Two recently evolved microRNAs enhance plant nitrogen starvation adaptation via regulation of glucosinolate synthesis.
Abstract
Nitrogen is an essential macronutrient required for plant growth and development. A number of genes respond to nitrogen starvation conditions. However, the functions of most of these nitrogen starvation-responsive genes are unclear. Our recent survey suggested that many microRNAs (miRNAs) are responsive to nitrogen starvation in Arabidopsis thaliana. Here, we identified a new miRNA (miR5090) from the complementary transcript of the MIR826 gene. Further investigation uncovered that both miRNA genes recently evolved from the inverse duplication of their common target gene, ALKENYL HYDROXALKYL PRODUCING2 (AOP2). Similar to miR826, miR5090 is induced by nitrogen starvation. By contrast, the AOP2 transcript level was negatively correlated with miR826 and miR5090 under nitrogen starvation. GUS-fused AOP2 expression suggested that AOP2 was posttranscriptionally suppressed by miR826 and miR5090. miRNA transgenic plants with significantly low AOP2 expression accumulated fewer Met-derived glucosinolates, phenocopying the aop2 mutants. Most glucosinolate synthesis-associated genes were repressed under nitrogen starvation conditions. Furthermore, miRNA transgenic plants with less glucosinolate displayed enhanced tolerance to nitrogen starvation, including high biomass, more lateral roots, increased chlorophyll, and decreased anthocyanin. Meanwhile, nitrogen starvation-responsive genes were up-regulated in transgenic plants, implying improved nitrogen uptake activity. Our study reveals a mechanism by which Arabidopsis thaliana regulates the synthesis of glucosinolates to adapt to environmental changes in nitrogen availability.
MicroRNAs (miRNAs) are a class of endogenous noncoding, small RNAs that regulate gene expression posttranscriptionally. miRNAs originate from primary miRNAs transcribed by RNA polymerase II. Dicer-like proteins in the nucleus orchestrate conversion of the primary miRNAs to precursor miRNAs and then to mature miRNAs (Chen, 2005; Voinnet, 2009). The mature miRNA duplexes are then methylated by HUA ENHANCER1 and exported to the cytoplasm by HASTY (the plant ortholog of exportin5), where they are incorporated into RNA-induced silencing complexes. In the RNA-induced silencing complex, miRNAs can either cleave the target mRNA or repress its translation through perfect, or almost perfect, complementary base pairing with its target sequences (Lu and Huang, 2008; Voinnet, 2009).
Plant miRNAs regulate many aspects of plant growth and development such as leaf morphogenesis (Palatnik et al., 2003), floral development, and the juvenile-to-adult transition (Wu et al., 2009). Recent reports revealed that several plant miRNAs are also involved in plant nutrient metabolism (Khraiwesh et al., 2012). Sulfate starvation induces miR395, which regulates sulfate assimilation and allocation by targeting ATP SULFURYLASEs (APS1, APS3, and APS4) and SULFATE TRANSPORTER2;1 in Arabidopsis thaliana (Liang et al., 2010). Phosphate deficiency upregulates miR399, which controls phosphate acquisition and root-to-shoot translocation by repressing PHOSPHATE2 (UBIQUITIN-CONJUGATING ENZYME24) (Chiou et al., 2006). Copper limitation induces miR397, miR398, miR408, and miR857, which regulate copper homeostasis by mediating the cleavage of genes encoding copper/zinc superoxide dismutases, copper chaperone for superoxide dismutase, and Laccases (Yamasaki et al., 2007; Abdel-Ghany and Pilon, 2008; Beauclair et al., 2010). During nitrogen deficiency, miR169 is down-regulated, whereas its targets nuclear factor Y subunit A family members are induced. Overexpression of miR169 results in the accumulation of less nitrogen than in the wild type, suggesting a role in impairing nitrogen uptake (Zhao et al., 2011). In addition, high-throughput sequencing of Arabidopsis thaliana miRNAs uncovered many miRNAs responsive to different nutrient-deficient conditions (Hsieh et al., 2009; Pant et al., 2009; Liang et al., 2012).
Nitrogen is a key component of many fundamental biological molecules such as nucleic acids, amino acids, proteins, and nitrogen-containing metabolites. Thus, plants must obtain sufficient nitrogen for normal growth and development (Peng et al., 2007; Wang et al., 2012). As sessile organisms, most plants absorb nitrogen from the soil through their roots. However, there is not always sufficient nitrogen in the soil because soil erosion, rainwater leaching, and microbial consumption have removed it. To cope with this nitrogen limitation, plants have evolved sophisticated mechanisms to adapt to inhospitable environments. These adaptation mechanisms include regulating nitrogen uptake system activity and modulating root system architecture (Peng et al., 2007; Tsay et al., 2011). Nitrogen uptake by plant roots involves multiple uptake systems (Vidal and Gutiérrez, 2008; Maathuis, 2009). Arabidopsis thaliana primarily acquires nitrogen in the form of NO3− using NO3− transporters from the NITRATE TRANSPORTER1 (NRT1) and NRT2 families. Some of them are induced by NO3− to ensure increased uptake when the substrate becomes available. The plant nitrogen status also affects NO3− uptake, with Gln acting as a negative feedback signal (Miller et al., 2008). NH4+ is another form of inorganic nitrogen that is taken up by a relatively large number of high- and low-affinity NH4+ transporters encoded by the AMMONIUM TRANSPORTER (AMT) family (Miller et al., 2009). However, the molecular mechanisms of nitrogen sensing, the nitrogen signaling network, and the developmental responses to nitrogen limitation are not well studied.
Glucosinolates are a group of plant secondary metabolites that are largely limited to species of the order Brassicales, which include nutritionally important Brassica spp. crops as well as the model plant Arabidopsis thaliana (Wittstock and Halkier, 2002). Glucosinolates are nitrogen-rich metabolites; therefore, nitrogen availability is crucial for their synthesis. The biosynthesis of glucosinolates includes three main processes: side chain elongation of amino acids, core structure formation, and modifications of the side chain (Grubb and Abel, 2006). ALKENYL HYDROXALKYL PRODUCING2 (AOP2) is responsible for the side chain modification of Met-derived glucosinolates (Kliebenstein et al., 2001; Grubb and Abel, 2006; Neal et al., 2010). Modifications of the glucosinolate side chain are particularly important because the structure of the side chain affects the biological activity of the glucosinolates and their degradation products (Hansen et al., 2007).
Although many nitrogen starvation-responsive miRNAs have been identified (Krapp et al., 2011; Liang et al., 2012), the functions for most of them are unclear under nitrogen starvation conditions. Our previous research revealed that miR826 is significantly induced by nitrogen starvation (Liang et al., 2012). Here, a novel miRNA (miR5090) was identified from the complementary transcripts of MIR826. Similar to miR826, miR5090 is also up-regulated by nitrogen starvation. Further investigation suggested that both miRNAs can improve the adaptation of Arabidopsis thaliana to nitrogen starvation by directly affecting the synthesis of Met-derived glucosinolates.
RESULTS
Identification of a Novel miRNA in Arabidopsis thaliana
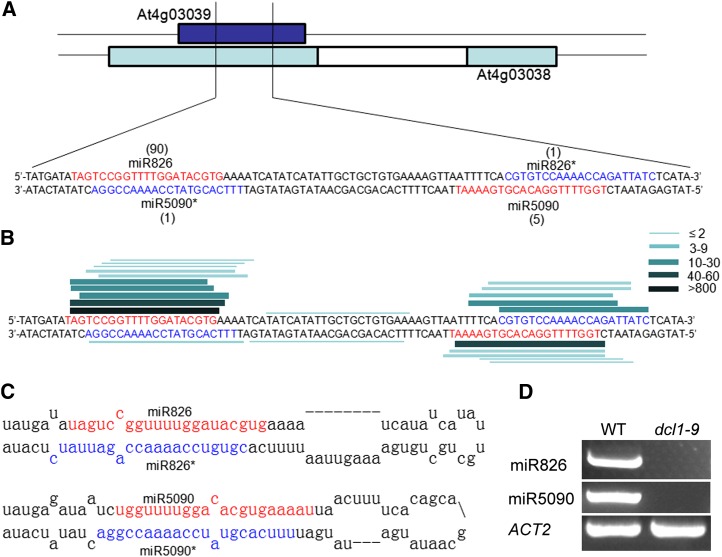
In our previous work (Liang et al., 2012), we used deep sequencing to analyze two small libraries derived from seedlings with or without nitrogen deprivation treatment. We found two small RNAs that completely matched with the complementary transcript (At4g03038) of the MIR826 gene (At4g03039; Fig. 1A). This transcript was annotated as another RNA. By prediction of its RNA secondary structure, a canonical miRNA precursor stem loop structure was produced. The two small RNAs perfectly correspond to the miRNA/miRNA* (the passenger strand) complex with 2-nucleotide 3′ overhangs (Fig. 1C). We speculated that At4g03038 is an miRNA-encoding gene. To investigate this hypothesis, we searched the Arabidopsis MPSS Plus database (http://mpss.udel.edu/at) for small RNA signatures that match with the stem loop sequence. We found that many small RNA reads are completely identical to the miR826 and mi826* sequences in the MIR826 gene, and that many small RNA reads match with the putative miRNA and miRNA* sequence in the At4g03038 transcript (Fig. 1B; Supplemental Fig. S1A). The combination of the stem loop structure and putative miRNA/ miRNA* sequence suggests that At4g03038 may be an authentic MIRNA gene. To confirm our hypothesis, we cloned the stem loop region downstream of the Cauliflower mosaic virus (CaMV) 35S promoter, and performed Agrobacterium tumefaciens-mediated transient expression experiments in Nicotiana benthamiana leaves. The antisense sequence of the putative miRNA was used as a probe to detect the expression of the miRNA by northern blotting. As shown in Supplemental Fig. S1B, compared with the empty vector with no signal detected, the CaMV 35S promoter-driven stem loop sequence accumulated a high abundance of mature small RNA sequence. To determine whether the small RNA products are DICER-LIKE1 (DCL1) dependent, we detected their abundance in the dcl1-9 mutant, revealing that these small RNAs are undetectable in the dcl1-9 mutant compared with the wild type (Fig. 1D). Analysis of the high-throughput sequencing data deposited in the Arabidopsis MPSS Plus database revealed that both miR826 and the novel miRNA sequences were undetectable in the dcl1-7 mutant, but were highly abundant in the wild type and dcl2/3/4 mutant (Supplemental Fig. S1C). Therefore, our results indicate that At4g03038 is an miRNA-encoding gene. This miRNA was designated as miR5090 according to the miRNA annotation system (Ambros et al., 2003).
