Abstract
Alternative splicing is the main mechanism of increasing the proteome diversity coded by a limited number of genes. It is well established that different tissues or organs express different splicing variants. However, organs are composed of common major cell types, including fibroblasts, epithelial, and endothelial cells. By analyzing large-scale data sets generated by The ENCODE Project Consortium and after extensive RT-PCR validation, we demonstrate that each of the three major cell types expresses a specific splicing program independently of its organ origin. Furthermore, by analyzing splicing factor expression across samples, publicly available splicing factor binding site data sets (CLIP-seq), and exon array data sets after splicing factor depletion, we identified several splicing factors, including ESRP1 and 2, MBNL1, NOVA1, PTBP1, and RBFOX2, that contribute to establishing these cell type–specific splicing programs. All of the analyzed data sets are freely available in a user-friendly web interface named FasterDB, which describes all known splicing variants of human and mouse genes and their splicing patterns across several dozens of normal and cancer cells as well as across tissues. Information regarding splicing factors that potentially contribute to individual exon regulation is also provided via a dedicated CLIP-seq and exon array data visualization interface. To the best of our knowledge, FasterDB is the first database integrating such a variety of large-scale data sets to enable functional genomics analyses at exon-level resolution.
Human genes are an assemblage of exons that can be differentially selected during splicing. Alternative splicing, which can produce splicing variants with different exonic content from a single gene, is the rule rather than an exception, as 95% of human genes generate several splicing variants (Kim et al. 2008; Hallegger et al. 2010; Kalsotra and Cooper 2011; Blencowe 2012; Kelemen et al. 2013). Alternative splicing relies on the combinatory action of splicing factors (e.g., SR and hnRNP proteins) that bind to exonic or intronic splicing regulatory sequences to either strengthen or inhibit splice site recognition by the splicing machinery, therefore enhancing or repressing the inclusion of alternative exons (Barash et al. 2010; Goren et al. 2010; Witten and Ule 2011). Similarly to how transcription factors control transcriptional programs by directing the expression of gene networks, splicing factors control splicing programs by regulating alternative splicing of co-regulated exons (Hartmann and Valcarcel 2009; Barash et al. 2010; Goren et al. 2010; Witten and Ule 2011). Alternative splicing is the main mechanism used to increase the proteome diversity coded by a limited number of genes, as the majority of alternative exons contains coding sequences (Kim et al. 2008; Hallegger et al. 2010; Kalsotra and Cooper 2011; Blencowe 2012; Kelemen et al. 2013). Because of the diversity generated by alternative splicing and the complexity of its regulation, functional genomics at exon-level resolution requires the development of new integrative bioinformatics approaches.
Functional genomics at exon-level resolution is necessary to better understand tissue-specific functions. Indeed, it is well established that different tissues (or organs) express different splicing variants (Bland et al. 2010; de la Grange et al. 2010; Hartmann et al. 2011; Llorian and Smith 2011; Barbosa-Morais et al. 2012; Merkin et al. 2012). The development of new technologies like splicing-sensitive microarrays and massive RNA sequencing fully establish that different tissues express different splicing programs as a consequence of the combinatorial actions of more-or-less tissue-specific splicing factors (Pan et al. 2008; Wang et al. 2008; Merkin et al. 2012). However, most organs are composed of common cell types, such as fibroblast and epithelial cells, which perform specific functions. Epithelial cells are tightly connected cells arranged in monolayer with several functions, such as protection, diffusion, secretion, absorption, and excretion, and establishing boundaries between compartments. Fibroblasts comprise the structural framework of tissues and synthesize the extracellular matrix, a supportive framework for epithelial cells. Unlike epithelial cells, fibroblasts can migrate as individual cells. Another important cell type is represented by endothelial cells that compose the endothelium, the thin layer of cells that lines the interior surface of blood vessels that supply tissues and organs with blood.
Recent large-scale analyses suggest that splicing programs may contribute to establishing cell type–specific functions. Indeed, the epithelial-to-mesenchymal transition (EMT) that corresponds to the trans-differentiation of epithelial to mesenchymal (fibroblast-like) cells relies not only on transcriptional programs but also on extensive changes in alternative splicing (Warzecha et al. 2009a, 2010; Shapiro et al. 2011; Venables et al. 2013). Splicing factors, including ESRP1 and 2 and RBFOX2 (RBM9), have been reported to play a key role in EMT (Warzecha et al. 2009a, 2010; Shapiro et al. 2011; Dittmar et al. 2012; Venables et al. 2013). Although a recent report indicates that several dozen genes are differentially spliced when comparing normal fibroblasts to epithelial cells isolated from colon and ovarian tissues (Venables et al. 2013), it is currently not known whether the major common cell types, namely fibroblast, epithelial, and endothelial cells, express specific splicing programs independently of their organ origin.
To address this question, we analyzed an ENCODE data set based on exon arrays performed on RNAs prepared from several dozen normal fibroblast, epithelial, and endothelial cells isolated from different tissues (Thurman et al. 2012). After extensive RT-PCR validation, we show that each major cell type expresses a specific splicing program independently of their tissue origin, suggesting a role of alternative splicing in establishing specific functions of common cell types shared by many tissues. Additionally, by analyzing several publicly available large-scale data sets related to these factors, we identified a set of splicing factors that coordinate these splicing programs. In order to provide the community with full support for alternative splicing analysis, we have made all of the analyzed data sets freely available through a user-friendly web interface named FasterDB, which describes all the known splicing variants of human and mouse genes as well as their splicing patterns across several dozens of normal cells, cancer cells, and tissues. Furthermore, FasterDB integrates several kinds of publicly available data sets, namely, splicing factor binding sites (CLIP-seq) and splicing factor depletion followed by exon-array analysis, to provide users with information regarding the splicing factors that potentially contribute to regulating each exon. To the best of our knowledge, FasterDB is the first database to integrate such a variety of data sets, thereby enabling functional genomics at exon-level resolution.
