Abstract
Background
V(D)J recombination takes place during lymphocyte development to generate a large repertoire of T- and B-cell receptors. Mutations in recombination-activating gene 1 (RAG1) and RAG2 result in loss or reduction of V(D)J recombination. It is known that different mutations in RAG genes vary in residual recombinase activity and give rise to a broad spectrum of clinical phenotypes.
Objective
We sought to study the immunologic mechanisms causing the clinical spectrum of RAG deficiency.
Methods
We included 22 patients with similar RAG1 mutations (c.519delT or c.368_369delAA) resulting in N-terminal truncated RAG1 protein with residual recombination activity but presenting with different clinical phenotypes. We studied precursor B-cell development, immunoglobulin and T-cell receptor repertoire formation, receptor editing, and B- and T-cell numbers.
Results
Clinically, patients were divided into 3 main categories: T−B− severe combined immunodeficiency, Omenn syndrome, and combined immunodeficiency. All patients showed a block in the precursor B-cell development, low B- and T-cell numbers, normal immunoglobulin gene use, limited B- and T-cell repertoires, and slightly impaired receptor editing.
Conclusion
This study demonstrates that similar RAG mutations can result in similar immunobiological effects but different clinical phenotypes, indicating that the level of residual recombinase activity is not the only determinant for clinical outcome. We postulate a model in which the type and moment of antigenic pressure affect the clinical phenotypes of these patients.
Key words: RAG deficiency, V(D)J recombination, B- and T-cell receptor repertoire, receptor editing, autoimmunity, next generation sequencing, immune repertoire analysis
Abbreviations used: BM, Bone marrow; CDR3, Complementary determining region 3; CID, Combined immunodeficiency; OS, Omenn syndrome; PB, Peripheral blood; RAG, Recombination-activating gene; RAGD, Recombination-activating gene deficiency; SCID, Severe combined immunodeficiency; TR, T-cell receptor; TRB, T-cell receptor β; TREC, T-cell receptor excision circle
Defects in V(D)J recombination result in a block in B- and T-cell differentiation because formation of immunoglobulin and T-cell receptors (TRs) is perturbed.1 This results in a combined immunodeficiency (CID) of B and T cells. V(D)J recombination is initiated by the recombination-activating gene (RAG) 1 and RAG2 proteins by creating double-stranded breaks in the immunoglobulin and TR loci. Subsequently, these breaks are processed and repaired by proteins involved in nonhomologous end joining. Thus far, genetic defects have been identified in the RAG1, RAG2, Artemis, ligase IV (LIG4), XLF (Cernunnos), and DNA-PKcs genes.2, 3, 4, 5, 6, 7, 8 The immunologic phenotypes and clinical presentations of these mutations are different, depending on the type of genetic defect (ie, null mutations or hypomorphic mutations with residual V[D]J recombination activity). Especially for the RAG genes, many different mutations have been described that give rise to residual activity of the mutated RAG protein.9 Different RAG mutations can result in a broad spectrum of clinical phenotypes, including severe combined immunodeficiency (SCID), RAG deficiency (RAGD) with skin inflammation and αβ T-cell expansion (classical Omenn syndrome [OS]), RAGD with skin inflammation but without T-cell expansion (incomplete OS), RAGD with maternofetal transfusion, RAGD with γδ T-cell expansion, late-onset SCID, RAGD with granulomas, and RAGD with CD4 cytopenia and thymus hypoplasia.9, 10 This broad spectrum of clinical phenotypes impedes timely recognition of RAGD and might thus delay treatment (hematopoietic stem cell transplantation).
In this study we selected 22 patients with RAGD with similar N-terminal truncating RAG1 mutations to study the effect of a similar mutation on the clinical phenotype. These patients could be divided into 3 main clinical phenotypes (ie, SCID, OS, and CID, which includes the other phenotypes). We studied whether key immunologic parameters (eg, precursor B-cell development, B- and T-cell numbers, and B- and T-cell repertoire) might explain the differences in clinical phenotypes.
Methods
Cell samples and flow cytometric immunophenotyping
Peripheral blood (PB), bone marrow (BM), and clinical data were obtained according to the guidelines of the Medical Ethics Committee of the Erasmus MC Rotterdam. Flow cytometric analysis was performed, as previously described.8, 11, 12
RAG analysis and in vitro V(D)J recombination assay
The RAG1 and RAG2 genes were amplified by means of PCR and sequenced, as previously described.13 The level of recombination activity of the RAG1 expression constructs was determined by using the recombination plasmid pDVG93, as described previously.10, 13 A TaqMan-based realtime quantitative (RQ)-PCR was used to measure RAG1 and RAG2 transcription levels in BM mononuclear cells, as described previously.14
T-cell receptor β analysis
T-cell receptor β (TRB) gene rearrangements were studied, as described previously.15
Sequence analysis of Vκ and Jκ genes
Vκ-Cκ junctions were amplified in a multiplex PCR by using primers specific for Vκ1-5 families (VκI: 5′-GTAGGAGACAGAGTCACCATCACT-3′, VκII: 5′-TGGAGAGCCGGCCTCCA-TCTC-3′, VκIII: 5′-GGGAAAGAGCCACCCTCTCCTG-3′, and VκIV: 5′-GGCGAGAGGGCC-ACCATCAAC-3′) and a Cκ primer (5′-ACTTTGGCCTCTCTGGATA-3′). PCR products were cloned in the pGEM-Teasy vector (Promega, Madison, Wis) and prepared for sequencing on the ABI Prism 3130 XL fluorescent sequencer (Applied Biosystems, Foster City, Calif). Obtained sequences were analyzed with the IMGT database (http://imgt.cines.fr/) to assign the Vκ and Jκ genes.16, 17 The productive and unique sequences were used to determine the frequency of the Vκ and Jκ genes.
Repertoire analysis with next-generation sequencing
The VH-JH junctions were amplified from post-Ficoll PBMCs in a multiplex PCR by using the VH1-6 FR1 and JH consensus BIOMED-2 primers.15 The primers were adapted for 454 sequencing by adding the forward A or reverse B adaptor, the “TCAG” key, and the multiplex identifier adaptor. PCR products were purified by using gel extraction (Qiagen, Valencia, Calif) and Agencourt AMPure XP beads (Beckman Coulter, Fullerton, Calif). Subsequently, the PCR concentration was measured with the Quant-it Picogreen dsDNA assay (Invitrogen, Carlsbad, Calif). The purified PCR products were sequenced on the 454 GS junior instrument according to the manufacturer's recommendations by using the GS junior Titanium emPCR kit (Lib-A), sequencing kit, and PicoTiterPlate kit (454 Life Sciences; Roche, Branford, Conn). By using the CLC genomic workbench software, the samples were separated based on their multiplex identifier sequence and trimmed, and reads with a quality score of less than 0.05 and less than 250 bp were discarded. The reads were uploaded to IMGT HighV-Quest software.18 Subsequently, these output files were uploaded to the custom Galaxy platform.19, 20, 21 Further processing was done in the R programming language22 to generate the tabular and graphic outputs. The complementary determining region 3 (CDR3) amino acid patterns were visualized with WebLogo (http://weblogo.berkeley.edu/).23, 24
Statistics
Differences in absolute numbers of lymphocyte subsets were analyzed by using the 2-tailed t test for independent samples (P < .05 was considered significant) in GraphPad Prism software (GraphPad Software, La Jolla, Calif).
