The endoplasmic reticulum binding protein BiP can function as a positive and negative modulator of cell death events in plant cells.
Abstract
The binding protein (BiP) has been demonstrated to participate in innate immunity and attenuate endoplasmic reticulum- and osmotic stress-induced cell death. Here, we employed transgenic plants with manipulated levels of BiP to assess whether BiP also controlled developmental and hypersensitive programmed cell death (PCD). Under normal conditions, the BiP-induced transcriptome revealed a robust down-regulation of developmental PCD genes and an up-regulation of the genes involved in hypersensitive PCD triggered by nonhost-pathogen interactions. Accordingly, the BiP-overexpressing line displayed delayed leaf senescence under normal conditions and accelerated hypersensitive response triggered by Pseudomonas syringae pv tomato in soybean (Glycine max) and tobacco (Nicotiana tabacum), as monitored by measuring hallmarks of PCD in plants. The BiP-mediated delay of leaf senescence correlated with the attenuation of N-rich protein (NRP)-mediated cell death signaling and the inhibition of the senescence-associated activation of the unfolded protein response (UPR). By contrast, under biological activation of salicylic acid (SA) signaling and hypersensitive PCD, BiP overexpression further induced NRP-mediated cell death signaling and antagonistically inhibited the UPR. Thus, the SA-mediated induction of NRP cell death signaling occurs via a pathway distinct from UPR. Our data indicate that during the hypersensitive PCD, BiP positively regulates the NRP cell death signaling through a yet undefined mechanism that is activated by SA signaling and related to ER functioning. By contrast, BiP’s negative regulation of leaf senescence may be linked to its capacity to attenuate the UPR activation and NRP cell death signaling. Therefore, BiP can function either as a negative or positive modulator of PCD events.
The binding protein (BiP) is an ER-resident molecular chaperone, which has been demonstrated to play a dynamic role in the regulation of various ER-supported processes in mammalian cells (for review, see Hendershot, 2004). BiP mediates the gating of the translocon pore, folding and assembly of nascent proteins, targeting of incorrectly folded proteins for degradation, contribution to ER calcium stores, perception of ER stress, and regulation of the unfolded protein response (UPR). Except for binding calcium, all of these BiP functions require the binding of the molecular chaperone to client proteins in an ATP-dependent manner.
Plant BiP has been demonstrated to display molecular chaperone activity and participate in protein folding and maturation. BiP associates detectably with normal storage proteins in an ATP-dependent manner (Gillikin et al., 1995; Vitale et al., 1995), interacts cotranslationally with rice (Oryza sativa) prolamin storage proteins (Li et al., 1993), and binds to exposed sites on phaseolin monomers but not to the trimeric form of the bean (Phaseolus vulgaris) protein (Foresti et al., 2003). Several other client proteins interact with plant BiP in an ATP-dependent manner, and thus a molecular activity for plant BiP has been extensively demonstrated (Brandizzi et al., 2003; Mainieri et al., 2004; Snowden et al., 2007; Park et al., 2010). However, the role of plant BiP in regulating signaling events that radiate from stress in the ER, as implicated by the protective functions of plant BiP under distinct stress conditions, is far less understood. These functions include the ability of BiP to attenuate ER stress (Leborgne-Castel et al., 1999; Alvim et al., 2001; Costa et al., 2008), to confer tolerance to drought in transgenic soybean (Glycine max) and tobacco (Nicotiana tabacum) plants (Alvim et al., 2001; Valente et al., 2009), to promote plant innate immunity (Wang et al., 2005), and to attenuate ER stress and osmotic stress-induced cell death (Reis et al., 2011).
The most well-characterized ER signaling pathway is the UPR, which is triggered by any condition that disrupts ER homeostasis and promotes an accumulation of unfolded proteins in the organelle lumen (Walter and Ron, 2011). In mammals, upon ER stress, the UPR is transduced by three distinct classes of ER transmembrane proteins: protein kinase RNA-like ER kinase, activating transcription factor6 (ATF6), and inositol-requiring protein1 (IRE1). Upon activation, these receptors act together to transiently attenuate protein synthesis, up-regulate ER folding capacity, and degrade misfolded proteins. BiP plays a pivotal role in regulating the activation status of protein kinase RNA-like ER kinase, ATF6, and IRE1 (Hendershot, 2004; Malhotra and Kaufman, 2007). In plants, the UPR is transduced by two classes of transmembrane receptors: the ATF6 orthologs basic Leucine zipper transcription factor17 (bZIP17) and bZIP28 and the IRE1 orthologs IRE1a and IRE1b in Arabidopsis (Arabidopsis thaliana; Eichmann and Schäfer, 2012; Iwata and Koizumi, 2012; Fanata et al., 2013; Reis and Fontes, 2013). BiP overexpression in tobacco and soybeans greatly inhibits the UPR, suggesting a role for BiP in regulating UPR activation (Leborgne-Castel et al., 1999; Alvim et al., 2001; Costa et al., 2008). Recently, plant BiP has been demonstrated to directly regulate the activation of the plant ER stress transducer bZIP28 (Srivastava et al., 2013).
In addition to the ER stress-specific signaling UPR, the ER accommodates the integration of multiple stress signals. One plant-specific, ER stress-shared response is ER and osmotic stress-integrated signaling, which converges on N-rich proteins (NRPs), a developmental and cell death (DCD) domain-containing protein, to transduce a cell death signal (Irsigler et al., 2007; Costa et al., 2008). As an integrated pathway, NRP/DCD-mediated cell death signaling is activated by either ER or osmotic stress but requires both signals for full activation (Reis and Fontes, 2012). Upon ER and osmotic stress, the expression of the transcriptional activator soybean early responsive to dehydration stress15 is induced, which then activates the promoters and expression of the NRP genes (DCD genes) NRP-A and NRP-B (Alves et al., 2011). An enhanced accumulation of NRPs up-regulates the cell death effector Glycine max NAC-domain containing protein81 (GmNAC81) (formerly designated GmNAC6; Faria et al., 2011) to induce a programmed cell death (PCD) event, which can be monitored by an induction of caspase-like activity, DNA fragmentation, chlorophyll loss, elevated peroxidation, and senescence-associated marker gene expression (Costa et al., 2008; Faria et al., 2011). Recently, we discovered another member of the GmNAC family, GmNAC30, which binds to GmNAC81 in the nucleus of plant cells to coordinately regulate the caspase1-like vacuolar processing enzyme gene expression, underlying a mechanism for the execution of the ER stress- and osmotic stress-induced cell death program (Mendes et al., 2013). The vacuolar processing enzyme (VPE) has been shown to trigger vacuolar collapse-mediated PCD in pathogenesis and development (Kinoshita et al., 1999; Hatsugai et al., 2004; Yamada et al., 2004). Recent studies have demonstrated that BiP negatively regulates the NRP-mediated cell death signaling pathway and manipulating BiP expression protects plants against drought (Valente et al., 2009; Reis et al., 2011). These previous studies evaluated the performance of soyBiPD-overexpressing transgenic lines exposed to different water deficit regimes. The BiP gene family is represented by at least four copies in the soybean genome (soyBiPA, soyBiPB, soyBiPC, and soyBiPD), and all of them have been shown to be induced by ER stressors (Kalinski et al., 1995; Cascardo et al., 2000, 2001). Among them, soyBiPD (Glyma05g36620.1) is the most well-characterized isoform and has been shown to protect plants against ER stress and dehydration (Alvim et al., 2001; Valente et al., 2009). The underlying mechanism of BiP-mediated increase in water stress tolerance is associated with its capacity to modulate the osmotic stress-induced NRP/DCD-mediated cell death response (Reis et al., 2011). However, whether BiP would also control PCD under developmental conditions remains unanswered.
As the gateway of antimicrobial protein and immune signaling component biosynthesis, the ER also functions as central regulator in the execution of immune responses in plants and animals. The ER participates in at least three different processes in plant innate immunity, and compelling evidence has linked BiP to all three ER-supported immunity functions (Eichmann and Schäfer, 2012). First, ER functions as a surveillance system of proper glycosylation and folding of immune signaling receptors (Li et al., 2009; Nekrasov et al., 2009; Saijo et al., 2009; Liebrand et al., 2012). Accordingly, overexpression of rice BiP3 regulates the rice disease resistance gene Xa21-mediated innate immunity by specifically regulating the processing and stability of the immune receptor (Park et al., 2010). Second, plant immunity depends on elevated secretory activity for the efficient production of immune proteins (Wang et al., 2005; Moreno et al., 2012). NONEXPRESSOR OF PR GENES1 (NPR1), the master regulator of salicylic acid (SA)-dependent systemic acquired resistance (SAR), coordinately controls the up-regulation of pathogenesis-related (PR) genes and UPR genes during SAR (Wang et al., 2005). BiP also participates in the establishment of efficient SAR (Wang et al., 2005). BiP2 silencing in Arabidopsis attenuates PR1 secretion, a marker of SAR, upon treatment with SA analogs and impairs resistance against bacterial pathogens. Finally, the ER has been demonstrated to participate in the hypersensitive response (HR), a PCD kind of host defense triggered in plant pathogen-incompatible interactions and in nonhost resistance (Ye et al., 2011; Xu et al., 2012). Reverse genetics and overexpression studies have revealed that BiP also participates in the pathogen-induced HR PCD, although with contrasting results. BiP2 silencing is associated with a delay in the establishment of nonhost HR PCD induced by Xanthomonas oryzae pv oryzae (Xu et al., 2012), whereas BiP overexpression alleviates cell death induced by ectopic expression of the Potato virus X TGBp3al protein (Ye et al., 2011).
Although BiP has been implicated in controlling cell death events in plant cells, whether BiP functions as a positive or negative modulator in HR PCD remains debatable. Additionally, an analysis of whether BiP-mediated regulation of cell death is linked to senescence under normal conditions is merited. Here, we employed soybean and tobacco transgenic lines with increased or suppressed BiP levels to elucidate the role of BiP as a mediator of development- and immunity-related cell death events.
RESULTS
Expression Profiles of BiP-Overexpressing Soybean Leaves
The soybean transgenic lines 35S::BIP4 and 35S::BiP2, which had been independently transformed with soyBiPD, have been previously demonstrated to overexpress a functional BiP and to accumulate different levels of BiP in the microsomal fractions (Valente et al., 2009; Supplemental Fig. S1, A and C). Therefore, we employed these transgenic lines to elucidate the BiP-induced transcriptome in soybean leaves grown under normal conditions.
Affymetrix GeneChipH Soybean Genome Arrays were used to determine the BiP-induced global variation of gene expression compared with wild-type leaves. Soybean leaves at the V3/V4 stage of development were harvested, and two biological replicates, consisting of a pool of leaves from three different soybean plants, were used for each treatment. The quality of the array was assessed by pairwise comparisons using MA plots (Supplemental Fig. S2A). Using criteria of a corrected P value less than 0.05 and log2-fold change greater than 1.5, we identified 310 genes that were differentially expressed in BiP-overexpressing leaves compared with control untransformed leaves (Supplemental Fig. S2B). A predominance of down-regulated genes (254) was observed over up-regulated genes (56). Functional annotation based on gene ontology and the David bioinformatics database (http://david.abcc.ncifcrf.gov/) revealed some interesting terms (Fig. 1A). Among the differentially expressed genes, we observed an overrepresentation of abiotic stress-responsive genes (13%) that largely predominated the down-regulated gene list (Supplemental Fig. S2D; Supplemental Tables S1 and S2). These down-regulated genes were represented by a large fraction of dehydration-induced genes and components of the antioxidant system. Therefore, under normal developmental conditions, BiP overexpression massively down-regulates components of the antioxidant system. Likewise, protein degradation and cell death-associated genes were mostly down-regulated in BiP-overexpressing lines (Supplemental Table S2, cell death and proteolysis), which may be consistent with the negative effects of BiP on stress-induced senescence or cell death programs (Valente et al., 2009; Reis et al., 2011).