Results
Identification of cell type–specific splicing programs independent of tissue origin
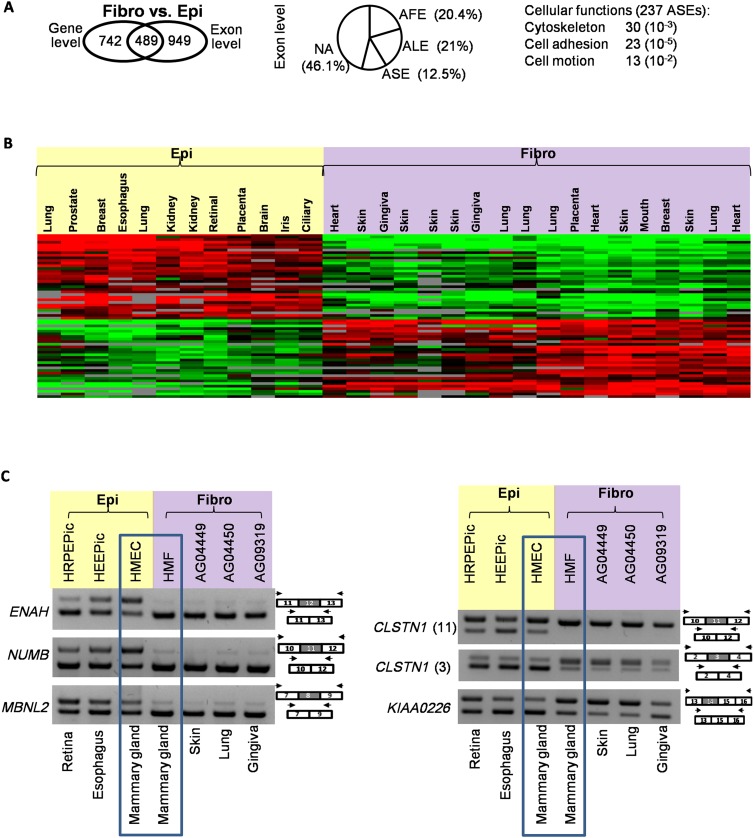
While a splicing program switch has been shown to occur in a few models of trans-differentiation of epithelial to mesenchymal (fibroblast-like) cells (Warzecha et al. 2009a, 2010; Shapiro et al. 2011; Dittmar et al. 2012; Venables et al. 2013), we tested whether epithelial cells and fibroblasts express a different splicing program independently of their tissue of origin. For this purpose, we analyzed an ENCODE data set corresponding to 18 and 12 normal fibroblast and epithelial cells, respectively, isolated from different organs (Supplemental Table S1). As shown in Figure 1A, fibroblast and epithelial cells express a transcriptome that differs both quantitatively (at the gene level) and qualitatively (at the exon level) (Supplemental Table S2). Focusing on variations at the exon level and using annotations based on known publicly available transcripts (see below), we observed that ∼20% of the cases corresponded to alternative first exons (AFE) and alternative last exons (ALE), while 12% of the cases corresponded to alternatively spliced exons (ASE) (Fig. 1A). About half of the cases were not annotated (NA), indicating potentially novel alternative exons. Remarkably, using the splicing index (SI), which represents the inclusion rate of the differentially spliced exons, fibroblasts and epithelial cells were clustered independently of their organ origin (Fig. 1B; Supplemental Fig. S1). Similar results were obtained by principal component analyses, which showed a clear clustering of the variables (cell lines) depending on their type (epithelial or fibroblast; Supplemental Fig. S3).
Figure 1.
Epithelial- and fibroblast-specific splicing variants. (A) Transcriptome analysis of fibroblasts (fibro) as compared to epithelial (epi) cells at both gene and exon levels. The number of genes differentially expressed at the gene and/or exon level when comparing both cell types is shown in the left panel. The middle panel indicates the classification of events corresponding to exon level variations. Differentially expressed exons were classified according to their annotation using publicly available transcripts: alternative first exon (AFE), alternative last exon (ALE), and alternative skipped exon (ASE). Not annotated (NA) corresponds to exons that do not correspond to any of the above-mentioned categories. The cellular functions of genes differentially spliced when comparing fibroblasts to epithelial cells are indicated in the right panel. (B) Heatmap presentation of the splicing index (SI) values for exons differentially spliced when comparing fibroblast to epithelial cells. Each line corresponds to a regulated exon, while each column corresponds to a specific cell. Green boxes (–1.5 < SI < 0) correspond to a low inclusion level in the cell as compared to all the others; red boxes (0 < SI < 1.5), high inclusion level; black boxes (SI = 0), no difference for exon inclusion between cells; and gray boxes, missing values. Exons were computationally split into several groups depending on their inclusion rate that correlates with the two major cell types. (C) RT-PCR validations using RNAs from fibroblasts and epithelial cells, as indicated.
As shown in Figure 1C (see also Supplemental Fig. S1), a large set of ASE events was validated by RT-PCR. Further demonstrating that a set of exons can be differentially spliced in major cell types even if they have the same tissue origin, the splicing pattern observed in HMEC cells, which are epithelial cells from the mammary gland, was different from the splicing pattern observed in HMF cells, which are fibroblast cells also from the mammary gland. Additionally, the splicing pattern observed in HMEC cells was similar to the splicing pattern observed in other epithelial cells from different tissues, and the splicing pattern observed in HMF cells was similar to the splicing pattern observed in other fibroblast cells from different tissues (Fig. 1C).
