conclude that the therapeutic nonequivalence of generic products of meropenem is due to greater susceptibility to DHP-I hydrolysis. These failing generics are compliant with USP requirements and would remain undetectable under current regulations.
INTRODUCTION
Generic medicines are essential for health care systems. According to industry estimates, the United States saved 1.07 trillion dollars between 2002 and 2011, more than a billion dollars every other day, basically by offering more than 10 generic versions of each compound (1). To achieve such goals, drug regulatory agencies (DRA) such as the U.S. Food and Drug Administration (FDA) must facilitate the licensing process while ensuring safety, efficacy, and quality. As a consequence, as long as the active pharmaceutical ingredient (API) falls within the expected range of potency, concentration, and serum pharmacokinetics (PK), any generic drug is labeled interchangeable and bioequivalent; efficacy and safety are assumed without direct proof (2). This policy on generic drugs, accepted and implemented worldwide, has one critical drawback: there is experimental evidence that bioequivalent generic antibiotics can be inferior in vivo with respect to the innovator (3–7), and therapeutic nonequivalence conveys clinical and microbiological failure (3, 4), increased mortality (3), and bacterial resistance (8). The catastrophic case of generic heparin tainted with oversulfated chondroitin sulfate exemplifies the universal importance of this problem (9, 10).
Our data from the neutropenic mouse thigh infection model, showing therapeutic nonequivalence of generic products of vancomycin commercialized in Colombia, ignited additional research with generics in the United States and France. Analytical chemistry data for six vancomycin generics obtained by FDA scientists demonstrated that factor B and total impurities made up 90 to 95% and 5 to 10%, respectively, which was in full compliance with the U.S. Pharmacopeia (11–13). Similarly, no significant differences were detected among six French vancomycin generics based on the rabbit endocarditis model used by scientists from the Pontchaillou University Hospital at Rennes (14). These studies were done several years after the innovator had abandoned antimicrobial business, selling its vancomycin production secrets to all interested parties, while our research took place before and after that decision (7, 8). The discontinuation of the innovator made vancomycin a less-than-ideal choice to study the mechanisms behind therapeutic nonequivalence, but the FDA reports showed that U.S. generics differed slightly in the amounts and variety of impurities and degradation products (11, 12). Although such differences do not violate the pharmacopeia, the fact is that vancomycin generics are not chemically identical. This might be relevant, because a thorough evaluation of a generic product of metronidazole established that therapeutic equivalence was the result of absolute chemical and biological identity with the innovator (15). Based on these findings and the fact that the differences between the innovator and “bioequivalent” generics that fail are seen exclusively in vivo, we hypothesized that so-called insignificant chemical differences can cause molecular instability that leads to therapeutic nonequivalence of certain generic antibiotics.
Carbapenems exhibit different degrees of susceptibility to dehydropeptidase I (DHP-I; EC 3.4.13.19), a mammalian zinc-dependent metalloenzyme that belongs to the M19 superfamily, which hydrolyzes dipeptides and dehydropeptides as well as beta-lactams of the trans-conformation, like thienamycin and its N-formimidoyl derivative, imipenem. The introduction of a 1-β-methyl substituent into the structure of thienamycin conferred enough stability against DHP-I to meropenem, ertapenem, and doripenem to dispense with the addition of DHP-I inhibitors, such as cilastatin (16). However, resistance of each carbapenem to hydrolysis depends on the DHP-I species. For instance, meropenem is stable to human and cavian DHP-I but very susceptible to the murine and leporid versions of the enzyme, while imipenem resistance to hydrolysis is exactly the opposite (17).
In order to compare the in vivo efficacies of meropenem products, we chose two animal species for the infection models, Cavia porcellus and Mus musculus. The first species is ideal because the cavian dehydropeptidase I (cDHP-I) and the human version of the enzyme (hDHP-I) have very low hydrolytic potencies against meropenem, while the murine enzyme (mDHP-I) is quite active, as demonstrated by the relative Vmax/Km ratios for this carbapenem: values of 1 for hDHP-I, 2.4 for cDHP-I, and 22.2 for mDHP-I (17).
With this approach, we demonstrate here that two of three DRA-licensed generics of meropenem are therapeutically nonequivalent due to greater susceptibility to DHP-I, while only minor chemical changes were found by liquid chromatography/mass spectrometry (LC/MS) analysis. These structural differences, hard to detect, innocent in appearance, and totally acceptable under current regulations, seem to be responsible for in vivo failure of generic meropenem.
MATERIALS AND METHODS
Study design.
The study design is illustrated in Fig. 1. The investigators were blind to the manufacturer of the study products until data analysis ended, except for conduct of LC/MS assays. For inclusion in this study, only bioequivalent generics were used, which implies pharmaceutical and pharmacokinetic equivalence with respect to the innovator. In addition, we required that all products have identical in vitro susceptibility testing profiles. Then, therapeutic equivalence with the innovator was determined by using the neutropenic mouse thigh (sepsis), brain (meningoencephalitis), and lung (pneumonia) infection models, as well as the neutropenic guinea pig soleus (sepsis) infection model.
FIG 1.
Flow chart for the project design.
Mechanistic studies included in vivo protection of meropenem (mouse models) with the DHP-I inhibitor cilastatin to measure the extent to which hydrolysis by mDHP-I was responsible for the lack of therapeutic equivalence and with high-performance LC with UV detection (HPLC-UV)/LC-MS analyses of the generic and innovator drugs to determine the kind of chemical differences causing therapeutic nonequivalence without affecting bioequivalence, looking at (i) the chemical entities within their formulations, (ii) the stability at room temperature of each pharmaceutical form in solution, and (iii) the susceptibility to hydrolysis by mDHP-I. Protocols were approved by the University of Antioquia Animal Care and Experimentation Ethics Committee.
Antibacterial and chemical agents.
Meropenem products were bought from local drugstores and prepared for experimental use as indicated by their makers (Table 1): the innovator, produced by Astra Zeneca (called iMer), and three generics produced by Vitalis (gMer-A), Procaps (gMer-B), and Farmionni (gMer-C). A reference standard (meropenem trihydrate; Sigma-Aldrich, St. Louis, MO) was employed for analytical chemistry only. Cilastatin was donated by Merck (Whitehouse Station, NJ).
TABLE 1.
Meropenem products, pharmaceutical form, license number, batch, and manufacturer
| Meropenem product (code) | Ampule presentationa | License no. | Batch(es) | Manufacturer |
|---|---|---|---|---|
| Analytical standard | Meropenem trihydrate powder, 500 mg | CAS no. 119478-56-7 | 106K0046V | Sigma-Aldrich, St. Louis, MO |
| Generic A (gMer-A) | Meropenem trihydrate powder + Na2CO3, 500 mg | INVIMA 2004M-0003819 | B5305032, B5305052, B040687, C100752, C010806, C050994 | Vitalis S.A.C.I., Bogotá, Colombia |
| Generic B (gMer-B) | Meropenem trihydrate powder + Na2CO3, 500 mg | INVIMA 2006M-0005526 | ES270220, ES230084 | Procaps S.A., Barranquilla, Colombia |
| Generic C (gMer-C) | Meropenem trihydrate powder + Na2CO3, 500 mg | INVIMA 2010M-0010487 | ES200261 | Farmioni (Procaps S.A.), Barranquilla, Colombia |
| Innovator (iMer) | Meropenem trihydrate powder + Na2CO3, 500 mg | INVIMA 2007M-006423-R1 | DY001, DF465, CS686, CN859, CH277, HF501, FF201 | AstraZeneca, Macclesfield, Cheshire, UK |
Only the analytical standard was devoid of the excipient, sodium carbonate (Na2CO3).
Bacterial strains.
Efficacy of meropenem was determined in vitro and in vivo against the wild-type (WT) clinical isolates Pseudomonas aeruginosa GRP-0019 and Klebsiella pneumoniae GRP-0107 and multidrug-resistant (MDR) P. aeruginosa GRP-0049 (resistant to all antipseudomonal drugs except carbapenems and polymyxins). Kocuria rhizophila ATCC 9341 was the agar-seeding strain for microbiological assays; K. pneumoniae ATCC 10031 and P. aeruginosa ATCC 27853 were the quality control organisms for susceptibility tests.
In vitro susceptibility testing.
The minimal inhibitory (MIC) and bactericidal (MBC) concentrations of meropenem against K. pneumoniae GRP-0107 (WT) and P. aeruginosa strains GRP-0019 (WT) and GRP-0049 (MDR) were determined by broth microdilution following CLSI protocols (18). All assays were performed at least twice; results were read for innovator and generic products simultaneously after 18 to 21 h of incubation, and their geometric means were compared by using the Kruskal-Wallis test (Prism 6; GraphPad Software Inc., La Jolla, CA). The quality control organisms during in vitro susceptibility testing were K. pneumoniae ATCC 10031 and P. aeruginosa ATCC 27853.
Microbiological assays (pharmaceutical equivalence).
Difco antibiotic medium 11 was the ideal diffusion agar for meropenem and was seeded with the indicator strain K. rhizophila ATCC 9341 and poured into a custom designed glass plate large enough to accommodate all samples simultaneously (19); this assay has been properly validated for this application (20). Standard curves (quintuplets) with 10 known concentrations of each product (doubling from 4 to 2,048 mg/liter) were obtained by linear regression and compared by curve-fitting analysis (CFA) to see if there were significant differences in their intercepts (concentration of the active pharmaceutical ingredient [API]) or the slopes (potency of the API). The CFA was computed by using Prism 5.0 (GraphPad Software, Inc., La Jolla, CA) (21).