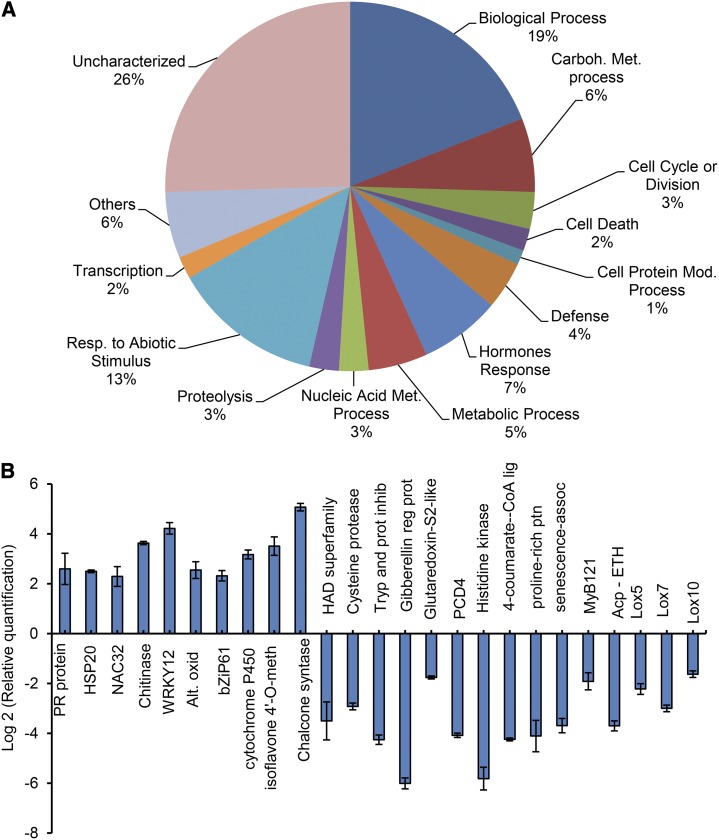
Figure 1.
Global variation of gene expression induced by soyBiPD overexpression. A, The pie chart illustrates the distribution of the differentially expressed genes across functional categories defined by the Gene Ontology Biological process. The numbers represent the frequency of genes in each category. B, The relative quantification of gene expression in transgenic plants compared with the wild type in a log2 scale (± sd, n = three biological replicates) determined by real-time PCR. The identities of each gene are presented in Supplemental Table S3.
Up-regulated defense and immune system-related genes clearly predominated in BiP-overexpressing lines. Most notable genes include the Bet vI gene family, PR genes, and lignin biosynthetic process genes, such as lignan, a chitinase class I, and a syringolide-induced protein (Durner et al., 1997; Broekaert et al., 1998; Jacobs et al., 1999; Lo et al., 1999; Slaymaker and Keen, 2004). These results are consistent with previous data, which suggest that BiP participates in plant immunity and enhanced PR protein synthesis (Jelitto-Van Dooren et al., 1999; Wang et al., 2005).
A set of flavonoid-related genes, which fall into the Biological Process/Metabolic Process category, was up-regulated in the BiP-overexpressing line. The O-methyltransferases, which participate in the biosynthesis of flavonoids and lignin, have been implicated in disease resistance in plants (Lam et al., 2007). Therefore, these flavonoid-related genes enhanced the list of biotic stress-responsive genes, which were up-regulated in the 35S::BiP4 line.
A representative subset of down- and up-regulated genes were selected to validate the microarray hybridization results by quantitative reverse transcription (qRT)-PCR (Fig. 1B). The qRT-PCR results perfectly matched the microarray hybridization results. Taken together, the global gene expression variation results indicated that in the BiP-overexpressing soybean leaves, cell death-associated genes are down-regulated and immune system components are up-regulated.
BiP-Overexpressing Lines Display Delayed Leaf Senescence under Normal Developmental Conditions
Because the gene expression profile was assessed under normal conditions and because BiP overexpression down-regulated genes associated with PCD, we examined whether BiP would regulate aging or leaf senescence under normal developmental conditions.
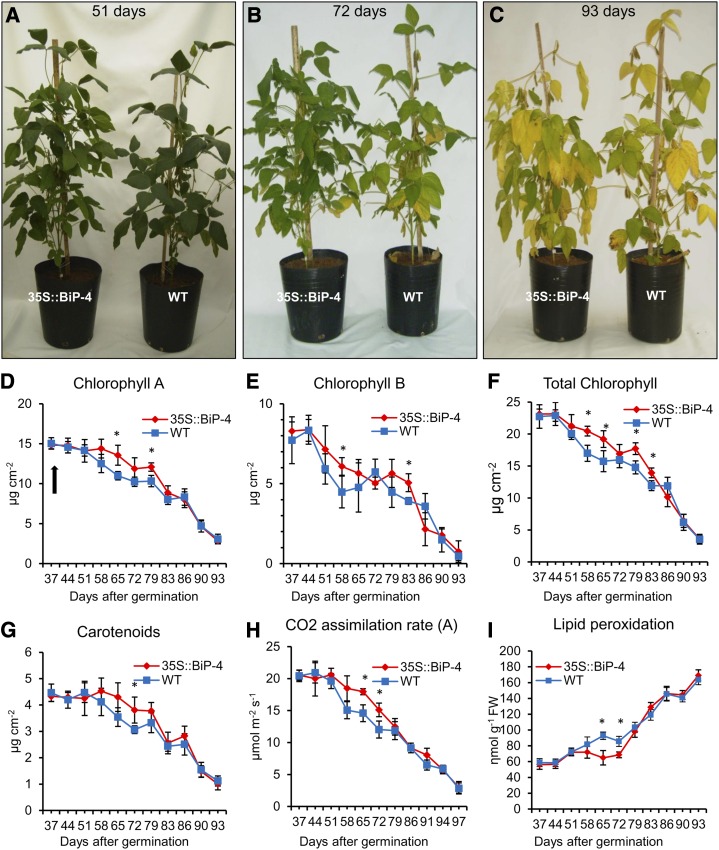
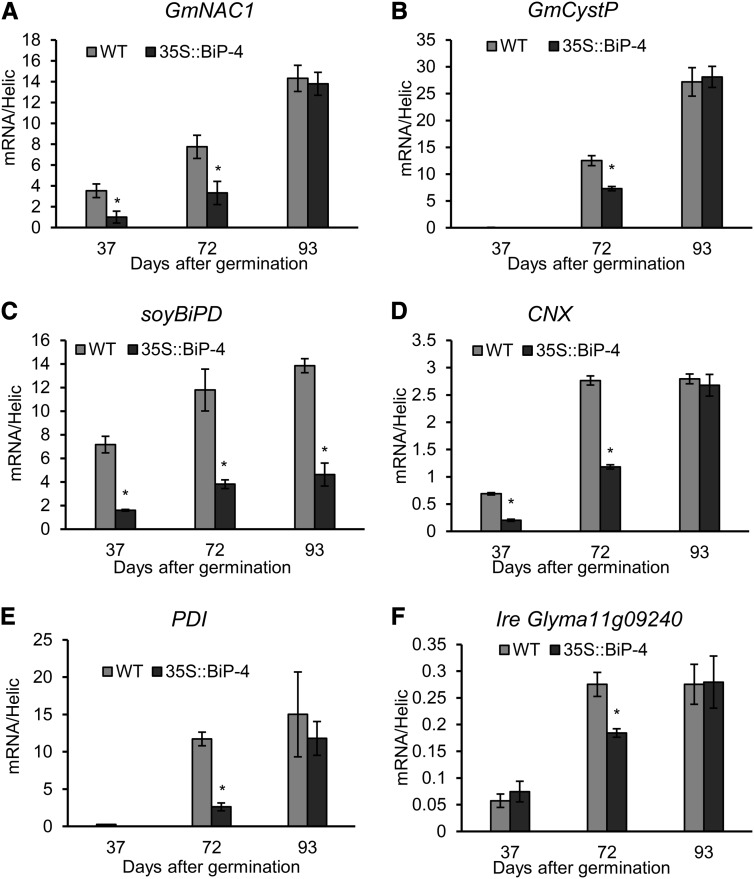
We observed that BiP-overexpressing plants flowered at similar rate as wild-type controls (approximately 37 d after germination [DAG] on average) but displayed delayed senescence after flowering, because the plants retained green leaves for a longer time (see Fig. 2, A–C for a comparison of leaf yellowing progression between 35S::BiP4 and the wild type at 51, 72, and 93 DAG). Accordingly, a clear difference in the quantitation of senescence-associated physiological markers between wild-type and 35S::BiP4 leaves was observed during 21 d of growth, between 58 to 79 DAG, which did not persist through later stages of development. For example, significant differences in the leaf pigment content, such as chlorophyll a (Fig. 2D), chlorophyll b (Fig. 2E), total chlorophyll (Fig. 2F), and carotenoids (Fig. 2G), between the wild-type and BiP-overexpressing lines were pronounced during this time window. At 65 and 72 DAG, the photosynthesis rate was significantly higher in BiP-overexpressing lines compared with wild-type, untransformed controls (Fig. 2H), whereas the malondialdehyde content, a product of senescence-associated lipid peroxidation, was lower in BiP-overexpressing lines (Fig. 2I). The expression of senescence-associated genes, such as GmNAC1 (Pinheiro et al., 2009) and Cys protease (GmCystP, Valente et al., 2009), was also monitored by qRT-PCR (Fig. 3, A and B), revealing the GmNAC1 and GmCystP transcript levels in wild-type and BiP-overexpressing lines displayed similar profiles; higher expression levels were observed at later developmental stages. However, at 72 DAG, the aging-associated induction of these genes was remarkably lower in BiP-overexpressing lines than in wild-type leaves. Collectively, these results indicate that senescence under normal greenhouse growth conditions is delayed in the BiP-overexpressing line. We also analyzed another independently transformed BiP-overexpressing line, 35S::BiP2, which accumulates a similar level of BiP (Supplemental Fig. S1C). We used an anti-BiP serum that cross reacts with a 28-kD polypeptide to probe the immunoblotting. This cross-reacting 28-kD polypeptide serves as an internal control for loading and thereby allows a quantitative comparison of BiP accumulation between the transgenic lines. The 35S::BiP2 line also displayed delayed senescence after flowering; the plants retained green leaves for a longer time (Supplemental Fig. S3A). During this time, the chlorophyll loss (103 DAG) and photosynthetic rate (78–103 DAG) were significantly lower in the transgenic line than in wild-type leaves, and the senescence-associated molecular markers GmNAC1 and GmCystP were induced to a lower extent in the 35S::BiP2 leaves (Supplemental Fig. S3, D and E). These results further indicate that BiP may modulate developmentally programmed leaf senescence. Because high secretory activity may also cause ER stress, we measured the extent of the UPR activation during development to correlate the BiP-mediated modulation of ER stress × senescence. We used the UPR marker genes, i.e. soyBiPD, calnexin (CNX), and protein disulfide isomerase (PDI), that were known to be induced at the RNA and protein levels by ER stress. We also selected two soybean IRE1 homologs, Glyma01g36200 and Glyma11g09240, using a sequence comparison with IRE1a and IRE1b from Arabidopsis and because they were induced by tunicamycin, a potent ER stressor (Supplemental Fig. S4), as additional ER stress-associated genes. All tested UPR marker genes were up-regulated during senescence, indicating that developmentally programmed leaf senescence activated the UPR in plants (Fig. 3, C–F; Supplemental Figs. S5A and S6, A–D). Consistent with a previous observation, BiP overexpression initiated a feedback regulation of endogenous BiP gene expression, as the level of senescence-mediated BiP induction was significantly lower in the 35S::BiP4 lines (Fig. 3C; Supplemental Fig. S6A; Leborgne-Castel et al., 1999; Costa et al., 2008). These results further confirm that BiP transcription is regulated via a feedback mechanism that involves monitoring BiP protein levels. BiP overexpression also attenuated the UPR activation during senescence, as the induction of the other UPR-associated marker genes (CNX, PDI, and IRE homologs) was significantly lower in BiP overexpressing than in the wild type. At a later developmental stage (93 DAG in35S::BiP4 and 110 DAG in 35S::BiP2), the BiP-mediated inhibition of the UPR was relieved, and the transcript levels of UPR marker genes reached wild-type levels. These results are consistent with previous observations, which demonstrated that the BiP modulation of UPR activation depends on the duration and intensity of the ER stress, where BiP inhibition is relieved with persistent stress (Reis et al., 2011).
