Abstract
Undiscovered side effects of drugs can have a profound effect on the health of the nation, and electronic health-care databases offer opportunities to speed up the discovery of these side effects. We applied a “medication-wide association study” approach that combined multivariate analysis with exploratory visualization to study four health outcomes of interest in an administrative claims database of 46 million patients and a clinical database of 11 million patients. The technique had good predictive value, but there was no threshold high enough to eliminate false-positive findings. The visualization not only highlighted the class effects that strengthened the review of specific products but also underscored the challenges in confounding. These findings suggest that observational databases are useful for identifying potential associations that warrant further consideration but are unlikely to provide definitive evidence of causal effects.
The increasing adoption of electronic health records (EHRs)1 and the availability of other data sources, such as administrative claims data2 and spontaneous adverse drug event reporting systems,3 promise a new era of medical discovery.4 One area that has shown concrete progress is pharmacovigilance.5 Adverse drug events represent a huge health and economic cost to the nation.6,7,8 It is simply not possible to detect all possible drug side effects in the drug-approval process because of small sample size, narrow study populations, and limited time course. Postmarket surveillance of drug safety—that is, pharmacovigilance—promises to detect important side effects as soon as possible to minimize the damage.
Before regulatory approval, while a drug is in development, randomized clinical trials represent the primary sources of safety information. Such experiments are generally regarded as the highest level of evidence, leading to an unbiased estimate of the average treatment effect.9 Unfortunately, most trials suffer from insufficient sample size and lack of applicability to reliably estimate the risk of other potential safety concerns for the target population.10,11 As a result, new evidence about safety is required even after a medical product is approved.
A number of techniques have been developed to infer drug side effects from large databases in the postapproval setting.12 Spontaneous adverse event reporting databases comprise voluntary reports of a suspected relationship between adverse effects following medical product exposure. As a result, these spontaneous databases present challenges in analysis, because there is no defined population from which to base the denominator when estimating reporting rates. The reports reflect a nonrandom sample from the total patients exposed and the total patients who have experienced the adverse event, but neither totals are reliably obtained. Disproportionality analysis methods for spontaneous adverse event reporting data were established as an approach to account for the lack of denominator by using the universe of all reports as a proxy to estimate the expected number of events that could be compared with the true observed count. Longitudinal observational health-care databases, such as administrative claims and EHRs, offer opportunity to define a population over time, enabling the estimation of background rates of events and drug utilization patterns, which can then be used as denominators for evaluating the strength of association between exposure and outcomes. However, retrospective observational database analyses suffer from a multitude of potential sources of bias due to the data capture process and heath-care delivery system. For example, it is common that the indication for a drug may bias the estimated association if it is associated with an increased risk of the outcome itself.13 Propensity score adjustment,14 self-controlled designs,12 and domain knowledge (e.g., indications)15 are commonly used to reduce confounding; however, health records have unreliable timing, and indications may be correlated so that a second indication may be confused with a side effect. Pharmacovigilance also presents the challenge of multiplicity, as there are >1,500 active ingredients in prescription medications and each requires monitoring for thousands of potential side effects; however, simultaneous evaluation of millions of statistical tests is likely to produce many false-positive findings due to chance alone. A number of techniques for addressing multiplicity, including false discovery rate analysis,16 have been suggested.
The consequence of dependencies, confounding, and other “noise” is an unacceptably high false-positive rate. The state of the art for pharmacovigilance on the Observational Medical Outcomes Partnership (OMOP)17 databases, which cover 140 million lives, produces areas under the receiver operating characteristic curve of almost 0.8.18 Even with a high threshold (relative risk > 2), which led to an average sensitivity of 0.28, the average specificity was only 0.87 and the average positive-predictive values reached only 0.51. Therefore, the discovery of an adverse event association through mining even very large databases cannot be used to directly infer actual risks. At best, the method generates a smaller pool of hypotheses that warrant further study. The volume of hypotheses when applied to all potential outcomes across the entire formulary of drugs, however, is likely to be in the hundreds or thousands.
