This work presents a systematic analysis of how VPS-C/HOPS, CCZ-1/SAND-1, and RAB-7, which have well-defined roles in lysosome formation, act in the biogenesis of Caenorhabditis elegans lysosome-related organelles. It identifies key molecular similarities and differences in trafficking to these homologous, yet distinct organelles.
Abstract
As early endosomes mature, the SAND-1/CCZ-1 complex acts as a guanine nucleotide exchange factor (GEF) for RAB-7 to promote the activity of its effector, HOPS, which facilitates late endosome–lysosome fusion and the consumption of AP-3–containing vesicles. We show that CCZ-1 and the HOPS complex are essential for the biogenesis of gut granules, cell type–specific, lysosome-related organelles (LROs) that coexist with conventional lysosomes in Caenorhabditis elegans intestinal cells. The HOPS subunit VPS-18 promotes the trafficking of gut granule proteins away from lysosomes and functions downstream of or in parallel to the AP-3 adaptor. CCZ-1 also acts independently of AP-3, and ccz-1 mutants mistraffic gut granule proteins. Our results indicate that SAND-1 does not participate in the formation of gut granules. In the absence of RAB-7 activity, gut granules are generated; however, their size and protein composition are subtly altered. These observations suggest that CCZ-1 acts in partnership with a protein other than SAND-1 as a GEF for an alternate Rab to promote gut granule biogenesis. Point mutations in GLO-1, a Rab32/38-related protein, predicted to increase spontaneous guanine nucleotide exchange, specifically suppress the loss of gut granules by ccz-1 and glo-3 mutants. GLO-3 is known to be required for gut granule formation and has homology to SAND-1/Mon1–related proteins, suggesting that CCZ-1 functions with GLO-3 upstream of the GLO-1 Rab, possibly as a GLO-1 GEF. These results support LRO formation occurring via processes similar to conventional lysosome biogenesis, albeit with key molecular differences.
INTRODUCTION
Many cell types contain multiple types of lysosomal compartments. Like the ubiquitous conventional lysosomes with which they typically coexist, lysosome-related organelles (LROs) are often acidified and produced through the action of the endocytic pathway (Marks et al., 2013; Yousefian et al., 2013). LROs represent a diverse array of compartments that are distinguished from each other by their cell type–specific morphology, composition, and function (Dell'Angelica et al., 2000). Defective formation of LROs can lead to the disease Hermansky–Pudlak syndrome (HPS). HPS patients and mouse HPS disease models have altered body pigmentation and blood clotting, resulting from aberrant biogenesis of LROs in melanocytes and platelets (Huizing et al., 2008). LROs are not restricted to vertebrates and are found in invertebrates, protozoa, and possibly plants (Raposo et al., 2007; Sohn et al., 2007; Docampo et al., 2010).
Gut granules are cell type–specific LROs found within Caenorhabditis elegans intestinal cells (Hermann et al., 2005). They contain birefringent and autofluorescent material, whose accumulation is coincident with their formation during embryogenesis (Laufer et al., 1980; Bossinger and Schierenberg, 1992). Gut granules function in zinc storage, limiting its toxicity (Roh et al., 2012), contain tryptophan derivatives of the kynurenine pathway (Coburn et al., 2013), and are linked to lipid metabolism (Mak et al., 2006; O'Rourke et al., 2009; O'Rourke and Ruvkun, 2013). Although gut granules share some characteristics and proteins with conventional lysosomes, the two organelles are distinct and coexist within intestinal cells (Clokey and Jacobson, 1986; Levitte et al., 2010; Hermann et al., 2012). Many of the factors mediating trafficking to C. elegans gut granules have evolutionarily conserved roles in the biogenesis of LROs in Drosophila and mammals (Hermann et al., 2005, 2012; Raposo et al., 2007).
Most of the molecular machinery known to guide LRO biogenesis is also used in the formation of conventional lysosomes. These include the AP-3 adaptor (Dell'Angelica, 2009), BLOC-1 (Di Pietro et al., 2006; Salazar et al., 2006; Setty et al., 2007), and HOPS tethering complexes (Zlatic et al., 2011a; Solinger and Spang, 2013), suggesting that the mechanisms promoting trafficking to LROs and lysosomes are likely to be similar. However, there are some factors, such as Rab32/38-related proteins (Ma et al., 2004; Hermann et al., 2005; Wasmeier et al., 2006), that likely function exclusively in the biogenesis of LROs. Knowing the similarities and differences between trafficking pathways leading to lysosomes and LROs will be key in understanding the biogenesis of LROs and the cellular processes that simultaneously generate these related, yet structurally and functionally distinct organelles.
Studies reveal an evolutionarily conserved mechanism for protein trafficking from early endosomes to lysosomes by which early endosomes, or early endosome subdomains (Rink et al., 2005; Vonderheit and Helenius, 2005), mature into late endosomes via the action of a SAND-1(Mon1)/CCZ-1 complex. This complex promotes both the inactivation and removal of RAB5 from early endosomes and the activation and recruitment of RAB7, a process called Rab conversion (Kinchen and Ravichandran, 2010; Nordmann et al., 2010; Poteryaev et al., 2010; Gerondopoulos et al., 2012). Active RAB7 recruits the HOPS tethering complex to late endosomes and lysosomes (Cabrera et al., 2010; Ostrowicz et al., 2010), promoting their heterotypic fusion to create endolysosomes (Caplan et al., 2001; Sriram et al., 2003; Richardson et al., 2004; Kinchen et al., 2008; Hickey and Wickner, 2010; Pols et al., 2013), which then mature into lysosomes via membrane budding (Pryor et al., 2000; Bright et al., 2005). HOPS also functions in the fusion of Golgi apparatus or early endosome–derived, AP-3–containing vesicles with endolysosomes and lysosomes (Darsow et al., 2001; Angers and Merz, 2009; Cabrera et al., 2010).
Parallels with lysosome biogenesis suggest that similar mechanisms function in trafficking to LROs. Mammalian paralogues of SAND-1 and CCZ-1, called HPS1 and HPS4 (Hoffman-Sommer et al., 2005; Kinch and Grishin, 2006), respectively, form BLOC-3 (Chiang et al., 2003; Martina et al., 2003; Nazarian et al., 2003), which functions in the formation of LROs (Oh et al., 1996; Suzuki et al., 2002). Rab7 is localized on immature and mature LROs (Jordens et al., 2006; Zhang et al., 2011; Ambrosio et al., 2012), and disrupting Rab7 activity can alter the distribution of LRO contents (Hirosaki et al., 2002; Kawakami et al., 2008; Hida et al., 2011). In various metazoan systems, mutations affecting the function of HOPS subunits cause reduced numbers of LROs (Sevrioukov et al., 1999; Hermann et al., 2005; Pulipparacharuvil et al., 2005; Sadler et al., 2005; Maldonado et al., 2006; Akbar et al., 2009; Thomas et al., 2011). AP-3 functions during the formation of LROs (Ooi et al., 1997; Simpson et al., 1997; Kantheti et al., 1998; Dell'Angelica et al., 1999; Feng et al., 1999; Mullins et al., 1999, 2000; Kretzschmar et al., 2000; Hermann et al., 2005) by interacting with the cytoplasmic domain of a subset of LRO-destined transmembrane proteins, directing their vesicular transport to LROs from early endosomes (Huizing et al., 2001; Theos et al., 2005; Setty et al., 2007).
Here we systematically analyze the role of genes promoting protein trafficking from early endosomes to lysosomes during gut granule biogenesis in C. elegans intestinal cells and document genetic interactions revealing their functional relationships. Our results suggest that the pathways constructing LROs and conventional lysosomes are likely similar; however, they highlight significant molecular differences.
RESULTS
Formation of autofluorescent and birefringent gut granules in endolysosomal trafficking mutants
To address whether the machinery that mediates protein trafficking from early endosomes to conventional lysosomes also affects trafficking to LROs, we used C. elegans mutants that lack or have strongly reduced activity of VPS-C/HOPS, CCZ-1, SAND-1/Mon1, and RAB-7. These mutants were analyzed for their effects on the formation of gut granules, which contain birefringent material and autofluorescent material and are acidified (Laufer et al., 1980; Clokey and Jacobson, 1986; Bossinger and Schierenberg, 1992). Defective gut granule biogenesis leads to loss of acidified intestinal organelles and loss and/or extracellular misaccumulation of the birefringent and autofluorescent material, which we refer to as a gut granule loss (Glo) phenotype (Hermann et al., 2005). Following standard C. elegans nomenclature, genes are given in lowercase italics and proteins are given in uppercase nonitalic.
VPS-C/HOPS.
VPS-C comprises VPS-11, -16, -18, and -33. Two additional subunits, VPS-39 and -41, associate with VPS-C to form HOPS (Nickerson et al., 2009; Balderhaar and Ungermann, 2013; Graham et al., 2013). C. elegans encodes a protein orthologous to each HOPS subunit (Solinger and Spang, 2013; Supplemental Figure S1), and there are mutants predicted to severely disrupt the function of each of the six HOPS subunits (Supplemental Figure S1). Our prior work showed that mutations disrupting vps-16 or vps-41 led to a Glo phenotype in adults (Hermann et al., 2005), and we found that additional mutant alleles and RNA interference (RNAi) targeting these genes led to a similar phenotype, further supporting their role in gut granule biogenesis (Figure 1 and Table 1). We investigated whether the four other subunits of VPS-C/HOPS were similarly required for gut granule biogenesis. Mutations in three other HOPS genes—vps-11, vps33.1, and vps-39—also led to a reduction in the number of autofluorescent compartments (Figure 1 and Table 1). Of these, vps-39 presented the weakest Glo phenotype, which was enhanced by vps-39(RNAi) (Table 1), suggesting that the vps-39 mutant allele retains function or the adult Glo phenotype can be maternally rescued. The vps-18(tm1125) mutant did not display an obvious alteration in adult gut granule number (Figure 1 and Table 1), although it did exhibit defective formation of gut granules in embryos (see later discussion).
FIGURE 1:
Autofluorescent compartments in adult intestinal cells. The 9-μm maximum-intensity projections, which span 50% the width of the intestine, of GFP channel autofluorescence centered on the lumen of the anterior intestine show that, in comparison to (A) wild type, the (B) vps-11, (C) vps-16, (E) vps-33, (F) vps-39, (G) vps-41, and (I) ccz-1 mutants exhibited reduced numbers of autofluorescent compartments. Mutations in (D) vps-18, (H) sand-1, and (J) rab-7 did not obviously alter the number of autofluorescent organelles. Black arrows denote representative autofluorescent compartments.
TABLE 1:
Gut granules in adults.
| Percentage of animals with the specified number of autofluorescent organelles in anterior intestinal cells | |||||||
|---|---|---|---|---|---|---|---|
| Genotype | 0 | 1–20 | 21–50 | 51–100 | 101–200 | >200 | n |
| Wild typea | 0 | 0 | 0 | 0 | 0 | 100 | 38 |
| VPS-C subunits | |||||||
| vps-11(ok1664) | 0 | 0 | 0 | 76 | 24 | 0 | 34 |
| vps-16(ok719) | 0 | 13 | 87 | 0 | 0 | 0 | 31 |
| vps-16(ok776) | 0 | 4 | 92 | 4 | 0 | 0 | 25 |
| vps-16(RNAi)b | 0 | 3 | 41 | 56 | 0 | 0 | 87 |
| spe-39(eb9) | 0 | 0 | 0 | 0 | 0 | 100 | 25 |
| spe-39(RNAi)b | 0 | 0 | 0 | 0 | 8 | 92 | 25 |
| vps-18(tm1125), 22°C | 0 | 0 | 0 | 0 | 0 | 100 | 37 |
| vps-18(tm1125), 25°C | 0 | 0 | 0 | 0 | 59 | 41 | 22 |
| vps-18(RNAi)b | 0 | 0 | 0 | 0 | 8 | 92 | 25 |
| vps-18(tm1125); vps-18(RNAi) | 0 | 0 | 19 | 62 | 19 | 0 | 21 |
| vps-33.1(ok2494) | 0 | 4 | 81 | 15 | 0 | 0 | 26 |
| vps-33.2(RNAi)b | 0 | 0 | 0 | 0 | 15 | 85 | 48 |
| vps-45(tm246)c | 0 | 0 | 0 | 0 | 0 | 100 | 22 |
| vps-45(RNAi)b | 0 | 0 | 0 | 0 | 0 | 100 | 31 |
| CORVET-specific subunit | |||||||
| vps-8(RNAi)b | 0 | 0 | 0 | 0 | 14 | 88 | 44 |
| HOPS-specific subunits | |||||||
| vps-39(ok2442) | 0 | 0 | 0 | 10 | 84 | 6 | 31 |
| vps-39(tm2253) | 0 | 0 | 0 | 10 | 90 | 0 | 31 |
| vps-39(RNAi)b | 0 | 0 | 0 | 0 | 46 | 54 | 37 |
| vps-39(tm2253); vps-39(RNAi)d | 0 | 33 | 67 | 0 | 0 | 0 | 21 |
| vps-41(ep402) | 0 | 0 | 64 | 36 | 0 | 0 | 22 |
| vps-41(ok3433) | 0 | 0 | 21 | 75 | 4 | 0 | 44 |
| vps-41(RNAi)b | 0 | 60 | 40 | 0 | 0 | 0 | 30 |
| HOPS regulators | |||||||
| ccz-1(ok2182) | 59 | 29 | 6 | 2 | 0 | 4 | 51 |
| ccz-1(t2129)d | 0 | 94 | 6 | 0 | 0 | 0 | 45 |
| ccz-1(t2129) +ccz-1::yfpe | 0 | 0 | 0 | 70 | 30 | 0 | 20 |
| ccz-1(RNAi)f | 0 | 0 | 0 | 5 | 90 | 5 | 21 |
| sand-1(ok1963) | 0 | 0 | 0 | 0 | 0 | 100 | 38 |
| sand-1(or552) | 0 | 0 | 0 | 0 | 0 | 100 | 25 |
| HOPS interactors | |||||||
| apt-6(ok429) | 0 | 72 | 25 | 3 | 0 | 0 | 32 |
| apt-7(tm920) | 0 | 83 | 17 | 0 | 0 | 0 | 30 |
| arl-8(tm2504)g | 0 | 0 | 0 | 0 | 0 | 100 | 20 |
| rab-7(ok511)h | 0 | 0 | 0 | 0 | 0 | 100 | 23 |
| rab-7(RNAi)i | 0 | 0 | 0 | 0 | 0 | 100 | 25 |
| Double mutants | |||||||
| apt-6(ok429); vps-18(tm1125)j | 100 | 0 | 0 | 0 | 0 | 0 | 45 |
| apt-7(tm920); vps-18(tm1125)j | 92 | 8 | 0 | 0 | 0 | 0 | 52 |
| apt-6(ok429); vps-41(ep402)k | 3 | 95 | 2 | 0 | 0 | 0 | 42 |
| apt-6(ok429); ccz-1(RNAi) | 75 | 25 | 0 | 0 | 0 | 0 | 20 |
| apt-7(tm920); ccz-1(RNAi) | 95 | 5 | 0 | 0 | 0 | 0 | 22 |
| apt-7(tm920); ccz-1(ok2182) | 100 | 0 | 0 | 0 | 0 | 0 | 25 |
| apt-6(ok429); rab-7(ok511)l | 0 | 50 | 50 | 0 | 0 | 0 | 24 |
| apt-7(tm920); rab-7(ok511)l | 0 | 62 | 38 | 0 | 0 | 0 | 21 |
| apt-6(ok429); rab-7(RNAi)l | 0 | 71 | 29 | 0 | 0 | 0 | 21 |
| apt-7(tm920); rab-7(RNAi)l | 0 | 87 | 13 | 0 | 0 | 0 | 23 |
| apt-7(tm920); sand-1(or552) | 0 | 96 | 4 | 0 | 0 | 0 | 28 |
| glo-2(zu455) | 0 | 46 | 54 | 0 | 0 | 0 | 63 |
| glo-2(zu455); rab-7(ok511)l | 0 | 52 | 48 | 0 | 0 | 0 | 27 |
| glo-2(tm592) | 0 | 0 | 0 | 31 | 69 | 0 | 29 |
| glo-2(tm592); rab-7(ok511)l | 0 | 0 | 0 | 41 | 59 | 0 | 22 |
| glo-3(kx90) | 0 | 33 | 67 | 0 | 0 | 0 | 21 |
| glo-3(kx90); rab-7(ok511)l | 3 | 90 | 7 | 0 | 0 | 0 | 30 |
| pgp-2(kx48) | 0 | 0 | 0 | 60 | 40 | 0 | 20 |
| pgp-2(kx48); rab-7(ok511)l | 0 | 27 | 73 | 0 | 0 | 0 | 30 |
All strains were grown at 22˚C unless otherwise noted. Adult-stage animals were analyzed using fluorescence microscopy with a GFP or DAPI filter set and were scored for the number of autofluorescent organelles within the anterior intestine located between the pharynx and vulva.