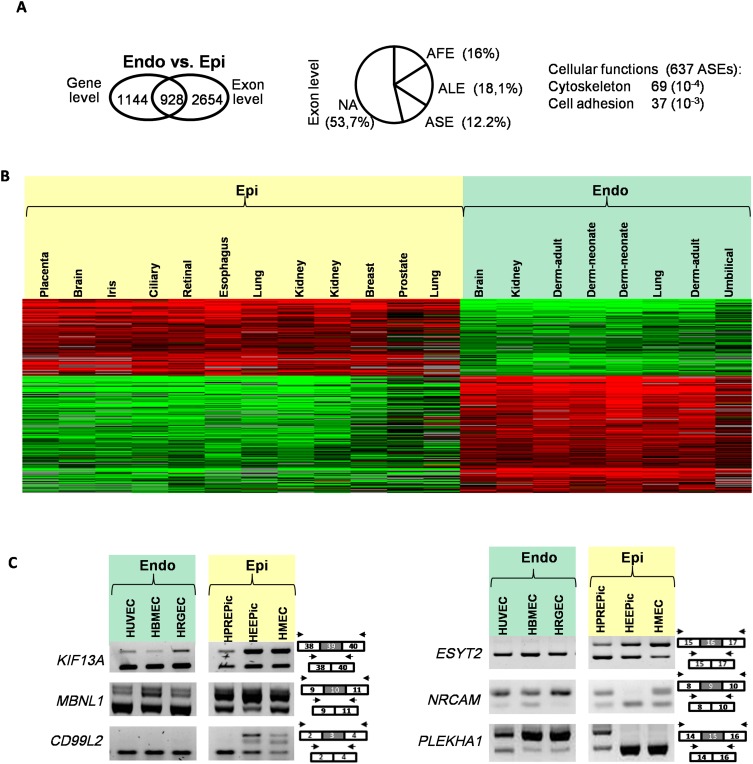
Endothelial cells are also a general cell type present in all organs, as they form blood vessels that supply organs with blood. Because of some functional similarities, endothelial cells are often compared to epithelial cells although they have different embryonic origins. It is not known whether endothelial cells express a specific splicing program or not. As shown in Figure 2, A and B, endothelial cells differ from epithelial cells in both transcriptional and splicing programs (Supplemental Table S3); this was additionally validated by RT-PCR (Fig. 2C; Supplemental Fig. S2). Likewise, endothelial cells differ from fibroblasts in their transcriptional and splicing programs (Supplemental Table S4).
Figure 2.
Epithelial- and endothelial-specific splicing variants. (A) Same as in Figure 1A but comparing endothelial (Endo) to epithelial (Epi) cells. (B) Same as in Figure 1B but comparing endothelial (Endo) to epithelial (Epi) cells. (C) RT-PCR validations using RNAs from endothelial and epithelial cells, as indicated.
A gene ontology (GO) annotation analysis revealed that genes regulated at the splicing level (i.e., ASE) may contribute to cell type–specific cellular programs, as they were mainly involved in cytoskeleton, cell adhesion, and motion, which are the main features distinguishing these cell types (Figs. 1A, 2A).
To further challenge a model in which each of the three major cell types (fibroblast, epithelial, and endothelial) is characterized by specific splicing programs independently from their tissue of origin, we compared each cell type to the other two (Supplemental Tables S5–S7). Remarkably, we identified a set of alternative cassette exons whose splicing index allowed a perfect clustering of each major cell type (Fig. 3A; Supplemental Fig. S4). Thus, we identified a set of exons that are differentially spliced across three major cell types independently of their tissue origin, as validated by RT-PCR (Fig. 3B; Supplemental Table S1). Similar results were obtained using alternative first or last exons (AFE or ALE, respectively), demonstrating that major cell types can differ by the exonic content (ASE, AFE, and ALE) of the transcripts they express (Supplemental Figs. S5, S6).
Figure 3.
Cell type–specific splicing programs. (A) Heatmap presentation of the SI values for exons differentially spliced across fibroblast, endothelial, and epithelial cells. Exons were computationally split into several groups depending on their inclusion rate in the three major cell types. Every brace corresponds to a group (names are indicated on the left); their exons are surrounded by a yellow rectangle on the heatmap. The different categories of exons are indicated as follows: epithelial-included (EPI+), epithelial-skipped (EPI–), endothelial-included (ENDO+), endothelial-skipped (ENDO–), fibroblast-included (FIBRO+), and fibroblast-skipped (FIBRO–) exons. (B) Inclusion rate of selected exons measured after RT-PCR using cells from different origins. The gene symbol and exon position are indicated. Inclusion rates are given for epithelial cells compared to both fibroblast and endothelial cells (upper panel), fibroblasts compared to both epithelial and endothelial cells (middle panels), and endothelial cells compared to both fibroblast and epithelial cells (lower panel).