Figure 1.
Identification of miR5090. A, Precursor sequences of miR826 and miR5090 with a tally of reads (from the nitrogen starvation library) mapping to the hairpins. B, Lines denote the sequences mapping to the miRNA precursors. The thickness and color of the lines correspond to the number of total reads representing each small RNA species in the Arabidopsis MPSS Plus database. C, The predicted miRNA precursors. D, RT-PCR analysis of miRNAs in the wild type (WT) and dcl1-9 mutant.
MIR826 and MIR5090 Are Recently Evolved miRNAs Originating in Arabidopsis thaliana Genomes by Duplication of AOP2
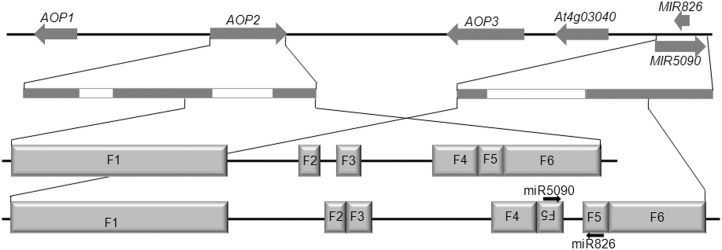
miR826 was identified as a recently evolved miRNA, which is only present in Arabidopsis thaliana. To investigate whether miR5090 is conserved in other plant species, we searched the miRBase database (http://www.mirbase.org) for potential homologs of miR5090. However, no miR5090 homolog was found. We further searched the National Center for Biotechnology Information nucleotide database (http://blast.ncbi.nlm.nih.gov) for highly similar sequences of miR5090 by BLASTN, and found that miR5090 only matches with the Arabidopsis thaliana AOP2 gene. Thus, the miR5090 sequence is specifically present in Arabidopsis thaliana. When the stem loop region sequence of miR5090 was used in a BLAST search for similar Arabidopsis thaliana transcripts, the only significant result was AOP2, which showed 89% similarity over a region of 278 nucleotides (E value = 4e−52) that extends well beyond the 21-nucleotide-long miR5090 pairing site. Target prediction (Dai and Zhao, 2011) results also indicated that AOP2 is the candidate target of miR5090. Allen et al. (2004) suggested that new miRNAs evolve through inverted duplication of target gene sequences. Therefore, we speculated that the MIR5090 gene originates from a duplication of its target, AOP2. To confirm our hypothesis, we compared the genome sequences of AOP2 and MIR5090. As shown in Figure 2, six regions (F1–F6) with a high similarity (>90%) are shared by AOP2 and MIR5090. Apparently, the MIR5090 gene is a duplicate of the AOP2 gene 3′ terminal region containing two exons and one intron. In contrast with AOP2, the MIR5090 gene contains an extra inverted F5 fragment (Fig. 2), which results in the stem loop structures in miR826 and miR5090 precursors (Fig. 1C). We also compared their flanking sequences and found no significant similarity. In the Arabidopsis thaliana chromosome, MIR826, MIR5090, and three AOP genes exist in a cluster (Fig. 2). By contrast, only two AOP genes were found in Arabidopsis lyrata, from which Arabidopsis thaliana is thought to be derived (Yogeeswaran et al., 2005). This suggested that MIR826 and MIR5090 are recently evolved miRNAs.
Figure 2.
Extended homology between miRNA genes and the AOP2 gene, suggestive of common origin. Three AOP genes and two miRNA genes are closely linked. The white bars and black bars indicate introns and exons. F1 to F6 correspond to the homologous sequences between the AOP2 gene and the miRNA genes.
AOP2 Is the Common Target of miR826 and miR5090
The AOP2 gene has been identified as the target of miR826 (Rajagopalan et al., 2006; Liang et al., 2012), which encodes a 2-oxoglutarate-dependent dioxygenase that is involved in glucosinolate biosynthesis. Interestingly, target prediction results suggested that AOP2 is also the target of miR5090. As shown in Figure 3A, the target site of miR5090 is shifted by 5 nucleotides from that of miR826. Usually, the target cleavage mediated by a plant miRNA occurs in the 10th nucleotide from the 5′ end of the miRNA (Allen et al., 2005). We identified two AOP2 cleavage sites by a 5′ RACE experiment from nitrogen-starved Arabidopsis thaliana seedlings, both of which are identical to the cleavage sites retrieved from degradome data (German et al., 2008; Liang et al., 2012) and perfectly match with the 10th nucleotides of miR826 and miR5090, respectively (Fig. 3A). Thus, these two cleavage products might result from cleavage by miR826 and miR5090. To further investigate whether both miRNAs mediate the putative cleavage of AOP2, we performed an Agrobacterium tumefaciens-mediated transient assay by coexpressing the miRNA genes (35S:MIR826 or 35S:MIR5090) with AOP2 (35S:AOP2) in N. benthamiana leaves. The results showed that AOP2 mRNA levels decreased to 10% or 30% when coexpressing with miR826 or miR5090 compared with the control level, respectively (Fig. 3C). To further confirm whether the cleavage of AOP2 occurs in the predicted target sites, several synonymous substitutions were introduced to generate miR826- and miR5090-resistant versions of AOP2 (35S:mAOP2; Fig. 3B). When miR826 or miR5090 were coexpressed with mAOP2, the mRNA levels of AOP2 were not affected. These results confirmed that AOP2 is the common target of miR826 and miR5090.
Figure 3.
AOP2 is the common target of miR826 and miR5090. A, Predicted miRNA/target duplex. Vertical arrows indicate the target cleavage positions. The number indicates the number of corresponding cleavage products from 5′ RACE experiment. B, Synonymous nucleotide substitutions in miRNA binding sites of AOP2. C, Cleavage of AOP2 transcripts by miR826 and miR5090 in planta. Constructs harboring the wild type or mutated AOP2 driven by the 35S promoter were coagroinoculated with the 35S:MIR826 or 35S:MIR5090 constructs in tobacco leaves. Empty vector was used as a negative control. Total RNAs were extracted after a 3-d inoculation and examined by qRT-PCR. The Student' t test indicated that the values marked by one asterisk are significantly different from the corresponding wild-type value (P < 0.01; n = 3).