aWild type had >200 at both 22 and 25°C.
bRNAi was carried out in the rrf-3(pk1426) RNAi–sensitive strain (Simmer et al., 2003). F33E2.4(RNAi) targeting a gene not involved in gut granule formation did not alter number of autofluorescent compartments in rrf-3(-).
cvps-45(tm246) animals were grown at 22˚C from early larvae to adulthood before scoring.
dvps-39(tm2253)/nT1[qIs51] animals were placed on RNAi plates as embryos and grown for two generations before being scored. Only animals lacking GFP, which are vps-39(-)/vps-39(-), were analyzed. F33E2.4(RNAi) targeting a gene not involved in gut granule formation did not alter number of autofluorescent compartments in these animals (unpublished data).
eDpy progeny of dpy-11(e224) ccz-1(t2129)/nT1[qIs51] were scored. dpy-11(e224) ccz-1(t2129); jcpEx2[ccz-1::yfp; myo-2::gfp] animals expressing GFP were scored. Non–GFP-expressing animals, which lack jcpEx2, were Glo (unpublished data).
fRNAi was carried out in the N2 background. F33E2.4(RNAi) targeting a gene not involved in gut granule formation did not alter number of autofluorescent compartments.
gNon–GFP-expressing arl-8(-)/arl-8(-) progeny of arl-8(tm2504)/nT1[qIs51] were scored.
hNon–GFP-expressing rab-7(-)/rab-7(-) progeny of rab-7(ok511)/mIn1[mIs14] were scored.
iAdults scored had prominent, enlarged, refractile structures visible with DIC microscopy in the most proximal oocytes, which are indicative of rab-7(-).
junc-4(e120), which did not alter the Glo phenotype, was present in the background.
kvps-41(-)/vps-41(-) adult progeny of vps-41(-)/+; apt-6(-)/apt-6(-) parents were identified by the accumulation of apoptotic corpses in the germline and arrested embryos in the uterus.
lrab-7(-)/rab-7(-) adult progeny of rab-7(-)/+; glo(-)/glo(-) parents were identified by the refractile structures visible with DIC microscopy in the most proximal oocytes, which are indicative of rab-7(-).
The VPS-C/HOPS subunit mutants that exhibited an adult Glo phenotype also displayed maternal effect embryonic lethality, producing progeny that arrest at a stage in embryogenesis before the formation of gut granules, thus preventing an analysis of their effect on gut granules biogenesis in embryos (unpublished observations). We therefore used RNAi, which often leads to a partial knockdown of gene function, to assess the role of VPS-C/HOPS genes in embryonic gut granule biogenesis. Birefringent material, which can be visualized with polarization microscopy, is restricted to, and first becomes apparent within, gut granules at the bean stage, midway through embryogenesis (Laufer et al., 1980). This marker continues to specifically accumulate in gut granules as embryos elongate through the 1.5-fold to the pretzel stage (Hermann et al., 2005; Figure 2A). We found that RNAi targeting vps-11, -16, -33.1, -39, and 41 led to an embryonic Glo phenotype (Figure 2, K–O, and Table 2).
FIGURE 2:
Analysis of birefringent and acidified gut granules in embryonic intestinal cells. Embryos were analyzed with polarization microscopy to detect birefringent material, stained with LysoSensor Green, and analyzed with fluorescence microscopy to detect acidified compartments. (A, F) Wild-type, (C, H) sand-1, and (D, I) rab-7 mutant embryos displayed birefringent and acidified gut granules (white arrows). (B, G) ccz-1 and (E, J) vps-18 mutants lacked acidified and birefringent gut granules and mislocalized birefringent material into the intestinal lumen (black arrows). (K–O) RNAi targeting subunits of the VPS-C/HOPS complex in an rrf-3(pk1426) RNAi–sensitive strain caused a reduction in the number of birefringent organelles (white arrows) and sometimes led to misaccumulation of birefringent material in the intestinal lumen (black arrows). (P, Q) Birefringent material (white arrows) was present within the intestinal cells of vps-18 mutants at 15°C and was distributed throughout the embryo at 25°C. The intestine is flanked by black arrowheads in A–P. Pretzel-stage embryos are shown in A–C, E, and K–P; 1.5-fold-stage embryos are shown in D and F–J. A terminally arrested pre–bean stage vps-18 mutant embryo is shown in Q. Embryos are ∼50 μm in length. (R) Individual vps-18 mutant embryos were shifted from 15 to 22°C at the indicated stage, and on reaching threefold stage or later, were analyzed using polarization microscopy. Embryos exhibiting <20 birefringent granules were scored as having a reduced number of granules. E2, E4, and E8 refer to the number of intestinal cells in the intestinal primordium; early-bean-stage embryos are E16 before the apical migration of intestinal nuclei. Bean and 1.5-fold stages refer to the body morphology. The number of embryos scored is indicated in parentheses near each data point. Unshifted vps-18 mutants grown at 15°C exhibited a Glo phenotype only 9% of the time (n = 33). Wild-type embryos that underwent similar temperature shifts always displayed >50 birefringent granules.
TABLE 2:
Gut granules in embryos.
| Genotype | Percentage lacking birefringence in intestinal cellsa | Percentage with 1–20 birefringent granules in intestinal cellsa | Percentage with 21–50 birefringent granules in intestinal cellsa | Percentage with >50 birefringent granules in intestinal cells | n |
|---|---|---|---|---|---|
| Wild typeb | 0 | 0 | 0 | 100 | 43 |
| apt-6(ok429) | 0 | 77 (58) | 23 (23) | 0 | 31 |
| apt-7(tm920) | 0 | 100 (81) | 0 | 0 | 27 |
| ccz-1(ok2182) | 100 (14) | 0 | 0 | 0 | 58 |
| ccz-1(t2129)c | 100 (38) | 0 | 0 | 0 | 90 |
| ccz-1(t2129) + ccz-1::yfpc | 0 | 5 | 71 | 24 | 21 |
| glo-3(kx90) | 44 (17) | 56 (35) | 0 | 0 | 84 |
| pgp-2(kx48) | 0 | 33 | 67 (67) | 0 | 21 |
| rab-7(ok511)d | 0 | 33 | 67 | 0 | 15 |
| rab-7(RNAi)e | 2 | 27 | 68 | 2 | 41 |
| sand-1(ok1963) | 0 | 0 | 0 | 100 | 51 |
| sand-1(or552) | 0 | 0 | 0 | 100 | 49 |
| vps-8(ok2912)f | 0 | 0 | 6 | 94 | 32 |
| vps-11(RNAi)g | 0 | 92 (28) | 8 | 0 | 40 |
| vps-16(RNAi)g | 0 | 100 (83) | 0 | 0 | 47 |
| vps-18(tm1125), 15oC | 0 | 6 | 58 | 36 | 33 |
| vps-18(tm1125), 22oC | 100 (68) | 0 | 0 | 0 | 106 |
| vps-18(tm1125) + vps-18::gfp, 22°C | 0 | 0 | 0 | 100 | 36 |
| vps-33.1(RNAi)g | 0 | 82 (38) | 18 | 0 | 48 |
| vps-33.2(RNAi)g | 0 | 0 | 5 | 95 | 37 |
| vps-39 (RNAi)g | 2 | 72 (6) | 26 | 0 | 47 |
| vps-41(RNAi)g | 0 | 58 (8) | 40 | 2 | 40 |
| vps-45(tm246) | 0 | 0 | 0 | 100 | 51 |
| vps-45(RNAi)g | 0 | 0 | 6 | 94 | 35 |
| Double mutants | 0 | ||||
| apt-6(ok429); vps-18(tm1125)h | 85 (66) | 15 (11) | 0 | 0 | 94 |
| apt-7(tm920); vps-18(tm1125)h | 88 (53) | 12 (8) | 0 | 0 | 74 |
| apt-7(tm920); sand-1(or552) | 0 | 82 (11) | 5 | 3 | 38 |
| apt-7(tm920); ccz-1(ok2182) | 100 (22) | 0 | 0 | 0 | 23 |
| apt-6(ok429); rab-7(ok511)i | 0 | 83 (35) | 17 | 0 | 23 |
| apt-7(tm920); rab-7(ok511)i | 0 | 96 (44) | 8 | 0 | 25 |
| glo-3(kx90); rab-7(RNAi)e | 57 (9) | 43 (23) | 0 | 0 | 131 |
| pgp-2(kx48); rab-7(ok511)i | 0 | 100 (20) | 0 | 0 | 20 |
All strains were grown at 22˚C unless otherwise noted. Threefold- and later-stage embryos were analyzed using polarization microscopy and scored for the presence and localization of birefringent material in the intestine.
aPercentage that also mislocalized birefringent material into the intestinal lumen are in parentheses.
bThe phenotype of wild type was not altered by growth at 15°C (unpublished data).
cDpy progeny of dpy-11(e224) ccz-1(t2129)/nT1[qIs51] were isolated, and their progeny were scored. dpy-11(e224) ccz-1(t2129); jcpEx2[ccz-1::yfp; myo-2::gfp] embryos expressing GFP were scored. Non–GFP-expressing animals, which lacked jcpEx2, were often Glo (not shown).
dNon–GFP-expressing rab-7(-)/rab-7(-) adults from rab-7(ok511)/mIn1[mIs14] parents were isolated, and their embryonic progeny were scored.
eRNAi was carried out in an otherwise wild-type background. F33E2.4(RNAi) targeting a gene not involved in gut granule formation did not alter the number of birefringent compartments in the indicated strain. All embryos scored displayed numerous enlarged refractile structures visible with DIC microscopy, indicative of rab-7(-).
fNon-GFP vps-8(-)/vps-8(-) progeny of vps-8(ok2912)/nT1[qIs51 GFP] were scored.
gRNAi was carried out in the rrf-3(pk1426) RNAi–sensitive strain (Simmer et al., 2003). F33E2.4(RNAi) targeting a gene not involved in gut granule formation did not alter number of birefringent compartments in rrf-3(-).
hunc-4(e120), which did not alter the Glo phenotype, was present in the background.
iEmbryonic progeny of rab-7(-)/rab-7(-); glo(-)/glo(-) parents were scored.
The vps-18(tm1125) mutant, in contrast to other VPS-C/HOPS mutants, is temperature sensitive for growth, being viable between 15 and 22°C and displaying embryonic lethality at 25°C (Xiao et al., 2009). This allele leads to temperature-sensitive defects in cell corpse degradation due to inhibition of lysosome formation and function (Xiao et al., 2009). vps-18 mutant adults did not display a significant Glo phenotype at any temperature (Figure 1D and Table 1). They do, however, show temperature-sensitive defects in the formation of embryonic gut granules (Figure 2E and Table 2). At 15°C, pretzel-stage vps-18 mutant embryos displayed a weak Glo phenotype (Table 2 and Figure 2P). At 22°C, vps-18 mutant embryos displayed a strong and fully penetrant Glo phenotype (Table 2 and Figure 2E), which was rescued by the introduction of vps-18(+)::gfp (Table 2). At 25°C, many vps-18 mutant embryos arrested before elongation, and 76% (n = 33) of these contained birefringent material distributed throughout the embryo (Figure 2Q), suggestive of altered formation or trafficking of birefringent material, which is normally restricted to intestinal cells (Hermann et al., 2005).
We carried out temperature-shift experiments to determine when vps-18 activity promotes the formation of birefringent gut granules. vps-18 mutant embryos cultured at the permissive temperature were shifted to the nonpermissive temperature during early to mid embryogenesis and then scored for the presence and localization of birefringent material at the pretzel stage. Consistent with VPS-18 functioning directly in the formation of gut granules, we found that nearly all of the vps-18 mutant embryos upshifted before the early bean stage, when birefringent gut granules first become apparent (Laufer et al., 1980), lacked or displayed a significant reduction in the number of birefringent intestinal granules (Figure 2R).
The vps-18(tm1125) allele introduces a premature stop codon predicted to truncate the C-terminal 760 amino acids of VPS-18, which contains the α-solenoid, coiled-coil, and RING finger domains (Xiao et al., 2009; Supplemental Figure 1), suggesting that this allele may completely disrupt vps-18(+) activity. However, we found that application of vps-18 RNAi to vps-18(tm1125) mutants led to a significant decrease in the number of autofluorescent granules (Table 1). Thus the vps-18(tm1125) mutant allele exhibits significant activity in promoting adult gut granule formation. In contrast, vps-18(tm1125) mutant embryos exhibited a fully penetrant Glo phenotype at 22°C (Table 2), suggesting that vps-18 function is severely reduced at this stage. We therefore carried out our analyses of vps-18 activity in embryos.
We first addressed whether the defect in gut granule formation exhibited by the vps-18 mutant extended beyond just the generation and localization of birefringent material. Gut granules are the major acidified compartments within embryonic intestinal cells and are stained by LysoSensor Green, whose accumulation and fluorescence depend on an acidic pH (Levitte et al., 2010). Whereas LysoSensor Green marked wild-type gut granules, organelles within vps-18 mutant intestinal cells were not stained, indicating a general defect in gut granule biogenesis (Figure 2, F and J).
In metazoans, there are alternate VPS-16 and VPS-33 subunits that associate with VPS-C/HOPS (Zlatic et al., 2011a; Solinger and Spang, 2013), which are necessary for the formation of some LROs (Lo et al., 2005; Urban et al., 2012). In C. elegans, SPE-39 is homologous to VPS-16, and VPS-33.2 and VPS-45 are homologous to VPS-33.1 (Solinger and Spang, 2013). Mutations in spe-39 or vps-45 and RNAi targeting spe-39, vps-33.2, or vps-45 did not lead to an adult Glo phenotype (Table 1). The Arf-like GTPase Arl8 physically interacts with VPS41 in mammals, and mutation of the C. elegans orthologue arl-8 phenocopies vps-18 mutants (Xiao et al., 2009; Nakae et al., 2010; Garg et al., 2011), suggesting that it functions with the HOPS complex. However, we did not see any effect of an arl-8 mutation on the formation of autofluorescent gut granules (Table 1). In yeast, the VPS-C subunits can also associate with VPS-3 and VPS-8, instead of VPS-39 and VPS-41, to generate CORVET (Peplowska et al., 2007). There is no obvious VPS-3 homologue in C. elegans (Solinger and Spang, 2013). Mutations or RNAi targeting vps-8/(C42C1.4) did not lead to a Glo phenotype (Tables 1 and 2). On the basis of the genes exhibiting a Glo phenotype when altered by mutation or targeted by RNAi, we conclude that the canonical HOPS complex, rather than the CORVET or a VPS-C/HOPS subcomplex, functions in the formation of gut granules.
sand-1/Mon1 and ccz-1.
Current models for membrane trafficking to conventional lysosomes in C. elegans point to a role for a complex of SAND-1 and CCZ-1 as regulators of early endosome–to–late endosome maturation and RAB-5–to–RAB-7 conversion (Kinchen and Ravichandran, 2010; Nieto et al., 2010; Poteryaev et al., 2010). Mammalian genomes encode three distinct proteins—Mon1a, Mon1b, and HPS1—that are homologous to SAND-1 and two distinct proteins—Ccz1 and HPS4—that are homologous to CCZ-1. HPS1 and HPS4 function in the formation of LROs (Oh et al., 1996; Suzuki et al., 2002). The genome of C. elegans encodes only single proteins that are highly homologous to SAND-1/Mon1 and CCZ-1.