The same microbiological assay was applied to determine the serum drug concentration of meropenem for characterization of single-dose mouse pharmacokinetics (PK). The standard curves were identical to those described above but made with the innovator product; the concentrations (4 to 2,048 mg/liter) were selected to span the levels attainable in mice after single-dose PK and multiple-dose pharmacodynamics (PD) experiments (see below). Plates were incubated for 18 h at 37°C under an aerobic atmosphere, and the same researcher measured the zones of inhibition sizes (diameters) for all assays with an electronic caliper. The diameters of the unknowns were interpolated against the standard curve to obtain the respective concentrations.
Single-dose serum drug pharmacokinetics in neutropenic mice infected in the thigh with P. aeruginosa GRP-0019 (bioequivalence).
Meropenem generic products were studied simultaneously with the innovator at three dose levels, 10, 40, and 160 mg/kg of body weight. Two hours after infection, two groups of animals (one for the innovator and one for the generic to be compared in each experiment) received a single subcutaneous injection (0.2 ml) containing the dose of meropenem to be tested. Data for each dose and product were obtained from two subgroups of 3 female mice bled by retroorbital puncture at each of the following data points: 5, 10, 15, 30, 45, and 60 min after dosing. To prevent hypotension due to excessive sampling, we intercalated both subgroups so that no animal was bled more than three times. Serum was separated by blood centrifugation at 10,000 × g during 5 min and stored immediately at −80°C. Once all samples had been obtained, we analyzed them in one microbiological assay. The PK parameters for maximal (Cmax) and minimal (Cmin) drug concentrations, half-life of elimination (t½), and area under the concentration-time curve (AUC) were obtained by noncompartmental analysis (WinNonlin 5.2; Pharsight Corp., Mountain View, CA). The AUCall of the generic products, expressed as a percentage of the innovator product and compared by analysis of variance (ANOVA), implied bioequivalence when it ranged between 80% and 125%, as accepted by DRA everywhere.
Pharmacodynamics: animal infection models.
Several aspects were common to the four animal models employed: (i) the experimental protocols were reviewed and approved by the University of Antioquia Animal Care and Experimentation Ethics Committee; (ii) specific-pathogen-free (SPF)-certified guinea pigs and murine-pathogen-free (MPF)-certified mice were bred and raised in separate environments within a high-tech facility under strict microbiological barriers (One-Cage Micro-Isolator systems; LabProducts Inc., Seaford, DE) until they reached the age and weight specified for each model; (iii) the Swiss albino murine strains Udea:ICR(CD-1) and Udea:ICR(CD-2) and the cavian Hartley strain Udea:Crl(HA) were used in the process; (iv) profound neutropenia was induced with intraperitoneal injections of cyclophosphamide 4 days (150 mg/kg) and 1 day (100 mg/kg) before the infection (22); (v) to prepare the bacterial inoculum, we had specific protocols for each pathogen and they were designed to obtain vigorous cell samples in the log phase of growth (23) (P. aeruginosa and K. pneumoniae were grown in BBL Trypticase soy broth [Becton, Dickinson & Co., Sparks, MD] at 37°C in an aerobic atmosphere, and broth cultures were adjusted to the inoculum size required for each model); (vi) treatment with meropenem products was always simultaneous (generic and innovator) and administered every 3 h (q3h) by subcutaneous injections of 200 μl per dose, while untreated controls received sterile saline injections via an identical volume and dosing schedule; (vii) each PD experiment generated a 24-h dose-response curve encompassing at least five total doses that extended from minimal effect (at doses as low as 2.5 mg/kg/day) to maximum effect (at doses as high as 2,560 mg/kg/day), with the dose ranges selected to answer the questions at hand; (viii) untreated controls in the same numbers as the treated subgroups were euthanized by cervical dislocation immediately after bacterial inoculation (hour −2), to confirm the inoculum size, or at the time of starting (hour 0) or ending (hour 24) therapy, although control mice left to the end of the experiment were usually found dead or premortem in all four models; meropenem-treated animals were also euthanized at hour 24; (ix) the target organ (whole thighs, lungs, brains, or solei muscles, depending on the model) was dissected aseptically, homogenized, serially diluted, plated in duplicate on solid medium (Trypticase soy agar), and incubated at 37°C for 18 to 24 h; (x) the target organs were washed at least twice to eliminate any remnant meropenem, and carryover detection was part of the protocol in every experiment (24), although it was not found even with the highest dose used (2,560 mg/kg/day, administered q3h); (xi) after colonies were harvested and counted for each organ, data were registered in an Excel database as the log10 CFU per gram of tissue (log10 CFU/g); (xii) the limit of detection was 2.0 log10 CFU/g, and therefore any organ with 0 colonies was entered in the database as 100 CFU/g; (xiii) to calculate the net antibacterial effect in 24 h, the number of CFU/g remaining in the target organ after 24 h of treatment was subtracted from the number of CFU/g that grew in the untreated control; (xiv) to calculate the time above the MIC (T>MIC) for any dose and strain, the innovator's PK parameters of the absorption constant (ka), clearance (CL), volume of the central compartment (Vc), distribution clearance (CLd), and volume of the peripheral compartment (Vp), as well as their respective coefficients of variation (CV), were computed under a parametric population PK model with the SADAPT-TRAN program (25, 26). In the Results section, these data are expressed as the fraction (percent) of the dosing interval (q3h) that meropenem serum levels remained above the MIC (%T>MIC). The particularities of each animal model are summarized as follows.
Neutropenic mouse thigh infection model (sepsis).
The neutropenic mouse thigh infection model is probably the animal model that affords the greatest contributions to antimicrobial PD (27); it is well known, and details for this model have been described elsewhere (5–7). For this model, we employed 6-week-old, 23- to 27-g female mice; infection was induced by intramuscular injection of 100 μl of the freshly prepared bacterial inoculum in each thigh (P. aeruginosa GRP-0019 or GRP-0049; 6.0 log10 CFU/ml). Treatment started 2 h postinfection, and there were two mice in each dose subgroup.
Neutropenic mouse brain infection model (bacterial meningoencephalitis).
For the neutropenic mouse brain infection model, we employed 6-week-old, 23- to 27-g female mice that were inoculated in the brain by retroorbital injection with 10 μl of a freshly prepared culture of P. aeruginosa GRP-0019 or GRP-0049. Treatment started 2 h postinfection, included three mice per dose subgroup, and ended 24 h later when animals were euthanized.
Neutropenic mouse lung aerosolized infection model (bacterial pneumonia).
Besides the target organ, the neutropenic mouse lung aerosolized infection model differs from all others in that it allows 38 instead of 26 h between infection and sacrifice; for inoculation, 6-week-old, 23- to 27-g female mice were exposed for 45 min to an aerosol containing K. pneumoniae GRP-0107 (9.0 log10 CFU/ml). Treatment started 14 h postinfection with three mice per dose subgroup and ended 24 h later, when animals were sacrificed.
Neutropenic guinea pig soleus infection model (sepsis).
Four- to 10-week-old male or female guinea pigs weighing on average 500 g (range, 250 to 800 g) were inoculated in the middle of their shanks (soleus muscle) with a bacterial suspension of P. aeruginosa GRP-0019 containing 6.0 log10 CFU/ml in a 100-μl volume. Production of SPF guinea pigs is very expensive and never as straightforward as with MPF mice; to collect enough individuals for one experiment, it is necessary to use any sex and size and to treat each animal according to its actual weight by subcutaneous injections of meropenem q3h (as described above).
Inhibition of mDHP-I with cilastatin.
Since meropenem is susceptible to hydrolysis by mDHP-I, we designed experiments with murine infection models (thigh, lung, and brain) to compare the impact of cilastatin on the PD of generics and the innovator, using meropenem-to-cilastatin (M:C) ratios of 1:0, 1:1, 1:3, and 1:5 (in mg/kg/day).
Statistical analysis of PD data.
A detailed description of the methods used for statistical analysis of PD data has already been published (7). In short, data from dose-effect experiments were modeled by nonlinear regression (NLR) and fitted to the Hill equation to produce significant pharmacodynamic parameters (PDP) for each product. The sigmoid dose-response model is described by the Hill equation, E = [(Emax) × DN]/(ED50N + DN), where E is the net antibacterial effect (in log10 CFU/g) after 24 h of treatment, D is the meropenem dose (in mg/kg/day), Emax is the maximum effect (in log10 CFU/g), the ED50 is the effective dose needed to achieve 50% of the Emax (in mg/kg/day), and N is Hill's slope (it correlates with the affinity of the drug for its target). Emax, ED50, and N are the primary PDP obtained by a least-squares NLR, but the nature of the data made the secondary PDP bacteriostatic dose (BD) much more meaningful than the ED50, and therefore it was used instead. Their magnitudes were compared by CFA with the SigmaPlot 12 program (Systat Software, San Jose, CA) and Prism 5.0 (21, 28). A thorough analysis of NLR results was done for every experiment, based not only on the significance of the PDP but also on the adjusted coefficient of determination (adjR2) and the standard error of the estimate (Sy|x) of each regression (SigmaPlot 12). When NLR diagnostics pointed toward any problem, like multicollinearity of the PDP (variance inflation factor [VIF], ≥4.0), violation of the NLR assumptions, or unexplained outliers, the causes were identified (if possible) and new experiments were performed with a design that addressed the origin of the problems. To be compared by CFA, all NLR were required to have statistically significant coefficients devoid of serious multicollinearity (VIF, ≥7.0) and had to respect the regression assumptions of normality and constant variance (homoscedasticity); no outlier was eliminated unless induced by a human error that was clearly stated in the experiment's log. Serious multicollinearity is usually defined as a VIF of ≥10, because it implies a very strong covariance (R2, 0.9) of the coefficient involved, but the actual impact of multicollinearity in any NLR is more dependent on the research problem under scrutiny, and it is our experience that any VIF above 7.0 implies a serious problem, either structural or sample based. In case of mild multicollinearity (VIF, <7.0) in one of the products, its regression was compared only if the affected PDP was highly significant (P < 0.0001) and its standard error was negligible (28).