The Transcript Abundance of miR826 and miR5090 Is Negatively Correlated with That of AOP2 in Response to Nitrogen Starvation
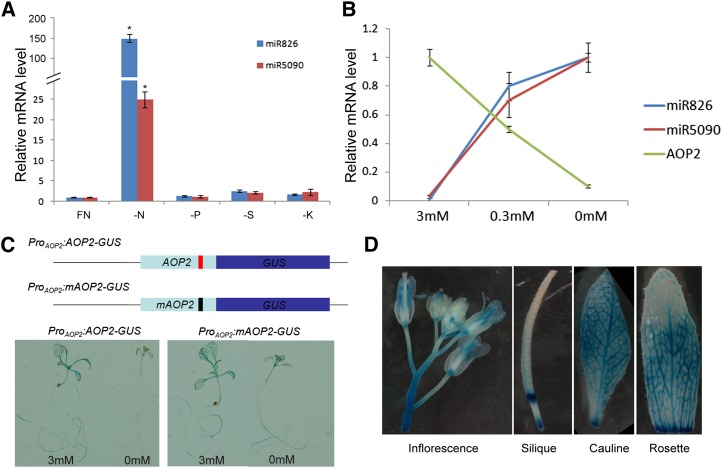
We previously found that miR826 transcript abundance was particularly high in the nitrogen deprivation library, and the miR5090 transcript was only found in the nitrogen deprivation library (Liang et al., 2012). To investigate whether both miRNAs are specifically responsive to nitrogen deprivation, we tested the expressions of miR826 and miR5090 in seedlings grown under different nutrient deprivation conditions. In contrast with the slight change caused by sulfur, potassium, or phosphorus deprivation, both miR826 and miR5090 were strongly induced by nitrogen starvation (Fig. 4A), indicating that miR826 and miR5090 were specifically up-regulated by nitrogen starvation. miRNAs suppress their target transcripts; therefore, the expression of miRNAs is usually inversely correlated with that of their targets. We thus detected the transcript levels of miR826, miR5090, and AOP2 transcripts in 10-d-old seedlings grown on medium supplemented with different nitrogen concentrations (nitrogen sufficient, 3 mm; nitrogen low, 0.3 mm; and nitrogen free, 0 mm). As expected, both miR826 and miR5090 expression levels increased with the decrease in nitrogen concentration, whereas AOP2 showed the reverse trend (Fig. 4B). Similar results were observed when roots and shoots were examined separately (Supplemental Fig. S2). These results suggested that AOP2 expression is negatively correlated with the expressions of miR826 and miR5090.
Figure 4.
AOP2 is regulated posttranscriptionally. A, Expression of miR826 and miR5090 under different nutrient deficiencies. FN, Full nutrient; K, potassium; N, nitrogen; P, phosphorus; S, sulfur. The Student’s t test indicated that the values marked by one asterisk are significantly different from the corresponding wild-type value (P < 0.01; n = 3). B, Expressions of miRNAs and AOP2 under nitrogen starvation. A and B, Ten-day-old seedlings were used for RNA extraction. The qRT-PCR analysis was repeated for three biological replicates, each of which consisted of three technical replicates. The error bars represent the sds from triplicate samples. C, GUS staining under nitrogen starvation conditions. The scheme represents two reporters with wild-type or mutated AOP2. Ten-d-old seedlings were used for GUS staining. D, GUS staining under different tissues and organs. The 2-month-old ProAOP2:AOP2-GUS reporter line was used for GUS staining.
However, we did not know whether the reduction in AOP2 expression directly resulted from the induction of miRNAs under nitrogen starvation conditions. To uncover the regulation of AOP2 by miRNAs, we prepared two reporters. First, the wild-type AOP2 complementary DNA (cDNA) was fused in frame with a GUS gene driven by the 2.9-kb genomic region upstream of the start codon of AOP2 (Fig. 4C). This wild-type reporter (ProAOP2:AOP2-GUS) would allow us to monitor AOP2 transcriptional and posttranscriptional regulation. In agreement with the quantitative reverse transcription PCR (qRT-PCR) results, ProAOP2:AOP2-GUS was strongly expressed in roots and shoots under nitrogen-sufficient conditions, whereas GUS staining was very weak under nitrogen-free conditions (Fig. 4C). To reveal the effect of miRNAs on the expression of AOP2, we prepared a second reporter, with mutated AOP2 (mAOP2) (Fig. 4C). Compared with nitrogen-sufficient conditions, the expression of ProAOP2:mAOP2-GUS was down-regulated only moderately under nitrogen-free conditions (Fig. 4C). GUS staining in the whole plant indicated that AOP2 was ubiquitously expressed in inflorescence, silique, cauline, and rosette leaves (Fig. 4D). These results demonstrated that under nitrogen starvation, the expression of AOP2 is suppressed by miRNAs at the posttranscriptional level.
Overexpression of miR826 or miR5090 Suppresses the Accumulation of the AOP2 Transcript
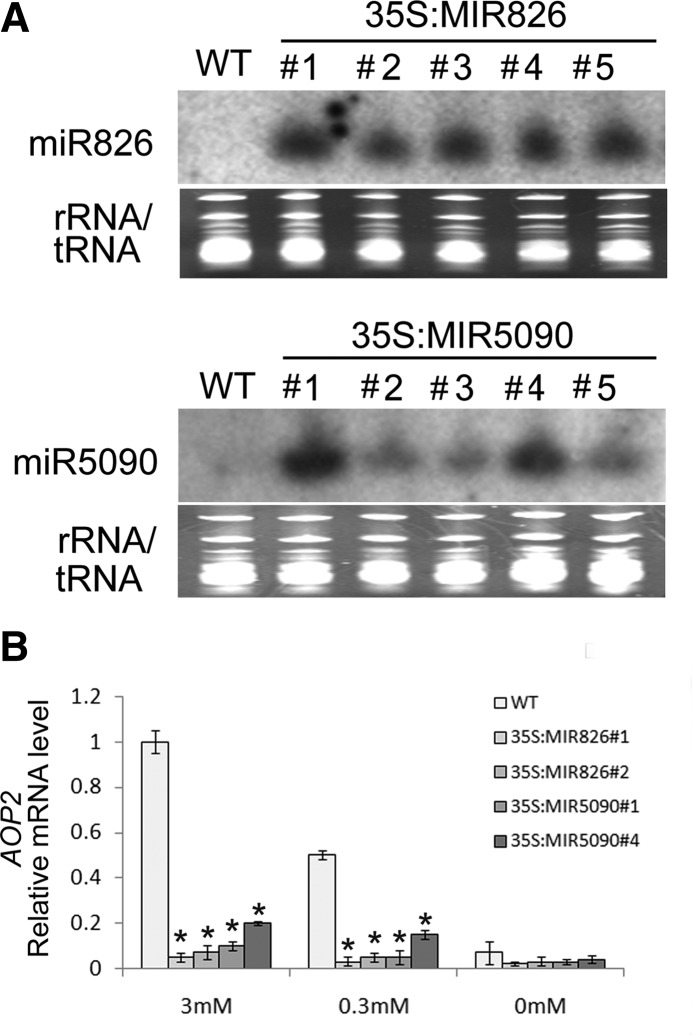
To further understand the functions of miR826 and miR5090, we generated transgenic plants overexpressing miR826 or miR5090. Twelve and eight independent transgenic lines for 35S:MIR826 and 35S:MIR5090 were obtained, respectively. We determined the miRNA expression levels of five independent lines for each genotype. The antisense DNA sequences of the two miRNAs were labeled with 32P and used as probes to detect the expression of these two miRNAs using northern blotting. Compared with the wild type, all of the transgenic plants we examined produced high quantities of mature miRNAs (Fig. 5A). Further analysis suggested that AOP2 transcript levels were dramatically down-regulated in the transgenic plants (Fig. 5B). When subjected to different nitrogen starvation conditions, the transcript abundance of AOP2 in transgenic plants remained low compared with the wild type (Fig. 5B). The overproduction of miR826 or miR5090 correlated well with the decreased AOP2 mRNA level in transgenic plants, suggesting that miR826 and miR5090 suppress AOP2 mRNA abundance.
Figure 5.
Expressions of miR826, miR5090, and AOP2 in wild-type (WT) and transgenic plants. A, Leaves from 4-week-old plants were used for RNA extraction. Ribosomal RNA/tRNA staining is shown as a loading control. Twenty micrograms of total RNA was used for northern blotting. Numbers 1 to 5 represent different transgenic lines. B, Expression of AOP2. Plants were grown for 10 d on MS medium with the indicated nitrogen concentrations. RNA was isolated from whole seedlings. The qRT-PCR analysis was repeated for three biological replicates, each of which consisted of three technical replicates. The error bars represent the sds from triplicate samples. A Student’s t test indicated that the values marked by one asterisk are significantly different from the corresponding wild-type value (P < 0.01; n = 3).