High-visibility drug market withdrawals, such as that of rofecoxib,19 have led investigators to assess when its side effects could have been discovered according to various databases.20,21,22 Retrospective assessments of the early appearance of a signal are common in the literature but are misleading as the investigation focuses on a single “known” signal rather than establishing the context of looking for these signals across an entire set of exposures and outcomes: these studies fail to account for the potential false-positive rate that would occur if the same method were similarly applied to all other drugs for the same outcome. Schuemie et al. have shown substantial risk of both false-positives and false-negative results when establishing decision thresholds near the effect size where rofecoxib signaled.23 Removing all drugs from the market whose relative risk confidence interval exceeds one or some other threshold is likely to cause more harm than good.
At this point in time, the only possible approach is to manually review and prioritize generated lists of hypotheses. Experts' domain knowledge of pharmacology, physiology, and health care may help in addressing issues such as confounding between indications and side effects. In the past, we have used bar plots12 and forest plots23 to better visualize and interpret pharmacovigilance results, but those approaches fall short because they convey no domain knowledge (indication and structure).
Genome-wide association studies identify relevant genetic changes associated with disease states from among the thousands to millions of potential sites. The typical visualization of these associations shows the statistical significance (–log P value) of the target sites compared with all others, where the sites are organized by their placement in the genome (see for example, Ikram et al.).24 The organization places sites within genes near each other and places sites that are genetically linked near each other. The visualization approach was adopted for clinical associations in the so-called phenome-wide association studies.25 These are an inverse of a genome-wide study, in which a single genetic locus is compared with all possible phenotypes. It is organized by clinical system, often using the International Classification of Diseases, 9th Revision, Clinical Modification26 for organizing the phenotypes so that those affecting similar systems are colocated.
Using an approach based on genome- and phenome-wide association studies, we propose a “medication-wide association study” (MWAS), in which each side effect is compared with all drugs available for comparison. We organize the drugs by the Anatomical Therapeutic Chemical Classification System,27 which groups drugs both by the organ system on which they act and by their therapeutic characteristics and chemical structure. We applied a self-controlled case series (SCCS) analysis to 6 years' data from two observational health-care databases—the Truven MarketScan Commercial Claims and Encounters (CCAE) administrative claims database with 46.5 million lives, and the GE Centricity EHR database with 11.2 million lives18—and four clinically important side effects: acute myocardial infarction, acute liver failure, acute renal failure, and upper gastrointestinal ulcer. We plotted drugs for which we had ground truth of either known side effects or known lack of side effects according to appropriately powered studies.
Results
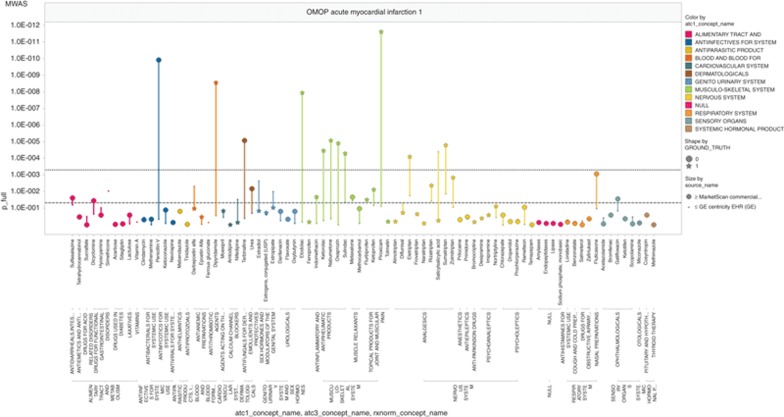
Figure 1 shows the four side-effect plots for the Truven MarketScan CCAE database. For myocardial infarction, a number of true associations (star markers) are above the threshold of P < 0.05, but there appears to be a class-specific tendency to display (e.g., anti-inflammatory) or not display (e.g., psychoanaleptics) an effect. Negative controls (circle markers) show P values almost as extreme as the true associations. For acute liver failure, the results are similar, with some classes with known effects displaying it and others not, and with a false-positive as high as the highest true-positives. Acute renal failure is similar. Upper gastrointestinal ulcer performs better with few notable false-positives.
Figure 1.