HPLC-UV quantitative assays of standard curves and enzymatic degradation: apparatus and chromatographic conditions.
The chromatographic system consisted of an HPLC quaternary pump (HP1100 column oven; Hewlett-Packard, Waldbronn, Germany) that was used and connected to a UV detector of the same series. Data analysis was performed using the manufacturer's software. Separation was achieved at 20°C by using a Satisfaction C18 Luna column (250 mm by 4.6 mm; 5 μm). The mobile phase consisted of a mixture of 0.1 M buffered phosphate (pH 6.8 to 7.0) and acetonitrile at a ratio of 90:10 delivered at 0.5 ml/min in an isocratic elution. Each sample was run for 15 min. The autosampler temperature was kept at 8°C, and the injection volume was 50 μl. The detection wavelength was set at 298 nm.
Sample preparation and solutions.
The pharmaceutical forms of meropenem products (iMer and gMer-A) were used for method development with the HPLC-UV, and a meropenem analytical standard (Sigma) was used as the external control. Methanol and HPLC-grade water were used for the respective dilutions in the optimization of conditions and analysis. For the standard curves, solutions were prepared by dissolving each product in sterile deionized water to generate a stock concentration of 5,000 mg/liter. HPLC-grade mobile-phase solvents (buffer phosphate and acetonitrile) were employed (29).
Standard curves.
The concentration of the API was calculated for each meropenem product by using the peak area ratios to model a validated standard curve by linear regression with Prism 5.0 software. The standard curves of all generics and the innovator were compared by CFA against the validated curve of the external meropenem standard (Sigma); generics were also compared against the innovator. The linear regression parameters (slope and intercept) were analyzed in the same form described above for the data obtained from microbiological assays; each curve was run at least twice.
HPLC quantification of meropenem hydrolysis by mDHP-I.
The mDHP-I enzyme was obtained from murine renal tissue following the methodology described by Campbell et al. (30); for kidney extraction, male MPF mice of the strain Udea:ICR(CD-2) were sacrificed and processed immediately. Fresh tissue aliquots (5 g) were homogenized, suspended to 5 ml with 4°C sterile normal saline, and centrifuged (13,000 × g) for 30 min. The supernatant was centrifuged 5 times, and the product obtained was a partially purified mDHP extract that was kept at 4°C for no more than 4 h. Three concentrations (125, 250, and 500 mg/liter) of each meropenem product and one concentration (500 mg/liter) of imipenem-cilastatin (Merck, Whitehouse Station, NJ; used as an mDHP-protected control) were mixed with 125, 250, or 500 μl, respectively, of the mDHP extract in a 1:1 volume ratio. This mix was incubated at 37°C for 4.5 h, and simultaneous samples were taken from each enzymatic reaction mixture (iMer, gMer-A, and imipenem-cilastatin) at 5, 30, 60, 90, 120, and 150 min. A separate group of meropenem samples at identical concentrations was incubated without the mDHP extract, as degradation controls. The samples were stored at −20°C until use in the HPLC quantification assay (no more than 6 h). For the assay, samples were cleaned of tissue particles by filtration through 0.22-μm devices, and the equipment was cleaned with the mobile phase for 15 min before starting with the next product. All samples of each product were run in duplicate at the same time, with the specific standard curve to quantify the antibiotic concentration determined at each point by interpolation. The degradation kinetics of gMer-A and iMer were compared by CFA (Prism 5.0).
Quantification of antibacterial activity after enzyme degradation of meropenem.
The microbiological assay described above was applied to determine the residual concentrations and potency of the API of meropenem products exposed to the mDHP extract (31). Standard curves of each meropenem product (freshly prepared duplicates ranging from 8 to 1,000 mg/liter) were used for references and plated simultaneously with the duplicate samples exposed to mDHP extract. Assays with iMer and gMer-A were performed simultaneously, zone sizes were measured as described above, problem concentrations were obtained by interpolation from the standard curves, and degradation kinetics of both products were compared by CFA (Prism 5.0).
Dynamic qualitative assay with LC/MS.
Analytical chemistry data were obtained with an Agilent 1100 liquid chromatograph coupled to a mass spectrometer electrospray ionization VL system. At the stationary phase, a Thermo Scientific Hypersil Gold analytical column (150 mm by 4.6 mm; 5 μm) was employed, with each meropenem product having its own column. We used the single-ion monitoring (SIM) mode [M+H] to obtain the chromatogram, and the scan mode to gather the mass spectra, with a range of 100 to 1,000 m/z. The mobile phase consisted of A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile), with A at 90% and B at 10%, 0.5 ml/min as the flow rate, and a total run time of 10 min (32). Working solutions for all studies were prepared by serial dilution of the stock solution (5,000 mg/liter).
In a first step, to compare the chromatograms (SIM mode) and the mass spectra (scan mode), all preparations for reference material (Sigma) and pharmaceutical formulations (iMer and gMer-A) were freshly prepared in deionized water at the moment of analysis by using a 250-mg/liter meropenem concentration. The mobile phase was kept running in the equipment for 15 min prior to sampling, and a blank sample was run after each product. In a second step, to compare the influence of storage time on the chromatograms and the mass spectra of iMer and gMer-A, samples with the same preparation and concentration methods were maintained at ambient temperature (25°C) during 48 h for mass and abundance evaluations at 6, 24, and 48 h. We repeated each run at least twice.
RESULTS
To facilitate navigation through the data, Fig. 1 depicts a flow chart of the experimental procedures.
In vitro susceptibility tests.
The geometric means of the minimal inhibitory (MIC) and bactericidal (MBC) concentrations of generics (gMer-A, gMer-B, and gMer-C) and innovator meropenem (iMer) were virtually identical against the three strains of P. aeruginosa of this study, producing the following data (in mg/liter): 1.0 (MIC) and 2.0 (MBC) against GRP-0019 (WT), 2.0 and 4.0 for the MDR strain GRP-0049, and data always within the CLSI range (0.25 to 1.0) for the quality control strain ATCC 27853. Against K. pneumoniae GRP-0107 (WT), the MIC and MBC were the same (0.06 mg/liter for all products except iMer, for which the MBC was 0.07 mg/liter), while for the quality control K. pneumoniae ATCC 10031, the MIC and MBC were 0.06 mg/liter for all products except gMer-B, for which the MBC was 0.07 mg/liter (exact P > 0.99, Kruskal-Wallis test).
In vivo PD: general results after comparing the three generics (gMer-A, gMer-B, and gMer-C) with the innovator (iMer) in four animal models infected with different pathogens.
Bacterial growth in untreated controls for the different models was steady, predictable, and reproducible, inducing sepsis (cavian soleus and murine thigh infection), meningoencephalitis (brain infection), or pneumonia (lung infection) in all subjects assigned to each animal model (Table 2). If left untreated, all models caused death for 100% of the animals within 24 h.
TABLE 2.
Bacterial inocula size ranges for untreated controls in the different animal models employed in this studya
| Bacterial pathogen | Neutropenic animal model | Size range of inoculum (log10 CFU/g of tissue) |
Growth in 24 h (log10 CFU/g of tissue) | |
|---|---|---|---|---|
| Start of treatment (h 0) | End of treatment (h 24) | |||
| P. aeruginosa GRP-0019 (WT) | Mouse thigh | 5.87–6.43 | 8.58–9.29 | 2.17–3.42 |
| Mouse brain | 4.29–4.98 | 8.91–9.73 | 3.97–5.26 | |
| Guinea pig soleus | 5.94–6.56 | 8.73–9.21 | 2.65–2.94 | |
| P. aeruginosa GRP-0049 (MDR) | Mouse thigh | 6.0–5.78 | 8.42–8.83 | 2.64–2.83 |
| Mouse brain | 4.27–4.46 | 8.68–8.99 | 4.31–4.72 | |
| K. pneumoniae GRP-0107 (WT) | Mouse lung | 6.70–7.04 | 9.58–10.08 | 2.66–3.38 |
Standard deviations for each value were omitted for the sake of clarity.
Enzymatic hydrolysis of meropenem by DHP-I in the murine models (mDHP-I) was manifested by one or several of these PD problems: (i) dramatic loss of antibacterial effect, (ii) increased experimental variance represented by a high Sy|x that diminished the statistical power for detection of less-effective products, (iii) violation of NLR assumptions, such as homoscedasticity or normality, which prevented a reliable statistical comparison, because those assumptions are essential for a valid fit of the data to the PD model, and (iv) diverse degrees of multicollinearity of the primary PDP in some experiments with an acceptable fit. These effects of mDHP-I were seen in all products, but the degree of meropenem hydrolysis varied between them with the animal model experiment used, the infected organ, the dose range of each experiment, the infecting strain and, as expected, the meropenem-to-cilastatin (M:C) ratio.