Results
Residual RAG1 activity in patients with N-terminal truncating RAG1 mutations
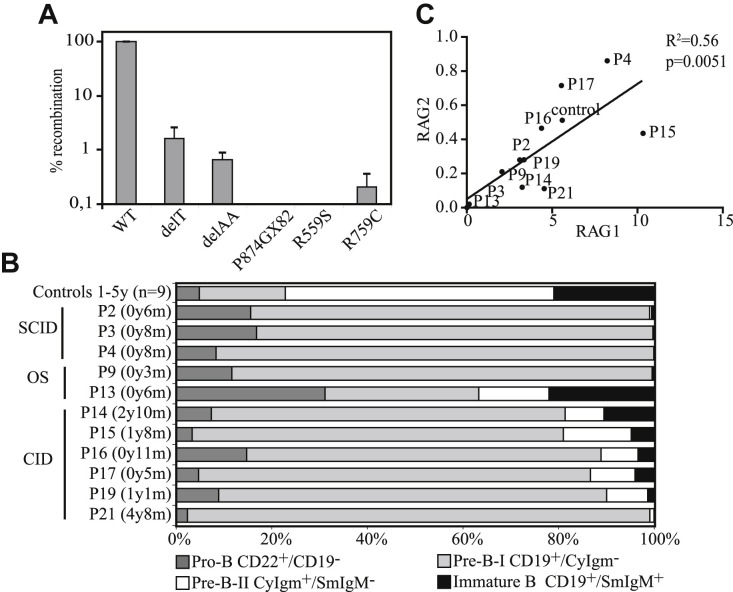
Over the past 10 years, we identified one of the 2 mutations resulting in N-terminal truncating RAG1 mutations in 22 patients (Table I, Table II ). These c.519delT (hereafter abbreviated as delT) and c.368_369delAA (hereafter abbreviated as delAA) mutations have been described before in several patients.13, 25, 26, 27, 28, 29 They were found to be hypomorphic13, 27 because translation can be reinitiated from the alternative start site methionine 202 (M202) or M183, resulting in an N-terminal truncated RAG1 protein13 with the same (comparable) residual RAG1 activity (<5% compared with wild type; Fig 1 , A).13 Sixteen patients were homozygous for the delAA or delT mutation, and 6 patients were compound heterozygous (Table I). Three RAG1 mutations found on the second allele were also analyzed in the in vitro recombination assay, showing no residual RAG1 activity (Fig 1, A). In addition, we determined the presence of polymorphisms in the RAG1 gene because these might influence the recombination activity of RAG1. The only polymorphism found was p.Arg249His, which was shown not to affect recombination activity.2
Table I.
Clinical data of patients with RAGD
| Onset of infections (mo) | Age at diagnosis (mo) | Infections | Respiratory tract infections | Autoimmunity/erythroderma | Hepatomegaly | Splenomegaly | Lymphadenopathy | |
|---|---|---|---|---|---|---|---|---|
| SCID | ||||||||
| P1 | 3 | |||||||
| P2 | 6 | 6 | BCG | No | ITP | No | No | No |
| P3 | 8 | 8 | Pneumonia and upper airway infections | |||||
| P4 | 6 | 8 | BCG | Mild | ||||
| OS | ||||||||
| P5 | 0 | 0.5 | Erythroderma | Yes | Yes | Yes | ||
| P6a | 0 | 0.5 | Recurrent pneumonia | Erythroderma | Yes | No | Yes | |
| P7a | 0 | 0.5 | CMV | No | Erythroderma | Yes | Yes | Yes |
| P8 | 0 | 3.5 | CMV, Candida species, MRSE | Severe pneumonia | Erythroderma | |||
| P9 | 0 | 4 | Erythroderma | Yes | No | Yes | ||
| P10 | 1 | 1 | Erythroderma | Yes | ||||
| P11 | 1.5 | 2 | Erythroderma | Yes | ||||
| P12 | 1 | 8 | BCG | Recurrent pneumonia | Erythroderma | Yes | Yes | Yes |
| P13 | 3 | 6 | Candida species, Mycobacterium bovis, coronavirus, rhinovirus | Recurrent upper and lower airway infections | Erythroderma, AIHA, ITP | Yes | No | No |
| CID | ||||||||
| P14b | 9 | 30 | CMV, Candida species | Recurrent bronchopneumonia | Yes | Yes | No | |
| P15b | 9 | 18 | CMV | Recurrent bronchopneumonia | Yes | Yes | No | |
| P16c | 1 | 11 | CMV | Chronic rhinitis | No | No | No | |
| P17 | 4 | 6 | CMV, BCG | Pneumonia | Yes | Yes | Yes | |
| P18 | 18 | 60 | CMV, BCG, rhinovirus | Yes | AIHA, ITP | No | No | No |
| P19 | 3 | 13 | Candida species | Chronic rhinitis and bronchitis | AIHA | No | No | No |
| P20 | 24 | 48 | AIHA | |||||
| P21 | 13 | 60 | Candida species, aspergillosis | Recurrent pneumonias, bronchitis | AIHA | No | No | No |
| P22c | 0 | 17 | Recurrent pneumonias, bronchitis | No | No | No | ||
Footnote symbols “a,” “b,” and “c” indicate relatives.
AIHA, Autoimmune hemolytic anemia; CMV, cytomegalovirus; ITP, idiopathic thrombocytopenic purpura; MRSE, methicillin-resistant staphylococcus epidermis.
Table II.
Immunologic data of patients with RAGD
| delT | delAA | Other | CD3+ T cells, absolute (× 10E9/L) | CD4+ T cells, absolute (× 10E9/L) | CD8+ cells, absolute | CD45RA (%) | γδT cells (%) | CD19+ cells, absolute (× 10E9/L) | NK cells, absolute (× 10E9/L) | |
|---|---|---|---|---|---|---|---|---|---|---|
| SCID | ||||||||||
| P1 | Homozygous | 0.06 (1.4-8.0) | 0.04 (0.9-5.5) | 0.01 (0.4-2.3) | 21.8 | 0.03 (0.6-3.1) | 0.08 (0.1-1.4) | |||
| P2 | Heterozygous | p.P874GfsX82 | 0.1 (2.4-6.9) | 0.06 (1.4-5.1) | 0.01 (0.6-2.2) | 32.1 | 24.4 | 0.01 (0.7-2.5) | 0.4 (0.1-1.0) | |
| P3 | Homozygous | 0.3 (1.6-6.7) | 0.06 (1.0-4.6) | 0.3 (0.4-2.1) | 0 (0.6-2.7) | 0.5 (0.2-1.2) | ||||
| P4 | Heterozygous | p.R559S | 0.3 (1.6-6.7) | 0.2 (1.0-4.6) | 0.04 (0.4-2.1) | 7.7 | 10 | 0 (0.6-2.7) | 0.1 (0.2-1.2) | |
| OS | ||||||||||
| P5 | Homozygous | 20.1 (2.3-7.0) | 7.56 (1.7-5.3) | 12.75 (0.4-1.7) | 4 | 0 (0.6-1.9) | 2.59 (0.2-1.4) | |||
| P6a | Homozygous | 3.7 (2.3-6.5) | 3.1 (1.5-5.0) | 0.4 (0.5-1.6) | 7.2 | 5.2 | 0.03 (0.6-3.0) | 0.8 (0.1-1.3) | ||
| P7a | Homozygous | 36 (2.3-6.5) | 10.7 (1.5-5.0) | 24.9 (0.5-1.6) | 3.1 | 1 | 0.03 (0.6-3.0) | 0.4 (0.1-1.3) | ||
| P8 | Homozygous | 3.93 (2.3-6.5) | 1.45 (1.5-5.0) | 2.19 (0.5-1.6) | 21.4 | 24.4 | 0.02 (0.6-3.0) | 0.88 (0.1-1.3) | ||
| P9 | Homozygous | 1.84 (2.3-6.5) | 1.48 (1.5-5.0) | 0.3 (0.5-1.6) | 4.6 | 3 | 0.004 (0.6-3.0) | 1.64 (0.1-1.3) | ||
| P10 | Heterozygous | p.R737H | 3.3 (2.3-7.0) | 0.32 (1.7-5.3) | 2.97 (0.4-1.7) | 0.3 | 0.1 | 0 (0.6-1.9) | 0.34 (0.2-1.4) | |
| P11 | Heterozygous | p.R559S | 4.33 (1.6-6.7) | 4 (1.0-4.6) | 0.27 (0.4-2.1) | 0.01 (0.6-2.7) | 0.93 (0.2-1.2) | |||