We found that ccz-1(t2129) and ccz-1(ok2182), two different mutant alleles of ccz-1 that lack or have severely reduced activity (Nieto et al., 2010), respectively, similarly resulted in the loss of birefringent gut granules and mislocalization of birefringent material into the intestinal lumen (Figure 2B and Table 2). Consistent with altered gut granule formation, ccz-1 mutant embryos lacked LysoSensor Green–stained compartments (Figure 2G). ccz-1 mutants also lacked or had substantially reduced numbers of autofluorescent gut granules in adults (Figure 1I and Table 1). The Glo phenotypes exhibited by the ccz-1(t2129) mutant were rescued by the introduction of ccz-1(+)::yfp (Tables 1 and 2). In contrast to ccz-1 mutants, the strong loss-of-function sand-1(or552) mutant and the likely null sand-1(ok1963) mutant (Poteryaev et al., 2007) did not exhibit a loss of birefringent, acidified, or autofluorescent gut granules (Figures 1H and 2, C and H, and Tables 1 and 2), suggesting that SAND-1 does not play a significant role in gut granule biogenesis.
rab-7.
C. elegans codes for a single RAB-7–related protein, which is orthologous to Rab7 in other organisms (Gallegos et al., 2012). In C. elegans, the SAND-1/CCZ-1 complex promotes association of RAB-7 with endosomal membranes (Poteryaev et al., 2007; Kinchen and Ravichandran, 2010; Nieto et al., 2010), likely by acting as a RAB-7 guanine nucleotide exchange factor (GEF; Nordmann et al., 2010; Gerondopoulos et al., 2012). The HOPS complex is an effector of RAB-7 in yeast and possibly other organisms (Nickerson et al., 2009; Epp et al., 2011; Zlatic et al., 2011a). Thus RAB-7 might act as a functional link between CCZ-1 and HOPS during trafficking to gut granules. If this is the case, rab-7 mutants should exhibit defects in gut granule biogenesis similar to ccz-1 and VPS-C/HOPS mutants.
The rab-7(ok511) null mutant exhibits maternal effect embryonic lethality and arrests at a stage after gut granules are normally formed (Yu et al., 2008), which allowed us to investigate the role of RAB-7 in the biogenesis of gut granules. Whereas the number of autofluorescent gut granules was not substantially reduced in rab-7 mutants (Figure 1J and Table 1), at the L4 stage their diameter was reduced by 65% relative to wild type (Supplemental Figure S2). The number of birefringent gut granules in rab-7 mutants was modestly reduced (Table 2); however, unlike ccz-1 and vps-18 mutants, birefringent material was never detected in the intestinal lumen (Figure 2D and Table 2). The average diameter of gut granules in 1.5-fold–stage embryos increased from 0.7 μm in wild type to 1.2 μm in rab-7 mutants (Supplemental Figure S2). Gut granules in rab-7 mutant embryos were stained by LysoSensor Green (Figure 2I), suggesting that they are acidified. Together these results indicate that loss of rab-7 function only subtly affects gut granule formation.
Protein trafficking to gut granules
The loss or misaccumulation of gut granule contents might result from specific defects in the formation/trafficking of birefringent or autofluorescent material or more generally from alterations in gut granule biogenesis. In addition, alterations in protein trafficking to gut granules might not obviously affect the localization of these materials.
To address these possibilities, we analyzed the localization of gut granule proteins in the endolysosomal trafficking mutants. CDF-2 and PGP-2 are multipass transmembrane proteins, which at steady state exclusively localize to the gut granule (Figure 3, A and B; Davis et al., 2009; Hermann et al., 2012). The single-pass membrane-associated protein LMP-1 localizes to both gut granules and conventional lysosomes (Hermann et al., 2012). LMP-1 transport to gut granules depends on the activity of the AP-3 adaptor complex, whereas CDF-2::green fluorescent protein (GFP) and PGP-2 can be trafficked to gut granules independently of AP-3 (Hermann et al., 2012). We carried out trafficking studies in 1.5-fold embryos, a stage when both gut granules and these proteins are initially being synthesized within the 20 newly polarized intestinal epithelial cells (Hermann et al., 2005).
FIGURE 3:
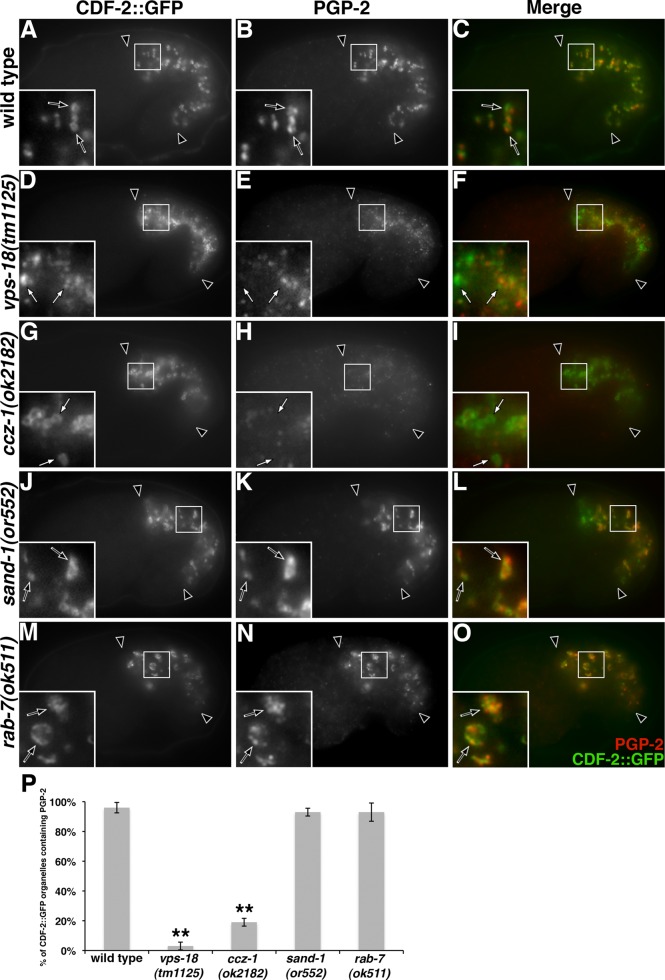
The distribution of CDF-2::GFP and PGP-2 is altered in vps-18 and ccz-1 mutants. Anti–PGP-2 antibodies and ectopically expressed CDF-2::GFP colocalized at gut granules in (A–C) wild type and (J–L) sand-1 and (M-O) rab-7 mutants (black arrows within insets). (D–F) CDF-2::GFP–containing organelles in vps-18 mutants lacked PGP-2 staining (white arrows in insets). (G–I) The majority of CDF-2::GFP–labeled compartments in ccz-1 mutants lacked anti–PGP-2 staining (white arrows in insets). In A–O, 1.5-fold-stage embryos are shown, black arrowheads flank the intestine, and the insets are 5 μm wide. (P) For each genotype, at least 25 randomly selected CDF-2::GFP–containing intestinal compartments in five different 1.5-fold-stage embryos were scored for the presence of anti-PGP-2 signals. The mean is plotted, and error bars represent the 95% confidence limit. A one-way analysis of variance (ANOVA) comparing each mutant to wild type was used to calculate p values (**p ≤ 0.001).
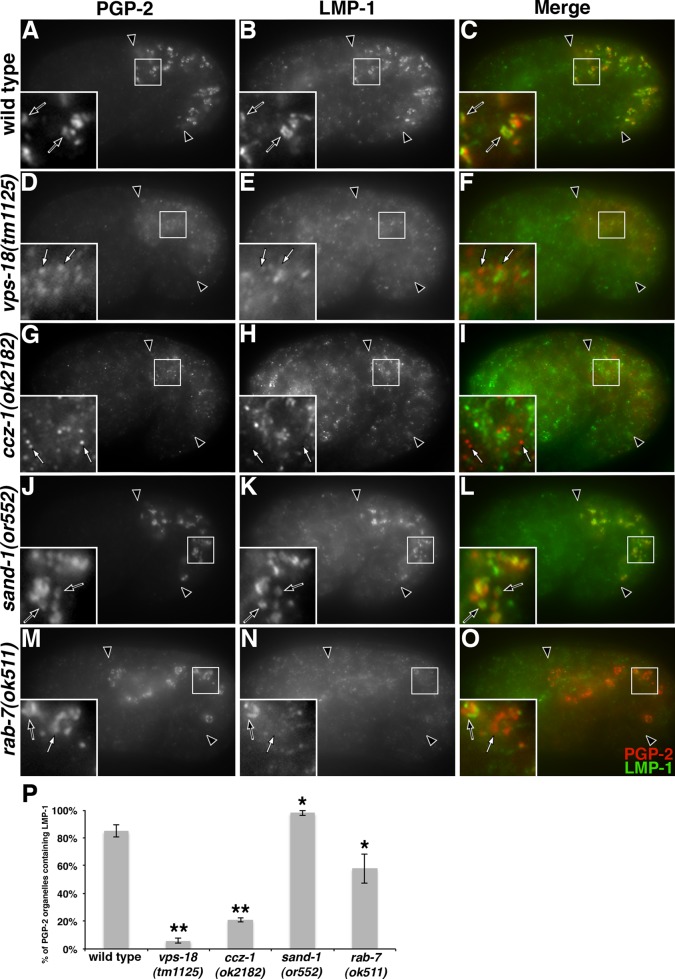
Similar to their effects on the localization of birefringent and autofluorescent material, the colocalization of CDF-2::GFP and PGP-2 at gut granules was not disrupted in sand-1 and rab-7 mutants (Figure 3, J–P). sand-1 mutants did not substantially reduce the localization of LMP-1 to gut granules containing CDF-2::GFP and PGP-2 (Figures 4, J–L, and 5, J–L). In contrast, in rab-7 mutant embryos, LMP-1 levels were significantly reduced and many CDF-2::GFP and PGP-2 compartments lacked detectable LMP-1 (Figures 4, M–P, and 5, M–P). These data indicate that gut granules are formed in sand-1 and rab-7 mutants. However, RAB-7 activity promotes the normal accumulation of LMP-1 at the gut granule.
FIGURE 4:
The distribution of PGP-2 and LMP-1 is altered in vps-18, ccz-1, and rab-7 mutants. Antibodies recognizing PGP-2 and LMP-1 colocalized at gut granules in (A–C) wild type and (J–L) sand-1 mutants (black arrows in insets). (D–I) PGP-2–stained organelles in vps-18 and ccz-1 mutants lacked LMP-1 staining (white arrows within insets). (M–O) In rab-7 mutants, LMP-1 staining was weakly present on some (black arrows in insets) but not all (white arrows in insets) anti-PGP-2–marked gut granules. In A–O, 1.5-fold-stage embryos are shown, black arrowheads flank the intestine, and insets are 5 μm wide. (P) For each genotype, at least 25 randomly selected PGP-2–stained intestinal compartments in five different 1.5-fold-stage embryos were scored for the presence of LMP-1 staining. The mean is plotted, and error bars represent the 95% confidence limit. A one-way ANOVA comparing each mutant to wild type was used to calculate p values (*p ≤ 0.05, **p ≤ 0.001).
FIGURE 5:
The distribution of CDF-2::GFP and LMP-1 is altered in vps-18, ccz-1, and rab-7 mutants. Nearly all CDF-2::GFP–containing organelles were marked by anti–LMP-1 antibodies in (A–C) wild-type and (J–L) sand-1 mutant cells (black arrows in insets). In (D–F) vps-18, (G–I) ccz-1, and (M–O) rab-7 mutants, a subset of CDF-2::GFP–marked organelles contained anti–LMP-1 staining (black arrows in insets), although most lacked LMP-1 (white arrows in insets). In A–L, 1.5-fold-stage embryos are shown, and in M–O, a late-bean-stage embryo is shown. In A–O, black arrowheads flank the intestine and insets are 5 μm wide. (P) For each genotype, at least 25 randomly selected CDF-2::GFP–containing intestinal compartments in five different 1.5-fold-stage embryos were scored for the presence of LMP-1 staining. The mean is plotted, and error bars represent the 95% confidence limit. A one-way ANOVA comparing each mutant to wild type was used to calculate p values (*p ≤ 0.05, **p ≤ 0.001).
Indicative of a significant defect in gut granule biogenesis, the distribution and colocalization of gut granule proteins were dramatically altered by mutations in vps-18 and ccz-1. PGP-2 and CDF-2::GFP colocalized at gut granules in wild-type 1.5-fold-stage embryos (Figure 3, A–C and P). In vps-18 mutants, CDF-2::GFP was enriched on organelles that did not contain PGP-2 (Figure 3, D–F and P). In ccz-1 mutants, CDF-2::GFP was localized to compartments that typically lacked PGP-2, which could be only weakly detected on intestinal organelles (Figure 3, G–I and P). LMP-1, which normally colocalized with PGP-2 on gut granules in wild type (Figure 4, A–C and P), rarely colocalized with PGP-2 in both vps-18 and ccz-1 mutants (Figure 4, D–I and P). CDF-2::GFP compartments in wild type contained LMP-1 (Figure 5, A–C and P), whereas a reduced proportion of CDF-2::GFP compartments contained LMP-1 in vps-18 and ccz-1 mutants (Figure 5, D–I and P).
Our results indicate that one or more gut granule–associated proteins are mislocalized in embryos deficient for VPS-18 or CCZ-1 function. PGP-2 exhibited weak staining in vps-18 and ccz-1 mutants, and LMP-1 normally associates with both gut granules and lysosomes. We therefore examined the localization of CDF-2::GFP in strains with altered VPS-18 and CCZ-1 activity. In vps-18 mutant embryos, CDF-2::GFP was often associated with organelles located near the apical surface of intestinal cells (compare Figure 6, A and D), which is where conventional lysosomes and early endosomes are typically localized (Hermann et al., 2012). In contrast, CDF-2::GFP–containing compartments were distributed throughout the cytoplasm of ccz-1 mutants (Figure 3G). In wild-type and both mutant strains, CDF-2::GFP did not accumulate on RAB-5–marked early endosomes (Supplemental Figure S3). Of note, CDF-2::GFP–containing organelles in vps-18 mutants were often marked by RAB-7, unlike ccz-1 mutants and wild type (Figure 6, A–I and S). RAB-7 was often associated with endolysosomes marked by LMP-1::GFP (Figure 7, F–H, and Supplemental Figure S2). Consistent with CDF-2::GFP being mislocalized to lysosomes in vps-18 mutants, CDF-2::GFP colocalized with an mCherry-tagged form of the lysosomal glucosylceramidase F11E6.1a (Levitte et al., 2010; Figure 6, M–O and T). CDF-2::GFP did not appreciably associate with F11E6.1a::mCherry in wild-type or ccz-1(-) mutant embryos (Figure 6, P–R and T). The identity of CDF-2::GFP compartments in ccz-1 mutants is unclear, as they lack markers for gut granules, early endosomes, late endosomes, and lysosomes, whereas disrupting VPS-18 activity results in gut granule proteins being missorted toward conventional lysosomes.
FIGURE 6:
CDF-2::GFP is mislocalized to endolysosomes in vps-18 mutants. (A–C, G–I) Endogenous RAB-7 detected with anti–RAB-7 antibodies was not associated with CDF-2::GFP–containing organelles in wild type or ccz-1 mutants (white arrows in insets). (D–F) In vps-18 mutants, anti–RAB-7 staining was often localized to CDF-2::GFP–marked compartments (black arrows in insets). The lysosomal hydrolase F11E6.1a::mCherry did not appreciably localize to CDF-2::GFP–marked compartments in (J–L) wild type or (P–R) ccz-1 mutants (white arrows in insets). (M–O) In vps-18 mutants, F11E6.1a::mCherry often colocalized with CDF-2::GFP–containing compartments (black arrows in insets). In A–R, single optical sections of 1.5-fold-stage embryos are shown, black arrowheads flank the intestine, and insets are 5 μm wide. In A–F, white arrowheads denote the apical surface of intestinal cells. (S, T) For each genotype, at least 25 randomly selected CDF-2::GFP–containing intestinal compartments in five different 1.5-fold-stage embryos were scored for the presence of RAB-7 or F11E6.1a::mCherry. The mean is plotted, and error bars represent the 95% confidence limit. A one-way ANOVA comparing each mutant to wild type was used to calculate p values (*p ≤ 0.05, **p ≤ 0.001).