Table 3 summarizes detailed results of the NLR shown in each figure, i.e., the values of the respective PDP, their statistical quality, and the NLR diagnostics. The daily dose of meropenem was administered every 3 h to optimize the PD index; the %T>MIC in dose-response experiments spanned from no effect to maximal effect. In the following paragraphs, the data generated by each model are presented in this order: a subtitle with a general description of the animal model (clinical syndrome and bacterial strains), followed by the relevance of the model, number of experiments done, the figure and table showing the respective data, results, and observations stemming from the results.
TABLE 3.
Primary pharmacodynamics parameters and mathematical quality of respective nonlinear regressions obtained from the dose-response relationships of the different animal modelsa
| Bacterial pathogen | Model | M:C ratio | Meropenem formb | Emax (mean Δlog10 CFU/g at 24 h ± SEM) | BD (mean mg/kg/day ± SEM) | N (mean Hill's slope ± SEM) | PDP with multicollinearity | P value (CFA) | T>MIC for BD (%) |
|---|---|---|---|---|---|---|---|---|---|
| P. aeruginosa GRP-0019 | Mouse thigh | 1:0 | iMer | NSc | NS | 0.769 ± 0.287 | All | NAd | NA |
| gMer-B | NS | NS | 0.673 ± 0.242 | All | |||||
| gMer-C | NS | NS | 0.645 ± 0.252 | All | |||||
| P. aeruginosa GRP-0019 | Mouse thigh | 1:0 | iMer | 6.358 ± 0.202 | 105.50 ± 8.07 | 2.079 ± 0.297 | None | 0.3220 | 67.9 |
| gMer-A | 6.028 ± 0.160 | 121.20 ± 7.36 | 2.948 ± 0.463 | None | 76.3 | ||||
| P. aeruginosa GRP-0019 | Mouse thigh | 1:1 | iMer | 5.929 ± 0.267 | 33.26 ± 2.53 | 3.025 ± 0.713 | None | 0.4150 | 19.5 |
| gMer-A | 6.480 ± 0.267 | 37.65 ± 2.34 | 2.243 ± 0.351 | None | 21.3 | ||||
| P. aeruginosa GRP-0019 | Mouse thigh | 1:3 | iMer | 6.935 ± 0.196 | 44.57 ± 2.32 | 1.861 ± 0.199 | None | 0.2310 | 23.8 |
| gMer-A | 7.019 ± 0.236 | 47.12 ± 2.55 | 1.520 ± 0.158 | None | 24.7 | ||||
| P. aeruginosa GRP-0049 | Mouse thigh | 1:1 | iMer | 6.836 ± 0.532 | 85.74 ± 5.17 | 1.303 ± 0.166 | Emax, BD | NA | NA |
| gMer-A | 6.465 ± 0.187 | 62.62 ± 2.31 | 1.857 ± 0.154 | None | 18.7 | ||||
| P. aeruginosa GRP-0049 | Mouse thigh | 1:3 | iMer | 5.827 ± 0.164 | 60.60 ± 3.03 | 3.290 ± 0.472 | None | 0.9863 | 18.2 |
| gMer-A | 5.797 ± 0.151 | 59.65 ± 2.79 | 3.529 ± 0.480 | None | 18.0 | ||||
| P. aeruginosa GRP-0049 | Mouse thigh | 1:5 | iMer | 5.812 ± 0.187 | 54.66 ± 3.22 | 3.102 ± 0.502 | None | 0.9661 | 16.8 |
| gMer-A | 5.904 ± 0.177 | 54.47 ± 2.98 | 3.225 ± 0.493 | None | 16.8 | ||||
| P. aeruginosa GRP-0019 | Mouse brain | 1:0 | iMer | 6.482 ± 0.212 | 66.25 ± 7.19 | 1.444 ± 0.236 | None | 0.0020 | 31.5 |
| gMer-B | 6.931 ± 0.295 | 58.88 ± 7.56 | 1.167 ± 0.235 | None | 28.7 | ||||
| gMer-C | 5.900 ± 0.104 | 44.29 ± 3.35 | 3.140 ± 0.415 | None | 23.7 | ||||
| P. aeruginosa GRP-0019 | Mouse brain | 1:0 | iMer | 6.427 ± 0.520 | 41.07 ± 4.07 | 0.689 ± 0.179 | All | NA | NA |
| gMer-A | 5.916 ± 0.318 | 44.03 ± 5.15 | 1.115 ± 0.279 | Emax, BD | NA | ||||
| P. aeruginosa GRP-0019 | Mouse brain | 1:1 | iMer | 7.164 ± 0.207 | 50.21 ± 2.81 | 0.846 ± 0.105 | None | NA | 25.7 |
| gMer-A | 6.605 ± 0.167 | 60.73 ± 4.37 | 1.279 ± 0.173 | Emax, N | NA | ||||
| P. aeruginosa GRP-0019 | Mouse brain | 1:3 | iMer | 6.537 ± 0.168 | 81.94 ± 6.86 | 2.057 ± 0.245 | None | <0.0001 | 41.5 |
| gMer-A | 7.279 ± 0.167 | 120.20 ± 5.80 | 1.709 ± 0.125 | None | 75.8 | ||||
| P. aeruginosa GRP-0049 | Mouse brain | 1:1 | iMer | 5.874 ± 0.2373 | 121.6 ± 10.7 | 3.676 ± 0.827 | None | NAe | NA |
| gMer-A | 6.311 ± 0.3355 | 96.11 ± 9.67 | 2.164 ± 0.4129 | None | 25.0 | ||||
| P. aeruginosa GRP-0049 | Mouse brain | 1:3 | iMer | 6.498 ± 0.113 | 87.64 ± 3.92 | 1.797 ± 0.126 | None | 0.0002 | 23.5 |
| gMer-A | 6.498 ± 0.363 | 136.80 ± 13.3 | 1.599 ± 0.246 | None | 32.4 | ||||
| K. pneumoniae GRP-0107 | Mouse lung | 1:0 | iMer | NS | NS | 0.722 ± 0.288 | All | NA | NA |
| gMer-A | NS | NS | 0.784 ± 0.259 | All | NA | ||||
| K. pneumoniae GRP-0107 | Mouse lung | 1:0 | iMer | 5.577 ± 0.310 | 13.91 ± 2.46 | 0.843 ± 0.166 | None | 0.1265 (iMer vs gMer-B)f | 92.9 |
| gMer-B | 5.764 ± 0.526 | 26.69 ± 6.10 | 0.771 ± 0.182 | Emax, BD | NA | ||||
| gMer-C | 5.906 ± 0.913 | NS | 0.614 ± 0.195 | All | NA | ||||
| K. pneumoniae GRP-0107 | Mouse lung | 1:3 | iMer | 5.916 ± 0.614 | 5.37 ± 1.25 | 0.875 ± 0.386 | Emax, N | NA | NA |
| gMer-A | 6.774 ± 0.955 | 9.89 ± 1.37 | 0.780 ± 0.237 | All | NA | ||||
| K. pneumoniae GRP-0107 | Mouse lung | 1:5 | iMer | 5.137 ± 0.260 | 9.05 ± 1.04 | 1.719 ± 0.453 | None | NA | 85.0 |
| gMer-A | NS | NS | 0.395 ± 0.152 | All | NA | ||||
| P. aeruginosa GRP-0019 | Guinea pig soleus | 1:0 | iMer | 6.485 ± 0.094 | 11.53 ± 0.40 | 3.331 ± 0.351 | None | <0.0001 | NDg |
| gMer-A | 6.642 ± 0.345 | 21.60 ± 2.55 | 1.444 ± 0.274 | None | ND | ||||
| P. aeruginosa GRP-0019 | Guinea pig soleus | 1:0 | iMer | 6.463 ± 0.184 | 10.99 ± 1.10 | 1.438 ± 0.257 | None | <0.0001f | ND |
| gMer-B | 6.349 ± 0.162 | 11.75 ± 0.99 | 1.503 ± 0.239 | None | ND | ||||
| gMer-C | 7.238 ± 0.466 | 25.12 ± 2.84 | 1.082 ± 0.197 | Emax | ND |
The ranges of the adjR2 and Sy|x were 0.7 to 0.988 and 0.222 to 0.624, respectively (data not shown).
gMer-A, generic product Vitalis; gMer-B, generic product Procaps; gMer-C, generic product Farmioni.
NS, not significant.
NA, not applicable. Statistical comparison of this regression was not possible due to serious multicollinearity of the PDP in one or both products.
*, in this instance in which this parameter was not applicable, violation of normality and homoscedasticity assumptions (iMer) prevented statistical comparison.
In this experiment, a formal statistical comparison was indicated because only one product had multicollinearity and it was mild (highest VIF, <7.0).
ND, not done.
Neutropenic guinea pig soleus infection model (sepsis by the WT strain P. aeruginosa GRP-0019).
