Abstract
Objectives
Aberrant expression of SOX4 in endometrial cancer has been identified and partially was contributed to hypermethylation of miR-129-2. Other miRNAs are suspected to influence SOX 4 as well. The current study seeks to identify other hypermethylated miRNAs that regulate SOX4 in endometrial carcinomas.
Methods
Methylation levels of miRNA promoter regions were measured by combined bisulfite restriction analysis (COBRA) and pyrosequencing assays. Gene expression was determined by RT-qPCR. Methylation level of a miRNA locus was corrected with clinicopathologic factors for 252 gynecological specimens.
Results
In silico analysis identified 13 miRNA loci bound on the 3′-UTR of SOX4. Using COBRA assays, increased methylation of miR-203, miR-219-2, miR-596, and miR-618 was detected in endometrial cancer cells relative to those seen in a normal cell line and in normal endometrium. Transfection of a miR-203 mimic decreased SOX4 gene expression. Hypermethylation of miR-203 was detected in 52% of type I endometrioid endometrial carcinomas (n=131) but was not seen in any of 10 uninvolved normal endometria (P<0.001). Methylation status of miR-203 was significantly associated with microsatellite instability and MLH1 methylation in endometrial tumors (P<0.001). Furthermore, hypermethylation of miR-203 was found in endometrioid and clear endometrial subtype tumors, but not in cervical squamous cell and ovarian carcinomas.
Conclusions
Hypermethylation of miR-203 is a frequent event in endometrial carcinomas and is strongly associated with microsatellite instability and MLH1 methylation status. Thus, miR-203 methylation level might represent a marker for patients with endometrioid and clear endometrial sub-cancers.
Keywords: Endometrial carcinoma, DNA methylation, SOX4, miR-203
Introduction
The SRY-related high-motility group box 4 gene (SOX4) is overexpressed in a variety of cancers, including prostate, lung, bladder, breast, gastric, and endometrial cancers [1, 2]. Its known functions include the regulation of embryonic development and differentiation to determine cell fate, as well as cellular transformation, cell survival and metastasis [1], suggesting an oncogenic role for SOX4 in some solid malignancies. In order to account for this overexpression, various mechanisms have been explored, such as chromosome amplification and post-transcription by microRNAs (also known as miRNAs) [1]. miRNAs are small noncoding RNAs that have recently been gaining attention in their roles in gene regulation [3]. More than 2500 human mature miRNAs have been identified [4], each believed to have the potential to post-transcriptionally modulate the expression of multiple mRNA targets [3, 4]. miRNAs regulate mRNA expression by forming imperfect pairing at the 3′-end of untranslated regions (3′-UTRs) of a target locus, which inhibits translation and may even promote degradation of the target mRNA.
While it is expected that promoter hypomethylation could upregulate a specific oncogene, hypermethylation-mediated silencing of a miRNA could activate its oncogenic target [2, 5]. This post-transcriptional regulation allows miRNA to control many regular processes of the cell [3], possibly even tumorigenesis. Recent studies have demonstrated that some cancers are associated with downregulation or even complete chromosomal deletion of specific miRNAs. In 2010, frequent deletions of miR-15 and miR-16 were first found in cells from patients with chronic lymphocytic leukemia [6]. In this study these deletions appeared to be associated with cell cycle arrest and apoptosis. Downregulation of miRNAs in cancers frequently occurs via DNA methylation. In endometrial and gastric cancers, repression of miR-129-2 by DNA hypermethylation was found to be correlated with overexpression of SOX4 [2, 7]. Subsequent demethylation of miR-129-2 resulted in partial downregulation of SOX4 expression, in a manner analogous to the downregulation of tumor suppressors by promoter hypermethylation. In this study we investigate the methylation status of other miRNAs and their relationship to SOX4 in endometrial cancer in search of potential markers of this disease.
Materials and Methods
Gynecological specimens
A total of 252 tissue specimens were obtained, either from Washington University in St. Louis described in a previous report [2], or through the Cooperative Human Tissue Network (CHTN). The Washington University cohort contained 131 tumors of type I endometrioid endometrial carcinomas (EEC). The CHTN cohort included 10 cancer-free normal, 23 EEC each paired with an adjacent normal, 17 EEC, 37 non-endometrioid endometrial carcinomas (NEEC), 24 ovarian tumors, and 10 cervical squamous cell carcinomas. Patient characteristics are summarized in Supplementary Table 1. All participants consented to both the molecular analyses and any follow-up studies, and the protocols were approved by the Human Studies Committees at Washington University in St. Louis, the Ohio State University, and Medical College of Wisconsin. Tumor specimens and adjacent normal tissues were collected from primary endometrial carcinomas at the time of hysterectomy. Normal control tissues were procured from women undergoing hysterectomy for non-cancer-related causes. All specimens were evaluated by at least one pathologist, who confirmed the diagnoses from hematoxylin- and eosin-stained tissue sections. The presence of microsatellite instability (MSI) and MLH1 methylation status was determined and reported previously [2]. Standard methods were used to extract DNA from tumors, corresponding non-neoplastic tissues, and normal controls.