| P12 | Homozygous | 2.21 (2.3-6.5) | 1.34 (1.5-5.0) | 0.61 (0.5-1.6) | 0.07 (0.6-3.0) | 0.56 (0.1-1.3) | ||||
| P13 | Homozygous | 0.6 (2.4-6.9) | 0.6 (1.4-5.1) | 0.01 (0.6-2.2) | 4.8 | 2.9 | 0.07 (0.7-2.5) | 0.2 (0.1-1.0) | ||
| CID | ||||||||||
| P14b | Homozygous | 0.3 (0.9-4.5) | 0.1 (0.5-2.4) | 0.07 (0.3-1.6) | 35 | 49.5 | 0.4 (0.2-2.1) | 0.8 (0.1-1.0) | ||
| P15b | Homozygous | 0.5 (1.4-8.0) | 0.1 (0.9-5.5) | 0.2 (0.4-2.3) | 38.9 | 64.1 | 0.4 (0.6-3.1) | 2.9 (0.1-1.4) | ||
| P16c | Homozygous | 0.16 (1.6-6.7) | 0.07 (1.0-4.6) | 0.02 (0.4-2.1) | 26.9 | 46.3 | 0.09 (0.6-2.7) | 0.32 (0.2-1.2) | ||
| P17 | Homozygous | 2.7 (1.6-6.7) | 0.2 (1.0-4.6) | 1.5 (0.4-2.1) | 90.7 | 90.2 | 0.06 (0.6-2.7) | 0.7 (0.2-1.2) | ||
| P18 | Heterozygous | p.R759C | 0.53 (0.9-4.5) | 0.07 (0.5-2.4) | 0.12 (0.3-1.6) | 57.2 | 0.12 (0.2-2.1) | 1.32 (0.1-1.0) | ||
| P19 | Homozygous | 0.10 (1.6-6.7) | 0.01 (1.0-4.6) | 0.10 (0.4-2.1) | 97.5 | 0.04 (0.6-2.7) | 0.23 (0.2-1.2) | |||
| P20 | Homozygous | 0.77 (0.9-4.5) | 0.25 (0.5-2.4) | 0.24 (0.3-1.6) | 41.7 | 0.02∗ (0.2-2.1) | 0.18 (0.1-1.0) | |||
| P21 | Heterozygous | p.A444V | 0.12 (0.9-4.5) | 0.10 (0.5-2.4) | 0.06 (0.3-1.6) | 14.4 | 0.001 (0.2-2.1) | 0.25 (0.1-1.0) | ||
| P22c | Homozygous | 1.97 (1.6-6.7) | 0.42 (1.0-4.6) | 1.50 (0.4-2.1) | 42 | 0.29 (0.6-2.7) | 0.72 (0.2-1.2) | |||
Numbers in parentheses indicate normal values. Footnote symbols “a,” “b,” and “c” indicate relatives.
NK, Natural killer.
Under rituximab treatment.
Fig 1.
RAG expression and precursor B-cell compartment. A, Recombination activity of the c.519delT (delT), c.delA368/A369 (delAA), p.P874GX82, p.R559S, and p.R759C RAG1 mutations was compared with wild-type (WT) RAG1. Only the delT and the delAA RAG1 mutations result in low levels of residual recombination activity. B, Composition of the precursor B-cell compartment in control subjects (n = 9), 3 patients with the “classical” SCID phenotype, 2 patients with OS, and 6 patients with CID. C, Relative RAG1 expression levels correlated to RAG2 expression in all the analyzed RAG patients, as determined by using RQ-PCR.
N-terminal truncating RAG1 mutations result in a spectrum of clinical phenotypes
Although all patients had similar RAG1 mutations, resulting in the same N-terminal truncation of the RAG1 protein, the clinical phenotypes varied substantially. The patients could be divided into 3 main clinical phenotypes: “classical” T−B− SCID (n = 4), OS (n = 9), and CID (n = 9, Table I, Table II). The patients with “classical” SCID were defined as having low B- and T-cell numbers and age at diagnosis before the first year of life. The patients with OS all had generalized and pronounced erythroderma. The patients with CID were given a diagnosis after the first year of life and had greater than 14% γδ T cells or normal levels of T cells (P17 and P22). Despite the same N-terminal truncation of RAG1 in the 22 patients, the range of clinical phenotypes strongly suggests that factors other than residual RAG1 activity contribute to the clinical phenotype.
All clinical phenotypes show a block in precursor B-cell development
RAGD results in a block in the precursor B-cell differentiation in BM at the B-cell stages during which V(D)J recombination of the immunoglobulin genes takes place.11 The relative distribution of pro-B, pre-BI, pre-BII, and immature B cells was assessed in BM from 11 of 22 patients to investigate precursor B-cell differentiation. In healthy children pro-B and pre-BI cells constitute 20% to 25% of the precursor B cells (Fig 1, B). All patients with “classical” SCID and OS, except P13, showed a complete block before the pre–BII-cell stage (Fig 1, B), whereas most of the patients with CID had a leaky block with greater than 10% pre-BII and immature B cells (Fig 1, B). RAG1 and RAG2 transcription levels were determined in the BM mononuclear cells to exclude that differences in RAG1 transcription levels caused these difference in precursor B-cell composition. It is known that RAG1 and RAG2 transcription levels are correlated, and that RAG1 and RAG2 levels in BM mononuclear cells depend on the number of cells expressing RAG (pre-BI and pre-BII cells).30 In all 11 studied patients, the RAG1 transcription level was correlated to RAG2 (Fig 1, C), indicating that the differences in severity of the precursor B-cell block were not caused by differences in expression of RAG1. B-cell numbers in PB were undetectable or very low in most patients, except P15, who had normal levels (Table II). Correlating the percentage of pre-BII and immature cells in BM with the number of peripheral B cells showed that only patients with greater than 10% pre-BII and immature B cells in BM (P13, P14, P15, P16, and P17) had detectable B cells in PB. Collectively, these data indicate that most patients with CID have a milder block in the precursor B-cell composition and that only patients with a leaky block have detectable B cells in the PB.