Figure 2.
Slightly delayed leaf senescence in 35S::BiP4 lines. A to C, Developmental performance of soybean plants after flowering. Wild-type (WT) and 35S::BiP4 soybean lines were grown under greenhouse conditions and were photographed at 51 DAG (A), at 72 DAG (B), and at 93 DAG (C). D to F, Chlorophyll loss during the progression of leaf senescence. The content of chlorophyll a (D), chlorophyll b (E), and total chlorophyll (F) was determined over time in the leaves of wild-type and 35S::BiP4 lines during leaf senescence. The arrow indicates the start of flowering. G, The carotenoids content during leaf senescence in wild-type and 35S::BiP4 leaves. H, CO2 assimilation during leaf senescence in wild-type and 35S::BiP4 leaves. I, Lipid peroxidation induced by leaf senescence in wild-type and 35S::BiP4 leaves. The leaf lipid peroxidation was monitored by determining the level of thiobarbituric acid-reactive compounds and expressed as the malondialdehyde content. The bars indicate the confidence intervals, and the asterisks indicate significant differences of P ≤ 0.05 when comparing the transgenic and nontransgenic lines on the same observation day.
Figure 3.
Gene expression analysis of senescence and cell death-associated genes in 35S::BiP4 transgenic plants under normal developmental conditions. Total RNA was isolated from wild-type (WT) and 35S::BiP4 leaves at 37, 72, and 93 DAG, and the induction of GmNAC1 (A), Cys protease, GmCystP (B), soyBiPD (C), CNX (D), PDI (E), and the IRE1 homolog Glyma11g09240 (F) was monitored by qRT-PCR. The bars indicate the confidence interval (P < 0.05, n = 3), and the asterisks indicate significant differences between wild-type and transgenic plants.
The BiP-Mediated Delay of Leaf Senescence Is Associated with an Attenuation of NRP-Mediated Cell Death Signaling
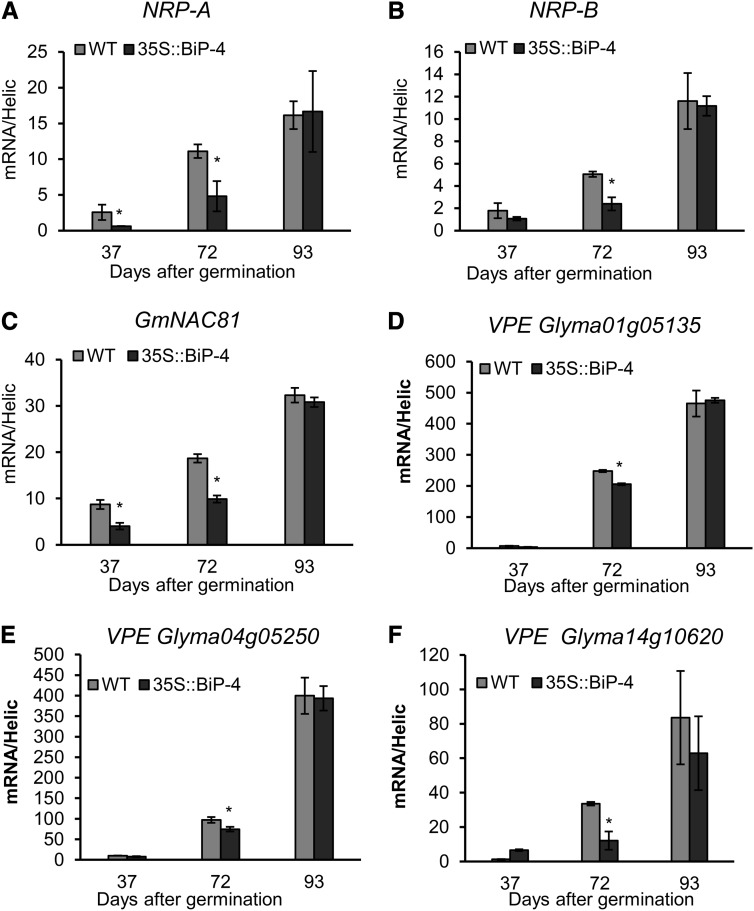
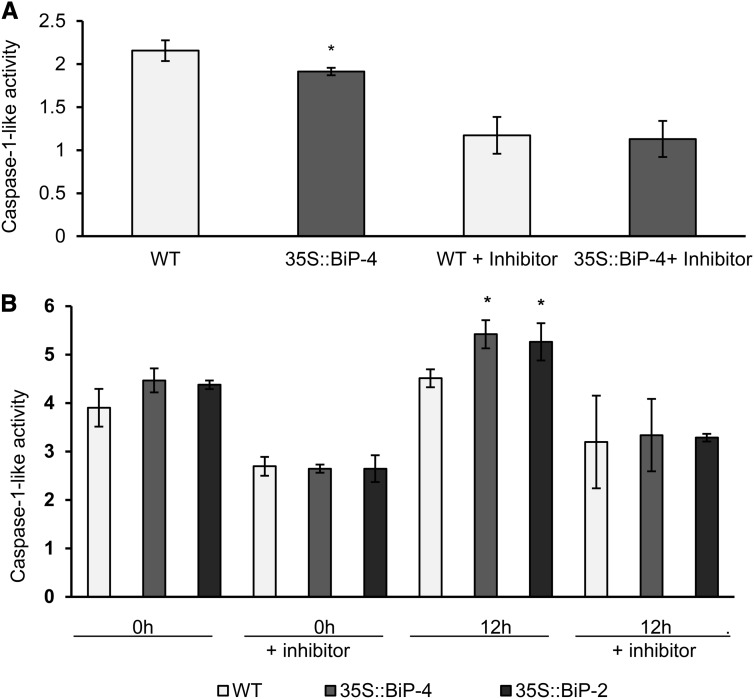
As BiP modulates the stress-induced NRP-mediated cell death response, we examined whether this pathway is also activated during developmental programmed leaf senescence. First, we assessed whether the expression of the components of this pathway, NRP-A, NRP-B, GmNAC-6, and VPE, are up-regulated during natural leaf senescence (Fig. 4). Under normal developmental conditions, all the tested components of the stress-induced NRP-mediated cell death signaling were up-regulated at the onset of leaf senescence and displayed increased transcript levels as leaf senescence progressed (Fig. 4). In BiP-overexpressing lines, the NRP-mediated signaling pathway was significantly attenuated at 72 DAG, but this attenuation did not persist through later stages of development. Likewise, in 35S::BiP2 line, the NRP-mediated signaling pathway was attenuated at 92 DAG but not at later stages of development (Supplemental Fig. S6). Because VPE displays caspase1-like activity (Tyr-Val-Ala-Asp proteolytic acivity [YVADase] activity) that is associated with the execution of PCD (Hara-Nishimura et al., 2005), we monitored the induction of YVADase in wild-type and 35S::BIP4 leaves undergoing senescence as additional evidence of NRP-mediated cell death signaling activation (Fig. 5A). At 72 DAG, YVADase activity was significantly lower in 35S::BiP4 leaves than in wild-type leaves, which is consistent with an inhibitory role of BiP in NRP-mediated cell death signaling. The inclusion of the YVADase-specific inhibitor N-Ac-Tyr-Val-Ala-Asp-CHO (Ac-YVAD-CHO) reduced enzyme activity to basal levels. Collectively, these results closely paralleled the BiP inhibition of the UPR and the BiP attenuation of leaf senescence. The BiP-mediated inhibition of NRP-mediated cell death signaling likely accounts, at least in part, for the attenuation of leaf senescence displayed by BiP-overexpressing lines.
Figure 4.
BiP overexpression attenuates mediators of the ER and osmotic stress-induced cell death response during development. Total RNA was isolated from wild-type (WT) and 35S::BiP4 leaves at 37, 72, and 93 DAG, and the induction of the indicated genes was monitored by qRT-PCR, as follows: NRP-A (A), NRP-B (B), GmNAC81 (C), VPE (Glyma01g05135; D), VPE (Glyma04g05250; E), and VPE (Glyma14g10620; F). The expression values were obtained using the comparative cycle threshold (2−ΔCT) method and the endogenous control helicase. The bars indicate a confidence interval (P < 0.05, n = 3), and the asterisks indicate significant differences between wild-type and transgenic plants.
Figure 5.
Caspase1-like (YVADase) activity in soybean leaves. A, BiP overexpression attenuates caspase1-like activity in leaves. Caspase1-like activity was determined from total protein of wild-type (WT) and 35S::BiP4 leaves at 72 DAG in the presence and absence of the specific inhibitor Ac-YVAD-CHO. B, Enhanced caspase1-like activity displayed by 35S::BiP4 and 35S::BiP2 inoculated leaves compared with wild-type leaves. Soybean wild-type, 35S::BiP4, and 35S::BiP2 leaves were inoculated with P. syringae pv tomato, and caspase1-like (YVADase) activity was determined at the postinoculation time as indicated in the figure. The inclusion of the caspase1-specific inhibitor reduced enzyme activity. The bars indicate the confidence interval based on Student’s t test (P < 0.05, n = 3), and the asterisks indicate significant differences between wild-type and transgenic plants.
In the BiP-Overexpressing Lines, the Onset of Nonhost Hypersensitive PCD Is Accelerated
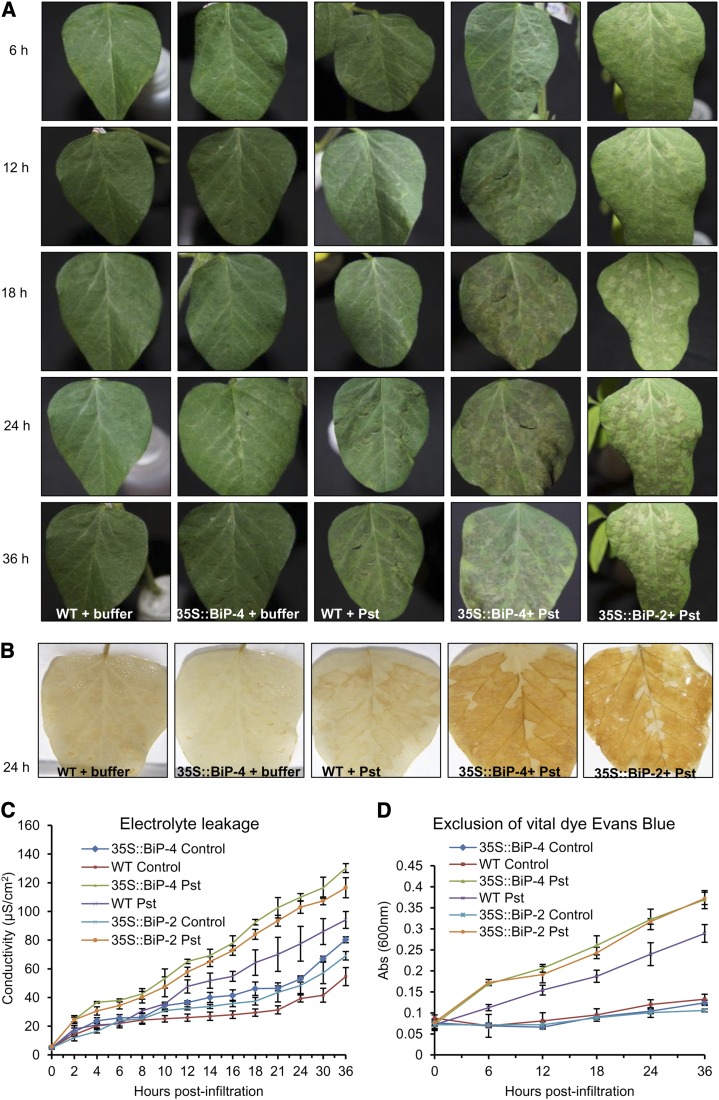
Although BiP overexpression down-regulated cell death-associated genes, we observed a clear predominance of up-regulated defense and immune system-related genes in BiP-overexpressing lines. Those genes included PR genes, which are readouts of HR, a form of PCD elicited in nonhost or incompatible interactions that is characterized by the rapid death of plant cells at the site of pathogen infection. Therefore, it was of interest to assess whether BiP overexpression would affect HR elicited by nonhost-pathogen interactions. As expected, inoculating soybean seedlings with Pseudomonas syringae pv tomato triggered a rapid cell death response, which was phenotypically visible at 24 h postinoculation (Fig. 6A). However, BiP-overexpressing lines displayed a cell death phenotype at 12 h postinoculation, and more severe necrotic lesions persisted throughout the experiment compared with inoculated wild-type leaves. Cell death was measured by H2O2 production in the inoculated leaves, and cell damage was monitored using electrolyte leakage from disk leaves and an exclusion of Evans blue vital dye (Fig. 6). H2O2 from oxidative bursts is known to drive PCD at challenged sites. A significantly enhanced oxidative burst was induced in wild-type inoculated leaves; however, much more H2O2 accumulated in 35S::BiP4 inoculated leaves at 24 h postinoculation (Fig. 6B). Electrolyte leakage was induced in wild-type leaves at 12 h postinoculation, and the values continuously increased with the progression of the HR response (Fig. 6C). The 35S::BiP4 inoculated leaves exhibited an enhanced conductivity from electrolyte leakage throughout the experiment compared with wild-type leaves. Likewise, the intensity of Evans blue dye stained cells, an indicator of cell death, was greater in 35S::BiP4 inoculated leaves throughout the experiment (Fig. 6D). We also monitored HR PCD induction by assaying the parallel induction of HR molecular markers, including PR1 and PR5, which are frequently used to examine local activation of defense mechanisms during HR, and Cys protease, a cell death-associated marker. The expression of these HR markers progressively increased with the HR response progression (Fig. 7, A–C). Although PR1 and PR5 induction has been associated with SAR, these genes are induced by many different stressors, including elicitor and wounding (van Loon and van Strien, 1999; van Loon et al., 2006). Consistent with the profiles of the other analyzed HR markers, the induction of PR1, PR5, and GmCystP by Pseudomonas spp. was much greater in 35S::BiP4 lines than in inoculated wild-type leaves at all experimental time points (Fig. 7, A–C). Collectively, these results establish that BiP-overexpressing lines displayed enhanced or accelerated PCD induced by HR.