Medication-wide association study (MWAS) analyses in Commercial Claims and Encounters (CCAE) database for (a) acute myocardial infarction, (b) acute liver injury, (c) acute kidney injury, and (d) upper gastrointestinal bleeding. Y-axis displays P values on the negative log scale. X-axis displays all the drugs studied for a given outcome, grouped by the Anatomical Therapeutic Chemical classification system. OMOP, Observational Medical Outcomes Partnership.
Figure 2 displays the P-value plots across the negative controls for each of the four outcomes. In all the cases, the proportion of tests with P < 0.05 is substantially higher than the 5% expected, indicating that these observational analyses do not satisfy the standard assumptions of independent and unbiased estimators.
Figure 2.
P-value plots for negative controls, trellised by outcome. Y-axis lists the P value for each drug–outcome pair and X-axis shows the percentile of the negative control drugs which have a P value at or below that P value. The black dashed line indicates the 45° line, which should approximate the P-value curves if the statistical tests were independent and unbiased. CCAE, Commercial Claims and Encounters; OMOP, Observational Medical Outcomes Partnership.
Figure 3 compares the results for CCAE and the GE Centricity database. For each drug, a line connects the results for the two databases, with the larger marker representing the CCAE database. In general, the CCAE P values are lower in value and therefore higher on the MWAS plots, likely because the database has a larger sample size and more complete data capture of health service utilization. The combination of the two databases does not appear, however, to help distinguish between positive and negative controls.
Figure 3.
Comparison between Commercial Claims and Encounters (CCAE) and GE databases of medication-wide association study (MWAS) analyses for acute myocardial infarction. Y-axis displays P values on the negative log scale. X-axis displays all the drugs studied for a given outcome, grouped by the Anatomical Therapeutic Chemical classification system. OMOP, Observational Medical Outcomes Partnership.
Discussion
Observational health-care databases are commonly used for evaluating specific hypotheses about potential drug safety issues, but only recently has the research community sought to systematically explore these data to proactively identify safety signals. In 2007, the US Congress passed the Food and Drug Administration Amendment Act, which required the Food and Drug Administration to establish a “postmarket risk identification and analysis system” with access to >100 million lives of electronic health-care data.28 In response, the Food and Drug Administration established the Sentinel Initiative, which has made progress toward developing a national data infrastructure, but has not yet conducted medication-wide analyses to identify potential safety concerns.29 Our work illustrates a proof-of-concept approach for signal generation that can enable standardized surveillance of specific health outcomes of interest across all medical products.
Our MWAS visualizations demonstrate both the opportunity and challenge of pharmacovigilance in these large health-care databases. Most of the signals identified in these analyses were positive controls that we would hope a system would detect, and the majority of negative controls failed to yield statistically significant false-positive associations. This performance reflects the previously documented predictive value of up to 0.8.18
Nevertheless, for each outcome, we observed a large number of drugs known not to have side effects that did have significant statistical associations. Conversely, many drugs known to have effects do not signal despite the large size of the database. All the four plots in Figure 1 contain false-positives (circles) above the Bonferroni-corrected threshold of ~0.0005, and three of the four have false-positives at the most significant P values. Therefore, the false-positives are not due to testing multiple hypotheses and we must consider sources of error such as confounding. For example, the very strong signal for hydrochlorothiazide causing acute renal failure may be due to its common coprescription in patients with renal impairment. The self-controlled design used in this analysis is only one of several alternative approaches that can be considered. While the SCCS explicitly addresses time-invariant confounding factors, such as gender, race, and genetics, it does not control for time-varying factors other than concomitant medication exposure. Other study design approaches include a new user cohort design, which uses an active comparator as a referent and estimates event rates during the time following initiation of treatment, and the case–control design, which compares exposure rates during the time before outcome incidence and compares with exposure rates among matched patients who did not experience the outcome. We present the results from the SCCS because this design has been demonstrated in OMOP's experiments to have higher predictive accuracy and lower bias than these alternative approaches.18 Future work should be considered to determine how best to combine results across multiple analyses to improve our understanding of the effects of medical products.