FIGURE 7:
Analysis of LMP-1::GFP trafficking. (A–E) Mutations in (B) vps-18, (C) ccz-1, and to a lesser extent (E) rab-7 led to an increased number of LMP-1::GFP–marked organelles. (R) The total number of LMP-1::GFP compartments was quantified in the four cells that compose Int2 and Int3 (marked by black arrows in A–E) in five different 1.5-fold-stage embryos of each genotype. The mean is plotted, and error bars represent the 95% confidence limit. A one-way ANOVA comparing each mutant to wild type was used to calculate p values (*p ≤ 0.05, **p ≤ 0.001). (F–H) Relative to wild type, a higher proportion of LMP-1::GFP compartments in (I–K) vps-18 mutants contained RAB-7 antibody signals, and a decreased proportion of LMP-1::GFP compartments in (L–N) ccz-1 and sand-1 mutants contained RAB-7 (in the insets, black arrows denote LMP-1::GFP compartments marked by RAB-7 antibodies and white arrows label compartments lacking a RAB-7 signal). (S) For each genotype, at least 25 randomly selected LMP-1::GFP–containing intestinal compartments in five different 1.5-fold-stage embryos were scored for the presence of signal from the RAB-7 antibody. The mean is plotted, and error bars represent the 95% confidence limit. A one-way ANOVA comparing each mutant to wild type was used to calculate p (*p ≤ 0.05, **p ≤ 0.001). In A–E, 1-μm maximum-intensity projections, and in F–Q, single optical sections, of 1.5-fold-stage embryos are shown. The white arrowhead in B denotes the apical cell membrane. In images showing embryos, the black arrowheads flank the intestine. The insets are 5 μm wide.
Formation of conventional endolysosomes
In addition to analyzing the effects of the endolysosomal trafficking pathway mutants on gut granule biogenesis, we also examined their role in controlling the number and identity of conventional endolysosomes in the embryonic intestinal primordium. LMP-1::GFP marks conventional lysosomes that are localized near the intestinal cell apical domain of 1.5-fold embryos (Hermann et al., 2012). Wild-type embryos contained on average 12 LMP-1::GFP compartments in the four intestinal cells that make up intestinal rings 2 and 3, whereas vps-18 mutants displayed fourfold more, which were enriched near the apical cell surface (Figure 7, A, B, and R). Relative to wild type, ccz-1 mutants had a twofold increase in the number of LMP-1::GFP compartments, which were not restricted to the apical domain (Figure 7, C and R). The number of LMP-1::GFP organelles was only slightly altered in sand-1 and rab-7 mutants (Figure 7, D, E, and R).
Owing to effects of vps-18 and ccz-1 mutations on the number and localization of LMP-1::GFP–containing compartments, we analyzed the identity of LMP-1::GFP–containing compartments using antibodies to RAB-5 and RAB-7. LMP-1::GFP did not accumulate on RAB-5 marked early endosomes in wild type or in vps-18 and ccz-1 mutants (Supplemental Figure S3, J–R and T). Of note, unlike wild type, vps-18 mutant embryos displayed an increase in the proportion of LMP-1::GFP compartments that also contained RAB-7 (Figure 7, F–K and S), suggesting that loss of VPS-18 function leads to an excess of late endosomes or lysosomes, consistent with known roles of HOPS promoting heterotypic fusion of late endosomes with lysosomes and homotypic fusion of lysosomes (Nickerson et al., 2009; Balderhaar and Ungermann, 2013; Solinger and Spang, 2013). In contrast, LMP-1::GFP organelles in ccz-1 and sand-1 mutants did not contain RAB-7 (Figure 7, L–Q and S), supporting prior observations that CCZ-1 and SAND-1/Mon1 act in conventional lysosome biogenesis by promoting the association of RAB-7 with endolysosomal membranes (Poteryaev et al., 2007; Kinchen and Ravichandran, 2010; Nieto et al., 2010). Our observations also suggest that LMP-1::GFP trafficking is more sensitive to the loss of CCZ-1 than SAND-1 or alternatively that defects in gut granule biogenesis in ccz-1 mutants, which are not present in sand-1 mutants, exacerbate effects on LMP-1::GFP localization, leading to the dramatic difference in LMP-1::GFP compartment number and subcellular localization in these two mutants
Interactions between AP-3 and endolysosomal trafficking factors in gut granule biogenesis
The HOPS complex in yeast mediates the docking of AP-3–containing vesicles at the lysosome-like vacuole through direct interactions of AP-3 and a short region of VPS41, which is conserved in C. elegans VPS-41 (Angers and Merz, 2009; Cabrera et al., 2010). In mammalian cells, HOPS subunits colocalize and physically interact with AP-3 subunits (Zlatic et al., 2011b). The AP-3 adaptor complex has evolutionarily conserved roles in trafficking to LROs (Dell'Angelica, 2009); however, the functional relationship between AP-3 and HOPS in the biogenesis of LROs has not been investigated.
If VPS-C/HOPS activity is restricted to the same step as AP-3 during trafficking to gut granules, then AP-3 and VPS-C/HOPS mutants should have similar defects in gut granule biogenesis, and the mutants should not exacerbate each other's phenotypes. Inconsistent with this model, vps-18 mutant embryos completely lacked birefringent intestinal granules, whereas they were always present in null mutants of apt-6 and apt-7, which encode AP-3 β- and μ-subunits, respectively (Table 2). Moreover, the majority of AP-3; vps-18 double-mutant embryos exhibited a complete loss of birefringent gut granules, resembling the vps-18 mutant (Table 2). Single VPS-C/HOPS and AP-3 mutants similarly displayed reduced numbers of autofluorescent gut granules (Table 1). However, the vps-18 mutation, which did not exhibit decreased numbers of autofluorescent compartments on its own at 22°C, led to a complete loss of autofluorescent gut granules when introduced into both apt-6 and apt-7 mutant backgrounds (Table 1). A vps-41 mutation also enhanced the adult Glo phenotype of the apt-6 mutant (Table 1). Together these genetic interactions suggest that VPS-C/HOPS and AP-3 function at distinct steps in the biogenesis of gut granules.
We also examined whether CCZ-1 has functions distinct from AP-3 in trafficking to gut granules. The application of ccz-1(RNAi), which led to a moderate decrease in the number of autofluorescent gut granules on its own, significantly enhanced the Glo phenotype of apt-6 and apt-7 mutants (Table 1). Similarly, autofluorescent and birefringent compartments, which were always present in AP-3 mutants, were absent in an apt-7; ccz-1 double mutant (Tables 1 and 2). These results point to CCZ-1 having activities that are distinct from those of AP-3.
We extended our analyses of the interactions between mutations disrupting the function of AP-3, VPS-C/HOPS, and CCZ-1 by comparing their effects, alone or in combination, on the relative localization of two gut granule proteins, CDF-2::GFP and PGP-2. Whereas vps-18 mutants rarely displayed CDF-2::GFP compartments containing PGP-2 (Figures 3, D–F, and 8J), ∼25% of CDF-2::GFP compartments were marked by PGP-2 in apt-7 and ccz-1 single mutants (Figures 3, G–I, and 8, A–C and J). We found that CDF-2::GFP compartments were more likely to contain PGP-2 in apt-7; vps-18 and apt-7; ccz-1 double mutants than the single mutants (Figure 8, D–J). The ability of an AP-3 mutant to modify the effects of mutations in vps-18 or ccz-1 on the colocalization of CDF-2::GFP and PGP-2 suggests that AP-3 functions upstream of, or parallel to, VPS-C/HOPS and CCZ-1 in trafficking to the gut granule.
FIGURE 8:
Effects of disrupting vps-18 and ccz-1 function on gut granule protein trafficking in apt-7 mutants. Embryos expressing CDF-2::GFP were stained with anti–PGP-2 antibodies. (A–C) In apt-7 single mutants, CDF-2::GFP–labeled compartments were rarely marked by PGP-2 antibodies (white and black arrows in insets). (D–I) Relative to the single mutants, apt-7; vps-18 and apt-7; ccz-1 double mutants displayed an increased proportion of organelles that contained both CDF-2::GFP and PGP-2 (black arrows in insets). (J) For each genotype, at least 25 randomly selected CDF-2::GFP–containing intestinal compartments in five different 1.5-fold-stage embryos were scored for the presence of PGP-2 staining. The mean is plotted, and error bars represent the 95% confidence limit. A one-way ANOVA comparing the indicated genotypes was used to calculate p values (*p ≤ 0.05, **p ≤ 0.001). In A–I, 1.5-fold-stage embryos are shown, and black arrowheads flank the intestine. The insets are 5 μm wide.
We used AP-3 and other glo mutants that exhibit partial defects in gut granule biogenesis and are thus sensitized to reveal gene function in the pathway to investigate whether SAND-1/Mon1 and RAB-7 play roles in trafficking to gut granules not apparent in an otherwise wild-type background. We found that mutations in sand-1 did not alter the Glo phenotype of apt-7 mutant embryos or adults (Tables 1 and 2). Moreover, the codistribution of gut granule proteins was not significantly different between apt-7 single and apt-7; sand-1 double mutants (Supplemental Figure S4). Thus we find no evidence supporting a role for SAND-1 in gut granule formation. The number of autofluorescent gut granules in AP-3; rab-7 mutant embryos and adults was not obviously different from that in AP-3 mutants (Tables 1 and 2). Similarly, the few autofluorescent gut granules in glo-2 mutant adults and birefringent gut granules in glo-3 mutant embryos remained in double mutants with rab-7 (Tables 1 and 2). However, the introduction the rab-7 mutation weakly enhanced the Glo phenotypes of pgp-2 mutant adults and embryos and glo-3 mutant adults. Similar to our analysis of protein trafficking, our genetic studies point to RAB-7 selectively and subtly contributing to gut granule formation.
GLO-1(ΔG4) mutants suppress ccz-1(-) and glo-3(-)
Our work shows that CCZ-1 and VPS-C/HOPS promote gut granule formation. In the lysosome biogenesis pathway, CCZ-1 likely functions with SAND-1/Mon1 as a GEF to activate RAB-7, which, through its effector HOPS, mediates protein trafficking by promoting membrane tethering and fusion (Nickerson et al., 2009; Zlatic et al., 2011a; Balderhaar and Ungermann, 2013). We show that rab-7 mutants do not phenocopy ccz-1 mutants, suggesting the possibility that a different Rab protein functions downstream of CCZ-1 in trafficking to the gut granule. Recently HPS-4, one of two mammalian homologues of CCZ-1, was identified as a subunit of the GEF that activates Rab32 and Rab38 (Gerondopoulos et al., 2012). GLO-1, which is known to function in gut granule formation, is the C. elegans Rab GTPase most homologous to these proteins (Hermann et al., 2005).
We used an in vivo genetic approach to analyze whether CCZ-1 might function as an upstream activator of the GLO-1 Rab to promote gut granule formation. If CCZ-1 acts as a GEF to activate GLO-1 in the gut granule biogenesis pathway, then it is possible that ccz-1 mutants are Glo due to the GLO-1 Rab being in its inactive GDP-bound form. To examine this possibility, we generated three different point mutations in the G4 domain of GLO-1 that are predicted to weaken the molecular interactions between GLO-1 and guanine nucleotides (Figure 9S). Analogous mutations in Rab7, which is in the same Rab subfamily as GLO-1 (Klopper et al., 2012; Rojas et al., 2012), led to greatly increased rates of spontaneous guanine nucleotide exchange in vitro and the ability to bypass the necessity of its corresponding GEF in vivo (Kucharczyk et al., 2001; McCray et al., 2010; Nordmann et al., 2010; Cabrera and Ungermann, 2013). We collectively refer to the GLO-1(K130E), GLO-1(D132A), and GLO-1(I133F) mutations as GLO-1(ΔG4) (Figure 9S). The mutations were created in a rescuing GFP-tagged form of GLO-1 (Hermann et al., 2005), which was expressed under the control of the vha-6 intestinal cell–specific promoter (Oka et al., 2001; Pujol et al., 2001). When introduced into a glo-1(zu437) null mutant background (Hermann et al., 2005), two of the point mutants exhibited rescuing activity similar to wild-type GLO-1, whereas GLO-1(K130E) was partially active (Table 3). None of the GLO-1(ΔG4) mutants exhibited dominant Glo phenotypes when expressed in wild type (unpublished data). The majority of ccz-1 mutant young adults expressing GFP::GLO-1 exhibited a low number of autofluorescent compartments, similar to ccz-1 mutants alone (Figure 9, B and C, and Table 3). In contrast, expression of all of the GLO-1(ΔG4) mutants significantly increased the number of autofluorescent organelles in ccz-1 mutants, with the majority of GLO-1(D132A)–containing animals resembling wild type (Figure 9, D and E, and Table 3). Consistent with the compartments being gut granules, the GLO-1(D132A) and GLO-1(I133F) constructs, which exhibited the strongest suppression, restored acidified organelles in ccz-1 mutants (Figure 9, K–N). These results are consistent with CCZ-1 acting upstream of, or parallel to, the GLO-1 Rab to promote gut granule biogenesis.
FIGURE 9:
Expression of GFP::GLO-1(ΔG4) mutants leads to increased numbers of autofluorescent and acidified gut granules in ccz-1 and glo-3 mutants. Young adults were analyzed for the presence of autofluorescent material or the accumulation of the acidophilic dye acridine orange. (A, J) Wild type contained many autofluorescent and acridine orange–stained organelles. (B, C, F, G, K, L, O, and P) ccz-1 and glo-3 mutants contained few autofluorescent and acridine orange–stained gut granules, whose numbers did not increase upon the expression of gfp::glo-1 under control of the vha-6 promoter. (D, E, H, I, M, N, Q, and R) The addition of glo-1(D132A) or glo-1(I133F), which contain mutations in the G4 motif of GLO-1, led to increased numbers of autofluorescent and acridine orange–labeled compartments in ccz-1 and glo-3 mutants. Maximum-intensity projections of fluorescence signals spanning the entire depth of the anterior intestine are shown. White arrows denote autofluorescent or acridine orange–stained organelles. (S) Alignment of the G4 domain of C. elegans (Ce) GLO-1 and related Rab proteins from humans (Hs) and yeast (Sc), denoting the location of mutations predicted to alter guanine nucleotide binding. (T) Alignment of the amino-terminal region of C. elegans GLO-3 (Ce) and HPS-1 from D. melanogaster (Dm) and humans (Hs). Location of β-sheets and α-helices that compose the Longing domain in HPS1 proteins are present below the sequences. A consensus of GLO-3 secondary structural predictions for β-sheets (listed as E) and α-helices (listed as H) are positioned above the sequences.