Cell type–specific splicing programs are controlled by balanced expression of antagonist splicing factors
By focusing on splicing factors with a marked cell type–specific expression pattern (e.g., up-regulated in only one cell type and down-regulated in the other two; see Supplemental Table S8), we identified a set of splicing factors whose expression level allowed us to classify each cell type (Fig. 4A; Supplemental Fig. S7). In particular, ESRP1 had a higher level of expression in epithelial cells than in other cell types, as confirmed by RT-qPCR (Fig. 4A,B; Supplemental Table S1), as expected (Warzecha et al. 2009a, 2010; Dittmar et al. 2012). Interestingly, PTBP1 (PTB) and MBNL1, which have been suggested to play a role in angiogenesis (Pascual et al. 2006; Pen et al. 2007; Masuda et al. 2009), were enriched in endothelial cells (Fig. 4A,B; Supplemental Table S1). Finally, RBFOX2 (RBM9) and NOVA1 were enriched in fibroblasts (Fig. 4A,B). Interestingly, in addition to sharing many common target exons with NOVA1, RBFOX2 is up-regulated during EMT and plays a critical role in this process (Zhang et al. 2008; Venables et al. 2013). These results were confirmed by analyzing the expression level of these splicing factors using RNA-seq and gene expression array data sets (Supplemental Fig. S8).
Figure 4.
Cell type–specific expression of splicing factors. (A) Heatmap of splicing factor expression level. Each line represents a splicing factor, while each column represents a specific cell. The color of the square corresponds to the variation of the expression level of the splicing factor in each specific cell as compared to the others (green, less expressed in the cell; red, more expressed in the cell; and black, no difference). (B) RT-qPCR analysis of the expression level of ESRP1, PTBP1, MBNL1, RBFOX2, and NOVA1 in a collection of fibroblasts, epithelial and endothelial cells. (C) Spearman correlations between splicing factor expression level and the inclusion rate of epithelial-included (EPI+), epithelial-skipped (EPI–), endothelial-included (ENDO+), endothelial-skipped (ENDO–), fibroblast-included (FIBRO+), and fibroblast-skipped (FIBRO–) exons. Warm colors indicate positive correlation (e.g., a high exon inclusion level that correlates with a high splicing factor expression level), whereas cold colors indicate negative correlation (e.g., a low exon inclusion level that correlates with a high splicing factor expression level). Gray boxes indicate correlations that were discarded because of values that were not statistically significant or insufficient data available to compute correlations. (D) Summary table of epithelial-, fibroblast-, and endothelial-specific exons predicted to be regulated by the ESRP1, PTBP1, and RBFOX2 splicing factors using RNA-seq, exon array, and CLIP-seq data sets (see Supplemental Fig. S4 for more information). The number and percentage of exons predicted to be regulated by each splicing factor in each category are indicated. (E) Schematic representation of splicing factor binding site enrichment in several sets of exons differentially regulated across epithelial, endothelial, or fibroblast cells. Columns define regions in which the binding site searches were done. (F) RT-PCR analyses of the effect of depleting ESRP1, RBFOX2, or PTBP1 on alternative splicing of selected genes in the MCF-7 epithelial cell line, the MDA-MB-231 fibroblast-like cell line, or the HUVEC endothelial cell line, respectively. (G) Venn diagrams representing the number of epithelial-, fibroblast-, and endothelial-specific exons predicted to be regulated by ESRP1, PTBP1, and/or RBFOX2.
To go a step further, we focused on the PTBP1, ESRP1, and RBFOX2 splicing factors, since their expression levels allowed each major cell type to be clustered (Supplemental Fig. S7B) and since large-scale data sets corresponding to these factors are publicly available (Warzecha et al. 2009b; Xiao et al. 2009; Xue et al. 2009; Yeo et al. 2009; Katz et al. 2010; Llorian et al. 2010; Huelga et al. 2012). As shown in Figure 4C, ESRP1 and 2 expression levels positively and negatively correlated with the inclusion rate (e.g., SI) of a set of included (EPI+) and excluded (EPI–) exons, respectively. Inversely, ESRP1 and 2 expression levels negatively and positively correlated with the inclusion rate of a set of included (FIBRO+) and excluded (FIBRO–) exons, respectively. Meanwhile, RBFOX2 expression levels positively and negatively correlated with the inclusion rate of a set of included (FIBRO+) and excluded (FIBRO–) exons, respectively. Finally, PTBP1 expression levels also nicely correlated with the inclusion rate of different exons enriched in endothelial cells.
To further explore the contribution of these splicing factors in controlling cell type–specific splicing programs, we analyzed publicly available data sets of crosslinking/immunoprecipitation-sequencing (CLIP-seq) and/or exon array or RNA-seq analyses after RNAi for PTBP1, ESRP1, and RBFOX2 in human cell lines (Warzecha et al. 2009b; Xiao et al. 2009; Xue et al. 2009; Yeo et al. 2009; Katz et al. 2010; Llorian et al. 2010; Huelga et al. 2012). These analyses (Fig. 4D; Supplemental Table S8; see Supplemental Fig. S7 for the strategy used) and the RT-PCR validations (Fig. 4F; Supplemental Fig. S7) demonstrated that ESRP1/2, PTBP1/2, and RBFOX2 regulate a large subset within the cell type–specific alternative exons.
Additionally, ESRP1/2 binding sites were enriched in introns downstream from epithelial-included exons and upstream of epithelial-excluded exons (Fig. 4E; Supplemental Fig. S9; Supplemental Table S8), as expected from the previously reported ESRP splicing code (Warzecha et al. 2009a, 2010; Shapiro et al. 2011; Dittmar et al. 2012). Likewise, RBFOX2 and PTBP1 binding sites were differentially enriched when comparing fibroblast-included and -excluded exons or endothelial-included and -excluded exons, respectively (Fig. 4E; Supplemental Fig. S9; Supplemental Table S8), in agreement with the previously reported RBFOX2 and PTBP1 splicing codes (Xue et al. 2009; Llorian et al. 2010).