Transgenic Plants Mimic the Phenotypes of aop2 Mutants
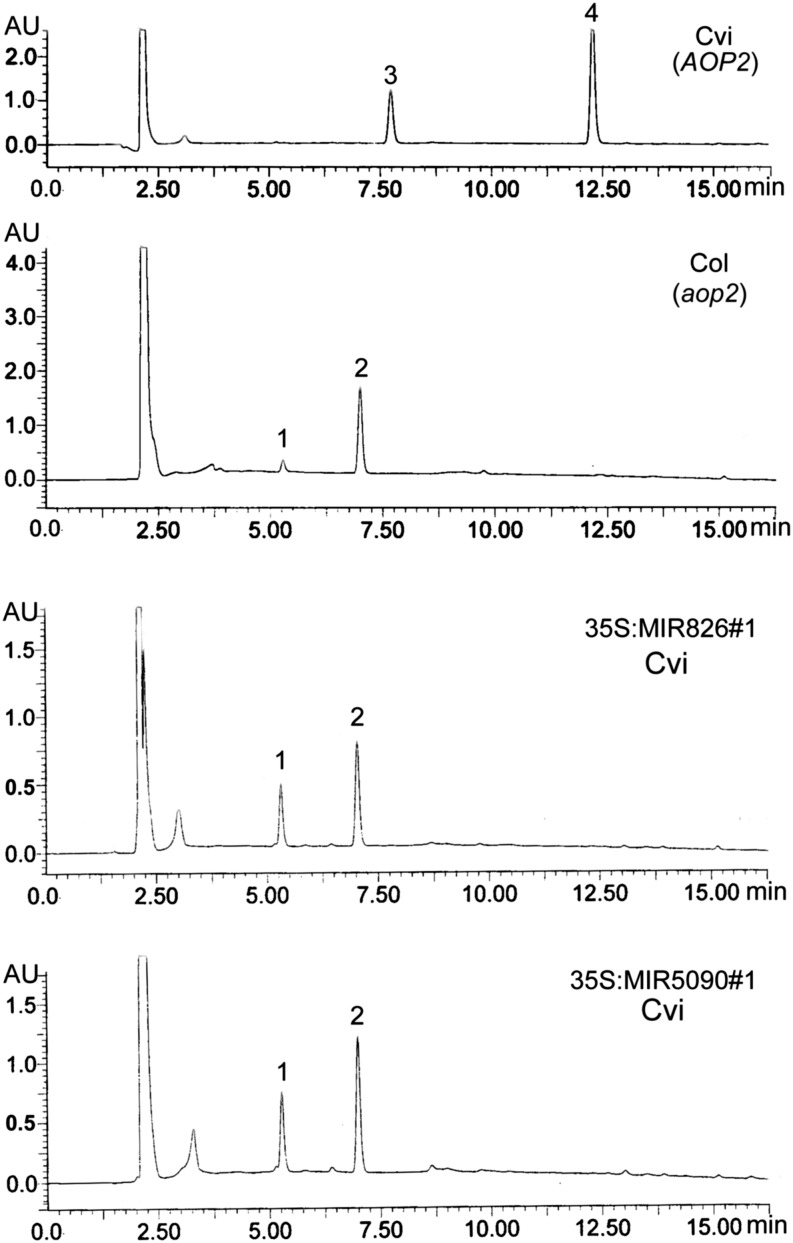
The AOP2 gene displays polymorphism across different Arabidopsis thaliana ecotypes (Kliebenstein et al., 2001). The ecotype Cape Verde Islands of Arabidopsis thaliana (Cvi) contains a functional AOP2 gene, whereas ecotype Columbia of Arabidopsis thaliana (Col) has a nonfunctional AOP2 gene resulting from a 5-bp deletion in its transcript. Therefore, all our experiments are performed in the Cvi ecotype. Given the significant reduction of AOP2 in transgenic plants, we expected that they could mimic the glucosinolate profiles of Col ecotype plants that contain a mutated aop2. Thus, we detected the glucosinolate profiles in miR826 and miR5090 transgenic plants to determine whether the composition of the glucosinolates had changed as a result of the down-regulation AOP2 expression. As shown in Figure 6, Cvi contains AOP2-catalyzed products 2-propenyl (7.7 min; peak 3) and 3-butenyl (12.2 min; peak 4) glucosinolates. By contrast, Col (a natural aop2 mutant) accumulated 3-methylsulfinylpropyl (5.2 min; peak 1) and 4-methylsulfinylbutyl (7.0 min; peak 2) glucosinolates, both of which are the substrates of AOP2 (Fig. 6). We then detected the glucosinolate profiles of miR826 and miR5090 transgenic plants: High levels of 3-methylsulfinylpropyl and 4-methylsulfinylbutyl glucosinolates, but not AOP2-catalyzed products, were detected. These results suggested that elevated miR826 or miR5090 expression suppresses the function of AOP2, resulting in the reduction of Met-derived glucosinolates.
Figure 6.
UPLC profile of glucosinolates from different plants. Peak 1 corresponds to 3-methylsulfinylpropyl glucosinolate. Peak 2 corresponds to 4-methylsulfinylbutyl glucosinolate. Peak 3 corresponds to 2-propenyl glucosinolate. Peak 4 corresponds to 3-butenyl glucosinolate.
Altered Expression of Genes Involved in Aliphatic Glucosinolate Synthesis
Glucosinolate biosynthesis involves three stages: 1) chain elongation of selected precursor amino acids (only Met and Phe), 2) formation of the core glucosinolate structure, and 3) secondary modifications of the amino acid side chain (Sønderby et al., 2010). AOP2 is a key regulator for glucosinolate synthesis, which is responsible for the secondary modifications of Met-derived glucosinolates that are the major components of aliphatic glucosinolates. Considering the down-regulation of AOP2 under nitrogen starvation conditions, we asked whether glucosinolate synthesis-associated genes would be affected by nitrogen starvation. Among the 31 genes (Table I), 19 decreased by 35% in the wild type under nitrogen-free conditions compared with that under nitrogen-sufficient conditions. Under both nitrogen-sufficient and nitrogen-deficient conditions, the down-regulated genes by nitrogen starvation always kept at lower levels in miRNA transgenic plants compared with the wild type. It suggested that both nitrogen starvation and AOP2 reduction caused down-regulation of glucosinolate synthesis-associated genes.
Table I. Relative expression of genes involved in aliphatic glucosinolate pathway.
| Gene Name | Gene Accession | Nitrogen-Sufficient Condition (3 mm) |
Nitrogen-Low Condition (0.3 mm) |
Nitrogen-Free Condition (0 mm) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild Type | 35S:MIR5090 | 35S:MIR826 | Wild Type | 35S:MIR5090 | 35S:MIR826 | Wild Type | 35S:MIR5090 | 35S:MIR826 | ||
| Side-chain elongation | ||||||||||
| BCAT3 | AT3G49680 | 1.00 | 0.97 | 0.70 | 0.93 | 0.76 | 0.68 | 0.85 | 0.78 | 0.65 |
| BCAT4 | AT3G19710 | 1.00 | 0.97 | 0.56 | 0.80 | 0.40 | 0.50 | 0.32 | 0.11 | 0.10 |
| BAT5 | AT4G12030 | 1.00 | 1.02 | 0.84 | 0.93 | 0.60 | 0.80 | 0.50 | 0.31 | 0.24 |
| LeuC | AT4G13430 | 1.00 | 0.90 | 0.73 | 1.25 | 0.87 | 0.77 | 0.90 | 0.67 | 0.61 |
| LeuD1 | AT2G43100 | 1.00 | 0.37 | 0.55 | 0.79 | 0.59 | 0.80 | 0.47 | 0.31 | 0.29 |
| LeuD2 | AT3G58990 | 1.00 | 1.00 | 0.67 | 1.06 | 0.79 | 0.77 | 0.61 | 0.51 | 0.33 |
| IPMDH1 | AT5G14200 | 1.00 | 1.02 | 0.63 | 0.87 | 0.61 | 0.66 | 0.47 | 0.36 | 0.25 |
| IPMDH2 | AT1G80560 | 1.00 | 0.69 | 0.69 | 0.99 | 1.08 | 1.21 | 0.91 | 0.89 | 1.07 |
| IPMDH3 | AT1G31180 | 1.00 | 0.59 | 0.41 | 0.91 | 0.63 | 0.74 | 0.62 | 0.33 | 0.43 |
| MAM1 | AT5G23010 | 1.00 | 0.73 | 0.51 | 0.68 | 0.43 | 0.55 | 0.60 | 0.18 | 0.12 |
| MAM3 | AT5G23020 | 1.00 | 0.13 | 0.17 | 0.19 | 0.14 | 0.11 | 0.41 | 0.16 | 0.18 |
| Core structure | ||||||||||
| CYP79F1 | AT1G16410 | 1.00 | 0.91 | 0.65 | 0.74 | 0.56 | 0.59 | 0.46 | 0.17 | 0.19 |
| CYP79F2 | AT1G16400 | 1.00 | 0.79 | 0.68 | 0.98 | 0.88 | 0.98 | 0.74 | 0.39 | 0.42 |
| CYP83A1 | AT4G13770 | 1.00 | 0.75 | 0.53 | 0.84 | 0.52 | 0.47 | 0.59 | 0.26 | 0.24 |
| GSTF11 | AT3G03190 | 1.00 | 0.91 | 0.79 | 0.95 | 0.62 | 0.77 | 0.37 | 0.25 | 0.26 |
| GSTU20 | AT1G78370 | 1.00 | 0.61 | 0.44 | 0.59 | 0.30 | 0.37 | 0.24 | 0.16 | 0.16 |
| GGP1 | AT4G30530 | 1.00 | 0.81 | 0.67 | 1.19 | 1.80 | 1.07 | 1.63 | 1.80 | 1.57 |
| SUR1 | AT2G20610 | 1.00 | 0.68 | 0.58 | 1.02 | 1.17 | 0.73 | 0.84 | 1.06 | 0.84 |
| UGT74C1 | AT2G31790 | 1.00 | 1.00 | 0.63 | 1.10 | 0.65 | 0.77 | 0.70 | 0.55 | 0.64 |
| ST5b | AT1G74090 | 1.00 | 0.94 | 0.80 | 1.02 | 0.83 | 0.66 | 0.76 | 0.72 | 0.57 |
| ST5c | AT1G18590 | 1.00 | 0.93 | 0.56 | 0.86 | 0.61 | 0.61 | 0.48 | 0.40 | 0.39 |
| Secondary modification | ||||||||||
| FMOGX-OX1 | AT1G65860 | 1.00 | 1.31 | 0.89 | 0.86 | 0.78 | 0.87 | 0.84 | 0.49 | 0.39 |
| FMOGX-OX2 | AT1G62540 | 1.00 | 1.53 | 1.08 | 2.32 | 1.94 | 2.95 | 2.97 | 2.24 | 4.23 |
| FMOGX-OX3 | AT1G62560 | 1.00 | 1.02 | 0.87 | 0.91 | 0.68 | 0.87 | 0.57 | 0.24 | 0.20 |
| FMOGX-OX4 | AT1G62570 | 1.00 | 1.01 | 0.92 | 1.98 | 2.70 | 2.67 | 7.37 | 5.98 | 5.35 |