Figure 6.
HR in soybean leaves induced by a nonhost-pathogen interaction. A, Leaf necrotic lesions induced by HR. Soybean wild-type (WT), 35S::BiP4, and 35S::BiP2 leaves were mock inoculated or inoculated with P. syringae pv tomato (Pst) and observed at intervals of 6, 12, 18, 24, and 36 h postinoculation. B, H2O2 production induced by HR. Wild-type, 35S::BiP2, and 35S::BiP4 leaves were stained with DAB 24 h after inoculation. C, Electrolyte leakage during HR. The conductivity from electrolyte leakage of mock- or P. syringae pv tomato-inoculated wild-type, 35S::BiP2, and 35S::BiP4 leaves was measured at the indicated periods postinoculation. D, Exclusion of Evans blue dye. The frequency of dead cells was quantified by the intensity of Evans blue staining of inoculated leaves as HR progresses. The bars represent a confidence interval based on Student’s t test (P < 0,05, n = 5).
Figure 7.
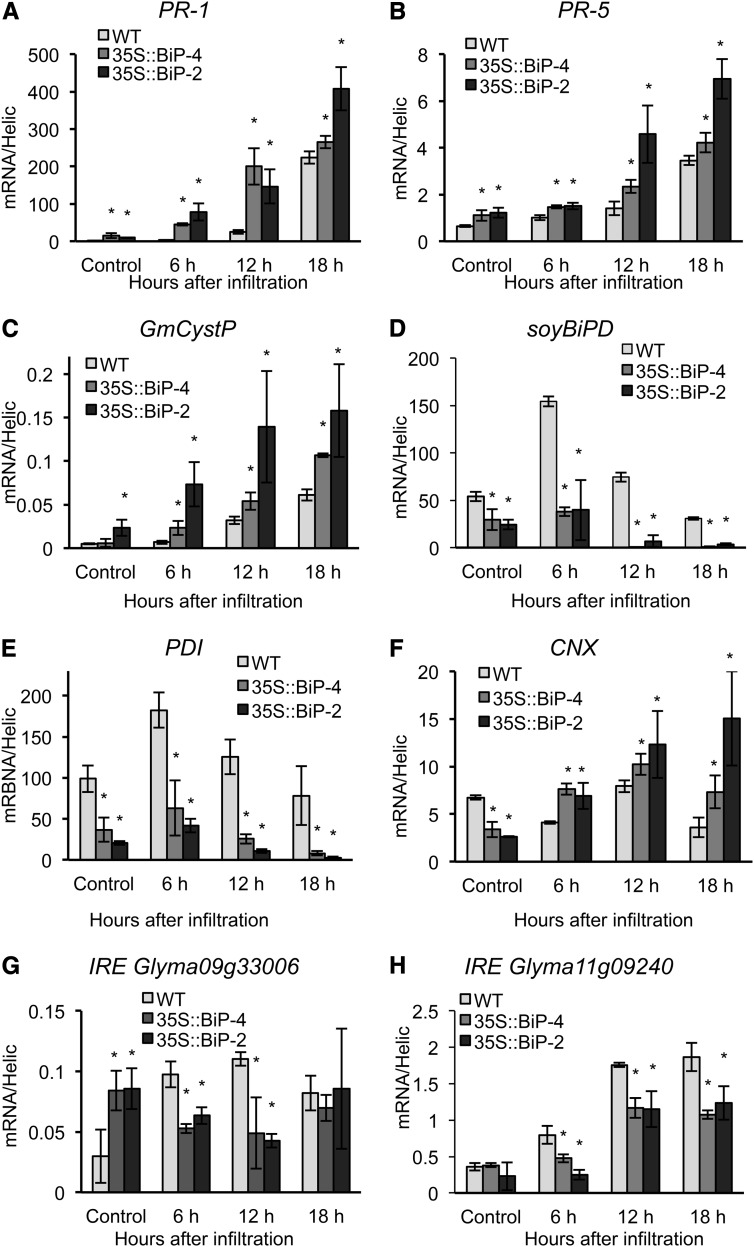
Induction of PR genes and UPR marker genes during HR elicited by P. syringae pv tomato. Total RNA was isolated from P. syringae pv tomato- or mock-inoculated wild-type (WT), 35S::BiP2 and 35S::BiP4 leaves, and the expression of the PR genes PR1 (A), PR5 (B), and Cys protease (GmCystP; C) was monitored by qRT-PCR. The induction of UPR marker genes, such as soyBiPD (D), PDI (E), CNX (F), the IRE1 homologs Glyma09g32970 (G), and Glyma11g09240 (H), was also monitored by qRT-PCR. The expression values were obtained using the 2−ΔCT method and a control helicase as the endogenous control. The bars indicate the confidence interval based on Student’s t test (P < 0.05, n = 3), and the asterisks indicate significant differences between wild-type and transgenic plants.
Under normal conditions, PR1 and PR5 were significantly induced in BiP-overexpressing lines compared with wild-type lines (see time zero in Fig. 7, A and B). Because PR1 and PR5 are SA-responsive gene markers, we examined whether BiP stimulated SA synthesis under normal conditions (Supplemental Fig. S7A). The SA levels, which were quantified in wild-type and 35S::BiP4 lines at the V3/V4 stage of development, were significantly higher in 35S::BiP4 lines. These results may indicate that higher SA accumulation in 35S::BiP4 lines represents a priming state that may be further amplified when SA signaling is biologically activated by biotrophic pathogens. Although we did not detect differences in jasmonic acid (JA) accumulation between the wild-type and 35S::BiP4 lines under normal conditions (Supplemental Fig. S7B), a higher SA accumulation in 35S::BiP4 may promote the antagonistic effect of SA on JA signaling, because several genes involved in JA synthesis were down-regulated by BiP overexpression (Supplemental Table S2).
UPR and NRP-Mediated Cell Death Signaling Are Induced by Nonhost-Pathogen Interactions and Antagonistically Modulated by BiP during HR
Next, we assessed whether the exogenous application of SA induced the UPR and the stress-induced NRP-mediated cell death signaling (Supplemental Fig. S8). As expected, the SA-responsive gene markers PR1 and PR5 and the cell death-associated gene markers GmNAC1 and GmCystP were efficiently induced by SA treatment (Supplemental Fig. S8, A–D). Except for the IRE homolog Glyma01g36200 (Supplemental Fig. S8), the other two IRE1 homologs and the other UPR gene markers, soyBiP, CNX, and PDI, were induced by exogenous SA application. Likewise, all the stress-induced NRP-mediated cell death signaling components, NRP-A, NRP-B, GmNAC81, and VPE genes, were efficiently induced by SA. The up-regulation of the UPR and NRP-mediated cell death signaling by SA prompted us to investigate whether these stress-induced pathways were induced by a Pseudomonas spp. invasion that triggers nonhost resistance in soybean.
The UPR marker genes soyBiP and PDI were induced by a Pseudomonas spp. invasion but displayed different induction kinetics than the PR genes (Fig. 7). In contrast to the more delayed response kinetics of the PR genes, the accumulation of soyBiP, PDI, and the IRE homolog Glyma09g32970 peaked as early as 6 h after infection. This is not surprising because the UPR has been reported as an early response to pathogen invasion in anticipation of PR synthesis along the ER (Jelitto-Van Dooren et al., 1999). Consistent with a role as a negative regulator of the UPR (Srivastava et al., 2013), BiP overexpression inhibited UPR activation triggered by Pseudomonas spp. invasion because the Pseudomonas spp.-mediated induction of UPR markers was attenuated in 35S::BiP4 lines (Fig. 7, D, E, G, and H). However, in BiP-overexpressing lines, CNX induction was stimulated further in response to a P. syringae pv tomato invasion (Fig. 7F). CNX is an integral membrane protein of ER, which shares a considerable homology with its luminal paralog calreticulin (CRT). Together, they constitute CNX/CRT cycle, which is crucial for proper folding of nascent proteins. Both lectins have also been shown to be involved in immunity. The chaperone function and the immune function of CRT have already been shown to be uncoupled (Qiu et al., 2012). Our results indicate that the immune function of CNX may predominate over its chaperone function as an UPR marker in nonhost resistance.
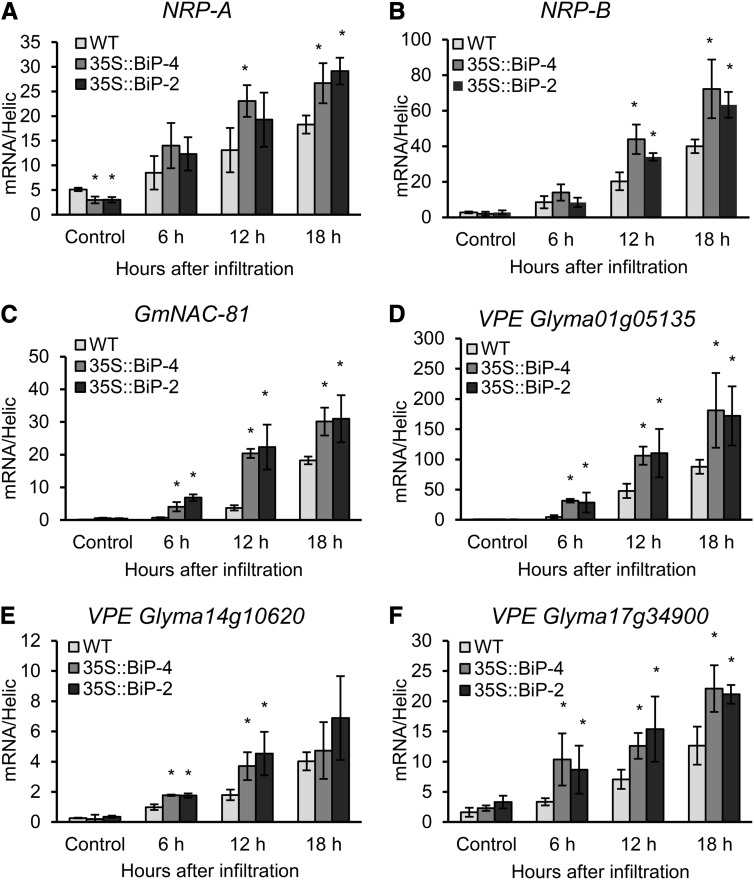
Additionally, the NRP-mediated cell death signaling was induced when the SA signaling was biologically activated by Pseudomonas spp. invasion (Fig. 8). In fact, all of the components of this cell death signaling, NRP-A, NRP-B, GmNAC81, and the VPE homologs, were effectively induced by Pseudomonas spp. infiltration and displayed similar induction kinetics to the SA-responsive PR genes. Moreover, BiP overexpression further stimulated the induction of NRP-mediated cell death signaling in the same manner as in the induction of the SA-responsive PR genes and CNX. The amplification of the HR-mediated cell death signal in 35S::BiP4 and 35S::BiP2 lines was also confirmed by an enhanced YVADase activity (caspase1-like activity) displayed by 35S::BiP4 and 35S::BiP2 inoculated leaves compared with inoculated wild-type leaves (Fig. 5B). These results indicate that the NRP-mediated signaling pathway was induced during the establishment of nonhost resistance and thus may participate in the execution of HR PCD. Furthermore, they suggest that the SA-mediated induction of the NRP-mediated cell death signaling may occur via a similar signaling branch as the induction of the PR genes, because they share similar induction kinetics and similar control mechanisms in response to pathogen invasion. In fact, BiP positively modulates both the PR genes and NRP-mediated cell death signaling pathway induction, which is antagonistic to its inhibition of the UPR in response to HR PCD.