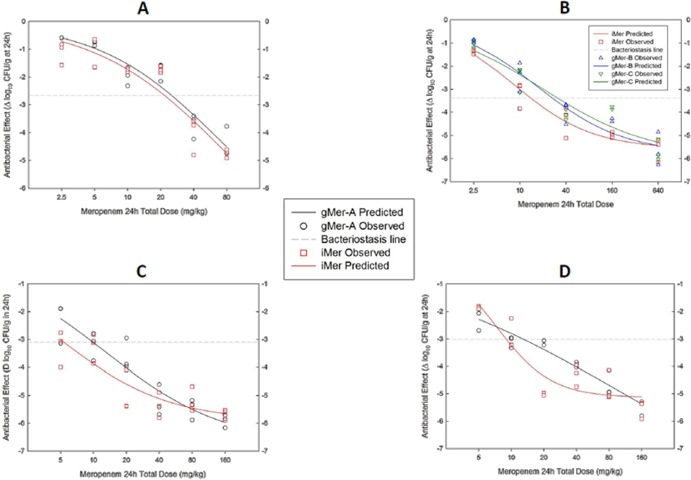
Of all mammalian dipeptidases studied so far, Cavia porcellus DHP-I (cDHP-I) has the closest Vmax/Km ratio to the human counterpart (ratios of 2.4 and 1, respectively), so it should not be necessary to protect meropenem with cilastatin when treating guinea pigs. The results from two individual experiments are illustrated in Fig. 2 and detailed in Table 3.
FIG 2.

Pharmacodynamics of three generics compared with the innovator of meropenem against the WT strain P. aeruginosa GRP-0019 (MIC, 1 mg/liter) in the neutropenic guinea pig soleus infection model (panels correspond to separate experiments). The hydrolytic activity of cDHP-I against meropenem was very close to that of hDHP-I, making this species useful to model the PD in a human-like environment and without protecting the antibiotic with cilastatin. All products fit the Hill equation without faults; gMer-A and gMer-C required 91% (A) and 122% (B) greater doses to attain bacteriostasis than iMer, respectively. gMer-B, on the other hand, was indistinguishable (P = 0.88) from iMer (B).
As expected, all products reached maximal effect and their NLR were impeccable (Table 3). This model showed that gMer-A (Fig. 2A) and gMer-C (Fig. 2B) were significantly less potent than iMer (P < 0.0001), while gMer-B was virtually identical to iMer (P = 0.88) (Fig. 2B). There were no differences in the Emax, but the magnitude of the difference in potency between products can be seen by comparing the BD (in mg/kg/day) of iMer (11.0 ± 1.1 [Fig. 2A] and 11.5 ± 0.4 [Fig. 2B]) and gMer-B (11.8 ± 0.99) with that of gMer-A (21.6 ± 2.6) and gMer-C (25.1 ± 2.8). It means that gMer-A required 91% and gMer-C 122% greater doses than iMer to stop the growth of P. aeruginosa GRP-0019 in the guinea pig, which is a human-like model in terms of DHP-I.
Neutropenic mouse thigh infection model (sepsis by P. aeruginosa, either WT strain GRP-0019 or MDR strain GRP-0049).
The hydrolytic activity of mDHP-I against meropenem is 22.2- and 9.25-fold greater than that of hDHP-I and cDHP-I, respectively (17). Within the murine models, the thigh is intermediate in terms of mDHP-I activity (approximately 6.4-fold greater than the brain and 469-fold lower than the lung); it has virtually no restrictions for the drugs to reach the site of infection and allows a thorough examination of the interaction between the dipeptidase, the carbapenem, and the DHP-I inhibitor cilastatin. All nine experiments with this model are detailed in Table 3, but only the data with the MDR strain are illustrated in Fig. 3 (five experiments).
FIG 3.
Pharmacodynamics of one generic product (gMer-A) compared with the innovator of meropenem against the MDR strain P. aeruginosa GRP-0049 (MIC, 2 mg/liter) in the neutropenic mouse thigh infection model. Against this organism, a 1:1 M:C ratio was insufficient to protect meropenem from mDHP-I hydrolysis, and it was necessary to accumulate data from three identical experiments to obtain a valid nonlinear regression for gMer-A; persistent multicollinearity of both products prevented their statistical comparison, but their PD profiles looked quite different (A). With a 1:3 M:C ratio, both products fit the Hill equation with a single experiment without faults, dropping the T>MIC to 18.1% (B); increasing the M:C ratio to 1:5 made both products even more potent (T>MIC, 16.8%), suggesting additional hydrolysis of meropenem caused by this strain, probably by an enzyme other than mDHP-I but still susceptible to cilastatin inhibition (C). Increasing concentrations of cilastatin made products identical, with overlapping PD curves (compare panel A with panels B and C).
Without cilastatin (M:C ratio, 1:0), very large doses (in mg/kg per day) of meropenem were required to generate a sigmoid dose-response curve against the WT strain, P. aeruginosa GRP-0019 (MIC, 1 mg/liter): 113.2 ± 5.6 (T>MIC of 72.2%) for bacteriostasis (BD) and 640 (T>MIC of 100%) for maximal effect (Emax). The addition of cilastatin (M:C ratio of 1:1) caused a 3.2-fold drop in the BD, 35.5 ± 1.8 mg/kg/day (T>MIC, 20.4%), but it did not drop further after increasing the M:C ratio to 1:3. Generics and innovator were indistinguishable without (P = 0.32) or with cilastatin (P = 0.23); therefore, each PDP was the same for all products (Table 3 provides the parameter values corresponding to the individual meropenem products, while values reported here come from unified regressions after finding no difference between products).
Pseudomonas aeruginosa GRP-0049 is resistant to all antibiotics except carbapenems and polymyxins, but the meropenem MIC and MBC (2 and 4 mg/liter, respectively) are only 1 dilution above those of the WT P. aeruginosa GRP-0019. Against this MDR strain, a 1:1 M:C ratio was insufficient to protect meropenem products from mDHP-I hydrolysis despite a dose range (20 to 640 mg/kg/day) spanning a T>MIC from 0% to 95.5%. Serious multicollinearity prevented comparison of meropenem products after three experiments under identical conditions (Fig. 3A). Increasing the M:C ratio to 1:3 (Fig. 3B) prevented mDHP-I hydrolysis, and both products gave excellent, identical regressions (P = 0.99). Therefore, their BD were the same: 60.1 ± 1.91 mg/kg/day (T>MIC, 18.1%). A 1:5 M:C ratio lowered the BD further, to 54.6 ± 2.04 mg/kg/day (T>MIC, 16.8%), again with identical PD profiles (P = 0.97) (Fig. 3C). The lower %T>MIC required for bacteriostasis of the MDR (18.1%) versus the WT strain (24.2%) indicated that the former is more susceptible to the meropenem-cilastatin combination than the latter, which could be explained if GRP-0049 were to express a cilastatin-inhibited dehydropeptidase (i.e., a dipeptidase of the M19 family).
Neutropenic mouse brain infection model (bacterial meningoencephalitis by P. aeruginosa, either the WT strain GRP-0019 or MDR strain GRP-0049).
Studies with rats have shown that DHP-I activity in the brain is negligible (33), approximately 6.4- and 3,000-fold less than in the muscle and the lungs, respectively (17). Since the fraction of cilastatin (16 to 66%) that reaches the cerebrospinal fluid (CSF) is about 3-fold that of meropenem (6 to 20%) in mammals with inflamed meninges (34–38), the site of infection in this model would have increased M:C ratios with respect to the actual proportions injected subcutaneously. It provides greater protection of meropenem without further increasing the dose of cilastatin and, therefore, is a more sensible model to determine therapeutic equivalence with the mouse. The results from six experiments are illustrated in Fig. 4 and detailed in Table 3.
FIG 4.
Pharmacodynamics of three generics compared with the innovator of meropenem against P. aeruginosa strains GRP-0019 (WT; A and B) and GRP-0049 (MDR; C and D) in the neutropenic mouse meningoencephalitis model. Without cilastatin, and using a dose-range encompassing the T>MIC from 12.4% to 100% (in serum) against GRP-0019, it was possible to obtain flawless fits for gMer-B, gMer-C, and iMer to the Hill model, demonstrating therapeutic equivalence of gMer-B and therapeutic nonequivalence of gMer-C (A). With cilastatin, only a 1:3 M:C ratio gave gMer-A a clean regression that, compared with iMer, required 47% more drug to achieve bacteriostasis (B). GRP-0049 was untreatable in this model without cilastatin (data not shown); with a 1:1 M:C ratio, gMer-A gave an accurate nonlinear regression but iMer did not (C), as seen in the thigh model with this MDR strain. A 1:3 M:C ratio gave an impeccable regression for both products, demonstrating that gMer-A required 56% more meropenem than iMer to attain bacteriostasis in vivo (D). These findings suggest that the innovator is preferentially hydrolyzed by GRP-0049, a strain that evolved in our hospital under the selective pressure of iMer (discussed in the text).
Against the WT strain, P. aeruginosa GRP-0019, a meropenem dose range (without cilastatin) from 20 to 1,280 mg/kg/day (%T>MIC range, 12 to 100%) revealed a significant difference (P = 0.002) in the PD of gMer-B, gMer-C, and iMer (Fig. 4A) due to an inferior bactericidal effect of gMer-C with respect to iMer: Emax, 5.90 ± 0.10 versus 6.69 ± 0.18 log10 CFU/g, respectively (P = 0.001). gMer-B was indistinguishable from iMer (P = 0.28) (Table 3). To compare gMer-A and iMer, we used a narrower dose range (20 to 640 mg/kg/day), but both products exhibited multicollinearity, indicating that even the very low activity of mDHP-I in the brain was enough to hydrolyze meropenem (Table 3). Addition of cilastatin in a 1:1 ratio protected iMer but not gMer-A (Table 3). A 1:3 M:C ratio gave excellent regressions for both products which, compared by CFA, demonstrated that gMer-A required a 46% greater dose than iMer to attain bacteriostasis (P < 0.0001) (Fig. 4B): BD = 120.2 ± 5.80 mg/kg/day (gMer-A) versus 81.9 ± 6.86 mg/kg/day (iMer) (T>MIC, 75.8% versus 41.5%, respectively).