| FMOGX-OX5 | AT1G12140 | 1.00 | 1.04 | 0.88 | 1.70 | 1.70 | 1.88 | 2.55 | 2.40 | 2.53 |
| GS-OH | AT4G03060 | 1.00 | 0.73 | 0.81 | 0.67 | 0.36 | 0.26 | 0.24 | 0.20 | 0.10 |
| AOP2 | AT2G25450 | 1.00 | 0.26 | 0.03 | 0.50 | 0.20 | 0.02 | 0.31 | 0.08 | 0.01 |
| Transcription factor | ||||||||||
| MYB28 | AT5G61420 | 1.00 | 1.48 | 1.19 | 0.78 | 0.54 | 0.86 | 1.00 | 0.90 | 0.65 |
| MYB29 | AT5G07690 | 1.00 | 1.09 | 0.78 | 0.84 | 0.59 | 0.61 | 0.39 | 0.19 | 0.16 |
| MYB76 | AT5G07700 | 1.00 | 0.93 | 0.80 | 0.61 | 0.32 | 0.36 | 0.27 | 0.24 | 0.10 |
| Unknown factor | ||||||||||
| AOP1 | 1.00 | 0.52 | 0.21 | 0.85 | 0.44 | 0.20 | 0.57 | 0.32 | 0.19 | |
miRNA Transgenic Plants Display Enhanced Tolerance to Nitrogen Starvation
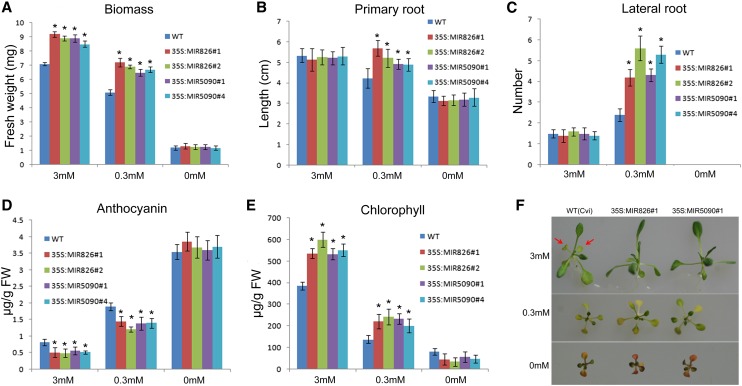
Considering the fact that miR826 and miR5090 are induced by nitrogen starvation, we asked whether they are involved in regulating the adaptation of Arabidopsis thaliana to nitrogen deficiency. Under nitrogen starvation conditions, plants often show physiological and developmental adaptation such as small stature, decreased primary root length, increased lateral root density, higher anthocyanin accumulation, and lower chlorophyll content (Maathuis, 2009; Tsay et al., 2011). Therefore, we evaluated the response of miRNA transgenic plants to nitrogen starvation. Wild-type and transgenic plants, which were grown on nitrogen-sufficient, nitrogen-low, and nitrogen-free agar medium, respectively, for 10 d, were used for the analysis. Under nitrogen-sufficient and nitrogen-low conditions, the fresh weight of transgenic plants was higher than that of the wild type (Fig. 7A). Although wild-type and transgenic plants displayed similar root systems under nitrogen-sufficient conditions, the transgenic plants generated longer primary roots and higher lateral root density than the wild type under nitrogen-low conditions (Fig. 7, B and C; Supplemental Fig. S3). With the reduction of nitrogen concentration, all plants produced more anthocyanin. However, under nitrogen-sufficient and nitrogen-low conditions, the anthocyanin concentration of transgenic plants was lower than in the wild type (Fig. 7D). By contrast, nitrogen starvation resulted in the reduction of chlorophyll concentration in all plants; however, under nitrogen-sufficient and nitrogen-low conditions, more chlorophyll was produced in the transgenic plants compared with the wild type (Fig. 7E). These results revealed that miRNA transgenic plants display enhanced tolerance to nitrogen limitation conditions, although no significant phenotypic difference was observed between wild-type and transgenic plants under nitrogen-free conditions. To determine whether transgenic plants were also tolerant to long-term nitrogen starvation, plants were grown on agar medium for 3 weeks. As shown in Figure 7F, wild-type plants generated more senescent leaves than transgenic plants under nitrogen-sufficient conditions. By contrast, transgenic plants were significantly bigger under nitrogen-low conditions than the wild type, despite similar senescent symptoms. We also determined the phenotypes of plant grown in soil and found that transgenic plants had a higher growth rate than wild-type plants (Supplemental Fig. S4). Total nitrogen measurement revealed that individual transgenic plants contained more nitrogen than individual wild-type plants, although their nitrogen concentrations were similar in both roots and shoots (Supplemental Fig. S5).
Figure 7.
Transgenic plants are less sensitive to nitrogen starvation. A, Biomass. Values are the means of three replicates of 20 plants. B, Primary root length. C, Lateral root density. D, Anthocyanin concentration. Shoots were harvested for analysis. E, Chlorophyll concentration. Shoots were harvested for analysis. A to E, Ten-d-old plants grown on plates were used for analysis. Error bars indicate the sds (n = 3). Asterisks indicate statistical significance at P < 0.01 compared with the wild type (Student’s t test). F, Three-week-old seedlings grown on medium with the indicated nitrogen concentrations.
Altered Responses to Nitrogen Limitation in Transgenic Plants
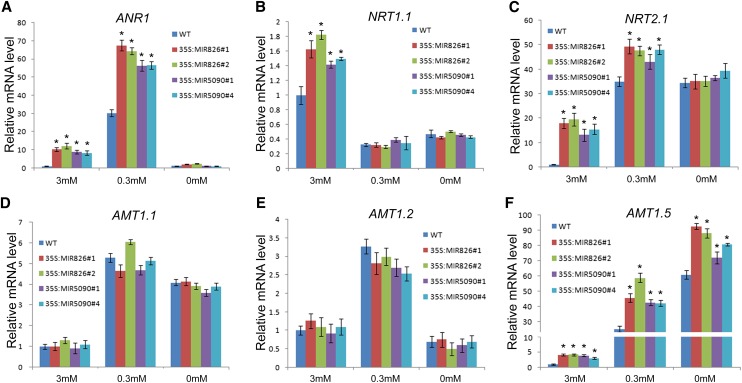
Considering the high biomass and lateral root density of transgenic plants, we speculated that transgenic plants might facilitate nitrogen uptake by the roots. An NO3−-triggered signaling pathway stimulates elongation of growing lateral roots (Zhang and Forde, 1998). This mechanism involves three genes: ARABIDOPSIS NITRATE-REGULATED1 (ANR1; Zhang and Forde, 1998), NRT1.1, and NRT2.1 (Remans et al., 2006a, 2006b). Expression analysis indicated that these genes were up-regulated in transgenic plants under nitrogen-sufficient conditions (Fig. 8, A and C). Under nitrogen-low conditions, the enhanced lateral root systems (Fig. 7C; Supplemental Fig. S3) of the transgenic plants correlated well with the elevated expression of ANR1 (Fig. 8A). NO3− and NH4+ are the two main forms of nitrogen nutrients in soils absorbed by plant roots. NRT2 transporters are responsible for NO3− uptake (Miller et al., 2009). Our results revealed that NRT2.1 expression was increased in transgenic plants under nitrogen-sufficient and nitrogen-low conditions (Fig. 8C), implying a stimulated nitrogen uptake activity. NH4+ uptake is attributed to AMT-type transporters (Yuan et al., 2007; Miller et al., 2009). We determined the expressions of AMT1.1, AMT1.2, and AMT1.5 (Fig. 8, D to F), demonstrating that only AMT1.5 was up-regulated in transgenic plants. Taken together, our results suggested that the nitrogen uptake system in transgenic plants may be improved, leading to enhanced tolerance to nitrogen limitation.