Figure 8.
Inoculation of soybean leaves with P. syringae pv tomato triggers NRP-mediated cell death signaling during the establishment of nonhost resistance. The total RNA was isolated from P. syringae pv tomato- or mock-inoculated wild-type (WT), 35S::BiP2, and 35S::BiP4 leaves, and induction of NRP-A (A), NRP-B (B), GmNAC81 (C), VPE (Glyma01g05135; D), VPE (Glyma14g10620; E), and VPE (Glyma17g34900; F) was monitored by qRT-PCR. Expression values were obtained as in Figure 7.
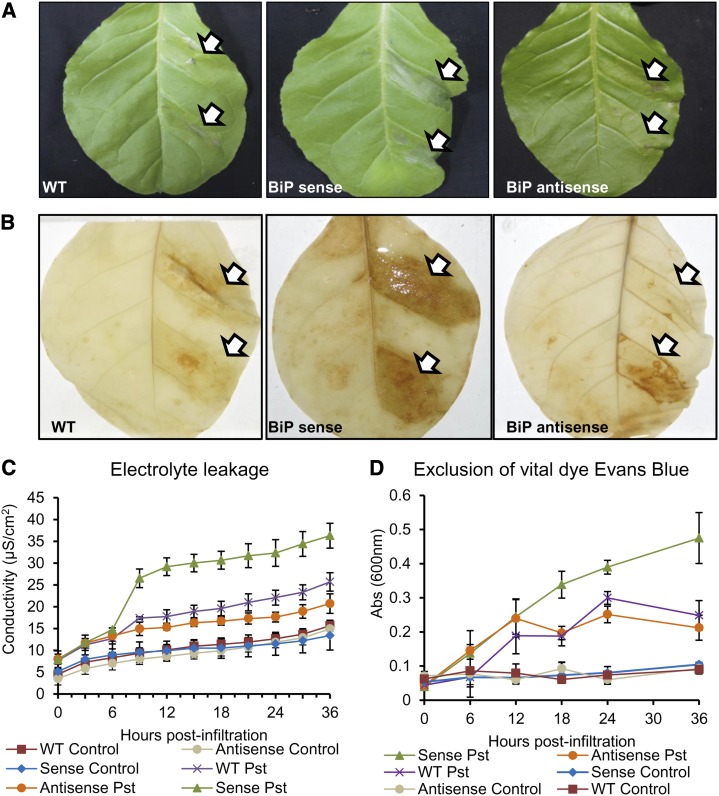
The relation between BiP mRNA abundance and the hypersensitive PCD response induced by nonhost-pathogen interactions was also investigated in tobacco, using lines with suppressed BiP expression (antisense lines AS1 and AS2) and lines with overexpressed BiP (sense lines S1 and S2; Alvim et al., 2001; Supplemental Fig. S1B). Tobacco leaves were also challenged with P. syringae pv tomato, and HR was examined 36 h postinoculation (Fig. 9). In transgenic sense lines with enhanced BiP accumulation, the leaf lesions were phenotypically more severe than in the wild-type or antisense lines (Fig. 9A). Likewise, H2O2 production was more pronounced in the sense lines, as judged by diaminobenzidine (DAB) staining (Fig. 9B). Cell death and cell damage induced by nonhost interactions were quantified by measuring the ion leakage and exclusion of Evans blue dye (Fig. 9, C and D). Similar to soybeans, the tobacco sense inoculated leaves exhibited an enhanced conductivity from electrolyte leakage and a greater intensity of stained cells with Evans blue dye throughout the experiment compared with wild-type leaves. These results confirmed that in BiP-overexpressing lines, the onset of HR PCD triggered by nonhost interactions is accelerated. Importantly, the cell death phenotype was associated with BiP levels, because the antisense lines with decreased BiP levels, when challenged with Pseudomonas spp., displayed decreased electrolyte leakage and dead cells at later stages postinoculation compared with wild-type leaves (Fig. 9, C and D, see 24 and 36 h postinoculation).
Figure 9.
HR elicited by nonhost-pathogen interactions in tobacco leaves with enhanced (sense) and suppressed (antisense) levels of BiP. A, The lesions on the tobacco leaves of wild-type (WT), sense, and antisense lines after 24 h of inoculation with a control MgCl2 buffer or with P. syringae pv tomato (Pst). B, DAB staining in wild-type, sense, and antisense leaves 24 h after inoculation with P. syringae pv tomato or a MgCl2 solution. C, The membrane integrity of leaf cells during the HR. The conductivity from electrolyte leakage of P. syringae pv tomato- and mock-inoculated wild-type, sense, and antisense leaves was measured at the indicated periods after inoculation. D, The exclusion of vital Evans blue dye from tobacco wild-type, sense, and antisense leaves during the HR. The bars represent the confidence interval (P < 0.05, n = 5).
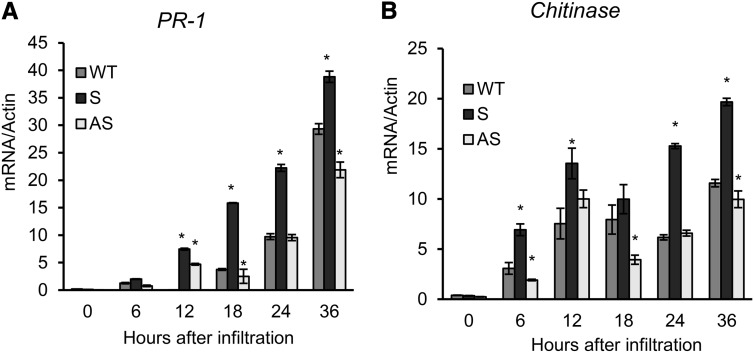
We also monitored the induction of HR PCD by assaying the parallel induction of pathogenesis-related genes, such as PR1, PR4, chitinase, and glucanase (Fig. 10). The SA-responsive marker genes PR1 (Fig. 10A) and chitinase (Fig. 10B) displayed a higher expression in sense leaves than in wild-type and antisense leaves inoculated with Pseudomonas spp. Remarkably, the induction of SA-responsive genes was less pronounced in the antisense line than in wild-type leaves at 36 h postinoculation, confirming that BiP suppression attenuated the HR triggered by nonhost-pathogen interactions. The BiP-mediated regulation of SA-responsive gene induction is consistent with our finding that BiP stimulates SA signaling activation.
Figure 10.
Time course of induction of PR genes by the nonhost interaction in tobacco leaves. Wild-type (WT), sense, and antisense tobacco leaves were inoculated with P. syringae pv tomato, and the induction of the PR genes, including PR1 and chitinase, was examined by qRT-PCR. The expression values were obtained using the 2−ΔCT method and actin as the endogenous control. The bars indicate the confidence interval (P < 0.05, n = 3), and the asterisks indicate significant differences between wild-type and transgenic plants.
DISCUSSION
Recent studies on ER stress-induced cell death have uncovered a relevant role for the molecular chaperone BiP in controlling cell death events in plant cells (Schröder and Kaufman, 2005; Valente et al., 2009; Reis et al., 2011). BiP has been demonstrated to control drought-induced cell death through the modulation of osmotic and ER stress NRP-mediated cell death signaling (Reis and Fontes, 2012). BiP has also been implicated in regulating PCD triggered by an immune response in plants and by ER stress specifically (Wang et al., 2005, 2007; Reis et al., 2011; Xu et al., 2012). Here, we employed soybean transgenic lines that overexpress a functional BiP to elucidate the role of plant BiP in developmentally programmed and biotic stress-induced cell death events. The BiP-induced transcriptome under normal conditions revealed a robust down-regulation of genes involved in developmental PCD and an up-regulation of genes involved in hypersensitive PCD triggered by nonhost plant-pathogen interactions. Consistent with the BiP-induced expression profile, we demonstrated several lines of evidence indicating that BiP can either negatively or positively modulate PCD events. Under normal conditions, the BiP-overexpressing lines display delayed leaf senescence and attenuated appearance of senescence-associated markers, such as chlorophyll loss, photosynthesis rate decrease, lipid peroxidation, and induction of senescence-associated genes. By contrast, we monitored hallmarks of HR PCD and demonstrated that enhanced BiP accumulation accelerated and amplified HR triggered by nonhost-pathogen interactions and that silencing BiP delayed the HR PCD. The BiP-mediated amplification of the HR PCD may be linked to the activation of the SA signaling because, even under normal conditions, the 35S::BiP4 line displayed an enhanced SA accumulation and an increased expression of SA-responsive marker genes, such as PR1, PR5, and PR10, compared with the wild type, which may serve as a priming state for further induction of SA signaling upon biological activation. Pseudomonas spp. infection induced a faster and stronger PR1 and PR5 gene expression in 35S::BiP4 inoculated leaves, which were associated with a more rapid appearance and amplification of hypersensitive PCD hallmarks, such as leaf necrotic lesions, H2O2 production, electrolyte leakage, and exclusion of Evans blue dye, as indicators of dead cells.
In this study, we demonstrated that the UPR was strongly activated in senescing leaves. This robust UPR activation may be due to an increased secretory activity of leaf cells during senescence. The BiP-overexpressing lines displayed an attenuated UPR activation at intermediate stages of leaf senescence (72 DAG), but this UPR inhibition was alleviated at later developmental stages when the induction of UPR gene markers (CNX, PDI, and IRE homologs) in 35S::BiP4 leaves approached the normal levels displayed by wild-type leaves. BiP overexpression in tobacco and soybeans was previously demonstrated to attenuate UPR activation by ER stress, suggesting a role for BiP in modulating UPR in plants (Leborgne-Castel et al., 1999; Alvim et al., 2001; Costa et al., 2008). A role of BiP as an UPR regulator in plants was further confirmed by recent data demonstrating that BiP regulates the activity to the UPR transducer bZIP28 (Srivastava et al., 2013). However, the BiP-mediated inhibition of UPR activation appears to depend on the duration and intensity of the ER stress, such that BiP inhibition is relieved during persistent stress (Reis et al., 2011). The results of this investigation are consistent with the hypothesis that BiP modulation of UPR is dependent on the ER processing capacity and secretory activity and might explain the controversy surrounding a potential role for BiP as a regulator of UPR in plants.