Immunoglobulin heavy chain combinatorial repertoire
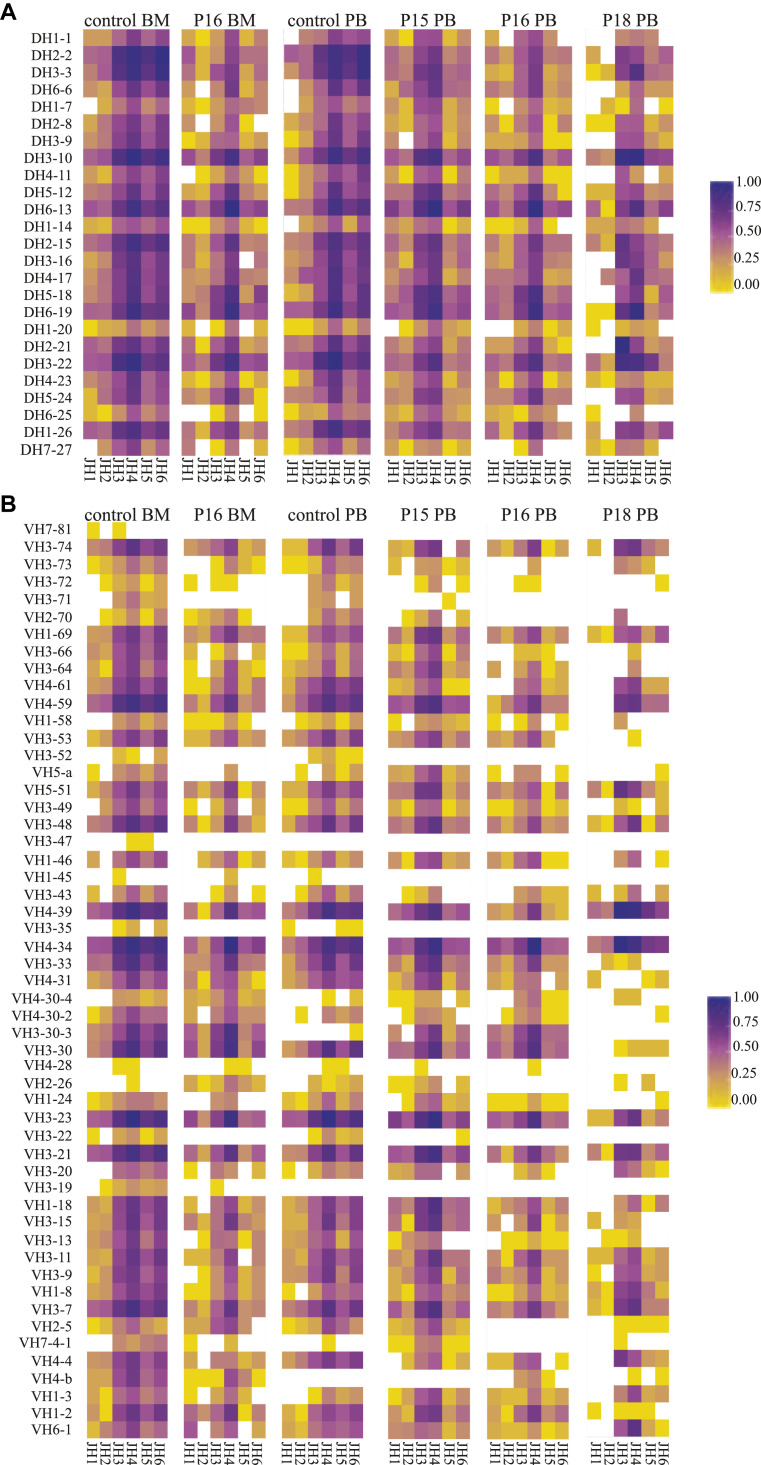
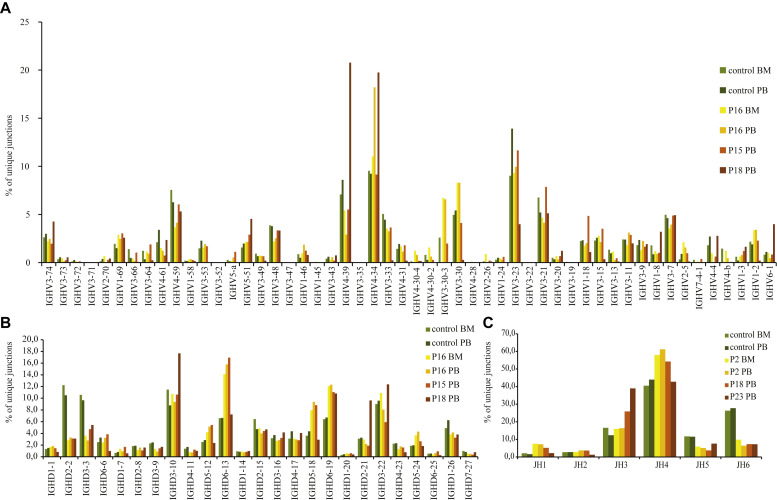
In those patients with detectable peripheral B cells, we studied the IGH V(D)J recombination repertoire. IGH gene rearrangements were amplified from mononuclear cells derived from PB, BM, or both and subsequently sequenced by using next-generation sequencing in healthy control subjects (PB and BM) and 3 patients with CID (P15, P16, and P18). The frequency of unique sequences in IGH genes was significantly lower in patients with RAGD than in control subjects (Table III ), which is a reflection of the low numbers of B cells present in PB. Despite the low recombination activity, IGHV, IGHD, and IGHJ gene use was not restricted (Fig 2 and see Fig E1 in this article's Online Repository at www.jacionline.org). Forty-eight of the 57 IGHV genes used in control subjects were identified in the patients with RAGD because were the 25 IGHD genes and all 6 IGHJ genes. IGHV, IGHD, and IGHJ gene uses were similar to those seen in control subjects, although some genes were used with different frequencies (Fig 2 and see Fig E1). Most strikingly, JH6 use was lower whereas JH4 use was higher compared with that seen in control subjects. The patients with RAGD had a significantly lower frequency (5.9% to 6.2% vs 20.9% to 24.3% in control subjects) of unproductive rearrangements (Table III), as reported previously.31 Unproductive rearrangements were defined as out-of-frame rearrangements or rearrangements with a stop codon. Therefore even though the patients with RAGD had reduced V(D)J recombination, leading to a limited TR and immunoglobulin repertoire, the IGH gene use was similar to that seen in control subjects without preferential use of the proximal or distal genes.
Table III.
Number of IGH sequences
| All sequences | Unique sequences | Unproductive | Productive | |
|---|---|---|---|---|
| Control BM | 35,472 | 18,241 (51.4) | 8,633 (24.3) | 26,839 (75.7) |
| P16 BM | 12,195 | 3,325 (27.3) | 1,629 (13.3) | 10,566 (86.7) |
| Control PB | 19,294 | 9,185 (61.2) | 4,030 (20.9) | 15,003 (77.8) |
| P15 PB | 16,826 | 7,706 (45.8) | 1,047 (6.2) | 15,779 (93.8) |
| P16 PB | 14,572 | 3,763 (25.8) | 896 (6.1) | 13,676 (93.9) |
| P18 PB | 25,100 | 3,730 (14.9) | 1,488 (5.9) | 23,612 (94.1) |
Numbers in parentheses indicate percentages. “Unproductive” refers to out-of-frame rearrangements or rearrangements containing a stop codon in the CDR3 region.
Fig 2.
Heat maps of the different combinations of immunoglobulin DH-JH (A) and VH-JH (B), as determined in the unique junctions (defined by the unique combination of VH, DH, JH, and nucleotide sequences of CDR3).