TABLE 3:
Effects of GLO-1(ΔG4) mutants on gut granule formation.
| Percentage of animals with the specified number of autofluorescent granules in anterior intestinal cells | |||||||
|---|---|---|---|---|---|---|---|
| Genotype | 0 | 1–20 | 21–50 | 51–100 | 101–200 | >200 | n (p value) |
| Wild type | 0 | 0 | 0 | 0 | 0 | 100 | 83 |
| apt-6(ok429) | 50 | 46 | 4 | 0 | 0 | 0 | 52 |
| apt-6(ok429) + gfp::glo-1 | 60 | 34 | 6 | 0 | 0 | 0 | 32 |
| apt-6(ok429) + gfp::glo-1(D132A) | 50 | 41 | 9 | 0 | 0 | 0 | 34 |
| apt-6(ok429) + gfp::glo-1(I133F) | 20 | 73 | 7 | 0 | 0 | 0 | 26* |
| ccz-1(ok2182)a | 19 | 67 | 14 | 0 | 0 | 0 | 99 |
| ccz-1(ok2182) + gfp::glo-1 | 6 | 44 | 42 | 8 | 0 | 0 | 53 |
| ccz-1(ok2182) + gfp::glo-1(K130E) | 0 | 17 | 46 | 18 | 12 | 7 | 60** |
| ccz-1(ok2182) + gfp::glo-1(D132A) | 0 | 0 | 0 | 0 | 28 | 72 | 25** |
| ccz-1(ok2182) + gfp::glo-1(I133F) | 0 | 4 | 32 | 24 | 32 | 8 | 25** |
| glo-1(zu437) | 100 | 0 | 0 | 0 | 0 | 0 | 35 |
| glo-1(zu437) + gfp::glo-1 | 0 | 0 | 0 | 0 | 100 | 0 | 52** |
| glo-1(zu437) + gfp::glo-1(K130E) | 0 | 0 | 56 | 44 | 0 | 0 | 72** |
| glo-1(zu437) + gfp::glo-1(D132A) | 0 | 0 | 0 | 0 | 100 | 0 | 71** |
| glo-1(zu437) + gfp::glo-1(I133F) | 0 | 0 | 0 | 0 | 100 | 0 | 68** |
| glo-2(zu455) | 43 | 55 | 2 | 0 | 0 | 0 | 67 |
| glo-2(zu455) + gfp::glo-1 | 39 | 51 | 10 | 0 | 0 | 0 | 31 |
| glo-2(zu455) + gfp::glo-1(D132A) | 32 | 65 | 3 | 0 | 0 | 0 | 34 |
| glo-2(zu455) + gfp::glo-1(I133F) | 29 | 62 | 7 | 2 | 0 | 0 | 45 |
| glo-3(kx94) | 67 | 18 | 5 | 5 | 4 | 1 | 85 |
| glo-3(kx94) + gfp::glo-1 | 51 | 41 | 8 | 0 | 0 | 0 | 37 |
| glo-3(kx94) + gfp::glo-1(D132A) | 1 | 0 | 18 | 17 | 24 | 40 | 68** |
| glo-3(kx94) + gfp::glo-1(I133F) | 7 | 32 | 3 | 3 | 13 | 38 | 52** |
| glo-4(ok623) | 100 | 0 | 0 | 0 | 0 | 0 | 33 |
| glo-4(ok623) + gfp::glo-1 | 100 | 0 | 0 | 0 | 0 | 0 | 30 |
| glo-4(ok623) + gfp::glo-1(K130E) | 100 | 0 | 0 | 0 | 0 | 0 | 61 |
| glo-4(ok623) + gfp::glo-1(D132A) | 100 | 0 | 0 | 0 | 0 | 0 | 64 |
| glo-4(ok623) + gfp::glo-1(I133F) | 100 | 0 | 0 | 0 | 0 | 0 | 63 |
| snpn-1(tm1892) | 86 | 14 | 0 | 0 | 0 | 0 | 43 |
| snpn-1(tm1892) + gfp::glo-1 | 60 | 40 | 0 | 0 | 0 | 0 | 35 |
| snpn-1(tm1892) + gfp::glo-1(D132A) | 59 | 41 | 0 | 0 | 0 | 0 | 34 |
| snpn-1(tm1892) + gfp::glo-1(I133F) | 63 | 34 | 3 | 0 | 0 | 0 | 38 |
All strains were grown at 22˚C unless otherwise noted. Individual young adult-stage animals were analyzed using fluorescence microscopy with a rhodamine filter set and were scored for the number of autofluorescent organelles within the anterior intestine located between the pharynx and vulva. The expression of GLO-1 was verified in all animals scored by the presence of GFP. A Fisher's exact test was used to compare a mutant expressing gfp::glo-1 to the same mutant expressing a gfp::glo-1(ΔG4) variant. **p ≤ 0.001, *p ≤ 0.05, lack of asterisk indicates p > 0.05.
aAll ccz-1(-) animals scored with or without gfp::glo-1 transgenes were ccz-1(-)/ccz-1(-) progeny of ccz-1(-)/+ adults. They were identified by containing within their uterus embryos with large refractile bodies visible with DIC microscopy.
We examined the specificity of the genetic interactions between the GLO-1(ΔG4) and ccz-1 mutants by testing whether the GLO-1(ΔG4) constructs altered the Glo phenotype of other mutants that disrupt gut granule biogenesis. It has been suggested that GLO-4 and its homologues act as a GEF for Rabs similar to GLO-1 (Ma et al., 2004; Hermann et al., 2005). However, the GLO-1(ΔG4) mutants did not suppress the Glo phenotype displayed by glo-4 mutants (Table 3). Similarly, apt-6 mutants were not rescued by introduction of GLO-1(ΔG4) constructs (Table 3), suggesting that the GLO-1(ΔG4) mutants do not generally restore gut granules in mutants with a partial Glo phenotype similar to ccz-1. Finally, we tested the ability of GLO-1(ΔG4) mutants to bypass the activity of BLOC-1, a complex that has evolutionarily conserved roles in LRO biogenesis, and found that the Glo phenotype resulting from mutations in two BLOC-1–encoding subunit genes, glo-2 and snpn-1, were not suppressed by GLO-1(ΔG4) constructs (Table 3).
CCZ-1 and its homologues are believed to function in a heterodimeric complex with a SAND-1/Mon1–related protein (Levine et al., 2013). Our results indicate that SAND-1 does not act with CCZ-1 to promote gut granule biogenesis. To identify other C. elegans genes that could function with CCZ-1, we performed PSI-BLAST searches with SAND-1/HPS1 family members from other species. Starting with Drosophila HPS1 (Lee et al., 2009), the fourth PSI-BLAST iteration led to the identification of C. elegans GLO-3, which shows low but significant similarity to human and Drosophila HPS1 proteins (Figure 9T). The region of highest homology with SAND-1/HPS1 family members contains a Longin domain (Kinch and Grishin, 2006), which consists of five β-strands sandwiched between α-helices (Filippini et al., 2001; Tochio et al., 2001). Longin domains are found in a variety of proteins that mediate membrane trafficking and notably are often present in Rab GEFs, where they mediate interactions with Rabs and other Longin domain–containing proteins (Collins et al., 2002; Kim et al., 2006; Yu et al., 2009; Marat et al., 2011; Levine et al., 2013; De Franceschi et al., 2014). Predictions of GLO-3 secondary structure suggest that GLO-3 has similarity to the first half of the Longin domain (Figure 9T).
glo-3 was identified in a genetic screen for factors that promote gut granule biogenesis, and glo-3 and ccz-1 mutants exhibit similar Glo phenotypes (Figure 9F; Rabbitts et al., 2008). We found that expression of two different GLO-1(ΔG4) mutants significantly increased the number of autofluorescent organelles in glo-3 mutants when compared with expression of wild-type GLO-1 (Figure 9, F–I, and Table 3). The GLO-1(ΔG4) constructs also increased the number of acidified organelles in glo-3 mutants (Figure 9O-R), suggesting that they lead to restoration of gut granules. These observations suggest that GLO-3 functions similarly to CCZ-1 and are consistent with both proteins acting upstream of the GLO-1 Rab.
DISCUSSION
HOPS function in LRO biogenesis
We showed that all six proteins predicted to compose the canonical HOPS complex in C. elegans are necessary for the formation of autofluorescent gut granules (Figure 1 and Tables 1 and 2). This requirement resembles that seen in Drosophila melanogaster, where disrupting the HOPS subunits Vps16A (Pulipparacharuvil et al., 2005), deep orange/Vps18 (Sevrioukov et al., 1999), carnation/Vps33A (Akbar et al., 2009), or light/Vps41 (Warner et al., 1998) leads to altered eye pigment, likely from the reduced number of LROs (pigment granules) present within the retinal epithelium (Lloyd et al., 1998). Similarly, in zebrafish, mutations in platinum/vps11 (Thomas et al., 2011), vps18 (Sadler et al., 2005; Maldonado et al., 2006), and lbk/vps39 (Schonthaler et al., 2008) disrupt LRO biogenesis, reducing the number of epidermal and retinal melanosomes. Only one HOPS subunit, Vps33a, has been shown to facilitate the formation of LROs in mammals; the buff/Vps33a partial-loss-of-function mutant exhibits reduced numbers of melanosomes and platelet dense granules in mice (Suzuki et al., 2002; Nguyen and Wei, 2004), both characteristics of HPS (Huizing et al., 2008). Mutations in HOPS subunit encoding genes have not been detected in HPS patients, likely due to their essential roles during mammalian embryogenesis (Messler et al., 2011; Aoyama et al., 2012; Peng et al., 2012).
Paralogues of VPS16 and VPS33 do not appear to function during C. elegans gut granule formation (Tables 1 and 2) or pigment granule biogenesis in other organisms (Matthews et al., 2005; Akbar et al., 2009). Instead, they act in the formation of different LROs, epidermal lamellar bodies, and platelet α-granules in mammals (Lo et al., 2005; Hershkovitz et al., 2008; Urban et al., 2012) and fibrous body-membranous organelles in C. elegans spermatocytes (Zhu and L'Hernault, 2003; Zhu et al., 2009). It is possible that the canonical HOPS complex (containing VPS11, VPS16a, VPS18, VPS33a, VPS39, VPS41) and an alternate HOPS-related complex (substituting Vps16b for Vps16a and Vps33b for Vps33a) function in the biogenesis of different types of LROs.
Unlike other HOPS-complex mutants in C. elegans, which are maternal effect lethal, the conditional vps-18(tm1125) mutant provided us with the opportunity to assess the function of VPS-18 during endolysosome and LRO biogenesis in the developing intestinal primordium. We found that loss of vps-18 activity resulted in a dramatic increase in LMP-1::GFP–containing conventional endolysosomes (Figure 7), similar to what was seen in coelomocytes of vps-18 mutants (Xiao et al., 2009). Drosophila epithelial cells lacking vps16A or vps33A activity also display increased number of endolysosomes (Pulipparacharuvil et al., 2005; Akbar et al., 2009), and compromising vps39 or vps41 activity in cultured mammalian cells leads to more late endosomes due to their impaired fusion with lysosomes (Pols et al., 2013). Disrupting HOPS-subunit function in C. elegans phagocytes leads to the accumulation of undegraded phagosomes, likely due to defects in the fusion of lysosomes with RAB-7–containing phagosomes (Kinchen et al., 2008; Xiao et al., 2009). Our data show that LMP-1::GFP compartments in vps-18 mutants more often contained RAB-7 than did those in wild type (Figure 7). This suggests that in the intestinal primordium, VPS-18 promotes the fusion of lysosomes with late endosomes, a well-documented function of the HOPS complex (Epp et al., 2011; Balderhaar and Ungermann, 2013; Solinger and Spang, 2013). Based on these phenotypes, it is likely that VPS-18 acts as part of the HOPS complex to direct the dynamics of late endosomal and lysosomal membranes in the embryonic intestine.
Our work strongly suggests that VPS-18 mediates gut granule formation due to its activity as a HOPS subunit. Analysis of five distinct gut granule markers indicates that vps-18 mutant embryos completely lack gut granules (Figures 2–5). The reduction in the number of autofluorescent and birefringent gut granules and misaccumulation of birefringent material by mutations or RNAi targeting all of the other HOPS-subunit-encoding genes (Figures 1 and 2 and Tables 1 and 2), including the HOPS-specific genes vps-39 and vps-41, point toward HOPS acting in the biogenesis of gut granules. Furthermore, loss of VPS-18 is predicted to lead to the disassembly of the HOPS complex (Plemel et al., 2011). The only known HOPS-independent function of VPS18 is as a subunit in the related CORVET complex (Nickerson et al., 2009; Balderhaar and Ungermann, 2013; Solinger and Spang, 2013). We did not detect an alteration in gut granule biogenesis when vps-8 activity was disrupted (Tables 1 and 2), which is predicted to specifically disrupt CORVET activity.
The HOPS and AP-3 complexes have evolutionarily conserved roles in protein trafficking to LROs, and their functional interactions are poorly understood (Dell'Angelica, 2009; Zlatic et al., 2011a). In Saccharomyces cerevisiae, which lacks LROs, HOPS mediates the tethering and consumption of Golgi-derived, AP-3–containing vesicles by promoting their fusion with conventional lysosomes (Darsow et al., 2001; Angers and Merz, 2009). This process is mediated by direct physical interactions between VPS41, associated with HOPS on the lysosomal membrane, and subunits of the AP-3 vesicle coat (Darsow et al., 2001; Angers and Merz, 2009; Cabrera et al., 2010). Our double-mutant analysis leaves open the possibility that HOPS functions downstream of AP-3, possibly in the same pathway, during trafficking to gut granules (Table 2 and Figure 8).
Our genetic studies also indicate that HOPS function in gut granule biogenesis extends beyond acting exclusively in the AP-3–trafficking pathway, as vps-18 mutants exhibit a stronger Glo phenotype than when AP-3 activity is abolished, and mutations in both vps-18 and vps-41 modify the Glo phenotype of AP-3 mutants (Tables 1 and 2). There are a number of ways that HOPS might promote gut granule biogenesis independent of AP-3. HOPS functions in the fusion of late endosomes and lysosomes by promoting the assembly and activity of soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) complexes, which mediate membrane fusion (Balderhaar and Ungermann, 2013; Pols et al., 2013; Solinger and Spang, 2013). SNAREs likely mediate trafficking to LROs (Ghiani et al., 2010; Tamura et al., 2011; Yatsu et al., 2013); however, the site of SNARE action and the specific membrane fusion events that they regulate during LRO biogenesis are unknown. HOPS activity has also been implicated in the maturation of early to late endosomes by functioning to inactivate Rab5 on early endosomes, as well as to activate and recruit Rab7 on nascent late endosomes (Wang et al., 2003; Rink et al., 2005; Nordmann et al., 2010; Poteryaev et al., 2010). Organelle maturation, possibly associated with Rab conversion (Hume et al., 2011), occurs during mammalian melanosome biogenesis (Sitaram and Marks, 2012). However, whether organelle maturation is a conserved feature of LRO biogenesis and whether HOPS directly facilitates this process are open questions.
Rab GTPase activity during gut granule formation
In yeast, Rab7 recruits HOPS and its associated tethering and membrane fusion activity (Price et al., 2000; Wurmser et al., 2000; Brett et al., 2008; Plemel et al., 2011). We found that HOPS mutants displayed stronger gut granule protein biogenesis and trafficking defects than did loss of RAB-7 (Figures 1–5), suggesting that HOPS activity in gut granule biogenesis only partially depends on RAB-7 and that HOPS interacts with another Rab or has Rab-independent activities.
Gut granules were formed in rab-7 mutants (Figures 1–3 and Tables 1– 2); however, their size was modestly altered (Supplemental Figure S2), and LMP-1 accumulation on nascent gut granules was disrupted (Figures 4 and 5). Similarly, Rab7 activity partially regulates the protein composition of melanosomes (Hida et al., 2011). Rab7 is associated with many different mammalian LROs (Jordens et al., 2006; Kawakami et al., 2008; Zhang et al., 2011; Ambrosio et al., 2012); however, we did not detect RAB-7 on gut granules at the stage when gut granule defects were observed (Figure 6 and Supplemental Figure S2), suggesting that RAB-7 exerts its effects indirectly or at an early point in the trafficking pathway to gut granules. In addition, the different effects of sand-1 and rab-7 mutants on gut granule formation (Figures 4 and 5) point to an LRO-related function of RAB-7 that is independent of SAND-1/Mon1–mediated activation.
C. elegans RAB-7 mediates the trafficking between early and late endosomes (Poteryaev et al., 2007), the fusion between lysosomes and cell corpse–containing phagosomes (Yu et al., 2008; Guo and Wang, 2010), and yolk degradation (Grant and Hirsh, 1999; Britton and Murray, 2004). In these processes, a complex of CCZ-1 and SAND-1 likely functions as a GEF to generate active GTP-bound RAB-7 (Poteryaev et al., 2007, 2010; Kinchen and Ravichandran, 2010; Nieto et al., 2010; Nordmann et al., 2010; Gerondopoulos et al., 2012; Yousefian et al., 2013). Given the subtle effects of rab-7 mutants on gut granules, it was surprising that ccz-1 mutants displayed significant defects in protein trafficking to gut granules (Figures 1–5 and Tables 1 and 2), which suggests that CCZ-1 functions to activate another Rab GTPase. In this case, CCZ-1 would be expected to function upstream of the Rab. In yeast, Ccz1 acts as a GEF for Rab7 in vitro, and predicted spontaneous nucleotide exchange mutations in Rab7 bypass the requirement for Ccz1 in lysosome biogenesis, providing a genetic indicator that Ccz1 acts upstream of Rab7 (Kucharczyk et al., 2001; Nordmann et al., 2010; Cabrera and Ungermann, 2013). Similar mutations in GLO-1, a Rab GTPase homologous to Rab32 and Rab38 and predicted to bypass GEF activity (McCray et al., 2010), restored gut granules in ccz-1 mutants (Figure 9 and Table 3). Recent in vitro studies demonstrate that mammalian BLOC-3, composed of HPS4 and HPS1, which are homologous to CCZ-1 and SAND-1/Mon1, respectively, functions as a GEF for Rab32 and Rab38 (Gerondopoulos et al., 2012). Mutations in HPS1 and HPS4 lead to Hermansky–Pudlak syndrome and display defects in the formation of LROs (Oh et al., 1996; Suzuki et al., 2002; Chiang et al., 2003; Martina et al., 2003; Nazarian et al., 2003; Gerondopoulos et al., 2012). Therefore, if CCZ-1 functions analogously to HPS4, it would be expected to act as a GEF for the Rab GLO-1.
sand-1 mutants did not substantially alter gut granule biogenesis (Figures 1–5 and Tables 1 and 2; Poteryaev et al., 2007), in contrast to what would be predicted if CCZ-1 and SAND-1 functioned together in a complex similar to HPS4 and HPS1. We demonstrate that C. elegans GLO-3 exhibits significant sequence similarity with human and Drosophila HPS1 and SAND-1/Mon1–related proteins (Figure 9). Of note, the region of highest homology is within the Longin domain of HPS1, which mediates the interaction with HPS4 in BLOC-3 (Carmona-Rivera et al., 2013), suggesting that GLO-3 functions with CCZ-1 in gut granule biogenesis. glo-3 mutants exhibit defects in gut granule biogenesis, and GLO-3::GFP localizes to the gut granule membrane (Rabbitts et al., 2008). Moreover, predicted spontaneous nucleotide exchange mutations in GLO-1 Rab restored gut granules in glo-3 mutants (Figure 9), consistent with GLO-3 and CCZ-1 functioning similarly in gut granule biogenesis. These results suggest the interesting possibility that CCZ-1 could have dual functions, acting with GLO-3 as part of a BLOC-3–like complex upstream of the Rab GLO-1 during trafficking to gut granules, and also acting with SAND-1 to activate RAB-7 during trafficking to conventional lysosomes. However, the genetic data leave open the possibility of alternate models in which CCZ-1, GLO-3, and GLO-1 have distinct functions unrelated to each other during gut granule protein trafficking.