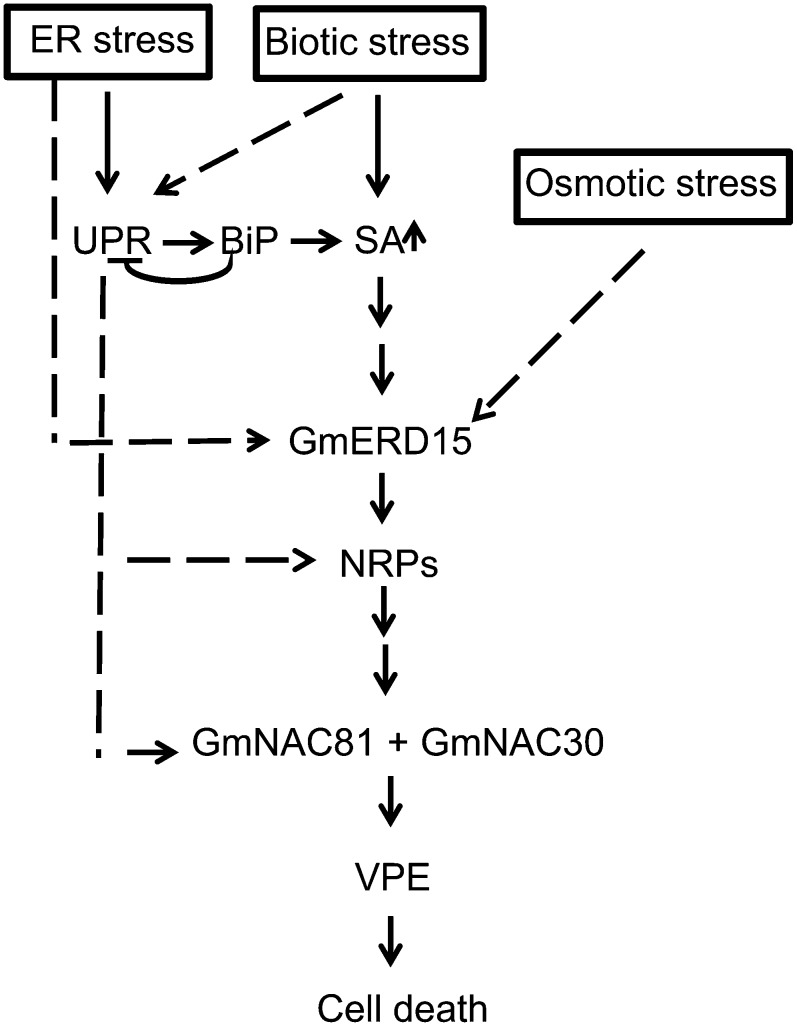
BiP modulates the propagation of an ER and osmotic stress-induced cell death signal by negatively regulating the expression and activity of NRP-mediated cell death signaling components (Reis et al., 2011). This interaction attenuates dehydration-induced cell death and promotes the adaptation of BiP-overexpressing lines to drought (Valente et al., 2009; Reis et al., 2011; Reis and Fontes, 2012). Here, we revealed that the stress-induced NRP-mediated cell death signaling also likely operates during developmentally programmed leaf senescence. We observed that all cell death pathway components, NRP-A, NRP-B, GmNAC81, and VPE homologs, were induced during leaf senescence. Furthermore, the GmNAC81-mediated induction of VPE during leaf senescence was associated with a specific increase in YVADase activity, demonstrating that a VPE-mediated PCD may be functional during leaf senescence. VPE is a Cys protease that exhibits caspase1-like activity and has been associated with Tobacco mosaic virus-induced hypersensitive cell death (Hatsugai et al., 2004) and more recently with developmental PCD (Hara-Nishimura et al., 2005). As an executioner of vacuolar collapse-mediated PCD, VPE acts as a processing enzyme to activate various vacuolar proteins, which participate in the disintegration of vacuoles, to initiate the proteolytic cascade in plant PCD (Hatsugai et al., 2004). We observed that BiP also attenuated NRP-mediated cell death signaling during leaf senescence by inhibiting the induction of pathway components, such as NRPs, GmNAC81, and VPE homologs, as well as YVADase activity. Therefore, BiP may delay leaf senescence by modulating NRP-mediated cell death signaling. The BiP-mediated negative modulation of NRP-mediated cell death signaling during leaf senescence may be linked to its function as an UPR regulator. In support of this hypothesis, we demonstrated that the BiP-mediated inhibition of the PCD signal was temporally coordinated with the BiP-mediated inhibition of the UPR, as it occurred at 72 DAG but not at later developmental stages. Furthermore, both GmNAC81 and NRP-B promoters harbor the UPR element CC(N)12CCACG (http://www.dna.affrc.go.jp/PLACE/signalscan.html; Higo et al., 1999), which was derived from the mammalian UPR cis-element CCAAT(N)9CCACG as a consensus sequence found in ER stress-inducible plant promoters (Martínez and Chrispeels, 2003). The putative UPR cis-element CCgta(N)9CCACG was found in the NRP-B promoter at position –200 and the sequence CCttc(N)9CCACG at position –383 in the GmNAC81 promoter, which could be activated by the UPR and hence be modulated by BiP (Fig. 11).
Figure 11.
BiP may modulate the NRP-mediated cell death signaling pathway by inhibiting UPR and stimulating SA signaling. The scheme illustrates the propagation of a cell death signal derived from prolonged ER, osmotic, and biotic stress through the NRP-mediated PCD signaling pathway. A broken arrow indicates an effect on gene expression, while a solid arrow indicates that the gene is an immediate downstream target. This investigation revealed that overexpression of BiP inhibits the UPR, the expression of NRPs, and GmNAC81 and as consequence attenuates cell death mediated by the NRP signaling as in leaf senescence. In response to a biotic stimulus, BiP positively modulates the SA signaling and activates the NRP-mediated cell death signaling.
In addition to being induced during leaf senescence, which was demonstrated by this investigation, and by ER and osmotic stress (Irsigler et al., 2007; Faria et al., 2011), the NRP-mediated cell death signaling is also induced by SA signaling. This induction was confirmed by an exogenous application of SA (Supplemental Fig. S8) and by the inoculation of soybean and tobacco leaves with P. syringae pv tomato, which incites a nonhost HR in these plant species. Under biological activation of SA signaling, BiP overexpression did not inhibit the induction of NRP-mediated cell death signaling but rather enhanced the expression of cell death pathway components and YVADase activity. The enhanced induction of NRP-mediated cell death signaling in BiP-overexpressing leaves paralleled an increased expression of PRs and the enhanced development of morphological and physiological hallmarks of HR PCD elicited by nonhost resistance. Therefore, in response to SA signaling, BiP induced further the NRP-mediated cell death signaling. By contrast, in the BiP-overexpressing lines, the induction of UPR marker genes in response to activated SA signaling was suppressed. These results are consistent with previous observation, which revealed an enhancement of the SA-mediated induction of UPR marker genes and a concomitant decrease in PR1 gene expression in Arabidopsis bip2 knockout lines (Wang et al., 2005).
SA plays a central role in plant defense signaling. It is required for the recognition of pathogen-derived components and subsequent establishment of local resistance in the infected region as well as for systemic resistance (Gaffney et al., 1993; Loake and Grant 2007). During SAR, there is a massive buildup of PR proteins in vacuoles and in the apoplast. Coordinated up-regulation of the protein secretory machinery is required to ensure proper folding, modification, and transport of PR proteins. Accordingly, the ER-resident gene BiP is induced before PR1 accumulation (Jelitto-Van Dooren et al., 1999; Wang et al., 2005; Fig. 6). NPR1, the master regulator of SA-dependent SAR, not only directly induces the PR genes, but also prepares the cell to secrete PR proteins by controlling the expression of the secretory machinery components and UPR genes (Wang et al., 2005). However, NPR1 regulates the expression of PR1 and UPR genes via interactions with distinct transcriptional factors (Zhang et al., 1999; Pajerowska-Mukhtar et al., 2012). We observed that BiP antagonistically modulates the SA-mediated induction of UPR genes and PR genes, which is coordinated with the induction of the NRP-mediated cell death response. Therefore, the SA-mediated induction of NRP cell death signaling likely occurs via a pathway distinct from the UPR, which contrasts with the coordinated induction of the NRP-mediated cell death response and the UPR by developmentally programmed leaf senescence.
In summary, the underlying mechanisms of BiP-mediated delay in leaf senescence or acceleration of HR PCD by the nonhost response may be associated, at least in part, with its capacity to modulate the NRP-mediated cell death response (Fig. 11). Although the BiP-mediated positive modulation of NRP cell death signaling occurs through a yet undefined mechanism that is activated by SA signaling and related to ER functioning, the BiP-mediated negative modulation of this pathway is likely associated with the capacity of the ER-resident molecular chaperone to regulate the UPR activation that occurs during leaf senescence.
MATERIALS AND METHODS
Plant Growth and Transgene Expression
Soybean (Glycine max ‘Conquista’) seeds transformed with soyBiPD under the control of the Cauliflower mosaic virus 35S promoter (Valente et al., 2009) as well as untransformed control seeds were germinated and grown in 3-L pots containing a mixture of soil and dung (3:1) in greenhouse conditions under natural conditions of light, a relative humidity of 65% to 85% and a temperature ranging from 35°C during the day to 15°C at night. At different periods and developmental stages, as indicated in the figure legends, the plant tissues were harvested, immediately frozen in liquid nitrogen, and stored at –80°C.
Transgenic lines were confirmed by PCR, and transgene accumulation was monitored by immunoblotting as previously described (Valente et al., 2009). Briefly, the total protein was extracted from untransformed or transformed soybean plant leaves and immunoblotted with an antibody prepared against an Escherichia coli-produced BiP carboxyl domain (anticarboxy BiP; Buzeli et al., 2002) at a 1:1,000 dilution and a goat anti-rabbit IgG alkaline phosphatase conjugate) at a 1:5,000 dilution. Alkaline phosphatase activity was assayed using 5-bromo-4-chloro-3-indolyl phosphate (Sigma) and p-nitro blue tetrazolium (Sigma).
RNA Extraction, Complementary DNA (cDNA) Synthesis, and mRNA Purification
The total RNA was extracted from leaves at the V3/V4 or V4 developmental stage, in which the third trifoliate leaf had fully expanded, using TRIzol (Life Technologies) and treated with RNase-Free DNase (Life Technologies). The RNA was quantified by spectrophotometry (Evolution 60 Thermo Scientific) and examined in a 1.3% (w/v) denaturing agarose gel that was stained with 0.1 μg mL–1 ethidium bromide. The total RNA was used for reverse transcription (RT)-PCR and microarray analyses.
RT-PCR assays were performed with 3 μg of total RNA, 5 µm of oligo(dT), 0.5 mm of deoxynucleoside triphosphates, and 1 Uracil of M-MLV reverse transcriptase (Life Technologies). For microarray analysis, mRNA was purified using the FastTrack 2.0 mRNA Isolation kit (Life Technologies) according to the manufacturer’s instructions, quantified by spectrophotometry (Evolution 60 Thermo Scientific) and electrophoresed in 1.5% (w/v) denaturing agarose gels in the presence of ethidium bromide.
Microarray Experiment and Data Analysis
For the microarray, RNA was extracted from untransformed wild-type and BiP-overexpressing soybean leaves at the V3 developmental stage. Three biological replicates corresponding to transformed (35S::BiP) and untransformed (wild-type) leaves were used to obtain two independent mRNA pools to hybridize duplicated chips for each sample. Hybridizations were performed on Affymetrix GeneChip Soybean Genome Arrays (http://affymetrix.com/index.affx) according to the manufacturer’s instructions and the following the steps: 1) double-stranded cDNA synthesis using the SuperScript One-Cycle cDNA Kit; 2) purification and cleaning of double-stranded cDNA with the cDNA Wash Buffer and Cleanup Spin Column; 3) synthesis of biotinylated complementary RNA (cRNA) using the IVT Labeling Kit; 4) amplification of cRNA using the IVT Master Mix, purification as in the second step, and quantification at 260 nm by spectrophotometry (Evolution 60 Thermo Scientific); and 5) fragmentation (hydrolysis) of biotinylated cRNA to 35 to 200 nucleotides. Then, the cRNA (15 µg) was hybridized to an Affymetrix Soybean Genome Array for 16 h according to the Affymetrix GeneChip protocol with buffers provided by the manufacturers. The arrays were washed in a GeneChip Fluidic Station 450 GCOS/Microarray Suite (GeneChip Operating Software) and stained using a GeneChip IVT Labeling Kit. The chips were scanned with a Gene Array Scanner 3000 (Affymetrix) at 3-µm-pixel resolution and a 570-nm wavelength, generating cell intensity files.