Selection of B cells is slightly impaired
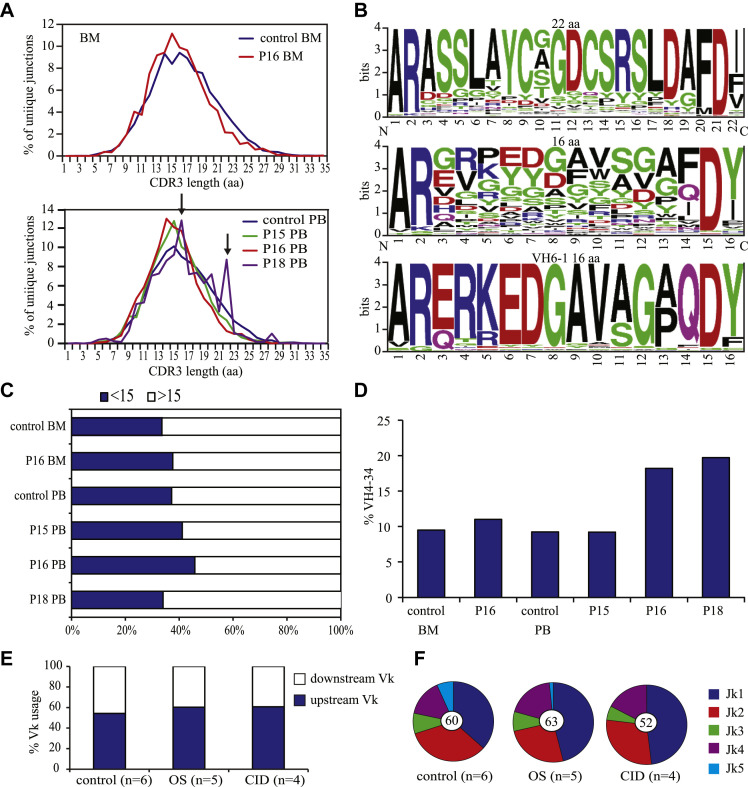
OS is characterized by autoimmune-like clinical features, including severe erythroderma, hepatosplenomegaly, and lymphadenopathy.32, 33 The immune dysregulation in patients with OS might be caused by the severe abnormalities of thymic architecture and impaired expression of autoimmune regulator and tissue-specific antigens.34, 35 In addition, hypomorphic Rag mouse models have shown a disturbance in B-cell tolerance.36, 37 In addition, patients with OS and also 1 patient with “classical” SCID and 4 patients with CID had autoimmunity, and all displayed idiopathic thrombocytopenic purpura, autoimmune hemolytic anemia, or both (Table I). Unfortunately, the thymic architecture and autoimmune regulator and tissue-specific antigen expression could not be studied in our patients, but we were able to evaluate 3 parameters in the IGH sequences that are associated with autoimmunity. These are characterized by long CDR3s, and the frequency of IGHV4-34, which is known to encode intrinsically self-reactive cold agglutinin antibodies that recognize carbohydrate antigens on erythrocytes.38, 39 The distribution of the CDR3 length of the unique junctions in BM and PB was similar to that seen in control subjects (Fig 3 , A), except patient 18, who seemed to have increased numbers of junctions with a CDR3 of 16 and 22 amino acids. These junctions with a CDR3 length of 22 amino acids displayed high similarity (Fig 3, B). No sequence similarity was found when all 16-amino-acid CDR3s were compared (Fig 3, B), but 18.3% of these junctions used IGHV6-1, and all these junctions had a highly similar CDR3 sequence (Fig 3, B), which suggests that they might recognize a common antigenic determinant. The frequency of long CDR3s (≥15 amino acids) was significantly lower in P15 and P16 (P < .0001) but not in P18 (Fig 3, C). The frequency of IGHV4-34 use was significantly higher in P16 (P < .0001) and P18 (P < .0001; Fig 3, D). From the 3 patients we analyzed, P18 had autoimmunity, which was reflected by the high frequency of IGHV4-34 use.
Fig 3.
Functional characteristics of IGH junctions. A, Functional characteristics of the IGH junctions were determined in 3 patients with RAGD in PB or BM. Distribution of CDR3 length frequencies in BM and PB was similar in control subjects and patients with RAGD; however, P18 had increased numbers of junctions, with a CDR3 length of 16 and 23 amino acids. B, Sequence logo showed no similarity of the 16-amino-acid CDR3s of P23 but high similarity of CDR3s of 16 amino acids using the IGHV6-1 gene and the 22-amino-acid CDR3s. C, The frequency of long CDR3s (≥15 amino acids) was decreased in P15 and P16. D,IGHV4-34 use was increased in P16 and P18. E and F, The percentage of IGKV and IGKJ genes was determined in 6 control subjects, 5 patients with OS, and 4 patients with CID. IGKV use was normal (Fig 3, E), but hardly any IGKJ5 gene was used (Fig 3, F).
In addition to selection against long CDR3s, B-cell tolerance is also generated by receptor editing of self-reactive B cells. These self-reactive B cells are induced to express the RAG proteins and edit their receptor light chains through available upstream Vκ and downstream Jκ genes to change the affinity of their receptors. Therefore the Vκ-Jκ junctions were amplified from 5 patients with OS and 4 patients with CID. The IGKV gene use was not significantly different from that seen in control subjects (Fig 3, E), but less IGKJ5 genes were used in the patients with RAGD (Fig 3, F). Therefore receptor editing seems partly affected, as deduced from the very low IGKJ5 use.
Difference between clinical phenotypes in absolute numbers of T cells but not in T-cell repertoire
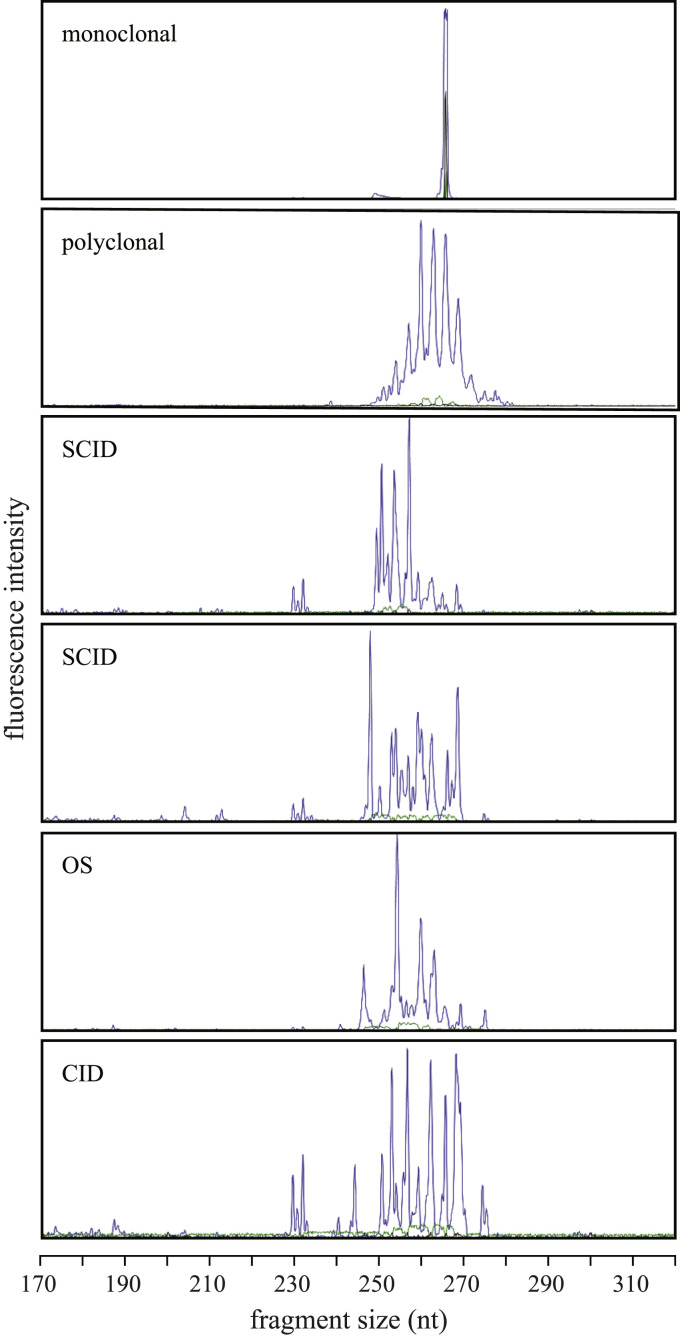
The hallmark of classical OS is an expansion of autologous T cells with an HLA-DR+CD45RO+ phenotype and an oligoclonal αβ T-cell repertoire.40 Consistent with this, most of the patients with OS had normal or increased CD3+ T-cell numbers; in addition, 2 patients with CID had normal numbers (P17 and P22), whereas all other patients had low absolute numbers of CD3+ T cells (Table II). Remarkably, many patients had high percentages (>14%) of γδ T cells, including 2 patients with “classical” SCID, 1 patient with OS, and 8 patients with CID (Table II). In addition, we determined the T-cell proliferation by determining the δREC-ψJα T-cell receptor excision circle (TREC)41 content per 50 ng of DNA in 3 patients with “classical” SCID, 7 patients with OS, and 5 patients with CID. In 11 patients TRECs were not detectable, and in the other 4 patients (P2, P5, P7, and P18), the number of TRECs/50 ng of DNA was less than 1 compared with 134 ± 75 TRECs/50 ng of DNA in control subjects (n = 7; age, 8 months to 11 years; data not shown), meaning that the T cells that were present in these patients showed extensive proliferation. Furthermore, the T-cell repertoire was determined by testing the TRB gene rearrangements in 2 patients with “classical” SCID, 3 patients with OS, and 2 patients with CID. In all patients the TRB repertoire was clearly restricted (Fig 4 ). Taken together, the T cells that were present in the patients with RAGD showed extensive proliferation and had a restricted TR repertoire.