Mechanism of gut granule biogenesis
Lysosomes and LROs, although distinct compartments that coexist in many cell types, share a general cellular mechanism of biogenesis. For both, a precursor to the fully developed compartment matures from another organelle. This is followed by retrograde removal of material from the pre-compartment, as well as the addition of other cargo via vesicular membrane transport. Integral to these processes are Rab GTPases, their regulators, and their effectors. During lysosome biogenesis, which is much better understood than LRO formation, the early endosome matures into a late endosome that receives material from other organelles, including AP-3 pathway vesicles and mature lysosomes (Luzio et al., 2007; Dell'Angelica, 2009; Marks et al., 2013). Rab7, its GEF CCZ-1/SAND-1, and the Rab-7 effector HOPS play key roles in organelle maturation and membrane docking and fusion (Balderhaar and Ungermann, 2013; Solinger and Spang, 2013).
The requirement for the GLO-1 Rab and AP-3 in gut granule biogenesis originally suggested that a process analogous to lysosome biogenesis was at work during the creation of this LRO (Hermann et al., 2005, 2012). Data presented here that HOPS and CCZ-1 function in gut granule formation further support this idea. Although we do not know the identity of the organelle that matures into a gut granule, there are precedents for pre-LROs emerging from the trans-Golgi network, early endosomes, or late endosomes (Marks et al., 2013). It is unlikely that gut granule precursors develop from lysosomes, since blocking lysosome biogenesis at the endolysosome stage, immediately before the formation of mature lysosomes, does not disrupt gut granule formation (Treusch et al., 2004; Campbell and Fares, 2010). SAND-1 and RAB-7 are required for normal maturation of early to late endosomes in C. elegans (Poteryaev et al., 2007, 2010). Thus the observations that sand-1 mutant embryos contained gut granules and rab-7 mutant embryos had nearly normal gut granules suggest that late endosomes do not significantly contribute to gut granule biogenesis and that early to late endosome maturation is not a major contributor to gut granule biogenesis. Gut granules are accessible to externally applied endocytic tracers (Clokey and Jacobson, 1986; Hermann et al., 2005), supporting the possibility that the pre–gut granule emerges from the early endosome rather than the trans-Golgi network. Possibly the CDF-2::GFP compartments that accumulate in ccz-1 mutants, which lack known gut granule and lysosomal proteins (Figures 2–5), represent a pre-LRO derived from the early endosome. In any case, our genetic studies suggest a model in which CCZ-1 and GLO-3 act as upstream activators of the GLO-1 Rab, which could function in the maturation of the pre–gut granule and recruit HOPS to regulate membrane fusion events between the pre-LRO and other organelles, such as those generated by AP-3. Additional studies are needed to test this idea and define the precise cellular and molecular processes that mediate gut granule formation. This work will not only provide insights into LRO biogenesis, but will also illuminate the key similarities and differences between lysosome and LRO biogenesis.
MATERIALS AND METHODS
Nematode strains and culture
C. elegans strains were grown at 22°C, unless noted, on NGM media seeded with OP50 bacteria as described (Stiernagle, 2006). N2 was used as the wild-type strain. In standard C. elegans nomenclature, gene names are written in lowercase italics and if appropriate followed by a mutant allele placed in parentheses, and proteins are written in all capitals. The following mutant alleles contained within the N2 background were used: apt-6(ok429), apt-7(tm920), arl-8(tm2504), ccz-1(ok2182), ccz-1(t2129), glo-1(zu437), glo-2(tm592), glo-2(zu455), glo-3(kx90), glo-3(kx94), glo-4(ok623), him-5(e1490), pgp-2(kx48), rab-7(ok511), sand-1(ok1963), sand-1(or552), snpn-1(tm1892), vps-8(ok2912), vps-11(ok1664), vps-16(ok719), vps-16(ok776), vps-18(tm1125), vps-33.1(ok2494), vps-39(ok2442), vps-39(tm2253), vps-41(ep402), vps-41(ok3433), and vps-45(tm246). The following transgenes were used: amIs2[cdf-2::gfp; unc-119(+)]I, amIs4[cdf-2::gfp; unc-119(+)], jcpEx2[ced-1p::ccz-1::yfp; unc-119(+); myo-2::GFP], kxEx148[F11E6.1::mCherry; Rol-6D], kxEx247[vha-6p::gfp::glo-1; Rol-6D], kxEx249[vha-6p::gfp::glo-1(K130E); Rol-6D], kxEx252[vha-6p::gfp::glo-1(D132A); Rol-6D], kxEx254[vha-6p::gfp::glo-1(I133F); Rol-6D], pwIs50[lmp-1p::lmp-1::gfp; unc-119(+)], pwEx31[lmp-1p::lmp-1::gfp; unc-119(+)], and yqEx115[vps-18p::vps-18::gfp; unc-76(+)]. The strains and alleles are described at WormBase (www.wormbase.org).
Genetic manipulations
Using standard genetic approaches, we found that the embryonic Glo phenotype of vps-18(tm1125) was recessively expressed and maternally and zygotically rescued. Introduction of a vps-18::gfp transgene whose expression is typically silenced in the germline (Kelly et al., 1997) rescued the embryonic Glo phenotype of vps-18(-), suggesting that embryonic expression is sufficient for vps-18(+) function in gut granule biogenesis. The presence of the vps-18(tm1125) deletion was verified by PCR and DNA sequencing. The Glo phenotype of ccz-1(ok2182) and ccz-1(t2129) embryos was recessively expressed and maternally and zygotically rescued. The adult Glo phenotype of these ccz-1 alleles, although recessive, was not maternally rescued. Introduction of a ccz-1::yfp transgene whose expression is controlled by the ced-1 promoter rescued the embryonic Glo phenotype of ccz-1(-), suggesting that embryonic expression is sufficient for ccz-1(+) function in gut granule biogenesis.
Genetic crosses were used to introduce and make homozygous the integrated cdf-2::gfp and lmp-1::gfp transgenes in different mutant backgrounds. Owing to linkage of pwIs50 and sand-1, sand-1(or552) was crossed into a strain containing the extrachromosomal pwEx31[lmp-1::gfp] transgene. The ccz-1(-) and vps-18(-) mutants were made homozygous after crossing into a kxEx148[F11E6.1::mCherry; Rol-6D]–containing strain. The mutations being examined for effects on protein trafficking were homozygous except for rab-7(ok511) and some ccz-1(ok2182)–containing strains, which were maintained as heterozygotes. Homozygous rab-7(-) and ccz-1(-) individuals were identified by the presence of large refractile organelles visible by differential interference contrast (DIC) microscopy.
apt(-); him-5(-) males were typically used to generate apt-6(-) and apt-7(-)–containing double mutants. From crosses of apt(-) with vps-18(-), resulting F2 adults that exhibited wild-type autofluorescence and corresponding F3 Glo embryos were isolated. These F3s were then cloned out to identify lines exhibiting the apt(-) Glo phenotype upon reaching adulthood. In crosses with vps-41(-) and apt-6(-), the apt-6(-) allele was marked by a closely linked dpy-5(-) mutation, and vps-41(-) was marked by a closely linked unc-6(-) mutation. Dpy Unc F2 offspring of the cross were isolated and scored. From crosses of apt(-) with ccz-1(-) and sand-1(-), resulting F2 progeny that exhibited the apt(-) adult Glo phenotype, which lacked early-stage embryos with large refractile organelles indicative of ccz-1(-) and sand-1(-), were isolated. F3 progeny were cloned out, and those producing embryos with refractile organelles were isolated. From crosses of apt(-) and other glo(-) mutants with rab-7(-), resulting F2 adult progeny exhibiting a Glo phenotype were isolated and allowed to self. Resulting F3 animals that laid rab-7(-) dead embryos were scored. In all cases multiple double-mutant lines were isolated and scored.
RNAi feeding protocol 1 (Kamath et al., 2001), using bacterial clones from the Ahringer RNAi library (available from Source Bioscience, Nottingham, United Kingdom) or, when these were not present for the gene being studied, the Vidal RNAi library (available from Thermo Scientific, Pittsburgh, PA), was used to knock down gene expression. In some cases rrf-3(pk1426) was used (Simmer et al., 2003). Briefly, gravid adults were bleached to isolate embryos, which were placed onto freshly seeded RNAi plates. When these embryos developed to adulthood, they and their resulting embryonic progeny were scored unless otherwise stated.
Generating GLO-1(ΔG4) mutants
The G4 domain of GLO-1 was altered using site-directed mutagenesis with QuikChange II (Agilent Technologies, Santa Clara, CA) and a vha-6p::gfp::glo-1::let-858 3′UTR plasmid as a template (Hermann et al., 2005). Primers p868, 5′CCAGCAATTTTACTTGCAAACGAGTGTGATATCGACAATAAAC3′, and p869, 5′GTTTATTGTCGATATCACACTCGTTTGCAAGTAAAATTGCTGG3′, were used to generate GLO-1(K130E).
Primers P870, 5′CTTGCAAACAAGTGTGCTATCGACAATAAACTTGGTGACG3′, and P871, 5′CGTCACCAAGTTTATTGTCGATAGCACACTTGTTTGCAAG3′, were used to generate GLO-1(D132A).
Primers P872, 5′CTTGCAAACAAGTGTGATTTCGACAATAACTTGGTGACG3′, and P873, 5′CGTCACCAAGTTATTGTCGAAATCACACTTGTTTGCAAG3′, were used to generate GLO-1(I133F). Underlined residues denote point mutations. For each mutagenized construct, the coding sequence of gfp::glo-1 was sequenced at the Oregon Health & Science University DNA Services Core facility (Portland, OR) to verify that only the desired point mutation had been introduced. The wild-type vha-6p::gfp::glo-1 plasmid and the three glo-1(ΔG4) mutant constructs were individually coinjected at 1 ng/μl with pRF4 [Rol-6D] at 100 ng/μl into glo-1(zu437). The resulting transmitting lines with similar expression of GFP::GLO-1 were characterized. Each of the extrachromosomal arrays was assessed for restoration of adult autofluorescent compartments and then moved via a genetic cross into a wild-type background. The arrays did not dominantly alter the number of autofluorescent compartments in wild type. The arrays were crossed from wild type into different glo-mutant backgrounds to generate the analyzed strains.
Microscopy
A Zeiss AxioImager.M2 microscope equipped with DIC, polarization, and fluorescence optics was used for all imaging (Zeiss, Thornwood, NY). Images were captured using an AxioCam MRm digital camera controlled with AxioVision 4.8 software. In all cases, optical Z-series through the regions of interest were captured, and, as indicated in the figure legends, individual optical sections or maximum intensity projections are presented.
L4-stage larvae and adults were immobilized on 3% agarose pads for imaging by mounting animals in 10 mM levamisole (Sigma-Aldrich, St. Louis, MO) or 0.1 μm polystyrene nanobeads (Polysciences, Warrington, PA). Autofluorescent gut granules were visualized with Zeiss 38 (GFP, excitation, BP 470/40; emission, BP 525/50), Zeiss 45 (mCherry; excitation, BP 560/40; emission, BP 630/75), or Zeiss 49 (4′,6-diamidino-2-phenylindole [DAPI]; excitation, G 365; emission, 445/50) filters. Acidic compartments within adults were stained with acridine orange (Sigma-Aldrich) as described (Hermann et al., 2005) and visualized with a Zeiss 45 filter. For each strain, a camera exposure time was identified that did not reveal endogenous autofluorescence in unstained animals, which was used to image acridine orange–stained compartments. ccz-1(ok2182)/+ animals containing gfp::glo-1 or gfp::glo-1(ΔG4) transgenes were maintained, and resulting homozygous ccz-1(-) adult progeny with the transgene, which were scored, were identified by both the presence of early-stage embryos with large refractile organelles indicative of ccz-1(-) and the intestinal expression of GFP. In all lines, ccz-1(-) animals lacking GFP expression exhibited a Glo phenotype.
Living embryos were mounted on 3% agarose pads in H2O or 1XM9 for imaging. To immobilize living pretzel-stage animals, excess respiring OP50 bacteria were added with the embryos to induce hypoxia. Within embryos, birefringent material was visualized with polarization optics, and autofluorescent compartments were visualized with a Zeiss 49 filter, LMP-1::GFP was visualized with a Zeiss 38 filter, and F11E6.1a::mCherry was visualized with a Zeiss 45 filter. The number of LMP-1::GFP–containing compartments was scored using Z-stacks spanning the entire intestine of 1.5-fold-stage embryos fixed with 100% MeOH to prevent the movement of organelles. LMP-1::GFP is enriched on the plasma membrane of intestinal cells, enabling the identification of the four cells that compose Int2 and Int3 (Leung et al., 1999) and the quantification of the number of LMP-1::GFP compartments contained within these cells. Acidified compartments within embryos were stained with LysoSensor Green (Invitrogen, Grand Island, NY) as described (Levitte et al., 2010) and imaged with a Zeiss 38 filter.
Embryos were fixed and stained with LMP-1 (Hadwiger et al., 2010), PGP-2 (Schroeder et al., 2007), RAB-5 (Audhya et al., 2007), and RAB-7 (Chen et al., 2010) antibodies as described (Leung et al., 1999). The specificity of these antisera using the staining protocol was confirmed in prior work. This protocol extracts the autofluorescent material present within embryonic gut granules. GFP fluorescence was used to visualize CDF-2::GFP and LMP-1::GFP in antibody-stained embryos. Because rab-7(-) causes maternal effect lethality, homozygous rab-7(-) embryos derived from homozygous rab-7(-) hermaphrodites were identified in mixed populations by their altered 1.5-fold body morphology and enlarged PGP-2 or CDF-2::GFP– marked gut granules. For colocalization studies, two-channel Z-stacks containing signals identifying different organelle associated proteins were acquired and analyzed. For each of the five 1.5-fold-stage embryos in each experiment, 25–35 organelles containing the protein of interest were randomly selected from within the intestine, and each was manually scored for the presence/colocalization of the other marker. If the signals marking the two proteins overlapped, they were scored as colocalizing. Statistical analysis, which is described in the figure legends, was carried out using Excel 2010 (Microsoft, Redmond, WA).
The images presented are representative of the data used to quantify number, size, and colocalization. The figures were prepared using Photoshop CS2 (Adobe, San Jose, CA), and only contrast was adjusted.
Bioinformatics
Default settings were used in PSI-BLAST searches (Altschul et al., 1997). The location of β-strands and α-helices in the Longin domain of HPS1 are from homology-based structural predictions (Kinch and Grishin, 2006). Secondary structure predictions of GLO-3 were generated using JPRED (Cole et al., 2008), PHD (Przybylski and Rost, 2002), and PSIPRED (Jones, 1999) algorithms.