Remarkably, a large number of fibroblast-, epithelial-, or endothelial-specific exons were predicted to be targeted by at least two splicing factors (Fig. 4G; Supplemental Table S8). Strikingly, as illustrated here for a few genes, some ESRP-regulated exons were regulated inversely by RBFOX2, while some RBFOX2-regulated exons were regulated inversely by PTBP1/2 (Supplemental Fig. S7). These data suggest that cell type–specific splicing programs may be controlled by a balanced expression of antagonist splicing factors. In other words, the splicing signature of a specific cell type likely depends on the combinatorial effects of both up- and down-regulated splicing factors, rather than by only up-regulated factors.
FasterDB: An integrative bioinformatics platform dedicated to alternative splicing analysis
In order to provide full support for performing functional genomics at exon-level resolution, the data sets presented above were organized in a freely available and user-friendly web interface. This novel database, named FasterDB (http://fasterdb.lyon.unicancer.fr/), provides researchers with information regarding the splicing variants generated for their genes of interest. Supplemental Figures S10–S21 demonstrate the use of FasterDB on the ENAH gene, which codes for the MENA protein that modulates cell adhesion and migration (Di Modugno et al. 2012) and which has an exon (exon 12) that is specifically included in epithelial cells (Figs. 1C, 3B). Thus, once the ENAH gene symbol is entered into the FasterDB search engine, FasterDB provides the description of the human and mouse genes as well as all known gene transcripts reported in public databases (Supplemental Fig. S10). Information regarding alternative use of exons, various features of exons and introns, exon conservation, UTRs, and miRNA binding site prediction can be obtained as well (Supplemental Figs. S11–S16).
FasterDB also provides information on the expression and splicing pattern of all protein-coding genes across a collection of 73 human cell lines (both normal and cancerous), 11 normal human tissues, nine mouse cell lines, and 11 normal mouse tissues corresponding to Affymetrix exon array data sets generated by The ENCODE Project Consortium and Affymetrix (Thurman et al. 2012). Clicking on “Expression” in the main toolbar (Supplemental Fig. S10B) reveals the relative level of gene expression across these panels (Supplemental Fig. S17) and provides the inclusion rate of any selected exon across the sample collection (Supplemental Fig. S18A), which can be helpful for selecting the adequate cellular model for functional studies of alternative splicing variants. To help with data mining, users can click on each cell line for a direct link to a dedicated visualization interface, named ELEXIR (Supplemental Figs. S18, S19).
Finally, FasterDB provides some clues about the splicing factors that might be involved in splicing regulation of their favorite gene. Indeed, the “Splicing factors” button in the main toolbar (Supplemental Fig. S10B) allows users to select different splicing factors and to access different kinds of information based on publicly available data sets (Hung et al. 2008; Warzecha et al. 2009b; Xiao et al. 2009, 2012; Xue et al. 2009; Yeo et al. 2009; Katz et al. 2010; Llorian et al. 2010; Wang et al. 2010; Grellscheid et al. 2011; Lebedeva et al. 2011; Mukherjee et al. 2011; Huelga et al. 2012; Lagier-Tourenne et al. 2012; Zarnack et al. 2013). For example, selecting hnRNPH/F on the “Splicing factors” screen (Supplemental Fig. S20) gives access to predicted binding motifs (Fig. 5A) as well as to a dedicated CLIP-seq data visualization interface (Supplemental Fig. S21) based on the data set generated by Huelga and Katz (Katz et al. 2010). From there, it is possible to zoom in and view the in cellulo binding sites of the selected splicing factor in the vicinity of the selected exon. For example, two hnRNPF binding sites were identified around ENAH exon 12, suggesting that hnRNPH/F might regulate ENAH splicing (Fig. 5B). To further challenge this possibility, the “Exon Arrays” button (Fig. 5A) gives access to Affymetrix exon array data obtained after hnRNPH/F depletion by Xiao and collaborators (Xiao et al. 2009). As shown in Figure 5C (upper panel), ENAH exon 12 probes appear red when comparing hnRNPH/F-depleted cells to control cells. This suggests that hnRNPH/F depletion favors exon inclusion, as validated by RT-PCR (Fig. 5C). Meanwhile, analysis of the ESRP data set generated by Warzecha et al. (2009b) predicts that ESRP depletion induces skipping of this exon (Fig. 5C, lower panel), as validated by RT-PCR (Fig. 4F). In sum, FasterDB will provide support for performing functional genomics at exon-level resolution by integrating publicly available large-scale data sets.
Figure 5.
Dedicated CLIP-seq data visualization web interface. (A) Localization of predicted hnRNPH/F binding motifs in the vicinity of the ENAH exon 12 and their link to splicing factor data sets. (B) Reads from a CLIP-seq experiment corresponding to hnRNPF binding sites in the vicinity of the ENAH exon 12. (C) Visualization of exon array probe intensities in hnRNPH/F- or ESRP1-depleted cells as compared to control cells. RT-PCR analysis of the ENAH exon 12 inclusion rate in hnRNPH/F-depleted or control cells.