If we group drugs by the organ system of their indications for each of the four side effects (drugs grouped by color in Figure 1), we found a tendency of the drugs to act similarly within groups. We found 28 groups where all drugs in the organ class were negative and no association was found and 5 in which there were drugs with known side effects and an association was found in more than half. Thus, 33 of 59 groups were handled well by the algorithm. In some cases, such as the positive effects of nonsteroidal anti-inflammatory drugs and acute myocardial infarction, the consistency of the findings supports the observation of a potential effect. There were 15 groups in which most or all of the known drugs with true side effects were missed, 2 groups in which a significant proportion of the drugs known not to have a side effect were found to have an association, 7 groups with a single spurious false-positive association, and 2 groups with a combination of a spurious association and incomplete or nearly complete identification of true side effects. For example, despite the known increased risk of acute liver injury after exposure to antivirals, the consistent lack of observed association could falsely lead to a conclusion that there is no effect. The tendency of drugs to act similarly within groups probably reflects biases due to the health-care process, because in most cases, the drugs within a group are not structurally similar. Despite the presence of these patterns, no single pattern appeared to reliably identify a drug as a true- or false-positive. For example, a single association within a group could be spurious or true, and a preponderance of associations within a group could represent accurate identification, a run of false-positives, or a combination.
Three of the graphs are notable for a lack of obvious confounding by indication. Drugs with an indication that was related to the side effects—cardiovascular for myocardial infarction, urologic for renal failure, and alimentary track for ulcer—did not produce false-positive associations, so the self-controlled study appeared to work in these cases. For acute liver failure, however, the false-positive findings observed for alimentary track drugs may be due in some way to the effects or treatment of liver failure.
One potential approach to addressing imperfect data is to combine evidence from disparate sources. Figure 3 shows two very different databases, derived from claims data and EHR data. Combining the two does not appear to help discriminate true signals from false ones; similar results were found for the other three side effects. We performed additional experiments with two additional databases and found that multiple approaches to synthesize evidence across databases failed to improve discrimination. These results suggest that different health-care databases may exhibit similar biases, such that pharmacovigilance activities may require information sources beyond observational data to support the evaluation of safety signals.
A P-value plot can be a useful test when each test can be considered as independent and unbiased.30 You can determine whether the number of significance tests is consistent with the unbiased, independence assumption by assessing whether the range of tests does not deviate from the 45% line. In the context of observational studies, we expect that results may be biased, and studies of the same outcome are likely correlated insofar as the sources of bias for a given outcome may be consistent across multiple drugs. This can be seen from the P-value plots of the negative controls (Figure 2), which show a disproportionate number of significant findings. For this reason, we argue that statistical significance using traditional P values or multiplicity-adjusted thresholds are insufficient, and instead rank-ordering effects based on P value, as we display in the MWAS plots, may be a more principled approach to triaging potential drug safety concerns.
The MWAS approach of systematic exploration of structured observational health-care claims and EHR databases is only one tool to complement other recent innovations toward improving the evidence base about the safety profile of medical products. LePendu et al. have demonstrated how natural language processing of free text in medical records can be used to draw inferences about potential drug–side effect relationships.31 Harpaz et al. recently measured the performance of new algorithms for data mining in spontaneous adverse event reporting data and demonstrated that disparate data may have differential performance across health outcomes of interest.32 Tatonetti et al.33,34 and Duke et al.35 have successfully demonstrated the potential to go beyond studying the main effects to also explore drug–drug interactions in the same data, and to integrate the results of observational analysis with other information sources, such as the published literature and chemical structure ontologies.
MWASs provide a structured approach for evaluating potential drug safety concerns across all products in a way that provides the necessary context for interpreting any one drug-safety question of interest. While these illustrations focus on a defined set of negative and positive control test cases for methodological purposes, we believe this graphical representation provides a consistent framework that can be applied to all drugs and outcomes as a means to assess the drug–outcome pairs for which we are still uncertain about the true extent of the potential relationship. That context involves understanding how unique a particular observation is by seeing how many other drugs yielded similar effects, and also involves seeing how consistent findings are with medical products that share similar characteristics. Further context is provided by evaluating an association through replication within two or more data sources. In this regard, the MWAS visualization using an SCCS analysis across multiple databases provides a framework that embodies several of the elements required for evaluating a potential causal effect, including strength of association, consistency, temporality, specificity, and coherence.36 Observational health-care data alone may not be sufficient to provide definitive evidence of any purported effect; however, systematic analysis of these data offers tremendous potential in providing credible evidence for advancing our understanding of the effects of medical products across large populations and a wide variety of products.