Figure 8.
Expression of nitrogen starvation-responsive genes. Roots of 10-d-old plants were used for expression analysis of ANR1 (A), NRT1.1 (B), NRT2.1 (C), AMT1.1 (D), AMT1.2 (E), and AMT1.5 (F). The qRT-PCR analysis was repeated for three biological replicates, each of which consisted of three technical replicates. The error bars represent the sds from triplicate samples. The Student’s t test indicated that the values marked by one asterisk are significantly different from the corresponding wild-type value (P < 0.01; n = 3).
DISCUSSION
Mineral nutrients are vital to plant growth and thus affect crop yield. Several miRNAs involved in nutrient homeostasis have been characterized such as miR169 for nitrogen (Zhao et al., 2011), miR395 for sulfur (Liang et al., 2010), miR397, miR398, miR408, and miR857 for copper (Abdel-Ghany and Pilon, 2008), miR399 for phosphorus (Chiou et al., 2006), and miR827 for nitrogen and phosphorus (Kant et al., 2011). Nitrogen, one essential macronutrient for plant growth, is often deficient in the environment where plants grow. Our previous research showed that many plant miRNAs are responsive to nitrogen starvation, among which miR826 was dramatically induced by nitrogen starvation (Liang et al., 2012). Here, we identified a novel miRNA gene (MIR5090) from the complementary transcript of MIR826. Like miR826, miR5090 was also significantly up-regulated by nitrogen starvation. Our results revealed that both miR826 and miR5090 regulate the adaptation of Arabidopsis thaliana to low nitrogen conditions by affecting glucosinolate synthesis.
MIR826 and MIR5090 Have Recently Evolved from Their Common Target, AOP2
More than 300 miRNAs were identified in Arabidopsis thaliana (miRBase database; http://www.mirbase.org), approximately one third of which are conserved across plant species. Generally, a conserved miRNA family contains more than one member. By contrast, there is only one member for miR826 and miR5090 families in Arabidopsis thaliana, implying that they are nonconserved miRNAs. In contrast with conserved miRNAs, miR826 and miR5090 are only identified in Arabidopsis thaliana. We could not find their potential orthologs in Arabidopsis lyrata, which shares over 80% of its miRNA genes with Arabidopsis thaliana (Fahlgren et al., 2010), implying that they are recently evolved miRNAs. To date, two hypotheses for the origins of miRNA have been proposed: One is the inverted duplication of the target (Allen et al., 2004), and the other is random sequence origin (Felippes et al., 2008). The DNA fragment containing the MIR826 and MIR5090 genes displays high similarity to the AOP2 gene (Fig. 2), indicating that they are evolved from an inverted duplication of the AOP2 sequence. Further sequence analysis revealed that a short DNA fragment (34 bp) is inversely duplicated, which contains the stem loop structure of two RNA transcripts (Pri-miR826 and Pri-miR5090). Three AOP genes are located in the same chromosome in a tandem manner in the Col ecotype (Fig. 2). The ecotype Landsberg erecta of Arabidopsis thaliana contains two AOP1 genes (AOP1.1 and AOP1.2) and one AOP3, but no AOP2. Both Col and Cvi contain one AOP1 gene and one AOP3 gene (in both ecotypes, AOP3 is transcriptionally silent due to natural variation; Kliebenstein et al., 2001). Thus, a recent local genome rearrangement event has caused the tandem repeat of AOP genes in Arabidopsis thaliana. Over time, mutational drift would lead to erosion of the extended similarities between the originating locus and the inverted repeat. However, the sequences of the MIR826 and MIR5090 genes show high similarity to a 3′ fragment of the AOP2 gene, implying that these two miRNAs evolved very recently. A similar example is the Arabidopsis thaliana-specific young miRNAs, miR161 and miR163, both of which are physically linked to their target loci and retain extended complementarity (Allen et al., 2004). Therefore, MIR826 and MIR5090 have recently evolved from the duplication of AOP2 genomic 3′ fragment.
AOP2 Is Down-Regulated by Nitrogen Starvation via miR826 and miR5090
As a suppressor of target genes, miRNAs often mediate the cleavage of their target transcripts (i.e. regulation occurs at the posttranscriptional level; Voinnet, 2009). Target prediction indicated that AOP2 is the common target of miR826 and miR5090. Our transient expression experiment revealed that both miRNAs are able to mediate the cleavage of AOP2 mRNAs. Moreover, the cleavage positions in AOP2 transcripts perfectly match with the predicted miRNA cleavage sites. When nucleotide mutations were introduced into the miRNA recognition sites, the mutated AOP2 mRNA became insensitive to miR826 and miR5090, suggesting that sequence complementarity of target sites are crucial for the regulation of AOP2 by miRNAs.
miRNA expression is inversely correlated with the expression of its target, unless that the miRNA and its target are expressed in different tissues or cell compartments (Voinnet, 2009). Upon a decrease in nitrogen concentration, expressions of miR826 and miR5090 were up-regulated, whereas AOP2 expression was down-regulated. This negative correlation implies that AOP2 expression is spatiotemporally consistent with its suppressors (miR826 and miR5090). The GUS reporter indicated that AOP2 is ubiquitously expressed in the whole plant (Fig. 4D). GUS expression from the wild-type reporter (ProAOP2:GUS-AOP2) was significantly repressed by nitrogen starvation, whereas that of mutated reporter (ProAOP2:GUS-mAOP2) was less affected by nitrogen starvation, indicating that AOP2 is suppressed by miR826 and miR5090 at the posttranscriptional level.
The genes from the same family are often targeted by miRNAs from different families. For example, miR160 and miR167 target ARF genes (Wang et al., 2005; Yang et al., 2006); miR397, miR408, and miR857 target Laccase genes (Abdel-Ghany and Pilon, 2008); and miR168 and miR403 target AGO genes (Allen et al., 2005). However, it does not happen frequently that one gene is targeted by more than one miRNA. In Arabidopsis thaliana, the CHX18 gene is targeted by miR780 and miR856 (Fahlgren et al., 2007). Jeong et al. (2011) demonstrated that miR156a and miR529a target SPL14 in rice (Oryza sativa). Our study established that miR826 and miR5090 can target the same target (AOP2). These two miRNAs may have different functions under different conditions, because, in addition to nitrogen starvation, the expression of AOP2 is also affected by light and dark (Neal et al., 2010). Alternatively, these two recently evolved miRNAs are currently undergoing evolutionary selection. As revealed by Fahlgren et al. (2007), MIRNA genes undergo relatively frequent birth and death, with only a subset being stabilized by integration into regulatory networks.
Overexpression of miR826 and miR5090 Results in Reduced Accumulation of Met-Derived Glucosinolates
Our results suggested that AOP2 is the common target of miR826 and miR5090. Compared with wild-type plants, AOP2 in miR826 and miR5090 transgenic plants is strongly down-regulated, regardless of the nitrogen status of the environment. Hence, it is expected that transgenic plants can phenocopy aop2 mutants. AOP2 functions in the side chain modification of Met-derived glucosinolates (Kliebenstein et al., 2001; Neal et al., 2010). Loss of function of AOP2 led to considerably lower accumulation of Met-derived glucosinolates than in an Arabidopsis thaliana ecotype with one functional AOP2 allele (Kliebenstein et al., 2001; Wentzell et al., 2007). Our results also confirmed that aop2 mutant accumulates much less alkenyl glucosinolates (Met-derived glucosinolates; Fig. 6). As expected, miRNA transgenic plants have a very similar glucosinolate profile to aop2 mutants. Therefore, overexpression of miRNAs is sufficient to reduce the accumulation of Met-derived glucosinolates. Our work provides a molecular tool for breeding of Brassica spp. vegetable crops with decreased levels of Met-derived glucosinolates, which has implications for production of functional foods enriched with particular glucosinolates.