Discussion
It is well established that different tissues or organs (e.g., liver or kidney) express different splicing variants (Pan et al. 2008; Wang et al. 2008; Bland et al. 2010; de la Grange et al. 2010; Hartmann et al. 2011; Llorian and Smith 2011; Barbosa-Morais et al. 2012; Merkin et al. 2012). However, all organs are composed of common major cell types like fibroblast, epithelial, and endothelial cells. In this report, we demonstrate that each major cell type expresses a specific splicing program independently of their organ origin (Figs. 1–3). It will be interesting to next determine whether a major cell type (e.g., endothelial cells) isolated from different tissues also expresses tissue-specific splicing programs. Looking at each column representing one specific cell type from a given organ in the clustering analyses (Figs. 1, 2; Supplemental Figs. S1–S3), the splicing pattern seems likely to integrate at least two levels of specificity, of cell type and tissue origin (Supplemental Fig. S22). Our observation has several consequences. For example, some tissue-specific splicing variants previously identified by comparing different tissues could in fact reflect the different relative proportions between common cell types in those tissues, as illustrated in Supplemental Figure S22. This could be particularly relevant for tumors that are often compared to normal control tissues to identify cancer-associated splicing variants. As many cancer cells derive from epithelial cells, some previously reported cancer-associated splicing variants could reflect epithelial cell enrichment in tumors as compared to normal tissues.
Remarkably, it has been previously shown that clustering cells based on global gene expression level also reveals that most cell lines cluster together rather than with their tissues of origin (Lukk et al. 2010). As similar results were obtained using alternative spliced exons (ASE) (Fig. 3) and alternative first or last exons (AFE or ALE, respectively) (Supplemental Figs. S5, S6), this demonstrates that major cell types not only differ by the set of genes they expressed but also by the exonic content (e.g., ASE, AFE, and ALE) of the transcripts produced by the expressed genes. Therefore, our data imply that understanding the function of cell types not only requires the characterization of expressed genes but also of splicing variants generated by these expressed genes. In this context, it is interesting to underscore that genes that are differentially spliced in fibroblasts, epithelial, and endothelial cells are often involved in cell–cell or cell–substrate interactions (Supplemental Fig. S23). This suggests that alternative splicing may play a role in modeling interactions with neighboring cells and with the microenvironment.
We also identified three splicing factors, ESRP1, PTBP1, and RBFOX2, which are likely to play a major role in setting up these cell type–specific splicing programs (Fig. 4). Obviously, other splicing factors may contribute as well (Supplemental Table S8). For example, the highly related hnRNPH1 and hnRNPF splicing factors, which are in the same family as ESRP1 and are also involved in EMT (Warzecha et al. 2009a, 2010; Shapiro et al. 2011; Dittmar et al. 2012), were shown to also have an enriched expression in epithelial cells (Fig. 4A; Supplemental Table S8). Of note, although NOVA1 plays a role in establishing a neuronal splicing program, we observed that NOVA1 expression was enriched in fibroblasts, similar to RBFOX2 (Supplemental Table S8). Interestingly, it has been shown that the targets of RBFOX2 and of NOVA1 often overlap (Zhang et al. 2008). Conducting splicing analysis in a cell type–specific manner might allow new cellular functions of splicing factors to be identified. In this context, our data suggest a role of the PTBP1 and MBNL1 splicing factors in endothelial cell biology (Supplemental Table S8). Interestingly, PTBP1 and MBNL1 have been suggested to play a role in angiogenesis (Pascual et al. 2006; Pen et al. 2007; Masuda et al. 2009).
Functional genomics now needs to be performed at an exonic resolution level, given the prevalence and importance of alternative splicing. This will require new integrative bioinformatic approaches to be developed that can handle the diversity of alternative exons and the complexity of alternative splicing regulation. In this setting, we have made the analyzed data sets used in this report freely available through the user friendly web interface of FasterDB, which describes all known splicing variants of human and mouse genes and their splicing pattern across several dozen normal and cancer cells and tissues, as well as information regarding which splicing factors contribute to individual exon regulation. The aim of this database is to help researchers identify the different splicing variants of their favorite genes as well as the tissue, cell type, or cell line in which they are expressed, in order to facilitate further functional and/or mechanistic studies. For example, when researchers identify a splicing variant in physiopathological conditions, FasterDB will be useful in deciding which cellular model might be suitable for functional analysis.
The second aim of FasterDB is to help researchers characterize the splicing factors regulating the identified splicing variants. To the best of our knowledge, FasterDB is the first database integrating large-scale data sets focused on splicing, including CLIP-seq and splicing-sensitive microarray data sets (Hung et al. 2008; Warzecha et al. 2009b; Xiao et al. 2009, 2012; Xue et al. 2009; Yeo et al. 2009; Katz et al. 2010; Llorian et al. 2010; Wang et al. 2010; Grellscheid et al. 2011; Lebedeva et al. 2011; Mukherjee et al. 2011; Huelga et al. 2012; Lagier-Tourenne et al. 2012; Zarnack et al. 2013). Our next goal will be to include other data sets, such as RNA-seq data sets. We also aim to label the protein domains coded by each alternatively spliced exon in order to help users predict potential functional consequences resulting from alternative splicing.
Methods
FasterDB core database
Human and mouse exons were collected from Ensembl (release 60, assemblies GRCh37 and NCBI m37) (Flicek et al. 2013) and aligned against the NCBI transcript database using MEGABLAST (v 2.2.25). These exons were aligned against genomic sequences to define their chromosomal coordinates and then clustered by genomic position to define seven major events (e.g., alternative first exon, alternative last exon, alternative 3′ splice site, alternative 5′ splice site, intron retention, exon deletion, and exon skipping). Scores were computed using MaxEntScan for each splice site. For each gene, a nonredundant repertory of untranslated regions was established using all corresponding transcripts. UTRs were more fully characterized by describing the motifs found in their sequences using PatSearch Tool (Grillo et al. 2003). miRNA binding sites were predicted using PITA, miRanda, and PicTar (John et al. 2004; Lewis et al. 2005; Kertesz et al. 2007). Conserved exons between human and mouse were identified by aligning each human exon against the exons of its orthologous mouse gene as provided by Ensembl. The “in silico PCR” tool is based on a multialignment of the transcript exons performed with ClustalW. FasterDB is built in Perl (v5.14.2) and runs on a Ubuntu server (v12.04.1) that hosts Apache (v.2.2.16) and MySQL (v5.1.49) servers. More information is available in Supplemental Material and can be downloaded from http://fasterdb.lyon.unicancer.fr.