The MDR P. aeruginosa GRP-0049 was untreatable without cilastatin in the meningoencephalitis model, despite a dose range from 20 to 640 mg/kg/day (%T>MIC, 0 to 95%). A 1:1 M:C ratio was enough for gMer-A to fit the Hill model, but not for iMer, which violated the normality and homoscedasticity assumptions (Fig. 4C; Table 3). With a 1:3 M:C ratio, both products generated excellent fits that were statistically different (P = 0.0002) by CFA due to nonequivalence of gMer-A, which required 56% more meropenem to attain bacteriostasis (Fig. 4D): BD, 136.8 ± 13.3 versus 87.6 ± 3.92 mg/kg per day, respectively (T>MIC, 32.4% versus 23.5%). As seen in the thigh model, the MDR P. aeruginosa GRP-0049 was more susceptible to the combination of meropenem with cilastatin than the WT GRP-0019 (iMer T>MIC values for bacteriostasis with a 1:3 M:C ratio: 23.5% and 41.5%, respectively). The most probable explanation for this interesting finding is that the MDR strain produces a M19 dipeptidase (i.e., a cilastatin-susceptible enzyme).
Neutropenic mouse lung aerosolized infection model (pneumonia by the WT strain K. pneumoniae GRP-0107; MIC, 0.06 mg/liter).
The high DHP-I activity characteristic of the lung hydrolyzed meropenem very efficiently, as reproduced in all five experiments with this model (Fig. 5 and Table 3). To achieve the maximal effect without cilastatin, iMer levels not only had to remain above the MIC 100% of the time (Fig. 5A) but also had to fluctuate between 30-fold (Cmin) and 897-fold of the MIC (Cmax) (Fig. 5B). Only one of the three generic products (gMer-B) demonstrated therapeutic equivalence in this model (P = 0.13); the other two (gMer-A and gMer-C) could not be compared with iMer because their PD profiles did not fit the Hill model (Table 3).
FIG 5.
Pharmacodynamics of three generics compared with the innovator of meropenem against the WT strain K. pneumoniae GRP-0107 (MIC, 0.06 mg/liter) in the neutropenic mouse pneumonia model. mDHP-I is very active in this tissue, providing a good model to test meropenem under a highly hydrolytic environment. Without cilastatin, a T>MIC range from 22.8% to 100% (2.5 to 80 mg/kg/day) was not enough to reach maximal efficacy, and the data did not fit the Hill equation (A). Increasing the dose to 640 mg/kg provided much higher serum concentrations of meropenem (30- to 897-fold above the MIC) during 100% of the dosing interval, allowing gMer-B and iMer to reach their maximal effect and fit the Hill equation (P = 0.13 for comparison by CFA); gMer-C could not be included in the analysis because it did not fit the model, suggesting greater susceptibility to mDHP-I (B). When the dose range was narrowed to 5 to 160 mg/kg (%T>MIC, 42.8 to 100%), the addition of cilastatin up to a 1:3 M:C ratio did not help gMer-A or iMer (C); under a 1:5 M:C ratio, only iMer generated a flawless regression, suggesting that gMer-A is more susceptible to mDHP-I (D).
Addition of cilastatin at a 1:1 (data not shown) or 1:3 M:C ratio (Fig. 5C) was not enough to protect iMer or gMer-A from mDHP-I hydrolysis. A 1:5 M:C ratio did protect iMer, but not gMer-A (Fig. 5D); it allowed the innovator to reach its maximal effect with 160 mg/kg (%T>MIC, 100%), a dose that provided serum drug concentrations ranging from 7-fold (Cmin) to 223-fold above the MIC (Cmax). It was not possible to attain a valid NLR for gMer-A; therefore, no formal statistical comparison with iMer could be performed (Fig. 5D).
Pharmaceutical equivalence.
The standard curves generated by microbiological assay showed that gMer-A, gMer-B, gMer-C, iMer, and the external control (meropenem USP; Sigma) are not different. All products gave the same results in terms of concentration (Pintercepts = 0.912) and potency (Pslopes = 0.997) of the API, demonstrating their pharmaceutical equivalence.
Single-dose serum drug pharmacokinetics (bioequivalence) in infected mice.
Table 4 shows the PK parameters for the generics and innovator, which exhibited identical linear PK in infected mice after subcutaneous administration of a single dose of 40 or 160 mg/kg (the values obtained after dosing 10 mg/kg were too close to the limit of detection [data not shown]). The AUCall for the generic products relative to iMer were 96.1 to 101.4% (at 40 mg/kg) and 95.0 to 96.9% (at 160 mg/kg), all within the 80% to 125% range accepted by DRA.
TABLE 4.
Single-dose serum PK parameters ± standard error computed by non compartmental analysis) of three generic and the innovator product of meropenem, after subcutaneous injection to neutropenic mice infected in the thighs with P. aeruginosa GRP-0019 (three dose levels, 10, 40 and 160 mg/kg, 6 mice per dose)*
| Meropenem product | Dose (mg/kg) | t1/2 (min) | C0 (mg/liter) | Cmax (mg/liter) | AUCall (min · mg/liter) | PAUCb |
|---|---|---|---|---|---|---|
| iMer | 40 | 24.82 ± 1.13 | 16.40 ± 0.81 | 14.90 ± 0.92 | 505.1 ± 15.06 | 0.565 |
| gMer-A | 23.50 ± 1.51 | 14.83 ± 0.58 | 14.47 ± 0.50 | 485.5 ± 15.83 | ||
| gMer-B | 22.68 ± 1.11 | 13.31 ± 3.38 | 15.85 ± 0.06 | 511.7 ± 8.49 | ||
| gMer-C | 22.92 ± 0.62 | 16.22 ± 0.73 | 15.44 ± 0.76 | 502.8 ± 11.74 | ||
| iMer | 160 | 14.66 ± 0.40 | 87.29 ± 6.17 | 77.32 ± 2.96 | 1964 ± 57.69 | 0.665 |
| gMer-A | 14.52 ± 0.44 | 82.69 ± 5.60 | 73.31 ± 5.60 | 1865 ± 87.74 | ||
| gMer-B | 15.07 ± 0.60 | 82.22 ± 3.50 | 76.29 ± 1.72 | 1904 ± 50.15 | ||
| gMer-C | 17.09 ± 2.97 | 80.35 ± 0.51 | 72.23 ± 3.88 | 1873 ± 36.41 |
PK parameters ± standard errors were computed by noncompartmental analysis of the three generic products and the innovator product meropenem, after subcutaneous injection with 10, 40, or 160 mg/kg into neutropenic mice (6 mice/dose) infected in the thigh with P. aeruginosa GRP-0019. The dose level of 10 mg/kg produced serum drug concentrations detectable only during the first time point, and therefore these data were not used and are not presented here.
P values for the comparison of the parameter AUCall were determined by using ANOVA.
Analytical chemistry: meropenem standard curves generated by using HPLC-UV.
Determination of API concentrations in the reference (Sigma), iMer, and gMer-A, gMer-B, and gMer-C products did not show differences in their intercepts (P = 0.99) or slopes (P = 0.99), demonstrating, as in the bioassay, pharmaceutical equivalence.
Quantifying of mDHP-I enzymatic degradation of meropenem by HPLC-UV and bioassay.
For HPLC, the remaining fraction of the API in each sample was obtained by peak area extrapolation from the standard curves. After 5 min of exposure to the enzyme, gMer-A had undergone faster kinetic degradation that continued to intensify for the first hour (Fig. 6). This difference was also evident as a remarkable decrease in the zone sizes of the bioassay with gMer-A, demonstrating the loss of antibacterial activity as the product was degraded (Table 5).
FIG 6.
Meropenem hydrolysis by mDHP-I: detection by HPLC-UV (left panels) and microbiological assay (right panels) of the remaining fraction of meropenem products with time after incubation at 37°C with mDHP-I extract for 4.5 h. The control curve (dotted) corresponds to meropenem incubated without DHP-I and shows the spontaneous degradation of the carbapenem. Both products were hydrolyzed by the enzyme, but gMer-A degraded much faster than iMer at all concentrations tested.
TABLE 5.
Quantification of enzymatic degradation of meropenem products by mDHP-I, determined by HPLC-UV and microbiological assaya
| Assay and reaction time (h) | Fraction of initial meropenem concn remaining (%) |
||||||
|---|---|---|---|---|---|---|---|
| Control | Initial 125 mg/liter |
Initial 250 mg/liter |
Initial 500 mg/liter |
||||
| iMer | gMer-A | iMer | gMer-A | iMer | gMer-A | ||
| HPLC-UV | |||||||
| 0.08 | 99 | 89 | 65 | 86 | 48 | 96.4 | 51 |
| 0.5 | 98.3 | 87 | 63 | 82 | 44 | 94 | 47 |
| 1.0 | 96 | 49 | 25 | 39 | 26 | 73 | 37 |
| 1.5 | 96 | 23 | 17 | 27 | 14 | 57 | 8 |
| 4.0 | 92 | 10 | 10 | 8 | 8.4 | 41 | 0 |
| 4.5 | 92 | 10 | 8 | 6 | 4 | 0 | 0 |
| Microbiological assay | |||||||
| 0.08 | 96 | 92 | 45 | 88 | 45 | 89 | 64 |
| 0.5 | 94 | 94 | 45 | 88 | 43 | 89 | 60 |
| 1.0 | 94 | 64 | 30 | 31 | 17 | 35 | 14 |
| 1.5 | 94 | 48 | 23 | 20 | 12 | 22 | 17 |
| 4.0 | 88 | 15 | 0 | 6 | 6 | 7 | 5 |
| 4.5 | 88 | 0 | 0 | 6 | 6 | 5 | 5 |
The enzymatic reaction was allowed to take place for 5 min at 37°C before taking the first sample for analysis; the control sample (iMer, 500 mg/liter) was incubated in sterile saline without the enzyme. The data are presented as the percentage of each initial concentration of meropenem (125, 250, or 500 mg/liter).