Fig 4.
TRB repertoire. TR spectratyping profiles of TRB gene rearrangements using the BIOMED-2 TRB tube B. The upper 2 panels show the monoclonal and the polyclonal controls. The patient panels SCID (P2 and P4), OS (P8), and CID (P21) show a restricted TRB repertoire.
Discussion
Many different RAG1 mutations have been reported to the RAG mutation database.42 Although most are null mutations, several have been described to result in residual recombinase activity.11, 13, 25, 27, 43 Previously, it was hypothesized that null mutations in RAG1 would result in “classical” T−B− SCID and that partial reduction of RAG activity would result in OS or an intermediate late-onset SCID or OS phenotype.28 Over the last few years, the spectrum of reported clinical phenotypes of RAGD has broadened and now also includes RAGD with γδ T-cell expansion, RAGD with skin inflammation but without T-cell expansion (incomplete OS), RAGD with granulomas, RAGD with maternofetal transfusion, and RAGD with CD4 cytopenia and thymus hypoplasia.9, 10 A few case reports have shown that the same RAG mutation can result in a different clinical phenotype.25, 29, 44, 45 This study is the first to report an in-depth immunobiological evaluation of 22 patients with RAGD with similar RAG1 mutations, resulting in the same N-terminal truncation of the RAG1 protein. These similar mutations result in 3 different clinical phenotypes, which indicates that a specific mutation does not predict a patient's clinical phenotype.
Because all patients had similar mutations, the residual RAG1 protein activity was expected to be comparable among all patients. The N-terminally truncated RAG1 protein is produced through translation starting from an alternative start site (M183 or M202), and hence the amount of protein is dependent on how efficiently these start sites are used. Because the RAG1 transcription level correlated with that of RAG2, we assume that all patients had similar expression of the mutant RAG1 protein (Fig 1, C). We cannot exclude that epigenetics and modifier genes accounted for small differences in RAG1 protein expression. Although a previous attempt to identify such modifier genes in human subjects was not successful,46 studies in mouse models could shed more light on the contribution of epigenetics and modifier genes.
In our cohort V(D)J recombination was not completely abolished but was strongly reduced because of the low residual activity of the RAG1 protein. Reduced V(D)J recombination was characterized by normal IGHV, IGHD, and IGHJ gene use, without preferential use of proximal or distal genes. However, as shown previously,31 the frequency of unproductive sequences was significantly lower than in healthy control subjects, indicating that the B cells in the patients with RAGD did not correct unproductive rearrangements by means of recombination of the second IGH allele.
As a consequence of the reduced V(D)J recombination, fewer B and T cells with a functional receptor can be produced. The proliferation of the lymphocytes is increased to compensate for low circulating B- and T-cell numbers. This idea is corroborated by the low numbers of TRECs in patients with RAGD. The increased proliferation of T cells might result in normal or increased T-cell counts, especially in the patients with OS; however, the corresponding TR repertoire in all the patients with RAGD remains restricted.
Most patients with RAGD showed clinical signs of immune dysregulation, such as erythroderma, lymphadenopathy, hepatosplenomegaly, idiopathic thrombocytopenic purpura, and autoimmune hemolytic anemia. B cells have been shown to contribute to the immune dysregulation seen in Rag mouse models.36, 37 Sera from these mice contained high-affinity anti–double-stranded DNA and tissue-specific autoantibodies, and B cells displayed impaired receptor editing. In addition, these mice had increased serum B cell–activating factor levels, which might rescue autoreactive B-cell clones. This increase in serum B cell–activating factor levels was also seen in patients with RAG-, Artemis-, and X-linked SCID.37 Similar to observations in mice, most patients with RAGD did not use the IGKJ5 gene, whereas IGKV gene use was normal. This suggests that receptor editing in this group of patients with RAGD was slightly impaired, which can either be a result of reduced recombination activity caused by the RAG1 mutation or by low B-cell numbers leading to reduced selection against autoreactive B cells. The IGH repertoire was investigated for long CDR3s and increased IGHV4-34 use, which are associated with autoreactive antibodies.47, 48 From the 3 patients with RAGD we analyzed, only P18 had autoimmunity, which was reflected by an increased VH4-34 gene use.
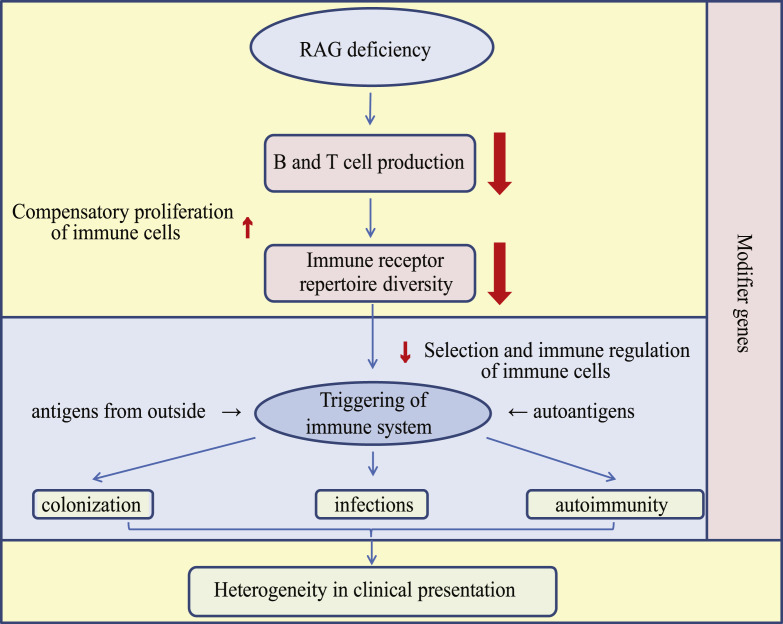
The patients divided into the 3 main clinical RAGD groups hardly differed in their immunobiological parameters, and consequently, we could not find any specific pattern that could explain the different clinical phenotypes. On the basis of our results and earlier reported data, we propose an explanatory model for the development of different clinical phenotypes in patients with RAGD with similar mutations (Fig 5 ). If RAGD results in reduced V(D)J recombination, low B- and T-cell numbers are produced with some (compensatory) clonal expansion. This expansion might increase the B- and T-cell numbers to even normal levels but does not change the limited repertoire. In such limited repertoire the selection against autoreactive cells is impaired. Provided the deficient immune system is not activated, patients with RAGD are asymptomatic. However, when the immune system will be activated by potentially a wide range of different (auto)antigens, the type of antigen and activated effector lymphocyte will have important consequences for the clinical phenotype. In addition, the impaired negative and positive selection of thymic lymphocytes and reduced number of regulatory T cells might result in autoimmunity when patients are exposed to autoantigens. This phenomenon can occur at any early stage, even in utero, as shown by the fact that patients with OS can have severe erythroderma already at birth, which is unlikely to be triggered by infections. Additionally, directly after birth, the skin and gastrointestinal tract become colonized by commensal bacteria, which can trigger the chronic diarrhea seen in most patients with RAGD. Key steps in the development of a certain clinical phenotype will be the B- and T-cell repertoire, the type of (auto)antigen exposure, the specificity of the antigen receptors and timing, the cell type involved in the immune activation, and the potential influence of genetic variations in modifier genes. Variability in any of these factors might eventually lead to different clinical phenotypes, despite a similar genetic defect.