Exon array data set analyses
Cell lines and tissues expression data were downloaded from the GEO and Affymetrix websites and corresponded to Affymetrix Exon Arrays data sets as listed in Supplemental Table S10. The preprocessing pipeline is described in more detail in the Supplemental Material (Part 2, Section 2.2, page 9). Briefly, low-quality and cross-hybridizing probes were removed, and signals were summarized at the gene level by using the median over all remaining probes. Three different ways of computing the inclusion/exclusion rates of a given exon were computed by measuring the ratio of (1) its expression level versus the gene expression level (NI); (2) its NI in the condition of interest relative to the NI in the control condition (global SI); or (3) its expression level versus the expression level of the flanked exons (local SI). Computation of the global and local SI is described in more detail in the Supplemental Material (Part 2, section 2.2, page 10). Identification of cell type–specific alternative exons was performed after computing local SI and selecting exons with local SI above 1.45 and with a P-value <0.05. Heatmaps were generated using the multiexperiment viewer application of the TM4 package. Hierarchical clustering of Mev4 was also used to cluster cell lines that have similar regulation. Input data contains the SI of each of the regulated ASEs in each of the analyzed cell lines. The SI was computed for each cell line in comparison with all the different types of cell lines. Functional enrichment and KEGG pathway mapping were done using DAVID (Huang da et al. 2009).
Elexir web interface
Elexir is a web application that allows users to choose an experiment within a set of stored exon array experiments. This interface was developed to allow the end user to easily browse and query the expression levels of one or more genes between conditions, with possible replicates for each condition. Different test conditions can be chosen for each experiment. A condition can be defined as a cell line, a tissue type, or a treatment. Paired or unpaired analyses can be done depending on sample relationships used for test and control conditions. The intensity report displays a schematic graph of the gene being analyzed with corresponding probes for each exon. The height of each bar represents the normalized probe intensity (in log2). The color reflects the ratio of probe intensity between test and control conditions. Green and red indicate that the probe intensity in the test condition is lower and higher, respectively, than the control condition. Information for each probe is given in the underlying descriptive table or can be easily obtained by clicking on the bar.
Splicing factor analyses
The regulation of cell type–specific ASEs by specific splicing factors was tested using three different sources: microarray data, literature RNA-seq, and CLIP-seq data sets. Chromosomal coordinates of exons regulated by ESRP1, PTBP1, and/or RBFOX2 identified by RNA-seq were retrieved from previously published work (Warzecha et al. 2009b; Xiao et al. 2009; Xue et al. 2009; Yeo et al. 2009; Katz et al. 2010; Llorian et al. 2010; Huelga et al. 2012). ASEs predicted to be regulated by a splicing factor using exon array or RNA-seq data sets were compared to ASEs predicted to be cell type–specific. For each cell type, we paid attention to the ASE regulation sense. For example, PTBP1 is up-regulated, while ESRP1 and RBFOX2 are down-regulated, in endothelial cells; thus, an endothelial ASE is predicted to be regulated by one of these factors if it is skipped upon PTBP1 depletion or included by ESRP1 or RBFOX2 depletion. To be considered as confident, CLIP hits must have been detected in the exon or within 100 nt upstream of or downstream from the exon.
Splicing factor binding sites were searched with the PatSearch Tool (Grillo et al. 2003) for the genomic sequences using previously defined splicing factor binding motifs (see Supplemental Table S8). A set of 1000 randomly selected alternative exons, and a set of 1000 randomly selected constitutive exons, were used as controls. Four regions were defined: 100 nt upstream of and downstream from the exon, the first 60 nt of the exon and the last 60 nt of the exon. For each region, a sliding window of a specific length was considered, and the enrichment score of the splicing factor was computed at each position as (Σ number of factor motifs at position X / total number of analyzed sequences) × 100. The total number of binding sites found in each region of each exon group was used to compute the standard deviation value.
Experimental validation
RNA from different cell types were purchased as indicated in Supplemental Table S1. Primers used for RT-PCR are described in Supplemental Table S9. RT-qPCR analyses were performed using primers described in Supplemental Table S9. Epithelial MCF-7, fibroblast-like MDA-MB-231 cells, and endothelial HUVEC cells were transfected using RNAiMax (Invitrogen) with control siRNAs or siRNAs against ESRP1 and 2, RBFOX2, or PTBP1 and 2 (Supplemental Table S9) 48 h before RNA extraction. Electrophoretic gels were analyzed with ImageJ software.
Data access
The FasterDB database is available at http://fasterdb.lyon.unicancer.fr/.
Supplementary Material
Acknowledgments
We thank the researchers who deposited their data sets in public libraries. We hope that our website will be welcomed by both users and the researchers who generated the data sets and that we did not forget to acknowledge any of those researchers in Supplemental Table S10 and on the FasterDB website. This work was supported by the Fondation pour la Recherche Médicale (FRM), INSERM Plan Cancer 2009-2013, Institut National du Cancer (INCa), Agence Nationale de la Recherche (ANR), and Association Française Contre les Myopathies (AFM). P.M. was supported by Lyon Science Transfert, J-P.V. and H.M. by FRM, F-O.D. by the Association pour la Recherche sur le Cancer, M.P.E. by AFM, S.S. by the Ligue National Contre le Cancer, L-C.T. and E.C. by INSERM “Plan Cancer 2009-2013.”