A CFA comparison of the linear regressions (concentration versus zone size) obtained from the microbiological assays confirmed widely divergent curves (P = 0.003) indicating that gMer-A was significantly more susceptible than iMer to mDHP-I hydrolysis.
Qualitative assay results with LC/MS.
In the SIM mode for fresh samples, the chromatograms of the reference (Sigma) and the pharmaceutical forms of iMer and gMer-A did not show differences in retention times or peak abundance levels of the analyte (API, m/z 384). In the scan mode (range, m/z 100 to 1,000) using fresh samples, gMer-A exhibited a different scan representation at retention time of 10.09 min, with one specific peak that had a main molecular mass of m/z 359 [M + 1] and was absent in the mass spectra of iMer; it probably corresponds to a three-sodium adduct accompanying a degradation product with a mass of m/z 295 (32). The two other major peaks were present in both products, with retention times of 2.9 and 4.8 min in gMer-A and 3.2 and 5.2 min in iMer (Fig. 7). When products were subjected to a dynamic test to determine their spontaneous degradation (29, 39, 40), changes appeared in the chromatograms as new peaks (degradation products) with simultaneous decreases in the peak of the API, but each product had different behavior with time. The peaks of the degradation products had a retention time of 2.8 (major masses, m/z 230.9 and 274) and 3.2 min (major mass, m/z 358.2) but were notably more abundant in gMer-A. Again, gMer-A had an additional peak (absent in the innovator), with a retention time of 12 min corresponding to a mass of m/z 359.1, which was also detected in fresh samples. Experiments were repeated at least 3 times (Fig. 8).
FIG 7.
LC/MS scan mode (range, m/z 100 to 1,000) of the pharmaceutical forms of one generic and the innovator of meropenem (fresh samples). The left (innovator) and right (generic) panels show above the spectrogram and, under it, the centroids graphs describing the composition masses of each peak numbered. There were no differences in the analyte signal (peak 2 in both panels), demonstrating that the active pharmaceutical ingredient (m/z 384) is present in both products at the same concentration, in compliance with current regulations. However, the generic product exhibited one additional peak, detected at 10 min (peak 3, right panel), with a main molecular mass of m/z 359 [M + 1] that was absent in the mass spectra of the innovator.
FIG 8.
LC/MS scan mode (range, m/z 100 to 1,000) for samples stored for 48 h at room temperature. Spectrograms for iMer (A) and gMer-A (B) are shown, demonstrating more abundance of degradation products in the pharmaceutical form of the generic, with different mass compositions between retention times of 3 and 6 min.
DISCUSSION
These data demonstrate that pharmaceutical equivalence and bioequivalence do not imply therapeutic equivalence of meropenem, as previously reported with gentamicin (5), ofloxacin (6), and vancomycin (7). Our results also provide a new principle applicable to any antimicrobial: there are subtle ways by which a bioequivalent generic can fail in vivo, ways so inconspicuous that they pass undetected under current regulations.
The great value of nonlinear regression is unquestionable, but it depends heavily on careful consideration of the regression diagnostics (28). Although it is okay to make general observations about an NLR with problems like multicollinearity or violation of the regression assumptions, it is unwise to draw conclusions about the hypothesis (like curve-fitting analysis) from faulty regressions, because the magnitudes and significance of the coefficients are ambiguous. For instance, multicollinearity among the distinct PDP was a common problem when meropenem hydrolysis was not completely blocked with cilastatin or when the dose-response range was too narrow to span the complete PD relationship, from no effect to maximal effect. This kind of multicollinearity is sample based (i.e., suboptimal experimental design) but can be misdiagnosed as structural (i.e., the intrinsic nature of the data does not fit the regression equation). The only solution for the first problem is optimization of the experimental design (i.e., new data), while structural multicollinearity disappears by fitting the correct equation to the same data. Here, taking the last path would lead to a fundamentally wrong conclusion, because all meropenem products fit the Hill equation perfectly well once the design was optimized (widening the dose range, protecting the carbapenem from mDHP-I with cilastatin, or using a mammal with human-like DHP-I activity). A similar approach must be taken toward any violation of the regression assumptions (28).
The MDR strain P. aeruginosa GRP-0049 required more cilastatin than the WT in the thigh and brain infection models, indicating bacterial expression of a dipeptidase of the M19 family (those inhibited by cilastatin); gene PA5389 (renamed cdhR) encodes the M19 dipeptidase CdhR, a transcriptional regulator of carnitine catabolic genes, which could be overexpressed in GRP-0049 (41, 42). CdhR seemed more active against the innovator, the only one meropenem product ever used in our hospital, where GRP-0049 evolved.
The LC/MS structure analysis of gMer-A suggested that nonequivalence is due to the presence of meropenem trisodium adducts leading to instability of the API and higher susceptibility to DHP-I hydrolysis. The greater susceptibility of gMer-A to DHP-I was demonstrated both in vitro and vivo, and although the data did not unveil the precise mechanism that makes it a better substrate than iMer for the enzyme, there are enough elements to formulate an educated guess. The fact that the differences in PD were easily uncovered by the model with the highest bacterial loads (brain) and undetectable in the murine model with the lowest growth (thigh) suggests a role for acidification at the site of infection. Although it is present for both products, the hydrogen released by gMer-A during the formation of trisodium adducts reduces the pH even further. Under mildly acidic pH, carbapenems are more susceptible to enzymatic hydrolysis, because the already strained double bond between carbons 2 and 3 of the beta-lactam ring becomes extremely unstable (43).
In conclusion, the subjacent mechanisms for in vivo failure of “bioequivalent” meropenem arise while manufacturing the API but are not detectable under current regulations. The solution to this serious problem demands the implementation of appropriate tools to discern effective from ineffective generics where their API are manufactured.
ACKNOWLEDGMENTS
This project was financed by Fundación Científica Rodrigo Vesga-Meneses and by the University of Antioquia's Sustainability Strategies 2011–2012 and 2013–2014.