Cell culture
Human endometrial cancer cell lines, AN3CA, HEC1A, Ishikawa, KLE, RL95-2 and SK-UT-1B, and a normal endometrial cell line, E6/E7, were used in this study [2]. For epigenetic studies, these cells were treated with 5-aza-2′-deoxycytidine (DAC, 5 μM, Sigma-Aldrich, St. Louis, MO) for 48 h and/or trichostatin A (TSA, 0.5 μM, Sigma-Aldrich) for 24 h. For transfection studies, Ishikawa cells were transfected with mature miRNA, and miRNA negative control #1 (Life Technologies, Grand Island, NY), using the Cell Line Nucleofector Kit (Lonza, Walkersville, MD) according to manufacturer’s instructions. DNA and RNA from treated and untreated cells were isolated as described previously [2].
Reverse transcription and quantitative PCR (RT-qPCR)
Total RNA (1 μg) underwent reverse transcription using Superscript III reverse transcriptase (Life Technologies). PCR was performed as described previously [2]. SOX4 primers were published previously (Table S2) [2], and TaqMan microRNA assay kits were obtained from Life Technologies. The relative expression of a gene in cells was determined by comparing the threshold cycle (Ct) of the gene against the Ct of housekeeping genes, GAPDH or U48.
Combined bisulfite restriction analysis (COBRA)
Genomic DNA (500 ng) was treated with sodium bisulfite using the EZ DNA Methylation kit from Zymo Research (Irvine, CA), following the manufacturer’s recommended protocol. COBRA was used to evaluate promoter methylation of miRNAs. Target sequences were amplified by PCR, and the products were digested with methylation-sensitive restriction enzymes, such as AciI (New England Biolabs, Ipswich, MA) to identify methylated alleles. Primer sequences are denoted in Table S3. Digested and non-digested PCR products were resolved on 2% agarose gels stained with ethidium bromide. Smaller DNA fragments digested by AciI were scored as “methylated” in a given sample.
Pyrosequencing
Pyrosequencing was performed using the PyroMark MD system (Qiagen, Valencia, CA) according to the manufacturer’s protocol. The oligonucleotide primers were purchased from Life Technologies, and used for the amplified region of miR-203 as follows: the forward primer, GTTTGGAGTTAGAGTTATAGTTAGG; the reverse biotinylated primer, CCCAACAACACTTAACTCTC; and the pyrosequencing primer, GATTAATTTAGGGGAGTTTA. Specific primers to detect methylation levels of miR-219-2, miR-335, miR-596 and miR-618 are denoted in Table S4. Methylation was quantified using the provided software (Qiagen).
Statistical analyses
The mRNA expression of endometrial cancer cell lines, and methylation levels of the tumors and adjacent normal endometrial tissues were compared by using the paired two-sample t-test. A P-value of <0.05 was considered to be significant. The marginal relationship between continuous methylation levels of miRNAs and a relevant categorical clinicopathologic covariate such as MSI was examined using the t-test for binary variables or ANOVA for categorical variables. All tests were two-sided, and all statistical analyses were performed using GraphPad Prism 5 software (GraphPad software, La Jolla, CA).
Results
Multiple miRNAs targeted on SOX4 exhibit hypermethylation in endometrial cancer cells
Using in silico analysis (miRanda database [4]), we identified 13 miRNA loci embedded in canonical CpG islands (Figs. 1A–B, S1A–B) located in the 5′-flanking regions, which play important roles on regulating gene expression. Among these loci, the expression of seven (miR-130b, miR-132, miR-191, miR-212, miR-335, miR-363 and miR-596) has been previously reported to be associated with cancer development and/or regulated by DNA methylation [8–13]. All 13 miRNA loci were evaluated by COBRA in six endometrial cancer cell lines (AN3CA, Ishikawa, HEC1A, KLE, RL95-2, and SK-UT-1B, Fig. 1B from left to right), and one (N) pooled sample derived from two noncancerous endometria (Fig. 1B). Methylation levels of miR-130b/301b, miR-191, miR-320 and miR-632 were not found in all of tested endometrial cancer cells. Promoter hypermethylation of miR-106a-363, miR-335, miR-203, miR-219-1, miR-219-2, miR-596, miR-618, and miR-1253 was detected in all or part of six endometrial cancer cell lines, as shown in Fig. 1B. However, because hypermethylation of miR-106a-363, miR-335 and miR-219-2 was also found in the normal sample (Fig. 1B), these sites were not evaluated further. Hypermethylation of miR-203 was found in five of six endometrial cancer cell lines, but not in the normal (N) endometrium, and not in the normal endometrial cell line (E6/E7), as analyzed by COBRA (Fig. 1C) and pyrosequencing (Figs. S1B–C). Based upon our COBRA analysis, along with other reports [5, 8, 13], miR-203, miR-219-2, miR-335, miR-596, and miR-618 were selected for further analysis, because these miRNAs were predominantly hypermethylated in cancer cell lines, but not in normal endometrium.