Supplementary Material
Acknowledgments
We are grateful to Juan Cabello, Barth Grant, Anne Spang, and Chonglin Yang for strains. We thank Peter Kennedy and Teri Peterson for assistance with statistical analysis. We acknowledge Lewis & Clark Bio361 students from 2006, 2010, and 2011 for carrying out the initial studies of ccz-1, sand-1, and vps-18 mutants. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Program P40 OD010440, the C. elegans Knockout Consortium, and the National Bioresource Project for C. elegans. This work was supported by the National Science Foundation (MCB-1120835), the James F. and Marion L. Miller Foundation, and the John S. Rogers Summer Research Program.
Abbreviations used:
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- Glo
gut granule loss
- HPS
Hermansky–Pudlak syndrome
- LRO
lysosome-related organelle
- SNARE
soluble N-ethylmaleimide–sensitive factor attachment protein receptor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-09-0521) on February 5, 2014.
REFERENCES
- Akbar MA, Ray S, Kramer H. The SM protein Car/Vps33A regulates SNARE-mediated trafficking to lysosomes and lysosome-related organelles. Mol Biol Cell. 2009;20:1705–1714. doi: 10.1091/mbc.E08-03-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio AL, Boyle JA, Di Pietro SM. Mechanism of platelet dense granule biogenesis: study of cargo transport and function of Rab32 and Rab38 in a model system. Blood. 2012;120:4072–4081. doi: 10.1182/blood-2012-04-420745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers CG, Merz AJ. HOPS interacts with Apl5 at the vacuole membrane and is required for consumption of AP-3 transport vesicles. Mol Biol Cell. 2009;20:4563–4574. doi: 10.1091/mbc.E09-04-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama M, Sun-Wada GH, Yamamoto A, Yamamoto M, Hamada H, Wada Y. Spatial restriction of bone morphogenetic protein signaling in mouse gastrula through the mVam2-dependent endocytic pathway. Dev Cell. 2012;22:1163–1175. doi: 10.1016/j.devcel.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Audhya A, Desai A, Oegema K. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol. 2007;178:43–56. doi: 10.1083/jcb.200701139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes—coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- Bossinger O, Schierenberg E. Transfer and tissue-specific accumulation of cytoplasmic components in embryos of Caenorhabditis elegans and Rabditis dolichura: in vivo analysis with a low-cost signal enhancement device. Development. 1992;114:317–330. [Google Scholar]
- Brett CL, Plemel RL, Lobingier BT, Vignali M, Fields S, Merz AJ. Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase. J Cell Biol. 2008;182:1141–1151. doi: 10.1083/jcb.200801001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright NA, Gratian MJ, Luzio JP. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr Biol. 2005;15:360–365. doi: 10.1016/j.cub.2005.01.049. [DOI] [PubMed] [Google Scholar]
- Britton C, Murray L. Cathepsin L protease (CPL-1) is essential for yolk processing during embryogenesis in Caenorhabditis elegans. J Cell Sci. 2004;117:5133–5143. doi: 10.1242/jcs.01387. [DOI] [PubMed] [Google Scholar]
- Cabrera M, et al. Phosphorylation of a membrane curvature-sensing motif switches function of the HOPS subunit Vps41 in membrane tethering. J Cell Biol. 2010;191:845–859. doi: 10.1083/jcb.201004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M, Ungermann C. Guanine nucleotide exchange factors (GEFs) have a critical but not exclusive role in organelle localization of Rab GTPases. J Biol Chem. 2013;288:28704. doi: 10.1074/jbc.M113.488213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Fares H. Roles of CUP-5, the Caenorhabditis elegans orthologue of human TRPML1, in lysosome and gut granule biogenesis. BMC Cell Biol. 2010;11:40. doi: 10.1186/1471-2121-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan S, Hartnell LM, Aguilar RC, Naslavsky N, Bonifacino JS. Human Vam6p promotes lysosome clustering and fusion in vivo. J Cell Biol. 2001;154:109–122. doi: 10.1083/jcb.200102142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Rivera C, Simeonov DR, Cardillo ND, Gahl WA, Cadilla CL. A divalent interaction between HPS1 and HPS4 is required for the formation of the biogenesis of lysosome-related organelle complex-3 (BLOC-3) Biochim Biophys Acta. 2013;1833:468–478. doi: 10.1016/j.bbamcr.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Jiang Y, Zeng S, Yan J, Li X, Zhang Y, Zou W, Wang X. Endocytic sorting and recycling require membrane phosphatidylserine asymmetry maintained by TAT-1/CHAT-1. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang PW, Oiso N, Gautam R, Suzuki T, Swank RT, Spritz RA. The Hermansky-Pudlak syndrome 1 (HPS1) and HPS4 proteins are components of two complexes, BLOC-3 and BLOC-4, involved in the biogenesis of lysosome-related organelles. J Biol Chem. 2003;278:20332–20337. doi: 10.1074/jbc.M300090200. [DOI] [PubMed] [Google Scholar]
- Clokey GV, Jacobson LA. The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech Ageing Dev. 1986;35:79–94. doi: 10.1016/0047-6374(86)90068-0. [DOI] [PubMed] [Google Scholar]
- Coburn C, et al. Anthranilate fluorescence marks a calcium-propagated necrotic wave that promotes organismal death in C. elegans. PLoS Biol. 2013;11:e1001613. doi: 10.1371/journal.pbio.1001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Darsow T, Katzmann DJ, Cowles CR, Emr SD. Vps41p function in the alkaline phosphatase pathway requires homo-oligomerization and interaction with AP-3 through two distinct domains. Mol Biol Cell. 2001;12:37–51. doi: 10.1091/mbc.12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DE, Roh HC, Deshmukh K, Bruinsma JJ, Schneider DL, Guthrie J, Robertson JD, Kornfeld K. The cation diffusion facilitator gene cdf-2 mediates zinc metabolism in Caenorhabditis elegans. Genetics. 2009;182:1015–1033. doi: 10.1534/genetics.109.103614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Franceschi N, Wild K, Schlacht A, Dacks JB, Sinning I, Filippini F. Longin and GAF domains: structural evolution and adaptation to the subcellular trafficking machinery. Traffic. 2014;15:104–121. doi: 10.1111/tra.12124. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC. AP-3-dependent trafficking and disease: the first decade. Curr Opin Cell Biol. 2009;21:552–559. doi: 10.1016/j.ceb.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Mullins C, Caplan S, Bonifacino JS. Lysosome-related organelles. FASEB. 2000;14:1265–1278. doi: 10.1096/fj.14.10.1265. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the b3A subunit of the AP-3 adaptor. Mol Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- Di Pietro SM, Falcon-Perez JM, Tenza D, Setty SR, Marks MS, Raposo G, Dell'Angelica EC. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol Biol Cell. 2006;17:4027–4038. doi: 10.1091/mbc.E06-05-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docampo R, Ulrich P, Moreno SN. Evolution of acidocalcisomes and their role in polyphosphate storage and osmoregulation in eukaryotic microbes. Philos Trans R Soc Lond B Biol Sci. 2010;365:775–784. doi: 10.1098/rstb.2009.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp N, Rethmeier R, Kramer L, Ungermann C. Membrane dynamics and fusion at late endosomes and vacuoles—Rab regulation, multisubunit tethering complexes and SNAREs. Eur J Cell Biol. 2011;90:779–785. doi: 10.1016/j.ejcb.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Feng L, et al. The beta3A subunit gene (Ap3b1) of the AP-3 adaptor complex is altered in the mouse hypopigmentation mutant pearl, a model for Hermansky-Pudlak syndrome and night blindness. Hum Mol Genet. 1999;8:323–330. doi: 10.1093/hmg/8.2.323. [DOI] [PubMed] [Google Scholar]
- Filippini F, Rossi V, Galli T, Budillon A, D'Urso M, D'Esposito M. Longins: a new evolutionary conserved VAMP family sharing a novel SNARE domain. Trends Biochem Sci. 2001;26:407–409. doi: 10.1016/s0968-0004(01)01861-8. [DOI] [PubMed] [Google Scholar]
- Gallegos ME, et al. The C. elegans Rab family: identification, classification and toolkit construction. PLoS One. 2012;7:e49387. doi: 10.1371/journal.pone.0049387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S, Sharma M, Ung C, Tuli A, Barral DC, Hava DL, Veerapen N, Besra GS, Hacohen N, Brenner MB. Lysosomal trafficking, antigen presentation, and microbial killing are controlled by the Arf-like GTPase Arl8b. Immunity. 2011;35:182–193. doi: 10.1016/j.immuni.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondopoulos A, Langemeyer L, Liang JR, Linford A, Barr FA. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol. 2012;22:2135–2139. doi: 10.1016/j.cub.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiani CA, Starcevic M, Rodriguez-Fernandez IA, Nazarian R, Cheli VT, Chan LN, Malvar JS, de Vellis J, Sabatti C, Dell'Angelica EC. The dysbindin-containing complex (BLOC-1) in brain: developmental regulation, interaction with SNARE proteins and role in neurite outgrowth. Mol Psychiatry. 2010;15:204–115. doi: 10.1038/mp.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SC, Wartosch L, Gray SR, Scourfield EJ, Deane JE, Luzio JP, Owen DJ. Structural basis of Vps33A recruitment to the human HOPS complex by Vps16. Proc Natl Acad Sci USA. 2013;110:13345–13350. doi: 10.1073/pnas.1307074110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Wang X. Rab GTPases act in sequential steps to regulate phagolysosome formation. Small GTPases. 2010;1:170–173. doi: 10.4161/sgtp.1.3.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger G, Dour S, Arur S, Fox P, Nonet ML. A monoclonal antibody toolkit for C. elegans. PLoS One. 2010;5:e10161. doi: 10.1371/journal.pone.0010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, et al. C. elegans BLOC-1 functions in trafficking to lysosome-related gut granules. PLoS One. 2012;7:e43043. doi: 10.1371/journal.pone.0043043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Schroeder LK, Hieb CA, Kershner AM, Rabbitts BM, Fonarev P, Grant BD, Priess JR. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2005;16:3273–3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershkovitz D, Mandel H, Ishida-Yamamoto A, Chefetz I, Hino B, Luder A, Indelman M, Bergman R, Sprecher E. Defective lamellar granule secretion in arthrogryposis, renal dysfunction, and cholestasis syndrome caused by a mutation in VPS33B. Arch Dermatol. 2008;144:334–340. doi: 10.1001/archderm.144.3.334. [DOI] [PubMed] [Google Scholar]
- Hickey CM, Wickner W. HOPS initiates vacuole docking by tethering membranes before trans-SNARE complex assembly. Mol Biol Cell. 2010;21:2297–2305. doi: 10.1091/mbc.E10-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida T, Sohma H, Kokai Y, Kawakami A, Hirosaki K, Okura M, Tosa N, Yamashita T, Jimbow K. Rab7 is a critical mediator in vesicular transport of tyrosinase-related protein 1 in melanocytes. J Dermatol. 2011;38:432–441. doi: 10.1111/j.1346-8138.2010.01004.x. [DOI] [PubMed] [Google Scholar]
- Hirosaki K, Yamashita T, Wada I, Jin HY, Jimbow K. Tyrosinase and tyrosinase-related protein 1 require Rab7 for their intracellular transport. J Invest Dermatol. 2002;119:475–480. doi: 10.1046/j.1523-1747.2002.01832.x. [DOI] [PubMed] [Google Scholar]
- Hoffman-Sommer M, Grynberg M, Kucharczyk R, Rytka J. The CHiPS domain—ancient traces for the Hermansky-Pudlak syndrome. Traffic. 2005;6:534–538. doi: 10.1111/j.1600-0854.2005.00301.x. [DOI] [PubMed] [Google Scholar]
- Huizing M, Helip-Wooley A, Westbroek W, Gunay-Aygun M, Gahl WA. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu Rev Genomics Hum Genet. 2008;9:359–386. doi: 10.1146/annurev.genom.9.081307.164303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Sarangarajan R, Stroval E, Zhou Y, Gahl WA, Boissy RE. AP-3 mediates tyrosinase but not TRP-1 trafficking in human melanocytes. Mol Biol Cell. 2001;12:2075–2085. doi: 10.1091/mbc.12.7.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume AN, Wilson MS, Ushakov DS, Ferenczi MA, Seabra MC. Semi-automated analysis of organelle movement and membrane content: understanding Rab-motor complex transport function. Traffic. 2011;12:1686–1701. doi: 10.1111/j.1600-0854.2011.01283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Jordens I, Westbroek W, Marsman M, Rocha N, Mommaas M, Huizing M, Lambert J, Naeyaert JM, Neefjes J. Rab7 and Rab27a control two motor protein activities involved in melanosomal transport. Pigment Cell Res. 2006;19:412–423. doi: 10.1111/j.1600-0749.2006.00329.x. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantheti P, et al. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron. 1998;21:111–122. doi: 10.1016/s0896-6273(00)80519-x. [DOI] [PubMed] [Google Scholar]
- Kawakami A, et al. Rab7 regulates maturation of melanosomal matrix protein gp100/Pmel17/Silv. J Invest Dermatol. 2008;128:143–150. doi: 10.1038/sj.jid.5700964. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Raunser S, Munger C, Wagner J, Song YL, Cygler M, Walz T, Oh BH, Sacher M. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127:817–830. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Doukoumetzidis K, Almendinger J, Stergiou L, Tosello-Trampont A, Sifri CD, Hengartner MO, Ravichandran KS. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen JM, Ravichandran KS. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 2010;464:778–782. doi: 10.1038/nature08853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinch LN, Grishin NV. Longin-like folds identified in CHiPS and DUF254 proteins: vesicle trafficking complexes conserved in eukaryotic evolution. Protein Sci. 2006;15:2669–2674. doi: 10.1110/ps.062419006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopper TH, Kienle N, Fasshauer D, Munro S. Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol. 2012;10:71. doi: 10.1186/1741-7007-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar D, Poeck B, Roth H, Ernst R, Keller A, Porsch M, Strauss R, Pflugfelder GO. Defective pigment granule biogenesis and aberrant behavior caused by mutations in the Drosophila AP-3beta adaptin gene ruby. Genetics. 2000;155:213–223. doi: 10.1093/genetics/155.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharczyk R, Kierzek AM, Slonimski PP, Rytka J. The Ccz1 protein interacts with Ypt7 GTPase during fusion of multiple transport intermediates with the vacuole in S. cerevisiae. J Cell Sci. 2001;114:3137–3145. doi: 10.1242/jcs.114.17.3137. [DOI] [PubMed] [Google Scholar]
- Laufer JS, Bazzicalupo P, Wood WB. Segregation of developmental potential in early embryos of Caenorhabditis elegans. Cell. 1980;19:569–577. doi: 10.1016/s0092-8674(80)80033-x. [DOI] [PubMed] [Google Scholar]
- Lee YS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung B, Hermann GJ, Priess JR. Organogenesis of the Caenorhabditis elegans intestine. Dev Biol. 1999;216:114–134. doi: 10.1006/dbio.1999.9471. [DOI] [PubMed] [Google Scholar]
- Levine TP, Daniels RD, Wong LH, Gatta AT, Gerondopoulos A, Barr FA. Discovery of new Longin and Roadblock domains that form platforms for small GTPases in Ragulator and TRAPP-II. Small GTPases. 2013;4:62–69. doi: 10.4161/sgtp.24262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitte S, Salesky R, King B, Coe Smith S, Depper M, Cole M, Hermann GJ. A Caenorhabditis elegans model of orotic aciduria reveals enlarged lysosome-related organelles in embryos lacking umps-1 function. FEBS J. 2010;277:1420–1439. doi: 10.1111/j.1742-4658.2010.07573.x. [DOI] [PubMed] [Google Scholar]
- Lloyd V, Ramaswami M, Kramer H. Not just pretty eyes: Drosophila eye color mutations and lysosomal delivery. Trends Cell Biol. 1998;8:257–259. doi: 10.1016/s0962-8924(98)01270-7. [DOI] [PubMed] [Google Scholar]