The cell intensity files were computationally analyzed using the R/Bioconductor package (Version 1.6/Version 2.6.0; Li and Wong, 2001). Normalization was performed using the Robust Multiarray Analysis method, which is available through the R/Bioconductor package for Affymetyrix arrays (Kerr et al., 2000). Microarray Quality Assessment was performed using functions from the Affy package along with functions from the R/Bioconductor package. To identify differentially expressed genes, we used R/Microarray ANOVA (http://churchill.jax.org/software/rmaanova.shtml), and the significance of differential gene expression was detected using F and Student’s t tests. The estimated P values were corrected by false discover rate and the resulting respective gene lists were classified according to the corrected P value and fold change. The genes were considered differentially expressed if they yielded a corrected P value of greater than 0.05 and a fold change greater than 1.5. The genes were annotated using the annotation Affymetrix file for the chip (http://www.affymetrix.com/support/technical/annotationfilesmain.affx), which were complemented with annotations from Soybase (http://www.soybase.org/) and Phytozome (http://www.phytozome.net/) databases.
qRT-PCR
All of the real-time PCR procedures, including tests, validations, and experiments, were conducted according to the recommendations of Applied Biosystems. Real-time RT-PCR reactions were performed on an ABI7500 instrument (Applied Biosystems) using cDNAs from the treatments, gene-specific primers (Supplemental Table S4), and the SYBR Green PCR Master Mix (Applied Biosystems). The amplification reactions were performed as follows: 10 min at 95°C, 40 cycles of 94°C for 15 s, and 60°C for 1 min. The soybean RNA helicase (Irsigler et al., 2007) and tobacco (Nicotiana tabacum ‘Havana’) RNA actin (Costa et al., 2008) were used as endogenous controls to normalize the real-time RT-PCR values. The gene expression was quantified using the 2−ΔCT (absolute quantification) or 2−ΔΔCT (relative quantification) method.
Physiological Measurements
The carotenoid, chlorophyll a, and chlorophyll b contents were determined as described by Wellburn (1994). Two 0.5-cm leaf discs were incubated with 5 mL of CaCO3-saturated dimethyl sulfoxide. After 12 h at room temperature, the total pigment content was determined spectrophotometrically at 480, 649.1, and 665.1 nm and expressed as μg cm–2.
Photosynthetic CO2 assimilation was measured in fully expanded leaves with a portable open-flow gas exchange system (LICOR 6400, Li-COR) under ambient CO2 concentrations, normal temperature conditions, and an artificial, saturating irradiance (1,000 µmol photons m–2 s–1 at the leaf level). All measurements were performed at 9 am.
The extent of leaf lipid peroxidation was estimated by measuring the malondialdehyde assay content, as described by Cakmak and Horst (1991). Approximately 0.150 g of leaves were homogenized with 2 mL of 0.1% (v/v) trichloroacetic acid. The mixture was centrifuged at 12,000g for 15 min. All steps were performed at 4°C. Supernatant aliquots (0.5 mL) were added to 1.5 mL 0.5% (v/v) thiobarbituric acid in 20% (v/v) trichloroacetic acid, and the samples were incubated at 90°C for 20 min. The reaction was stopped by incubation on ice, followed by centrifugation at 13,000g for 4 min. The absorbance of the supernatant was determined at 532 nm and subtracting from it the nonspecific A600. The malondialdehyde assay concentration was calculated using the molar absorptivity coefficient of 155 mm–1 cm–1.
Caspase1-Like (YVADase) Activity
Total protein was extracted from soybean leaves at different developmental stages or inoculated with Pseudomonas syringae pv tomato or mock-treated leaves. Caspase1-like activity was determined using the Caspase1 Colorimetric Assay Kit (Merck Millipore) according to the manufacturer’s instructions. The substrate was YVAD-pNitroaniline, and the inhibitor of caspase1 activity was the synthetic tetrapeptide Ac-YVAD-CHO supplied by the kit.
Total Protein Extraction from Leaves and Quantification
Total protein extracts were obtained as described by Görg et al. (1988) with some modifications. Approximately 200 mg of leaves from sense, antisense, and untransformed tobacco lines were crushed in liquid nitrogen, and the powder was homogenized with 10% (w/v) trichloroacetic acid in acetone containing 0.07% (v/v) 2-mercaptoethanol. The total protein was precipitated for 120 min and recovered by precipitation at 20,000g for 20 min. The pellet was washed twice with acetone containing 0.07% (v/v) 2-mercaptoethanol, and the residual acetone was removed under vacuum. The pellet was resuspended by ultrasonication in 8 m urea, 4% (v/v) Triton X-100, and 60 mm dithiothreitol. The total protein extract was stored at –20°C. The protein concentration was determined according to Bradford (1976) using bovine serum albumin as standard.
Bacterial Strains and Plant Inoculation
For the plant inoculation experiments, a BiP-overexpressing soybean transgenic line was used at the VC developmental stage (fully expanded unifoliate leaves) as well as an untransformed line and 3-week-old leaves of tobacco (sense, antisense, and the wild type; Alvim et al., 2001). P. syringae pv tomato was cultured at 28°C in King’s B (King et al., 1954) medium for 18 h. After centrifugation, the bacterial cultures were resuspended in 10 mm MgCl2. The leaves of transgenic and wild-type plants were inoculated with bacterial suspensions at an optical density of 600 nm of 0.2 (which corresponds to approximately 1 × 107 cells mL–1; Bogdanove et al., 1998) while applying a controlled pressure against the abaxial epidermis. Alternatively, the leaves were inoculated by aspersion of the bacterial suspension (1 × 107 cells mL–1), stored in a cloud chamber for 3 d, and transferred to a growth chamber at 28°C and 80% humidity with a 12-h photoperiod (Budde and Ullrich, 2000). The plant material was frozen in liquid nitrogen and stored at –80°C.
Determination of SA Contents by Ultra-Performance Liquid Chromatography (UPLC)- Mass Spectrometry Detection
The plant extracts were prepared as described previously (Forcat et al., 2008) with some modifications. Approximately 200 mg of fresh tissue were crushed in liquid nitrogen, transferred to a falcon tube, and added to 400 µL of 10% (v/v) methanol and 1% (v/v) acetic acid. The plant extract was sonicated (Ultrasonic Homogenizer 4610 Series, Cole-Parmer) twice at 30% power for 20 s on ice. After incubation for 30 min on ice, the extract was purified by centrifugation for 10 min at 13,000g and 4°C. The supernatant was carefully removed and reserved, whereas the pellet was reextracted as described. The supernatants were combined, centrifuged as before, and filtered (syringe filters, 0.45 µm, GHP Acrodisc).
Liquid chromatography (LC) was performed using a nanoAquity UPLC System (Waters) coupled to a binary pump and an autosampler. Ten microliters of the sample were separated by UPLC-mass spectrometry (MS) using a trap column (Waters) and a analytical ProteCol C18 5-Å 300 μm × 100 mm capillary column (SGE Analytical Science) operating at a flow rate of 3.0 μL min–1. The mobile phase buffers used for the gradient program were water with 0.1% (v/v) formic acid and acetonitrile with 0.1% (v/v) formic acid. The gradient program consisted of 5% (v/v) acetonitrile with 0.1% (v/v) formic acid for 5 min, linear ramping to 50% (v/v) acetonitrile with 0.1% (v/v) formic acid over 35 min, linear ramping to 95% (v/v) acetonitrile with 0.1% (v/v) formic acid over 10 min, holding at 95% (v/v) acetonitrile with 0.1% (v/v) formic acid for 10 min, ramping back to 5% (v/v) acetonitrile with 0.1% (v/v) formic acid over 5 min, and holding at 5% (v/v) acetonitrile with 0.1% (v/v) formic acid for 5 min. The eluted compounds were injected on line in anion trap mass spectrometer (Bruker Daltonics, Ion Trap amaZon) using a micro-electrospray ionization (ESI) needle. The mass spectrometer was operated in a multiple reaction mode (MRM) due to its high selectivity using precursor-to-product ion transitions. All of the UPLC-ESI MS and MS/MS parameters were optimized using individual standard solutions of SA at a concentration of 1 ng μL–1. Full scan data acquisition was performed by scanning from a mass-to-charge ratio of 60 to 300 at the positive mode, and the data were acquired for 70 min for each LC-MS/MS run. The UPLC-ESI-MS/MS assays were performed for three biological replicates. The data acquisition of both LC-MS instruments was managed using the Hystar package (Bruker Daltonics). In the product ion scan experiments, MS/MS product ions were produced by collision-activated dissociation of selected precursor ions. Thus, the combination of the parent mass and unique fragment ions was used to monitor SA selectively in crude plant extracts. MRM acquisition was performed by monitoring the 139/121. The mass spectrums were processed using a data analysis package (Bruker Daltonics), and the compound list was generated automatically using the optimized setting. The mass spectrum2 fragments were verified to confirm the detection of SA in the samples. Total ion chromatograms were used for quantitative analysis. The relative abundance of SA was normalized and expressed by integrating the total area of the chromatogram ion, which corresponds to SA, relative to the total area of all compounds observed in the MRM assays.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Ectopic expression of soyBiPD in transgenic plants.
Supplemental Figure S2. Number of differentially expressed genes in transformed plants (35S::BiP4) under normal developmental conditions.
Supplemental Figure S3. Delayed leaf senescence in 35S::BiP2 transgenic lines.
Supplemental Figure S4. Multiple alignment of Glyma01g36200, Glyma09g32970, Glyma11g09240, IRE1 (BAB63366), and IRE2 (BAB63367).
Supplemental Figure S5. Analysis of IRE Glyma01g36200 and VPE (Glyma17g34900) transcript accumulation during the development of transgenic and wild-type plants by qRT-PCR.
Supplemental Figure S6. Attenuated NRP/DCD-mediated cell death signaling and UPR activation displayed by the 35S::BiP2 line.
Supplemental Figure S7. Relative abundance of SA and JA in wild-type and 35S::BiP4 lines at the V3/V4 developmental stage.
Supplemental Figure S8. Induction of gene expression by the exogenous application of SA in wild-type soybean leaves.
Supplemental Table S1. Functional categorization of up-regulated genes in 35S::BiP4 compared with the wild type under normal developmental conditions.
Supplemental Table S2. Functional categorization of down-regulated genes in 35S::BiP4 lines.
Supplemental Table S3. Microarray-selected genes analyzed by qRT-PCR.
Supplemental Table S4. Gene-specific primers for qRT-PCR.