Loss of Function of AOP2 Causes Improved Nitrogen Starvation Adaptation in Arabidopsis thaliana
In our nitrogen starvation adaptation experiments, transgenic plants displayed significant growth advantages compared with the wild type. Corresponding to the phenotypes, the expressions of nitrogen starvation-responsive genes were altered in transgenic plants (Fig. 8). The increases in the expressions of nitrogen starvation-responsive genes may stimulate nitrogen uptake abilities of transgenic plants because the total nitrogen amount of individual transgenic plants was higher than that of individual wild-type plants (Supplemental Fig. S5). Under nitrogen-sufficient conditions, no morphological difference was observed for the roots of transgenic plants and the wild type. By contrast, under the nitrogen-low conditions, transgenic plants had longer primary roots and more lateral roots than the wild type. In agreement with root phenotypes, NRT2.1, a key regulator in root development under nitrogen-limited conditions (Remans et al., 2006b), increased in transgenic plants compared with the wild type. Therefore, in addition to the increased expression of nitrogen transporters, transgenic plants also enhanced their root systems for adaptation to nitrogen-low conditions. Despite having the same origin, the mature sequence of miR826 is different from that of miR5090. However, miR826 transgenic plants displayed phenotypes nearly identical to that of miR5090 transgenic plants. The only common feature for miR826 and miR5090 is that they have the same target gene, AOP2. Therefore, loss of function of AOP2 in transgenic plants causes the enhanced adaptation to nitrogen starvation. As a key enzyme in glucosinolate synthesis, AOP2 is the major regulator for aliphatic glucosinolate accumulation (Wentzell et al., 2007). An AOP2 null variant shows up to 3-fold fewer aliphatic glucosinolates compared with the functional AOP2 variant (Kliebenstein et al., 2001). In miRNA transgenic plants, the function of AOP2 was dramatically suppressed, which may reduce the consumption of nitrogen used for glucosinolate synthesis, thereby increasing the synthesis of nitrogen-containing metabolites that are necessary for plant growth and development under nitrogen starvation conditions. Meanwhile, nitrogen is one of the major components that are integrated into glucosinolates; therefore, it is possible that the reduction of glucosinolates promotes plants to acquire more nitrogen to meet the demand of glucosinolate synthesis. Correspondingly, nitrogen starvation also repressed most glucosinolate synthesis-associated genes (Table I). Under nitrogen-sufficient conditions, the expression levels of glucosinolate synthesis-associated genes were lower in miRNA transgenic plants than in the wild type, implying that the suppression of AOP2 by miRNAs mimics nitrogen starvation. Indeed, as shown in Figure 8, several nitrogen starvation-responsive genes were up-regulated in miRNA transgenic plants. A similar negative feedback regulation was reported, in which nitrogen influx increased when Gln synthesis was blocked (Rawat et al., 1999). Given that the loss of function of AOP2 caused the enhanced nitrogen starvation adaptation, we also compared mAOP2 transgenic plants with the wild type under nitrogen starvation conditions and found that they showed similar nitrogen starvation adaptation (data not shown), which implied that elevated AOP2 is not sufficient to raise nitrogen consumption. Plants have evolved diverse mechanisms to adapt to nitrogen starvation conditions. Apparently, reduction of nitrogen consumption and increased nitrogen absorption is an efficient strategy to maintain normal growth and development of plants when nitrogen is unavailable. Our work suggested that Arabidopsis thaliana plants have evolved new miRNAs that affect glucosinolate synthesis, leading to improved adaptation to nitrogen starvation conditions.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Cape Verde Islands (Cvi) was used as the wild-type plant in all of our experiments. The seeds were surface-sterilized with 20% bleach and washed three times with sterile water. Sterilized seeds were suspended in 0.1% agarose and plated on Murashige and Skoog (MS) medium. After vernalization for 2 d in the dark at 4°C, the plates were transferred to the culture room at 22°C under a 16-h light/8-h dark photoperiod. For determination of glucosinolate content, 7-d-old seedlings were planted in soil maintained in growth chambers at 22°C and 75% humidity under a 16-h light/8-h dark photoperiod. For observation of root phenotypes, seedlings were grown on vertical MS agar medium containing 0.8% agar. Nitrogen content in the medium was 3 mm (1 mm NH4NO3 and 1 mm KNO3), 0.3 mm (0.1 mm NH4NO3 and 0.1 mm KNO3), and nitrogen free, respectively. To evaluate the seed batch variation, homozygous T3 and T4 generation transgenic plants were respectively used for evaluation of phenotypes.
5′ RACE
Following the manufacturer’s instructions for the SMARTer RACE cDNA Amplification Kit (Clontech), 1 μg of total RNA isolated from seedlings grown on nitrogen-free MS medium was used for reverse transcription. Gene-specific primers (designed according to the protocol) and the UPM primer (provided by a kit) were used to conduct PCRs, and purified PCR products were sequenced.
Construct Generation
The putative promoters of AOP2 were amplified from Arabidopsis thaliana (Cvi) genomic DNA using primers Pro-AOP2-F and Pro-AOP2-R (Supplemental Table S1). The fused ProAOP2-GUS-(m)AOP2 was cloned into the pOCA28 vector containing a kanamycin resistance gene. The genomic sequences containing the stem loops of MIR826 or MIR5090 were used as synthetic precursor sequences. The sequences were amplified from Arabidopsis thaliana genomic DNA by PCR using primers PremiR826-F, PremiR826-R, PremiR5090-F, and PremiR5090-R (Supplemental Table S1). The PCR products of the precursor sequence were cloned into the pMD18-T vector (http://www.takara.com.cn; Takara) and confirmed by sequencing. The miR826 and miR5090 precursors were subcloned into the pOCA30 vector containing the CaMV 35S promoter. All of the constructs were then transformed into Agrobacterium tumefaciens strain GV3101. Arabidopsis thaliana transformation was performed by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected using 50 µg/mL kanamycin.
Histochemical GUS Staining
Plant samples were immersed immediately in 1.5 mL staining solution containing 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (Sigma) in 0.1 m sodium phosphate buffer (pH 7.3). The reaction was performed in the dark at 37°C until a blue-indigo color appeared. After the reaction, seedlings were rinsed in 0.1 m sodium phosphate buffer (pH 7.3). The samples were then rinsed twice in 70% ethanol to remove chlorophylls.
Gene Expression Analysis
For gene expression analysis, plants were grown on MS medium with the indicated nitrogen concentrations for 10 d.
Total RNA was extracted with the Trizol reagent (Invitrogen). Low-Mr RNAs were separated by electrophoresis through denaturing 15% polyacrylamide gels, and miRNA gel-blot hybridizations were performed as described previously (Liang et al., 2010). DNA oligonucleotides complementary to miR826 or miR5090 were end labeled with [α -32P] dATP using T4 polynucleotide kinase and used for hybridizations.
For real-time reverse transcription PCR (RT-PCR), 0.5 µg of total RNA was reverse transcribed using an oligo(dT)18 primer (Fermentas) in a 20-µL reaction mixture with RevertAid M-MuLV reverse transcriptase (Fermentas). After heat inactivation, a 1-µL aliquot was used for real-time qRT-PCR. All qRT-PCR analyses were performed using a Lightcycler FastStart DNA Master SYBR Green I kit on a Roche LightCycler real-time PCR machine, according to the manufacturer’s instructions. Real-time RT-PCR for miRNAs was detected by stem loop RT-PCR. To produce miRNA-fused stem loop cDNA, 0.5 μg of total RNA was used for the reverse transcription, with miRNA mature sequence-specific stem loop reverse transcription primers designed according to the stem loop RT-PCR protocol (Varkonyi-Gasic et al., 2007). ACTIN2 (AT3G18780) was used as a control for qRT-PCR. The primers used in qRT-PCR are listed in Supplemental Table S1.
Transient Expression in Nicotiana benthamiana
Constructs were transformed into Agrobacterium tumefaciens strain EHA105 and selected on Luria-Bertani medium containing rifampicin at 50 μg/mL and spectinomycin at 100 mg/L. Agrobacterium tumefaciens cells were then infiltrated into leaves of N. benthamiana. For coinfiltration experiments, equal volumes of an Agrobacterium tumefaciens culture containing 35S:MIR826, 35S:MIR5090, or vector (OD600 = 1.75) and 35S:(m)AOP2 (OD600 = 0.25) were mixed before infiltration into N. benthamiana leaves. Leaves were harvested 3 d after infiltration, and total RNA was extracted for small RNA blotting and real-time RT-PCR experiments.
Measurement of Chlorophyll Content
Chlorophyll contents were measured as described by Woodward and Bennett (2005). The pigments from leaves of 10-d-old seedlings were extracted with 5 mL of dimethylformamide for 24 h in the dark at 4°C, and the optical densities (OD664 and OD647) for each sample were measured. The chlorophyll content was calculated as follows: ([OD664 × 7.04] + [OD647 × 20.27]) ×5 ⁄sample weight (in grams) = micrograms chlorophyll/gram fresh weight.
Measurement of Anthocyanin Content
Ten-day-old seedlings grown on MS medium with the indicated nitrogen concentrations were used for anthocyanin analysis. Anthocyanin contents were measured as previously described (Rabino and Mancinelli, 1986). The pigments were extracted from leaves with 99:1 methanol:HCl (v/v) overnight at 4°C. The OD530 and OD657 for each sample were measured, and OD530 − 0.25 × OD657 was used to compensate for the contribution of chlorophyll and its products to the absorption at 530 nm.
Nitrogen Content Analysis
The shoots and roots of plants grown in soil for 4 or 5 weeks were separately harvested and dried at 65°C for 3 d. The samples were milled to a fine powder for nitrogen analysis. Nitrogen analysis was performed using a carbon and nitrogen analyzer (Vario MAX CN; Elementar Analysensysteme).