Footnotes
Published ahead of print 25 November 2013
REFERENCES
- 1.Generic Pharmaceutical Association 2012. Generic drug savings in the U.S., 4th annual ed. Generic Pharmaceutical Association, Washington, DC: http://www.ahipcoverage.com/wp-content/uploads/2012/08/2012-GPHA-IMS-GENERIC-SAVINGS-STUDY.pdf [Google Scholar]
- 2.WHO 1998. Marketing authorization of pharmaceutical products with special reference to multisource (generic) products: a manual for a drug regulatory authority. World Health Organization, Geneva, Switzerland: http://apps.who.int/prequal/info_general/documents/WHO_DMP_RGS_98_5_R.pdf [Google Scholar]
- 3.Mastoraki E, Michalopoulos A, Kriaras I, Mouchtouri E, Falagas ME, Karatza D, Geroulanos S. 2008. Incidence of postoperative infections in patients undergoing coronary artery bypass grafting surgery receiving antimicrobial prophylaxis with original and generic cefuroxime. J. Infect. 56:35–39. 10.1016/j.jinf.2007.09.011 [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez CA, Agudelo M, Cataño JC, Zuluaga AF, Vesga O. 2009. Potential therapeutic failure of generic vancomycin in a liver transplant patient with MRSA peritonitis and bacteremia. J. Infect. 59:277–280. 10.1016/j.jinf.2009.08.005 [DOI] [PubMed] [Google Scholar]
- 5.Zuluaga AF, Agudelo M, Cardeño JJ, Rodriguez CA, Vesga O. 2010. Determination of therapeutic equivalence of generic products of gentamicin in the neutropenic mouse thigh infection model. PLoS One 5(5):e10744. 10.1371/journal.pone.0010744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez CA, Agudelo M, Zuluaga AF, Vesga O. 2010. In vitro and in vivo comparison of the anti-staphylococcal efficacy of generic products and the innovator of oxacillin. BMC Infect. Dis. 10:153. 10.1186/1471-2334-10-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vesga O, Agudelo M, Salazar BE, Rodriguez CA, Zuluaga AF. 2010. Generic vancomycin products fail in vivo despite being pharmaceutical equivalents of the innovator. Antimicrob. Agents Chemother. 54:3271–3279. 10.1128/AAC.01044-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez CA, Agudelo M, Zuluaga AF, Vesga O. 2012. Generic vancomycin enriches resistant subpopulations of Staphylococcus aureus after exposure in a neutropenic mouse thigh infection model. Antimicrob. Agents Chemother. 56:243–247. 10.1128/AAC.05129-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gauzit R, Lakdhari M. 2012. Generic antibiotic drugs: is effectiveness guaranteed? Med. Mal. Infect. 42:141–148. 10.1016/j.medmal.2011.10.013 [DOI] [PubMed] [Google Scholar]
- 10.Blossom DB, Kallen AJ, Patel PR, Elward A, Robinson L, Gao G, Langer R, Perkins KM, Jaeger JL, Kurkjian KM, Jones M, Schillie SF, Shehab N, Ketterer D, Venkataraman G, Kishimoto TK, Shriver Z, McMahon AW, Austen KF, Kozlowski S, Srinivasan A, Turabelidze G, Gould CV, Arduino MJ, Sasisekharan R. 2008. Outbreak of adverse reactions associated with contaminated heparin. N. Engl. J. Med. 359:2674–2684. 10.1056/NEJMoa0806450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadwiger ME, Sommers CD, Mans DJ, Patel V, Boyne MT., II 2012. Quality assessment of U.S. marketplace vancomycin for injection products using high-resolution liquid chromatography–mass spectrometry and potency assays. Antimicrob. Agents Chemother. 56:2824–2830. 10.1128/AAC.00164-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nambiar S, Madurawe RD, Zuk SM, Khan SR, Ellison CD, Faustino PJ, Mans DJ, Trehy ML, Hadwiger ME, Boyne MT, II, Biswas K, Cox EM. 2012. Product quality of parenteral vancomycin products in the United States. Antimicrob. Agents Chemother. 56:2819–2823. 10.1128/AAC.05344-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.U.S. Food and Drug Administration Accessed 2 October 2012 FDA statement about product quality of parenteral vancomycin products, March 14, 2012. U.S. FDA, Rockville, MD: http://www.fda.gov/Drugs/DrugSafety/ucm295414.htm [Google Scholar]
- 14.Tattevin P, Saleh-Mghir A, Davido B, Ghout I, Massias L, Garcia de la Maria C, Miró JM, Perronne C, Laurent F, Crémieux AC. 2013. Comparison of six generic vancomycin products for treatment of methicillin-resistant Staphylococcus aureus experimental endocarditis in rabbits. Antimicrob. Agents Chemother. 57:1157–1162. 10.1128/AAC.01669-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agudelo M, Vesga O. 2012. Therapeutic equivalence requires pharmaceutical, pharmacokinetic, and pharmacodynamic identities: true bioequivalence of a generic product of intravenous metronidazole. Antimicrob. Agents Chemother. 56:2659–2665. 10.1128/AAC.06012-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. 2011. Carbapenems: past, present, and future. Antimicrob. Agents Chemother. 55:4943–4960. 10.1128/AAC.00296-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukasawa M, Sumita Y, Harabe ET, Tanio T, Nouda H, Kohzuki T, Okuda T, Matsumura H, Sunagawa M. 1992. Stability of meropenem and effect of 1 beta-methyl substitution on its stability in the presence of renal dehydropeptidase I. Antimicrob. Agents Chemother. 36:1577–1579. 10.1128/AAC.36.7.1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing, approved standard M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 19.Bennett JV, Brodie JL, Benner EJ, Kirby WMM. 1966. Simplified, accurate method for antimicrobial assay of clinical specimens. Appl. Microbiol. 14:170–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuluaga AF, Agudelo M, Rodriguez CA, Vesga O. 2009. Application of microbiological assay to determine pharmaceutical equivalence of generic intravenous antibiotics. BMC Clin. Pharmacol. 9:1. 10.1186/1472-6904-9-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glantz SA. 2002. Prime of biostatistics, 5th ed, p 230–297 McGraw-Hill, New York, NY [Google Scholar]
- 22.Zuluaga AF, Salazar BE, Rodriguez CA, Zapata AX, Agudelo M, Vesga O. 2006. Neutropenia induced in outbred mice by a simplified low-dose cyclophosphamide regimen: characterization and applicability to diverse experimental models of infectious diseases. BMC Infect. Dis. 6:55. 10.1186/1471-2334-6-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Restrepo AV, Salazar BE, Agudelo M, Rodriguez CA, Zuluaga AF, Vesga O. 2005. Optimization of culture conditions to obtain maximal growth of penicillin-resistant Streptococcus pneumoniae. BMC Microbiol. 5:34. 10.1186/1471-2180-5-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dankert J, Holloway Y, Joldersma W, Hess J. 1983. Importance of minimizing carry-over effect at subculture in the detection of penicillin-tolerant viridans group streptococci. Antimicrob. Agents Chemother. 23:614–616. 10.1128/AAC.23.4.614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulitta JB, Bingölbali A, Shin BS, Landersdorfer CB. 2011. Development of a new pre- and post-processing tool (SADAPT-TRAN) for nonlinear mixed-effects modeling in S-ADAPT. AAPSJ 13:201–211. 10.1208/s12248-011-9257-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Argenio DZ, Schumizky A, Wang X. 2009. ADAPT 5 user's guide: pharmacokinetic/pharmacokynamic systems analytical software. Biomedical Simulations Resource, Los Angeles, CA [Google Scholar]
- 27.Craig WA. 1998. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin. Infect. Dis. 26:1–10 [DOI] [PubMed] [Google Scholar]
- 28.Glantz SA, Slinker BK. 2001. Primer of applied regression and analysis of variance, 2nd ed. McGraw-Hill, New York, NY [Google Scholar]
- 29.Ozkan Y, Küçükgüzel I, Ozkan SA, Aboul-Enein HY. 2001. A rapid, sensitive high performance liquid chromatographic method for the determination of meropenem in pharmaceutical dosage form, human serum and urine. Biomed. Chromatogr. 15:263–266. 10.1002/bmc.68 [DOI] [PubMed] [Google Scholar]
- 30.Campbell BJ, Lin YC, Davis RV, Ballew E. 1966. The purification and properties of a particulate renal dipeptidase. Biochim. Biophys. Acta 118:371–386 [DOI] [PubMed] [Google Scholar]
- 31.Mendez AS, Weisheimer V, Oppe TP, Steppe M, Schapoval EE. 2005. Microbiological assay for the determination of meropenem in pharmaceutical dosage form. J. Pharm. Biomed. Anal. 37:649–653. 10.1016/j.jpba.2004.11.030 [DOI] [PubMed] [Google Scholar]
- 32.Sun C, Wu J, Pan Y. 2009. Characterization of novel hydrolysis products of carbapenems by electrospray ionization mass spectrometry. Rapid. Commun. Mass Spectrom. 23:3205–3212. 10.1002/rcm.4240 [DOI] [PubMed] [Google Scholar]
- 33.Kera Y, Liu Z, Matsumoto T, Sorimachi Y, Nagasaki H, Yamada RH. 1999. Rat and human membrane dipeptidase: tissue distribution and developmental changes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 123:53–58 [DOI] [PubMed] [Google Scholar]
- 34.Harrison MP, Moss SR, Featherstone A, Fowkes AG, Sanders AM, Case DE. 1989. The disposition and metabolism of meropenem in laboratory animals and man. J. Antimicrob. Chemother. 24(Suppl A):265–277 [DOI] [PubMed] [Google Scholar]
- 35.Dupuis A, Pariat C, Courtois P, Couet W, Bouquet S. 2000. Imipenem but not meropenem induces convulsions in DBA/2 mice, unrelated to cerebrospinal fluid concentrations. Fundam. Clin. Pharmacol. 14:163–165. 10.1111/j.1472-8206.2000.tb00406.x [DOI] [PubMed] [Google Scholar]
- 36.Dupuis A, Caillaud A, Pariat C, Courtois P, Couet W, Bouquet S. 2000. Comparative cerebrospinal fluid diffusion of imipenem and meropenem in rats. J. Pharm. Pharmacol. 52:1143–1149. 10.1211/0022357001774912. [DOI] [PubMed] [Google Scholar]
- 37.Maiques JM, Doménech A, Cabellos C, Fernández A, Ribes S, Tubau F, Gudiol F, Viladrich PF. 2007. Evaluation of antimicrobial regimens in a guinea-pig model of meningitis caused by Pseudomonas aeruginosa. Microbes Infect. 9:435–441. 10.1016/j.micinf.2006.12.013 [DOI] [PubMed] [Google Scholar]
- 38.Jacobs RF, Kearns GL, Brown AL, Longee DC. 1986. Cerebrospinal fluid penetration of imipenem and cilastatin (primaxin) in children with central nervous system infections. Antimicrob. Agents Chemother. 29:670–674. 10.1128/AAC.29.4.670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai SY, Hu CQ. 2005. Chromatographic determination of polymerized impurities in meropenem. J. Pharm. Biomed. Anal. 37:585–589. 10.1016/j.jpba.2004.11.023 [DOI] [PubMed] [Google Scholar]
- 40.Cielecka-Piontek J, Zajac M, Jelińska A. 2008. A comparison of the stability of ertapenem and meropenem in pharmaceutical preparations in solid state. J. Pharm. Biomed. Anal. 46:52–57. 10.1016/j.jpba.2007.08.024 [DOI] [PubMed] [Google Scholar]
- 41.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acid Res. 39(Database issue):D596–D600 http://www.pseudomonas.com/getAnnotation.do?locusID=PA5389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wargo MJ, Hogan DA. 2009. Identification of genes required for Pseudomonas aeruginosa carnitine catabolism. Microbiology 155:2411–2419. 10.1099/mic.0.028787-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Craig WA. 1997. The pharmacology of meropenem, a new carbapenem antibiotic. Clin. Infect. Dis. 24(Suppl 2):S266–S275 [DOI] [PubMed] [Google Scholar]