Methods
We conducted this analysis in two observational health-care databases, the Truven MarketScan CCAE administrative claims database and the GE Centricity EHR database.18 CCAE represents a privately insured population and captures inpatient and outpatient medical claims and pharmacy claims of multiple insurance plans. The database used in this analysis contained 46.5 million lives with >97.6 million patient-years of observation from 2003 to 2009. We defined periods of drug exposure based on pharmacy dispensing records and procedural administrations. The GE MQIC (Medical Quality Improvement Consortium) represents the group of providers who use the GE Centricity Electronic Medical Record and who contribute their data for secondary analytic use. The GE MQIC database reflects events in usual care, including patient problem lists, prescribing patterns and over-the-counter use of medications, and other clinical observations as experienced in the ambulatory care setting. GE contains 11.2 million lives with data from 1996 to 2008. Drug exposures were inferred from medication history and prescriptions written. For both databases, we applied standardized algorithms to define acute myocardial infarction, acute liver failure, acute renal failure, and upper gastrointestinal bleeding based on diagnosis codes on patient and outpatient medical claims.37
For each outcome, we identified a set of negative and positive controls. Ground truth was established based on systematic literature review and natural language processing of structured product labeling, with positive controls identified as drugs with Boxed Warnings or Precautions that are supported by published evidence with no conflicting published studies, and negative controls defined as drugs with no evidence suggesting an association in either labeling or literature.38 Drugs with inconsistent evidence were excluded. The MWAS plots shown in Figure 1 display the full set of negative and positive controls for each outcome that were tested as part of the OMOP experiment. The specific number of drugs varies by outcome; 118 drugs were studied for acute liver injury, 102 for acute myocardial infarction, 88 for acute renal failure, and 91 for gastrointestinal bleeding. Analyses were performed on RxNorm ingredient concepts. RxNorm concepts were classified using the Anatomical Therapeutic Chemical hierarchy only for presentation purposes, but this classification does not affect the effect estimation procedure. The RxNorm-to-Anatomical Therapeutic Chemical mapping is part of the OMOP vocabulary model and was created by and licensed from FirstDataBank. The entire OMOP vocabulary is publicly available online (http://omop.org/CDMvocabV4).
For each drug–outcome pair, we performed an SCCS analysis,39,40 which compares the event rate during time-at-risk with the rate during the time unexposed among patients who had at least one exposure and one outcome record. We defined time-at-risk as the all-time postexposure start, including the index date when treatment was initiated and continuing through the end of the patient's observation period. All time before starting the drug exposure is considered as the unexposed period. We included all occurrences of outcome. We applied a regularized implementation of the SCCS model,41 with the regularization parameter determined by crossvalidation, and we did multivariate adjustment for time-varying concomitant medications. The multivariate SCCS implementation uses all RxNorm ingredients as potential covariates in the model. Only those RxNorm ingredients which are observed in patients with exposure to the target drug and an occurrence of the event are actually fit within each model. Each analysis produced an incidence rate ratio, 95% confidence interval, and P value. The MWAS plot displays the P value on the negative log scale across all drugs for the same outcome. Drugs are grouped according to the Anatomical Therapeutic Chemical classification system. The source code of the SCCS implementation used to produce this analysis is publicly available online (http://omop.org/MethodsLibrary). The entire result set of all methods executions across a network of observational databases for all drug-outcome test cases is also publicly available online (http://omop.org/Research).
Only fully deidentified data sets were used in the study and only aggregate-level data are reported, so the review by Institutional Review Board was not required.
Author contributions
P.R., D.M., P.S., M.S., and G.H. wrote the manuscript. P.R., D.M, P.S., M.S., and G.H. designed the research. P.R., D.M., P.S, M.S., and G.H. performed the research. P.R., M.S., and G.H. analyzed the data. P.R., D.M., P.S., and M.S. contributed new reagents/analytical tools.
Conflict of interest
The authors declared no conflicts of interest.