Glucosinolate Extraction
Glucosinolate extraction was performed as described by Kliebenstein et al. (2001) and Neal et al. (2010). Leaves of 5-week-old wild-type and transgenic plants were harvested, freeze-dried, and then ground to powder. Then 250 mg of the powder was suspended in 5 mL of 70% methanol. After incubation at 70°C for 20 min, 1 mL Ba(OAc)2 (0.4 mol/L) was added and centrifuged at 3000 rpm for 5 min, and the supernatant was added to DEAE-Sephadex A25. The column was then washed twice with water and twice with 1 mL of 20 mm sodium acetate. Sulfatase solution (75 μL), prepared as described by Graser et al. (2000), was then added to the column and left to stand overnight. Desulfonated glucosinolates were eluted in 1-mL aliquots of deionized water and analyzed by ultra-performance liquid chromatography (UPLC)-tandem mass spectrometry.
UPLC-Tandem Mass Spectrometry Analysis of Glucosinolates
Glucosinolate samples were analyzed using a Waters ACQUITY UPLC system and Xevo TQ-S mass spectrometer. Samples (20 μΛ) were separated using an Agilent Zorbax SB-C18 column (4.6 × 250 mm i.d., 5 μm particle size) operated at 1 mL/min at 30°C using the following separation gradient described by Neal et al. (2010): solvent A: H2O; solvent B: MeCN: 1.5% to 5% (v/v) B (6 min), 5% to 7% (v/v) B (2 min), 7% to 21% (v/v) B (10 min), 21% to 29% (v/v) B (5 min), 29% to 57% (v/v) B (14 min), followed by a cleaning cycle: 57% to 93% (v/v) B (2 min), 5 min hold, 93% to 1. 5% (v/v) B (3 min), 6 min hold. Eluting compounds were monitored at 229 nm. The mass spectral analysis of glucosinolates was obtained with positive electrospray ionization (ESI) in multiple reaction monitoring mode. The ESI source was operated at 4 kV, and the sample cone was operated at 20 V. Nitrogen was used both as bath gas (100°C; 250 L/h) and nebulizing gas (15 L/h). ESI spectra were recorded in the mass range of a 100 to 800 mass-to-charge ratio. Mass spectra of 3-methylsulfinylpropyl, 4-methylsulfinylbutyl, 2-propenyl, and 3-butenyl glucosinolates were analyzed by detecting a single M+Na+ specific for the glucosinolate being tested. 2-propenyl glucosinolate in the samples was further identified with its standard purchased from Sigma-Aldrich.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At4g03060 (AOP2), At4g03038 (miR5090), and At4g03039 (miR826).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of small RNA sequences.
Supplemental Figure S2. Expression of miRNAs and AOP2 under nitrogen starvation.
Supplemental Figure S3. Phenotypes of transgenic plants grown on plates.
Supplemental Figure S4. Phenotypes of transgenic plants grown in soil.
Supplemental Figure S5. Nitrogen contents of wild-type and transgenic plants.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank the editor and two anonymous reviewers for their constructive comments, which helped us to improve the manuscript, the Biogeochemical Laboratory and Central Laboratory (Xishuangbanna Tropica Botanical Garden) for their assistance in the determination of nitrogen contents, and the Analytical Instrumentation Center (Kunming Institute of Botany) for UPLC-tandem mass spectrometry analysis of glucosinolates.
Glossary
- miRNA
microRNA
- miRNA*
the passenger strand
- CaMV
Cauliflower mosaic virus
- cDNA
complementary DNA
- qRT-PCR
quantitative reverse transcription PCR
- Cvi
ecotype Cape Verde Islands of Arabidopsis thaliana
- Col
ecotype Columbia of Arabidopsis thaliana
- MS
Murashige and Skoog
- RT-PCR
reverse transcription PCR
- UPLC
ultra-performance liquid chromatography
- ESI
electrospray ionization
References
- Abdel-Ghany SE, Pilon M. (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem 283: 15932–15945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36: 1282–1290 [DOI] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. (2003) A uniform system for microRNA annotation. RNA 9: 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauclair L, Yu A, Bouché N. (2010) microRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J 62: 454–462 [DOI] [PubMed] [Google Scholar]
- Chen XM. (2005) MicroRNA biogenesis and function in plants. FEBS Lett 579: 5923–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL. (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dai X, Zhao PX. (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39: W155-W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Jogdeo S, Kasschau KD, Sullivan CM, Chapman EJ, Laubinger S, Smith LM, Dasenko M, Givan SA, Weigel D, et al. (2010) MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22: 1074–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes FF, Schneeberger K, Dezulian T, Huson DH, Weigel D. (2008) Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA 14: 2455–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Graser G, Schneider B, Oldham NJ, Gershenzon J. (2000) The methionine chain elongation pathway in the biosynthesis of glucosinolates in Eruca sativa (Brassicaceae). Arch Biochem Biophys 378: 411–419 [DOI] [PubMed] [Google Scholar]
- Grubb CD, Abel S. (2006) Glucosinolate metabolism and its control. Trends Plant Sci 11: 89–100 [DOI] [PubMed] [Google Scholar]
- Hansen BG, Kliebenstein DJ, Halkier BA. (2007) Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J 50: 902–910 [DOI] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ. (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DH, Park S, Zhai J, Gurazada SGR, De Paoli E, Meyers BC, Green PJ. (2011) Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23: 4185–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Peng M, Rothstein SJ. (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Zhu JK, Zhu JH. (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T. (2001) Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Berthomé R, Orsel M, Mercey-Boutet S, Yu A, Castaings L, Elftieh S, Major H, Renou JP, Daniel-Vedele F. (2011) Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol 157: 1255–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, He H, Yu D. (2012) Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS ONE 7: e48951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Yang FX, Yu DQ. (2010) MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J 62: 1046–1057 [DOI] [PubMed] [Google Scholar]
- Lu XY, Huang XL. (2008) Plant miRNAs and abiotic stress responses. Biochem Biophys Res Commun 368: 458–462 [DOI] [PubMed] [Google Scholar]
- Maathuis FJ. (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12: 250–258 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Fan X, Shen Q, Smith SJ. (2008) Amino acids and nitrate as signals for the regulation of nitrogen acquisition. J Exp Bot 59: 111–119 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Shen Q, Xu G. (2009) Freeways in the plant: transporters for N, P and S and their regulation. Curr Opin Plant Biol 12: 284–290 [DOI] [PubMed] [Google Scholar]
- Neal CS, Fredericks DP, Griffiths CA, Neale AD. (2010) The characterisation of AOP2: a gene associated with the biosynthesis of aliphatic alkenyl glucosinolates in Arabidopsis thaliana. BMC Plant Biol 10: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu XL, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR. (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng MS, Bi YM, Zhu T, Rothstein SJ. (2007) Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol Biol 65: 775–797 [DOI] [PubMed] [Google Scholar]
- Rabino I, Mancinelli AL. (1986) Light, temperature, and anthocyanin production. Plant Physiol 81: 922–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat SR, Silim SN, Kronzucker HJ, Siddiqi MY, Glass AD. (1999) AtAMT1 gene expression and NH4+ uptake in roots of Arabidopsis thaliana: evidence for regulation by root glutamine levels. Plant J 19: 143–152 [DOI] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A. (2006a) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA 103: 19206–19211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A. (2006b) A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol 140: 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby IE, Geu-Flores F, Halkier BA. (2010) Biosynthesis of glucosinolates—gene discovery and beyond. Trends Plant Sci 15: 283–290 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Ho CH, Chen HY, Lin SH. (2011) Integration of nitrogen and potassium signaling. Annu Rev Plant Biol 62: 207–226 [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Gutiérrez RA. (2008) A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol 11: 521–529 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY. (2005) Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17: 2204–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Hsu PK, Tsay YF. (2012) Uptake, allocation and signaling of nitrate. Trends Plant Sci 17: 458–467 [DOI] [PubMed] [Google Scholar]
- Wentzell AM, Rowe HC, Hansen BG, Ticconi C, Halkier BA, Kliebenstein DJ. (2007) Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet 3: 1687–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA. (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7: 263–270 [DOI] [PubMed] [Google Scholar]
- Woodward AJ, Bennett IJ. (2005) The effect of salt stress and abscisic acid on proline production, chlorophyll content and growth of in vitro propagated shoots of Eucalyptus camaldulenis. Plant Cell Tissue Organ Cult 82: 189–200 [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Han SJ, Yoon EK, Lee WS. (2006) ‘Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells’. Nucleic Acids Res 34: 1892–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M. (2007) Regulation of copper homeostasis by micro-RNA in Arabidopsis. J Biol Chem 282: 16369–16378 [DOI] [PubMed] [Google Scholar]
- Yogeeswaran K, Frary A, York TL, Amenta A, Lesser AH, Nasrallah JB, Tanksley SD, Nasrallah ME. (2005) Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res 15: 505–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Loqué D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wirén N. (2007) The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19: 2636–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG. (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhao M, Ding H, Zhu JK, Zhang F, Li WX. (2011) Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol 190: 906–915 [DOI] [PMC free article] [PubMed] [Google Scholar]