Fig 5.
Model for development of clinical phenotype in patients with RAGD. RAGD results in reduced V(D)J recombination, leading to fewer B and T cells with a limited repertoire. In an attempt to compensate for low numbers, B and T cells start to proliferate, but the repertoire remains limited and imbalanced, so that selection and immune regulation are impaired. Most likely the type of antigenic stimulation together with the incomplete and imbalanced repertoire that has been developed will affect the eventual clinical phenotype with immune dysregulation problems.
In conclusion, this study clearly shows that the type of RAG1 mutation and the level of residual RAG1 recombinase activity are not the only determinants predicting the clinical phenotype, as previously assumed. The clinical outcome of an individual patient with RAGD depends on a complex interplay between the (limited) immune receptor repertoire, (auto)antigen exposure, the specificity of antigen receptors, and the timing and cell type involved in immune activation. Therefore the clinical outcome of patients with RAGD with similar mutations is extremely difficult to predict.
Clinical implications.
RAGD can result in a broad spectrum of clinical presentations, but the level of residual RAG activity is not always predictive for the clinical outcome.
Acknowledgments
We thank B. H. Barendregt and I. Pico-Knijnenburg for technical assistance, S. de Bruin-Versteeg for making the figures, D. Zessen for help with the repertoire analysis, and Professor A. J. Cant for discussion and advice.
Footnotes
Supported by grants from the foundation “Sophia Kinderziekenhuis Fonds” (grant 589 to H.I. and M.v.d.B.), the Dutch Organization for Scientific Research (NWO/ZonMw VIDI grant 91712323 to M.v.d.B.), and the IGA NT/13271, MH CZ–DRO, University Hospital Motol, Prague, Czech Republic (grant 00064203 to E.M.).
Disclosure of potential conflict of interest: H. IJspeert has received research support from Sophia Kinderziekenhuis Fonds (grant 589). G. J. Driessen has received research support from Baxter. I. Kondratenko has received consultancy fees from and is employed by Russian Children's Clinical Hospital and has received lecture fees from and has received payment for manuscript preparation from Russian National Scientific Medical University. M. van der Burg has received research support from ZonMW (Vidi grant 91712323). The rest of the authors declare that they have no relevant conflicts of interest.
Appendix
Fig E1.
Immunoglobulin heavy chain gene usage. Frequency of IGHV(A), IGHD(B), and IGHJ(C) gene use in control BM and PB and in P16 BM and PB, P15 PB, and P18 PB.
References
- 1.Schatz D.G. V(D)J recombination. Immunol Rev. 2004;200:5–11. doi: 10.1111/j.0105-2896.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- 2.Schwarz K., Gauss G.H., Ludwig L., Pannicke U., Li Z., Lindner D. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- 3.Moshous D., Callebaut I., de Chasseval R., Corneo B., Cavazzana-Calvo M., Le Deist F. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 4.Ahnesorg P., Smith P., Jackson S.P. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 5.Buck D., Malivert L., de Chasseval R., Barraud A., Fondaneche M.C., Sanal O. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 6.O'Driscoll M., Gennery A.R., Seidel J., Concannon P., Jeggo P.A. An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair (Amst) 2004;3:1227–1235. doi: 10.1016/j.dnarep.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 7.van der Burg M., Ijspeert H., Verkaik N.S., Turul T., Wiegant W.W., Morotomi-Yano K. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest. 2009;119:91–98. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Burg M., van Veelen L.R., Verkaik N.S., Wiegant W.W., Hartwig N.G., Barendregt B.H. A new type of radiosensitive T-B-NK+ severe combined immunodeficiency caused by a LIG4 mutation. J Clin Invest. 2006;116:137–145. doi: 10.1172/JCI26121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niehues T., Perez-Becker R., Schuetz C. More than just SCID—the phenotypic range of combined immunodeficiencies associated with mutations in the recombinase activating genes (RAG) 1 and 2. Clin Immunol. 2010;135:183–192. doi: 10.1016/j.clim.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Kuijpers T.W., Ijspeert H., van Leeuwen E.M., Jansen M.H., Hazenberg M.D., Weijer K.C. Idiopathic CD4+ T lymphopenia without autoimmunity or granulomatous disease in the slipstream of RAG mutations. Blood. 2011;117:5892–5896. doi: 10.1182/blood-2011-01-329052. [DOI] [PubMed] [Google Scholar]
- 11.Noordzij J.G., de Bruin-Versteeg S., Verkaik N.S., Vossen J.M., de Groot R., Bernatowska E. The immunophenotypic and immunogenotypic B-cell differentiation arrest in bone marrow of RAG-deficient SCID patients corresponds to residual recombination activities of mutated RAG proteins. Blood. 2002;100:2145–2152. [PubMed] [Google Scholar]
- 12.Noordzij J.G., Verkaik N.S., van der Burg M., van Veelen L.R., de Bruin-Versteeg S., Wiegant W. Radiosensitive SCID patients with Artemis gene mutations show a complete B-cell differentiation arrest at the pre-B-cell receptor checkpoint in bone marrow. Blood. 2003;101:1446–1452. doi: 10.1182/blood-2002-01-0187. [DOI] [PubMed] [Google Scholar]
- 13.Noordzij J.G., Verkaik N.S., Hartwig N.G., de Groot R., van Gent D.C., van Dongen J.J. N-terminal truncated human RAG1 proteins can direct T-cell receptor but not immunoglobulin gene rearrangements. Blood. 2000;96:203–209. [PubMed] [Google Scholar]
- 14.Boeckx N., Willemse M.J., Szczepanski T., van der Velden V.H., Langerak A.W., Vandekerckhove P. Fusion gene transcripts and Ig/TCR gene rearrangements are complementary but infrequent targets for PCR-based detection of minimal residual disease in acute myeloid leukemia. Leukemia. 2002;16:368–375. doi: 10.1038/sj.leu.2402387. [DOI] [PubMed] [Google Scholar]
- 15.van Dongen J.J., Langerak A.W., Bruggemann M., Evans P.A., Hummel M., Lavender F.L. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 16.Lefranc M.P. IMGT databases, web resources and tools for immunoglobulin and T cell receptor sequence analysis. Leukemia. 2003;17:260–266. doi: 10.1038/sj.leu.2402637. http://imgt.cines.fr. [DOI] [PubMed] [Google Scholar]
- 17.Lefranc M.P. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2003;31:307–310. doi: 10.1093/nar/gkg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alamyar E., Duroux P., Lefranc M.P., Giudicelli V. IMGT® tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- 19.Goecks J., Nekrutenko A., Taylor J., Galaxy T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blankenberg D., Von Kuster G., Coraor N., Ananda G., Lazarus R., Mangan M. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19:Unit 19.10.1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2013. [Google Scholar]