Study Highlights

Acknowledgments
G.H. was funded by a grant from the National Library of Medicine, “Discovering and applying knowledge in clinical databases” (R01 LM006910). P.R., M.S., P.S., and D.M. are research investigators for the Observational Medical Outcomes Partnership (OMOP). P.R., M.S., and P.S. are employees of Janssen Research and Development, and do not receive funding for their participation in OMOP. D.M. receives funding through the Foundation of the National Institutes of Health.
References
- Blumenthal D., Tavenner M. The “meaningful use” regulation for electronic health records. N. Engl. J. Med. 2010;363:501–504. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- Access to CMS Data & Application. < http://www.cms.gov/Research-Statistics-Data-and-Systems/CMS-Information-Technology/AccesstoDataApplication/index.html > Accessed 20 January 2013.
- US Food and Drug Administration (FDA). Adverse Event Reporting System (AERS). < http://www.fda.gov/cder/aers > Accessed 20 January 2013.
- Friedman C.P., Wong A.K., Blumenthal D. Achieving a nationwide learning health system. Sci. Transl. Med. 2010;2:57cm29. doi: 10.1126/scitranslmed.3001456. [DOI] [PubMed] [Google Scholar]
- World Health Organization. The importance of pharmacovigilance—safety monitoring of medicinal products. World Health Organization; Geneva; . < http://apps.who.int/medicinedocs/en/d/Js4893e/ > 2002. Accessed 20 January 2013. [Google Scholar]
- Lazarou J., Pomeranz B.H., Corey P.N. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998;279:1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- Classen D.C., Pestotnik S.L., Evans R.S., Lloyd J.F., Burke J.P. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA. 1997;277:301–306. [PubMed] [Google Scholar]
- Ahmad S.R. Adverse drug event monitoring at the Food and Drug Administration. J. Gen. Intern. Med. 2003;18:57–60. doi: 10.1046/j.1525-1497.2003.20130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins D., et al. GRADE Working Group Grading quality of evidence and strength of recommendations. BMJ. 2004;328:1490. doi: 10.1136/bmj.328.7454.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin J.A., Glasser S.C., Ellenberg S.S. Adverse event detection in drug development: recommendations and obligations beyond phase 3. Am. J. Public Health. 2008;98:1366–1371. doi: 10.2105/AJPH.2007.124537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller P.C., Evans S.J. A model for the future conduct of pharmacovigilance. Pharmacoepidemiol. Drug Saf. 2003;12:17–29. doi: 10.1002/pds.773. [DOI] [PubMed] [Google Scholar]
- Harpaz R., DuMouchel W., Shah N.H., Madigan D., Ryan P., Friedman C. Novel data-mining methodologies for adverse drug event discovery and analysis. Clin. Pharmacol. Ther. 2012;91:1010–1021. doi: 10.1038/clpt.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A.M. Confounding by indication. Epidemiology. 1996;7:335–336. [PubMed] [Google Scholar]
- Rosenbaum P.R., Rubin D.B. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:41–55. [Google Scholar]
- Wang X., Hripcsak G, Friedman C. Characterizing environmental and phenotypic associations using information theory and electronic health records. BMC Bioinformatics. 2009;10 suppl. 9:S13. doi: 10.1186/1471-2105-10-S9-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- Stang P.E., et al. Advancing the science for active surveillance: rationale and design for the Observational Medical Outcomes Partnership. Ann. Intern. Med. 2010;153:600–606. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- Ryan P.B., Madigan D., Stang P.E., Overhage J.M., Racoosin J.A., Hartzema A.G. Empirical assessment of methods for risk identification in healthcare data: results from the experiments of the Observational Medical Outcomes Partnership. Stat. Med. 2012;31:4401–4415. doi: 10.1002/sim.5620. [DOI] [PubMed] [Google Scholar]
- Arellano F.M. The withdrawal of rofecoxib. Pharmacoepidemiol. Drug Saf. 2005;14:213–217. doi: 10.1002/pds.1077. [DOI] [PubMed] [Google Scholar]
- Lependu P., Iyer S.V., Fairon C., Shah N.H. Annotation Analysis for Testing Drug Safety Signals using Unstructured Clinical Notes. J. Biomed. Semantics. 2012;3 suppl. 1:S5. doi: 10.1186/2041-1480-3-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein J.S., Sordo M., Kohane I.S., Mandl K.D. The tell-tale heart: population-based surveillance reveals an association of rofecoxib and celecoxib with myocardial infarction. PLoS ONE. 2007;2:e840. doi: 10.1371/journal.pone.0000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.S., et al. Early detection of adverse drug events within population-based health networks: application of sequential testing methods. Pharmacoepidemiol. Drug Saf. 2007;16:1275–1284. doi: 10.1002/pds.1509. [DOI] [PubMed] [Google Scholar]
- Schuemie M.J., et al. Using electronic health care records for drug safety signal detection: a comparative evaluation of statistical methods. Med. Care. 2012;50:890–897. doi: 10.1097/MLR.0b013e31825f63bf. [DOI] [PubMed] [Google Scholar]
- Ikram M.A., et al. Genomewide association studies of stroke. N. Engl. J. Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny J.C., et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Classification of Diseases, 9th Revision, Clinical Modification. < http://www.cdc.gov/nchs/icd/icd9cm.htm > Accessed 21 January 2013.