- Lo B, et al. Requirement of VPS33B, a member of the Sec1/Munc18 protein family, in megakaryocyte and platelet alpha-granule biogenesis. Blood. 2005;106:4159–4166. doi: 10.1182/blood-2005-04-1356. [DOI] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- Ma J, Plesken H, Treisman JE, Edelman-Novemsky I, Ren M. Lightoid and Claret: a Rab GTPase and its putative guanine nucleotide exchange factor in biogenesis of Drosophila eye pigment granules. Proc Natl Acad Sci USA. 2004;101:11652–11657. doi: 10.1073/pnas.0401926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak HY, Nelson LS, Basson M, Johnson CD, Ruvkun G. Polygenic control of Caenorhabditis elegans fat storage. Nat Genet. 2006;38:363–368. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- Maldonado E, Hernandez F, Lozano C, Castro ME, Navarro RE. The zebrafish mutant vps18 as a model for vesicle-traffic related hypopigmentation diseases. Pigment Cell Res. 2006;19:315–326. doi: 10.1111/j.1600-0749.2006.00320.x. [DOI] [PubMed] [Google Scholar]
- Marat AL, Dokainish H, McPherson PS. DENN domain proteins: regulators of Rab GTPases. J Biol Chem. 2011;286:13791–13800. doi: 10.1074/jbc.R110.217067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MS, Heijnen HF, Raposo G. Lysosome-related organelles: unusual compartments become mainstream. Curr Opin Cell Biol. 2013;25:495–505. doi: 10.1016/j.ceb.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Moriyama K, Bonifacino JS. BLOC-3, a protein complex containing the Hermansky-Pudlak syndrome gene products HPS1 and HPS4. J Biol Chem. 2003;278:29376–29384. doi: 10.1074/jbc.M301294200. [DOI] [PubMed] [Google Scholar]
- Matthews RP, Plumb-Rudewiez N, Lorent K, Gissen P, Johnson CA, Lemaigre F, Pack M. Zebrafish vps33b, an ortholog of the gene responsible for human arthrogryposis-renal dysfunction-cholestasis syndrome, regulates biliary development downstream of the onecut transcription factor hnf6. Development. 2005;132:5295–5306. doi: 10.1242/dev.02140. [DOI] [PubMed] [Google Scholar]
- McCray BA, Skordalakes E, Taylor JP. Disease mutations in Rab7 result in unregulated nucleotide exchange and inappropriate activation. Hum Mol Genet. 2010;19:1033–1047. doi: 10.1093/hmg/ddp567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messler S, et al. The TGF-beta signaling modulators TRAP1/TGFBRAP1 and VPS39/Vam6/TLP are essential for early embryonic development. Immunobiology. 2011;216:343–350. doi: 10.1016/j.imbio.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Mullins C, Hartnell LM, Bonifacino JS. Distinct requirements for the AP-3 adaptor complex in pigment granule and synaptic vesicle biogenesis in Drosophila melanogaster. Mol Gen Genet. 2000;263:1003–1014. doi: 10.1007/pl00008688. [DOI] [PubMed] [Google Scholar]
- Mullins C, Hartnell LM, Wassarman DA, Bonifacino JS. Defective expression of the mu3 subunit of the AP-3 adaptor complex in the Drosophila pigmentation mutant carmine. Mol Gen Genet. 1999;262:401–412. doi: 10.1007/s004380051099. [DOI] [PubMed] [Google Scholar]
- Nakae I, Fujino T, Kobayashi T, Sasaki A, Kikko Y, Fukuyama M, Gengyo-Ando K, Mitani S, Kontani K, Katada T. The Arf-like GTPase Arl8 mediates delivery of endocytosed macromolecules to lysosomes in Caenorhabditis elegans. Mol Biol Cell. 2010;21:2434–2442. doi: 10.1091/mbc.E09-12-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Falcon-Perez JM, Dell'Angelica EC. Biogenesis of lysosome-related organelles complex 3 (BLOC-3): a complex containing the Hermansky-Pudlak syndrome (HPS) proteins HPS1 and HPS4. Proc Natl Acad Sci USA. 2003;100:8770–8775. doi: 10.1073/pnas.1532040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Wei ML. Characterization of melanosomes in murine Hermansky-Pudlak syndrome: mechanisms of hypopigmentation. J Invest Dermatol. 2004;122:452–460. doi: 10.1046/j.0022-202X.2004.22117.x. [DOI] [PubMed] [Google Scholar]
- Nickerson DP, Brett CL, Merz AJ. Vps-C complexes: gatekeepers of endolysosomal traffic. Curr Opin Cell Biol. 2009;21:543–551. doi: 10.1016/j.ceb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C, Almendinger J, Gysi S, Gomez-Orte E, Kaech A, Hengartner MO, Schnabel R, Moreno S, Cabello J. ccz-1 mediates the digestion of apoptotic corpses in C. elegans. J Cell Sci. 2010;123:2001–2007. doi: 10.1242/jcs.062331. [DOI] [PubMed] [Google Scholar]
- Nordmann M, Cabrera M, Perz A, Brocker C, Ostrowicz C, Engelbrecht-Vandre S, Ungermann C. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol. 2010;20:1654–1659. doi: 10.1016/j.cub.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Oh J, Bailin T, Fukai K, Feng GH, Ho L, Mao JI, Frenk E, Tamura N, Spritz RA. Positional cloning of a gene for Hermansky-Pudlak syndrome, a disorder of cytoplasmic organelles. Nat Genet. 1996;14:300–306. doi: 10.1038/ng1196-300. [DOI] [PubMed] [Google Scholar]
- Oka T, Toyomura T, Honjo K, Wada Y, Futai M. Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. J Biol Chem. 2001;276:3309–33085. doi: 10.1074/jbc.M101652200. [DOI] [PubMed] [Google Scholar]
- Ooi CE, Moreira JE, Dell'Angelica EC, Poy G, Wassarman DA, Bonifacino JS. Altered expression of a novel adaptin leads to defective pigment granule biogenesis in the Drosophila eye color mutant garnet. EMBO J. 1997;16:4508–4518. doi: 10.1093/emboj/16.15.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol. 2013;15:668–676. doi: 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowicz CW, Brocker C, Ahnert F, Nordmann M, Lachmann J, Peplowska K, Perz A, Auffarth K, Engelbrecht-Vandre S, Ungermann C. Defined subunit arrangement and Rab interactions are required for functionality of the HOPS tethering complex. Traffic. 2010;11:1334–1346. doi: 10.1111/j.1600-0854.2010.01097.x. [DOI] [PubMed] [Google Scholar]
- Peng C, et al. Ablation of vacuole protein sorting 18 (Vps18) gene leads to neurodegeneration and impaired neuronal migration by disrupting multiple vesicle transport pathways to lysosomes. J Biol Chem. 2012;287:32861–32873. doi: 10.1074/jbc.M112.384305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell. 2007;12:739–750. doi: 10.1016/j.devcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Plemel RL, Lobingier BT, Brett CL, Angers CG, Nickerson DP, Paulsel A, Sprague D, Merz AJ. Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol Biol Cell. 2011;22:1353–1363. doi: 10.1091/mbc.E10-03-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols MS, ten Brink C, Gosavi P, Oorschot V, Klumperman J. The HOPS proteins hVps41 and hVps39 are required for homotypic and heterotypic late endosome fusion. Traffic. 2013;14:219–232. doi: 10.1111/tra.12027. [DOI] [PubMed] [Google Scholar]
- Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141:497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Poteryaev D, Fares H, Bowerman B, Spang A. Caenorhabditis elegans SAND-1 is essential for RAB-7 function in endosomal traffic. EMBO J. 2007;26:301–312. doi: 10.1038/sj.emboj.7601498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A, Seals D, Wickner W, Ungermann C. The docking stage of yeast vacuole fusion requires the transfer of proteins from a cis-SNARE complex to a Rab/Ypt protein. J Cell Biol. 2000;148:1231–1238. doi: 10.1083/jcb.148.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor PR, Mullock BM, Bright NA, Gray SR, Luzio JP. The role of intraorganellar Ca(2+) in late endosome-lysosome heterotypic fusion and in the reformation of lysosomes from hybrid organelles. J Cell Biol. 2000;149:1053–1062. doi: 10.1083/jcb.149.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybylski D, Rost B. Alignments grow, secondary structure prediction improves. Proteins. 2002;46:197–205. doi: 10.1002/prot.10029. [DOI] [PubMed] [Google Scholar]
- Pujol N, Bonnerot C, Ewbank JJ, Kohara Y, Thierry-Mieg D. The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J Biol Chem. 2001;276:11913–11921. doi: 10.1074/jbc.M009451200. [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Akbar MA, Ray S, Sevrioukov EA, Haberman AS, Rohrer J, Kramer H. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J Cell Sci. 2005;118:3663–3673. doi: 10.1242/jcs.02502. [DOI] [PubMed] [Google Scholar]
- Rabbitts BM, Kokes M, Miller NE, Kramer M, Lawrenson AL, Levitte S, Kremer S, Kwan E, Weis AM, Hermann GJ. glo-3, a novel Caenorhabditis elegans gene, is required for lysosome-related organelle biogenesis. Genetics. 2008;180:857–871. doi: 10.1534/genetics.108.093534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Marks MS, Cutler DF. Lysosome-related organelles: driving post-Golgi compartments into specialisation. Curr Opin Cell Biol. 2007:394–401. doi: 10.1016/j.ceb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SCW, Winistorfer SC, Poupon V, Luzio JP, Piper RC. Mammalian late vacuole protein sorting orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol Biol Cell. 2004;15:1197–1210. doi: 10.1091/mbc.E03-06-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Roh HC, Collier S, Guthrie J, Robertson JD, Kornfeld K. Lysosome-related organelles in intestinal cells are a zinc storage site in C. elegans. Cell Metab. 2012;15:88–99. doi: 10.1016/j.cmet.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas AM, Fuentes G, Rausell A, Valencia A. The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J Cell Biol. 2012;196:189–201. doi: 10.1083/jcb.201103008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132:3561–3572. doi: 10.1242/dev.01918. [DOI] [PubMed] [Google Scholar]
- Salazar G, et al. BLOC-1 complex deficiency alters the targeting of adaptor protein complex-3 cargoes. Mol Biol Cell. 2006;17:4014–4026. doi: 10.1091/mbc.E06-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonthaler HB, Fleisch VC, Biehlmaier O, Makhankov Y, Rinner O, Bahadori R, Geisler R, Schwarz H, Neuhauss SC, Dahm R. The zebrafish mutant lbk/vam6 resembles human multisystemic disorders caused by aberrant trafficking of endosomal vesicles. Development. 2008;135:387–399. doi: 10.1242/dev.006098. [DOI] [PubMed] [Google Scholar]
- Schroeder LK, Kremer S, Kramer MJ, Currie E, Kwan E, Watts JL, Lawrenson AL, Hermann GJ. Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle. Mol Biol Cell. 2007;18:995–1008. doi: 10.1091/mbc.E06-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, et al. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol Biol Cell. 2007;18:768–780. doi: 10.1091/mbc.E06-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukov EA, He JP, Moghrabi N, Sunio A, Kramer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol Cell. 1999;4:479–486. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson F, Peden AA, Christopoulou L, Robinson MS. Characterization of the adaptor-related complex, AP-3. J Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram A, Marks MS. Mechanisms of protein delivery to melanosomes in pigment cells. Physiology (Bethesda) 2012;27:85–99. doi: 10.1152/physiol.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn EJ, Rojas-Pierce M, Pan S, Carter C, Serrano-Mislata A, Madueno F, Rojo E, Surpin M, Raikhel NV. The shoot meristem identity gene TFL1 is involved in flower development and trafficking to the protein storage vacuole. Proc Natl Acad Sci USA. 2007;104:18801–18806. doi: 10.1073/pnas.0708236104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Spang A. Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 2013;280:2743–2757. doi: 10.1111/febs.12151. [DOI] [PubMed] [Google Scholar]
- Sriram VS, Krishnan KS, Mayor S. deep-orange and carnation define distinct stages in late endosomal biogenesis in Drosophila melanogaster. J Cell Biol. 2003;161:593–607. doi: 10.1083/jcb.200210166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. 2006. Maintenance of C. elegans. Wormbook Feb 11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, et al. Hermansky-Pudlak syndrome is caused by mutations in HPS4, the human homolog of the mouse light-ear gene. Nat Genet. 2002;30:321–324. doi: 10.1038/ng835. [DOI] [PubMed] [Google Scholar]
- Tamura K, Ohbayashi N, Ishibashi K, Fukuda M. Structure-function analysis of VPS9-ankyrin-repeat protein (Varp) in the trafficking of tyrosinase-related protein 1 in melanocytes. J Biol Chem. 2011;286:7507–7521. doi: 10.1074/jbc.M110.191205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos AC, et al. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol Biol Cell. 2005;16:5356–5372. doi: 10.1091/mbc.E05-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JL, Vihtelic TS, denDekker AD, Willer G, Luo X, Murphy TR, Gregg RG, Hyde DR, Thummel R. The loss of vacuolar protein sorting 11 (vps11) causes retinal pathogenesis in a vertebrate model of syndromic albinism. Invest Ophthalmol Vis Sci. 2011;52:3119–3128. doi: 10.1167/iovs.10-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochio H, Tsui MM, Banfield DK, Zhang M. An autoinhibitory mechanism for nonsyntaxin SNARE proteins revealed by the structure of Ykt6p. Science. 2001;293:698–702. doi: 10.1126/science.1062950. [DOI] [PubMed] [Google Scholar]
- Treusch S, Knuth S, Slaugenhaupt SA, Goldin E, Grant BD, Fares H. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc Nat Acad Sci USA. 2004;13:4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban D, et al. The VPS33B-binding protein VPS16B is required in megakaryocyte and platelet alpha-granule biogenesis. Blood. 2012;120:5032–5040. doi: 10.1182/blood-2012-05-431205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderheit A, Helenius A. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol. 2005;3:e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Stromhaug PE, Kauffman EJ, Weisman LS, Klionsky DJ. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J Cell Biol. 2003;163:973–985. doi: 10.1083/jcb.200308071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner TS, Sinclair DA, Fitzpatrick KA, Singh M, Devlin RH, Honda BM. The light gene of Drosophila melanogaster encodes a homologue of VPS41, a yeast gene involved in cellular-protein trafficking. Genome. 1998;41:236–243. [PubMed] [Google Scholar]
- Wasmeier C, Romao M, Plowright L, Bennett DC, Raposo G, Seabra MC. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J Cell Biol. 2006;175:271–281. doi: 10.1083/jcb.200606050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Chen D, Fang Z, Xu J, Sun X, Song S, Liu J, Yang C. Lysosome biogenesis mediated by vps-18 affects apoptotic cell degradation in Caenorhabditis elegans. Mol Biol Cell. 2009;20:21–32. doi: 10.1091/mbc.E08-04-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsu A, Ohbayashi N, Tamura K, Fukuda M. Syntaxin-3 is required for melanosomal localization of Tyrp1 in melanocytes. J Invest Dermatol. 2013;133:2237–2246. doi: 10.1038/jid.2013.156. [DOI] [PubMed] [Google Scholar]
- Yousefian J, Troost T, Grawe F, Sasamura T, Fortini M, Klein T. Dmon1 controls recruitment of Rab7 to maturing endosomes in Drosophila. J Cell Sci. 2013;126:1583–1594. doi: 10.1242/jcs.114934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Lin J, Jin C, Xia B. Solution structure of human zeta-COP: direct evidences for structural similarity between COP I and clathrin-adaptor coats. J Mol Biol. 2009;386:903–912. doi: 10.1016/j.jmb.2008.12.083. [DOI] [PubMed] [Google Scholar]
- Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6:e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu K, Robert KW, DeBolt KM, Hong N, Tao JQ, Fukuda M, Fisher AB, Huang S. Rab38 targets to lamellar bodies and normalizes their sizes in lung alveolar type II epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2011;301:L461–L477. doi: 10.1152/ajplung.00056.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu GD, L'Hernault SW. The Caenorhabditis elegans spe-39 gene is required for intracellular membrane reorganization during spermatogenesis. Genetics. 2003;165:145–157. doi: 10.1093/genetics/165.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu GD, Salazar G, Zlatic SA, Fiza B, Doucette MM, Heilman CJ, Levey AI, Faundez V, L'Hernault SW. SPE-39 family proteins interact with the HOPS complex and function in lysosomal delivery. Mol Biol Cell. 2009;20:1223–1240. doi: 10.1091/mbc.E08-07-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatic SA, Tornieri K, L'Hernault SW, Faundez V. Metazoan cell biology of the HOPS tethering complex. Cell Logist. 2011a;1:111–117. doi: 10.4161/cl.1.3.17279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatic SA, Tornieri K, L'Hernault SW, Faundez V. Clathrin-dependent mechanisms modulate the subcellular distribution of class C Vps/HOPS tether subunits in polarized and nonpolarized cells. Mol Biol Cell. 2011b;22:1699–1715. doi: 10.1091/mbc.E10-10-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.