- 23.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corneo B., Moshous D., Gungor T., Wulffraat N., Philippet P., Le Deist F.L. Identical mutations in RAG1 or RAG2 genes leading to defective V(D)J recombinase activity can cause either T-B-severe combined immune deficiency or Omenn syndrome. Blood. 2001;97:2772–2776. doi: 10.1182/blood.v97.9.2772. [DOI] [PubMed] [Google Scholar]
- 26.de Villartay J.P., Lim A., Al-Mousa H., Dupont S., Dechanet-Merville J., Coumau-Gatbois E. A novel immunodeficiency associated with hypomorphic RAG1 mutations and CMV infection. J Clin Invest. 2005;115:3291–3299. doi: 10.1172/JCI25178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santagata S., Gomez C.A., Sobacchi C., Bozzi F., Abinun M., Pasic S. N-terminal RAG1 frameshift mutations in Omenn's syndrome: internal methionine usage leads to partial V(D)J recombination activity and reveals a fundamental role in vivo for the N-terminal domains. Proc Natl Acad Sci U S A. 2000;97:14572–14577. doi: 10.1073/pnas.97.26.14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villa A., Sobacchi C., Notarangelo L.D., Bozzi F., Abinun M., Abrahamsen T.G. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Martinez L., Vazquez-Ortiz M., Gonzalez-Santesteban C., Martin-Nalda A., Vicente A., Plaza A.M. From severe combined immunodeficiency to Omenn syndrome after hematopoietic stem cell transplantation in a RAG1 deficient family. Pediatr Allergy Immunol. 2012;23:660–666. doi: 10.1111/j.1399-3038.2012.01339.x. [DOI] [PubMed] [Google Scholar]
- 30.van Zelm M.C., van der Burg M., de Ridder D., Barendregt B.H., de Haas E.F., Reinders M.J. Ig gene rearrangement steps are initiated in early human precursor B cell subsets and correlate with specific transcription factor expression. J Immunol. 2005;175:5912–5922. doi: 10.4049/jimmunol.175.9.5912. [DOI] [PubMed] [Google Scholar]
- 31.Ohm-Laursen L., Nielsen C., Fisker N., Lillevang S.T., Barington T. Lack of nonfunctional B-cell receptor rearrangements in a patient with normal B cell numbers despite partial RAG1 deficiency and atypical SCID/Omenn syndrome. J Clin Immunol. 2008;28:588–592. doi: 10.1007/s10875-008-9210-7. [DOI] [PubMed] [Google Scholar]
- 32.Ochs H.D., Davis S.D., Mickelson E., Lerner K.G., Wedgwood R.J. Combined immunodeficiency and reticuloendotheliosis with eosinophilia. J Pediatr. 1974;85:463–465. doi: 10.1016/s0022-3476(74)80445-2. [DOI] [PubMed] [Google Scholar]
- 33.Omenn G.S. Familial Reticuloendotheliosis with Eosinophilia. N Engl J Med. 1965;273:427–432. doi: 10.1056/NEJM196508192730806. [DOI] [PubMed] [Google Scholar]
- 34.Cavadini P., Vermi W., Facchetti F., Fontana S., Nagafuchi S., Mazzolari E. AIRE deficiency in thymus of 2 patients with Omenn syndrome. J Clin Invest. 2005;115:728–732. doi: 10.1172/JCI23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poliani P.L., Facchetti F., Ravanini M., Gennery A.R., Villa A., Roifman C.M. Early defects in human T-cell development severely affect distribution and maturation of thymic stromal cells: possible implications for the pathophysiology of Omenn syndrome. Blood. 2009;114:105–108. doi: 10.1182/blood-2009-03-211029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cassani B., Poliani P.L., Marrella V., Schena F., Sauer A.V., Ravanini M. Homeostatic expansion of autoreactive immunoglobulin-secreting cells in the Rag2 mouse model of Omenn syndrome. J Exp Med. 2010;207:1525–1540. doi: 10.1084/jem.20091928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter J.E., Rucci F., Patrizi L., Recher M., Regenass S., Paganini T. Expansion of immunoglobulin-secreting cells and defects in B cell tolerance in Rag-dependent immunodeficiency. J Exp Med. 2010;207:1541–1554. doi: 10.1084/jem.20091927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pascual V., Victor K., Lelsz D., Spellerberg M.B., Hamblin T.J., Thompson K.M. Nucleotide sequence analysis of the V regions of two IgM cold agglutinins. Evidence that the VH4-21 gene segment is responsible for the major cross-reactive idiotype. J Immunol. 1991;146:4385–4391. [PubMed] [Google Scholar]
- 39.Silberstein L.E., Jefferies L.C., Goldman J., Friedman D., Moore J.S., Nowell P.C. Variable region gene analysis of pathologic human autoantibodies to the related i and I red blood cell antigens. Blood. 1991;78:2372–2386. [PubMed] [Google Scholar]
- 40.de Saint-Basile G., Le Deist F., de Villartay J.P., Cerf-Bensussan N., Journet O., Brousse N. Restricted heterogeneity of T lymphocytes in combined immunodeficiency with hypereosinophilia (Omenn's syndrome) J Clin Invest. 1991;87:1352–1359. doi: 10.1172/JCI115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breit T.M., Verschuren M.C., Wolvers-Tettero I.L., Van Gastel-Mol E.J., Hahlen K., van Dongen J.J. Human T cell leukemias with continuous V(D)J recombinase activity for TCR-delta gene deletion. J Immunol. 1997;159:4341–4349. [PubMed] [Google Scholar]
- 42.RAG1 mutation database. Available at: http://www.uta.fi/imt/bioinfo/RAG1base. Accessed December 27, 2013.
- 43.Villa A., Santagata S., Bozzi F., Giliani S., Frattini A., Imberti L. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93:885–896. doi: 10.1016/s0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]
- 44.Chou J., Hanna-Wakim R., Tirosh I., Kane J., Fraulino D., Lee Y.N. A novel homozygous mutation in recombination activating gene 2 in 2 relatives with different clinical phenotypes: Omenn syndrome and hyper-IgM syndrome. J Allergy Clin Immunol. 2012;130:1414–1416. doi: 10.1016/j.jaci.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pasic S., Djuricic S., Ristic G., Slavkovic B. Recombinase-activating gene 1 immunodeficiency: different immunological phenotypes in three siblings. Acta Paediatr. 2009;98:1062–1064. doi: 10.1111/j.1651-2227.2009.01250.x. [DOI] [PubMed] [Google Scholar]
- 46.Haq I.J., Steinberg L.J., Hoenig M., van der Burg M., Villa A., Cant A.J. GvHD-associated cytokine polymorphisms do not associate with Omenn syndrome rather than T-B- SCID in patients with defects in RAG genes. Clin Immunol. 2007;124:165–169. doi: 10.1016/j.clim.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 47.Klonowski K.D., Primiano L.L., Monestier M. Atypical VH-D-JH rearrangements in newborn autoimmune MRL mice. J Immunol. 1999;162:1566–1572. [PubMed] [Google Scholar]
- 48.Wardemann H., Yurasov S., Schaefer A., Young J.W., Meffre E., Nussenzweig M.C. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]