Glossary
- ER
endoplasmic reticulum
- PCD
programmed cell death
- HR
hypersensitive response
- UPR
unfolded protein response
- SA
salicylic acid
- SAR
systemic acquired resistance
- qRT
quantitative reverse transcription
- DAG
d after germination
- YVADase
Tyr-Val-Ala-Asp proteolytic acivity
- YVAD
Tyr-Val-Ala-Asp
- Ac-YVAD-CHO
N-Ac-Tyr-Val-Ala-Asp-Chinese Hamster Ovary (Ac-YVAD-CHO)
- JA
jasmonic acid
- DAB
diaminobenzidine
- cDNA
complementary DNA
- RT
reverse transcription
- cRNA
complementary RNA
- UPLC
Ultra-Performance Liquid Chromatography; ESI, electrospray ionization
- LC
liquid chromatography
- MS
mass spectrometry
- ESI
electrospray ionization
- MRM
multiple reaction mode
- 2−ΔCT
comparative cycle threshold
- NRP
N-rich protein
References
- Alves MS, Reis PA, Dadalto SP, Faria JA, Fontes EPB, Fietto LG. (2011) A novel transcription factor, ERD15 (Early Responsive to Dehydration 15), connects endoplasmic reticulum stress with an osmotic stress-induced cell death signal. J Biol Chem 286: 20020–20030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvim FC, Carolino SMB, Cascardo JCM, Nunes CC, Martinez CA, Otoni WC, Fontes EPB. (2001) Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiol 126: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Kim JF, Wei ZM, Kolchinsky P, Charkowski AO, Conlin AK, Collmer A, Beer SV. (1998) Homology and functional similarity of an hrp-linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato. Proc Natl Acad Sci USA 95: 1325–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Hanton SL, DaSilva LLP, Boevink P, Evans D, Oparka K, Denecke J, Hawes C. (2003) ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J 34: 269–281 [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Van Parijs J, Allen AK, Peumans WJ. (1998) Comparison of some molecular, enzymatic and antifungal properties of chitinases from thorn-apple, tobacco and wheat. Physiol Mol Plant Pathol 33: 319–331 [Google Scholar]
- Budde IP, Ullrich MS. (2000) Interactions of Pseudomonas syringae pv. glycinea with host and nonhost plants in relation to temperature and phytotoxin synthesis. Mol Plant Microbe Interact 13: 951–961 [DOI] [PubMed] [Google Scholar]
- Buzeli RA, Cascardo JC, Rodrigues LA, Andrade MO, Almeida RS, Loureiro ME, Otoni WC, Fontes EPB. (2002) Tissue-specific regulation of BiP genes: a cis-acting regulatory domain is required for BiP promoter activity in plant meristems. Plant Mol Biol 50: 757–771 [DOI] [PubMed] [Google Scholar]
- Cakmak I, Horst JH. (1991) Effects of aluminium on lipid peroxidation, superoxid dismutase, catalase and peroxidade activities in root tips of soybean (Glycine max). Physiol Plant 83: 463–468 [Google Scholar]
- Cascardo JCM, Almeida RS, Buzeli RAA, Carolino SMB, Otoni WC, Fontes EPB. (2000) The phosphorylation state and expression of soybean BiP isoforms are differentially regulated following abiotic stresses. J Biol Chem 275: 14494–14500 [DOI] [PubMed] [Google Scholar]
- Cascardo JCM, Buzeli RAA, Almeida RS, Otoni WC, Fontes EPB. (2001) Differential expression of the soybean BiP gene family. Plant Sci 160: 273–281 [DOI] [PubMed] [Google Scholar]
- Costa MD, Reis PA, Valente MA, Irsigler AS, Carvalho CM, Loureiro ME, Aragão FJ, Boston RS, Fietto LG, Fontes EPB. (2008) A new branch of endoplasmic reticulum stress signaling and the osmotic signal converge on plant-specific asparagine-rich proteins to promote cell death. J Biol Chem 283: 20209–20219 [DOI] [PubMed] [Google Scholar]
- Durner J, Shah J, Klessig DF. (1997) Salicylic acid and disease resistance in plants. Trends Plant Sci 2: 266–274 [Google Scholar]
- Eichmann R, Schäfer P. (2012) The endoplasmic reticulum in plant immunity and cell death. Front Plant Sci 3: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanata WID, Lee SY, Lee KO. (2013) The unfolded protein response in plants: a fundamental adaptive cellular response to internal and external stresses. J Prot 93: 356–368 [DOI] [PubMed] [Google Scholar]
- Faria JA, Reis PA, Reis MT, Rosado GL, Pinheiro GL, Mendes GC, Fontes EPB. (2011) The NAC domain-containing protein, GmNAC6, is a downstream component of the ER stress- and osmotic stress-induced NRP-mediated cell-death signaling pathway. BMC Plant Biol 11: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcat S, Bennett MH, Mansfield JW, Grant MR. (2008) A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti O, Frigerio L, Holkeri H, de Virgilio M, Vavassori S, Vitale A. (2003) A phaseolin domain involved directly in trimer assembly is a determinant for binding by the chaperone BiP. Plant Cell 15: 2464–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Gillikin JW, Fontes EPB, Boston RS. (1995) Protein-protein interactions within the endoplasmic reticulum. Methods Cell Biol 50: 309–323 [PubMed] [Google Scholar]
- Görg A, Postel W, Gunther S. (1988) Two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 9: 531–546 [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Hatsugai N, Nakaune S, Kuroyanagi M, Nishimura M. (2005) Vacuolar processing enzyme: an executor of plant cell death. Curr Opin Plant Biol 8: 404–408 [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I. (2004) A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305: 855–858 [DOI] [PubMed] [Google Scholar]
- Hendershot LM. (2004) The ER function BiP is a master regulator of ER function. Mt Sinai J Med 71: 289–297 [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irsigler AS, Costa MD, Zhang P, Reis PA, Dewey RE, Boston RS, Fontes EPB. (2007) Expression profiling on soybean leaves reveals integration of ER- and osmotic-stress pathways. BMC Genomics 8: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N. (2012) Plant transducers of the endoplasmic reticulum unfolded protein response. Trends Plant Sci 17: 720–727 [DOI] [PubMed] [Google Scholar]
- Kalinski A, Rowley DL, Loer DS, Foley C, Buta G, Herman EM. (1995) Binding-protein expression is subject to temporal, developmental and stress-induced regulation in terminally differentiated soybean organs. Planta 195: 611–621 [DOI] [PubMed] [Google Scholar]
- Kerr MK, Mitchell M, Churchill GA. (2000) Analysis of variance for gene expression microarray data. J Comp Biol 7: 819–837 [DOI] [PubMed] [Google Scholar]
- King EO, Ward K, Raney DE. (1954) Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med 44: 301–307 [PubMed] [Google Scholar]
- Kinoshita T, Yamada K, Hiraiwa N, Kondo M, Nishimura M, Hara-Nishimura I. (1999) Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J 19: 43–53 [DOI] [PubMed] [Google Scholar]
- Jacobs AK, Dry IB, Robinson SP. (1999) Induction of different pathogenesis-related cDNA in grapevine infected with powdery mildew and treated with ethephon. Plant Pathol 48: 325–336 [Google Scholar]
- Jelitto-Van Dooren EPWM, Vidal S, Denecke J. (1999) Anticipating endoplasmic reticulum stress: a novel early response before pathogenesis-related gene induction. Plant Cell 11: 1935–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KC, Ibrahim RK, Behdad B, Dayanandan S. (2007) Structure, function, and evolution of plant O-methyltransferases. Genome 50: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Leborgne-Castel N, Jelitto-Van Dooren EPWM, Crofts AJ, Denecke J. (1999) Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell 11: 459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wong L. (2001) Emerging patterns and gene expression data. Genome Inform 12: 3–13 [PubMed] [Google Scholar]
- Li J, Zhao-Hui C, Batoux M, Nekrasov V, Roux M, Chinchilla D, Zipfel C, Jones JD. (2009) Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci USA 106: 15973–15978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu Y, Zhang DZ, Gillikin JW, Boston RS, Franceschi VR, Okita TW. (1993) Rice prolamine protein body biogenesis: a BiP-mediated process. Science 262: 1054–1056 [DOI] [PubMed] [Google Scholar]
- Liebrand TW, Smit P, Abd-El-Haliem A, de Jonge R, Cordewener JH, America AH, Sklenar J, Jones AM, Robatzek S, Thomma BP, et al. (2012) Endoplasmic reticulum-quality control chaperones facilitate the biogenesis of Cf receptor-like proteins involved in pathogen resistance of tomato. Plant Physiol 159: 1819–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SC, Hipskind JD, Nicholson RL. (1999) cDNA cloning of a sorghum pathogenesis-related protein (PR-10) and differential expression of defense-related genes following inoculation with Cochliobolus heterostrophus or Colletotrichum sublineolum. Mol Plant Microbe Interact 12: 479–489 [DOI] [PubMed] [Google Scholar]
- Loake G, Grant M. (2007) Salicylic acid in plant defence—the players and protagonists. Curr Opin Plant Biol 10: 466–472 [DOI] [PubMed] [Google Scholar]
- Mainieri D, Rossi M, Archinti M, Bellucci M, De Marchis F, Vavassori S, Pompa A, Arcioni S, Vitale A. (2004) Zeolin. A new recombinant storage protein constructed using maize γ-zein and bean phaseolin. Plant Physiol 136: 3447–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ. (2007) The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol 18: 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez IM, Chrispeels MJ. (2003) Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes GC, Reis PAB, Carvalho HH, Aragão JF, Fontes EBP. (2013) GmNAC6 and GmNAC30 integrate the ER stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc Natl Acad Sci USA 110: 19627–19632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, Chen Y, Brandizzi F, Dong X, Orellana A, et al. (2012) IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE 7: e31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, Lacombe S, Rougon A, Bittel P, Kiss-Papp M, Chinchilla D, et al. (2009) Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J 28: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar KM, Wang W, Tada Y, Oka N, Tucker CL, Fonseca JP, Dong X. (2012) The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr Biol 22: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, Han SW, Chen X, Ronald PC. (2010) Elucidation of XA21-mediated innate immunity. Cell Microbiol 12: 1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro GL, Marques CS, Costa MDL, Reis PAB, Alves MS, Carvalho CM, Fietto LG, Fontes EPB. (2009) Complete inventory of soybean NAC transcription factors: sequence conservation and expression analysis uncover their distinct roles in stress response. Gene 444: 10–23 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Xi J, Du L, Roje S, Poovaiah BW. (2012) A dual regulatory role of Arabidopsis calreticulin-2 in plant innate immunity. Plant J 69: 489–500 [DOI] [PubMed] [Google Scholar]
- Reis PAB, Fontes EPB. (2012) N-rich protein (NRP)-mediated cell death signaling: a new branch of the ER stress response with implications for plant biotechnology. Plant Signal Behav 7: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis PAB, Fontes EPB (2013) Cell death signaling from the endoplasmic reticulum in soybean. In JE Board, ed, A Comprehensive Survey of International Soybean Research-Genetics, Physiology, Agronomy and Nitrogen Relationships, Ed 1, Vol 1. InTech, Rijeka, pp. 261–271. [Google Scholar]
- Reis PAB, Rosado GL, Silva LA, Oliveira LC, Oliveira LB, Costa MD, Alvim FC, Fontes EPB. (2011) The binding protein BiP attenuates stress-induced cell death in soybean via modulation of the N-rich protein-mediated signaling pathway. Plant Physiol 157: 1853–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Häweker H, Dong X, Robatzek S, Schulze-Lefert P. (2009) Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J 28: 3439–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder M, Kaufman RJ. (2005) ER stress and the unfolded protein response. Mutat Res 569: 29–63 [DOI] [PubMed] [Google Scholar]
- Slaymaker DH, Keen NT. (2004) Syringolide elicitor-induced oxidative burst and protein phosphorylation in soybean cells, and tentative identification of two affected phosphoproteins. Plant Sci 166: 387–396 [Google Scholar]
- Snowden CJ, Leborgne-Castel N, Wootton LJ, Hadlington JL, Denecke J. (2007) In vivo analysis of the lumenal binding protein (BiP) reveals multiple functions of its ATPase domain. Plant J 52: 987–1000 [DOI] [PubMed] [Google Scholar]
- Srivastava R, Deng Y, Shah S, Rao AG, Howell SH. (2013) BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell 25: 1416–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente MA, Faria JA, Soares-Ramos JR, Reis PA, Pinheiro GL, Piovesan ND, Morais AT, Menezes CC, Cano MA, Fietto LG, et al. (2009) The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. J Exp Bot 60: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon L, van Strien E. (1999) The families of pathogenesis related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55: 85–97 [Google Scholar]
- van Loon LC, Rep M, Pieterse CMJ. (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Vitale A, Bielli A, Ceriotti A. (1995) The binding protein associates with monomeric phaseolin. Plant Physiol 107: 1411–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334: 1081–1086 [DOI] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X. (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wang S, Narendra S, Fedoroff N. (2007) Heterotrimeric G protein signaling in the Arabidopsis unfolded protein response. Proc Natl Acad Sci USA 104: 3817–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn AR. (1994) The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrometers of different resolution. J Plant Physiol 144: 307–313 [Google Scholar]
- Xu G, Li S, Xie K, Zhang Q, Wang Y, Tang Y, Liu D, Hong Y, He C, Liu Y. (2012) Plant ERD2-like proteins function as endoplasmic reticulum luminal protein receptors and participate in programmed cell death during innate immunity. Plant J 72: 57–69 [DOI] [PubMed] [Google Scholar]
- Yamada K, Shimada T, Nishimura M, Hara-Nishimura I. (2004) A VPE family supporting various vacuolar functions in plants. Physiol Plant 123: 369–375 [Google Scholar]
- Ye C, Dickman MB, Whitham SA, Payton M, Verchot J. (2011) The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol 156: 741–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fan W, Kinkema M, Li X, Dong X. (1999) Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci USA 96: 6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]