- WHO Collaborating Centre for Drug Statistics Methodology. Anatomical therapeutic chemical classification system: structure and principles. < http://www.whocc.no/atc/structure_and_principles/ > Accessed 21 January 2013.
- Food and Drug Administration Amendments Act of 2007. Public Law 110–85, 21 STAT. 823. 2007.
- Robb M.A., et al. The US Food and Drug Administration's Sentinel Initiative: expanding the horizons of medical product safety. Pharmacoepidemiol. Drug Saf. 2012;21 suppl. 1:9–11. doi: 10.1002/pds.2311. [DOI] [PubMed] [Google Scholar]
- Schweder T, Spjotvoll E. Plots of P-values to evaluate many tests simultaneously. Biometrika. 1982;69:493–502. [Google Scholar]
- LePendu P., et al. Pharmacovigilance using clinical notes. Clin. Pharmacol. Ther. 2013;93:547–555. doi: 10.1038/clpt.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz R., DuMouchel W., LePendu P., Bauer-Mehren A., Ryan P., Shah N.H. Performance of pharmacovigilance signal-detection algorithms for the FDA adverse event reporting system. Clin. Pharmacol. Ther. 2013;93:539–546. doi: 10.1038/clpt.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatonetti N.P., Ye P.P., Daneshjou R., Altman R.B. Data-driven prediction of drug effects and interactions. Sci. Transl. Med. 2012;4:125ra31. doi: 10.1126/scitranslmed.3003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatonetti N.P., Fernald G.H., Altman R.B. A novel signal detection algorithm for identifying hidden drug-drug interactions in adverse event reports. J. Am. Med. Inform. Assoc. 2012;19:79–85. doi: 10.1136/amiajnl-2011-000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke J.D., et al. Literature based drug interaction prediction with clinical assessment using electronic medical records: novel myopathy associated drug interactions. PLoS Comput. Biol. 2012;8:e1002614. doi: 10.1371/journal.pcbi.1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A.B. The environment and disease: association or causation. Proc. R. Soc. Med. 1965;58:295–300. doi: 10.1177/003591576505800503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Observational Medical Outcomes Partnership, Health Outcomes of Interest Library. < http://omop.org/HOI > Accessed 24 July 2013.
- Tisdale J., Miller D.Drug-Induced Diseases: Prevention, Detection, and Management2nd edn. (Bethesda, MD: American Society of Health-System Pharmacists; 2010 [Google Scholar]
- Whitaker H.J., Hocine M.N., Farrington C.P. The methodology of self-controlled case series studies. Stat. Methods Med. Res. 2009;18:7–26. doi: 10.1177/0962280208092342. [DOI] [PubMed] [Google Scholar]
- Whitaker H.J., Farrington C.P., Spiessens B., Musonda P. Tutorial in biostatistics: the self-controlled case series method. Stat. Med. 2006;25:1768–1797. doi: 10.1002/sim.2302. [DOI] [PubMed] [Google Scholar]
- Madigan D., Ryan P., Simpson S., Zorych I.Bayesian methods in pharmacovigilance. Bayesian Statistics 9.eds.Bernardo J.M.et al) (Oxford, UK Oxford University Press; 2011 [Google Scholar]