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.162933.113.
References
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ 2010. Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338: 1587–1593 [DOI] [PubMed] [Google Scholar]
- Bland CS, Wang ET, Vu A, David MP, Castle JC, Johnson JM, Burge CB, Cooper TA 2010. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res 38: 7651–7664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ 2012. An exon-centric perspective. Biochem Cell Biol 90: 603–612 [DOI] [PubMed] [Google Scholar]
- de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D 2010. Splicing factor and exon profiling across human tissues. Nucleic Acids Res 38: 2825–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Modugno F, Iapicca P, Boudreau A, Mottolese M, Terrenato I, Perracchio L, Carstens RP, Santoni A, Bissell MJ, Nistico P 2012. Splicing program of human MENA produces a previously undescribed isoform associated with invasive, mesenchymal-like breast tumors. Proc Natl Acad Sci 109: 19280–19285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Jiang P, Park JW, Amirikian K, Wan J, Shen S, Xing Y, Carstens RP 2012. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol Cell Biol 32: 1468–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. 2013. Ensembl 2013. Nucleic Acids Res 41: D48–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Kim E, Amit M, Vaknin K, Kfir N, Ram O, Ast G 2010. Overlapping splicing regulatory motifs–combinatorial effects on splicing. Nucleic Acids Res 38: 3318–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grellscheid S, Dalgliesh C, Storbeck M, Best A, Liu Y, Jakubik M, Mende Y, Ehrmann I, Curk T, Rossbach K, et al. 2011. Identification of evolutionarily conserved exons as regulated targets for the splicing activator tra2β in development. PLoS Genet 7: e1002390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo G, Licciulli F, Liuni S, Sbisa E, Pesole G 2003. PatSearch: A program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res 31: 3608–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallegger M, Llorian M, Smith CW 2010. Alternative splicing: Global insights. FEBS J 277: 856–866 [DOI] [PubMed] [Google Scholar]
- Hartmann B, Valcarcel J 2009. Decrypting the genome's alternative messages. Curr Opin Cell Biol 21: 377–386 [DOI] [PubMed] [Google Scholar]
- Hartmann B, Castelo R, Minana B, Peden E, Blanchette M, Rio DC, Singh R, Valcarcel J 2011. Distinct regulatory programs establish widespread sex-specific alternative splicing in Drosophila melanogaster. RNA 17: 453–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. 2012. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 1: 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LH, Heiner M, Hui J, Schreiner S, Benes V, Bindereif A 2008. Diverse roles of hnRNP L in mammalian mRNA processing: A combined microarray and RNAi analysis. RNA 14: 284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS 2004. Human MicroRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA 2011. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet 12: 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S 2013. Function of alternative splicing. Gene 514: 1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E 2007. The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284 [DOI] [PubMed] [Google Scholar]
- Kim E, Goren A, Ast G 2008. Alternative splicing and disease. RNA Biol 5: 17–19 [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur C, et al. 2012. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci 15: 1488–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N 2011. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell 43: 340–352 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Llorian M, Smith CW 2011. Decoding muscle alternative splicing. Curr Opin Genet Dev 21: 380–387 [DOI] [PubMed] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. 2010. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol 17: 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukk M, Kapushesky M, Nikkilä J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A 2010. A global map of human gene expression. Nat Biotechnol 4: 322–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Abdelmohsen K, Gorospe M 2009. RNA-binding proteins implicated in the hypoxic response. J Cell Mol Med 13: 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB 2012. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 338: 1593–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M Jr, Tuschl T, Ohler U, Keene JD 2011. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell 43: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Pascual M, Vicente M, Monferrer L, Artero R 2006. The Muscleblind family of proteins: An emerging class of regulators of developmentally programmed alternative splicing. Differentiation 74: 65–80 [DOI] [PubMed] [Google Scholar]
- Pen A, Moreno MJ, Martin J, Stanimirovic DB 2007. Molecular markers of extracellular matrix remodeling in glioblastoma vessels: Microarray study of laser-captured glioblastoma vessels. Glia 55: 559–572 [DOI] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB 2011. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 7: e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Brosseau JP, Gadea G, Klinck R, Prinos P, Beaulieu JF, Lapointe E, Durand M, Thibault P, Tremblay K, et al. 2013. RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol Cell Biol 33: 396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kayikci M, Briese M, Zarnack K, Luscombe NM, Rot G, Zupan B, Curk T, Ule J 2010. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol 8: e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP 2009a. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 33: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Shen S, Xing Y, Carstens RP 2009b. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol 6: 546–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP 2010. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J 29: 3286–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten JT, Ule J 2011. Understanding splicing regulation through RNA splicing maps. Trends Genet 27: 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Wang Z, Jang M, Nutiu R, Wang ET, Burge CB 2009. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nat Struct Mol Biol 16: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Tang P, Yang B, Huang J, Zhou Y, Shao C, Li H, Sun H, Zhang Y, Fu XD 2012. Nuclear matrix factor hnRNP U/SAF-A exerts a global control of alternative splicing by regulating U2 snRNP maturation. Mol Cell 45: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. 2009. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell 36: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH 2009. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol 16: 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, Reyes A, Anders S, Luscombe NM, Ule J 2013. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152: 453–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ 2008. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev 22: 2550–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.