Fig. 1.
miR-203 is a novel hypermethylated marker in endometrial cancer. (A) The diagram of predicted miRNA binding sites on SOX4 3′-UTR. (B) Summary of the methylation status by COBRA of thirteen miRNA regions in endometrial cancer cell lines (from left to right: AN3CA, Ishikawa, HEC1A, KLE, RL95-2 and SK-UT-1B) and one normal (N) pooled sample derived from two noncancerous endometria as a negative control. m: methylated; and u: unmethylated. (C) Hypermethylation of miR-203 in endometrial cancer cell lines, as revealed by COBRA analysis. E6/E7: normal endometrial cell line; SssI, methylated positive control; N: normal endometrium. +, AciI restriction enzyme added; and −, without AciI.
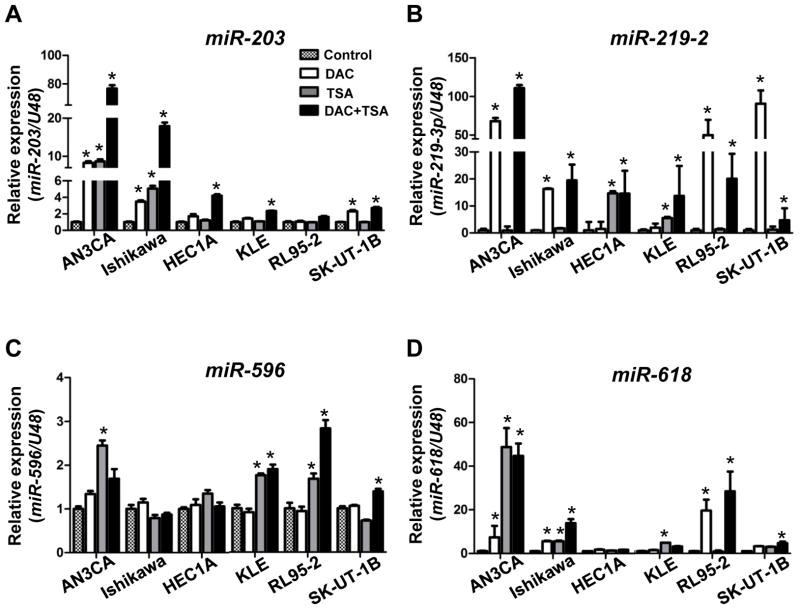
Because the hypermethylation of miR-203, miR-219-2, miR-335, miR-596, and miR-618 was observed in endometrial cancer cell lines (Fig. 1B) and the 5′-end of these loci have canonical CpG islands (Figs. S1B and S2–5, panels A), we examined whether these miRNAs can be reactivated in endometrial cancer cell lines. When these cancer cells were treated with a demethylating agent, DAC (5 μmol/L), a histone deacetylase inhibitor, TSA (0.5 μmol/L), or DAC and TSA in combination, reactivation of miR-203, miR-219-2, miR-596 and miR-618 was observed (Fig. 2). These results suggest that the silencing of miR-203, miR-219-2, miR-596, and miR-618 in endometrial cancer cells is mediated through promoter hypermethylation.
Fig. 2.
Relative expression levels of miR-203 (A), miR-219-2 (B), miR-596 (C), and miR-618 (D) in endometrial cancer cells after treatment with DAC and/or TSA. Gene expression was determined by RT-qPCR analysis and compared to untreated controls. U48 was used as an internal control gene. Error bar: SD; and *: P<0.05 compared with untreated control of the same cell type.
Methylation miR-203 is significantly correlated with microsatellite instability (MSI) status and MLH1 methylation
To determine whether hypermethylation of miRNAs mentioned above is presented in primary endometrial specimens, we performed pyrosequencing or MassARRAY (Supplementary Methods) analysis. The methylation status of miR-219-2, miR-335, miR-596, and miR-618 was examined. Because these miRNAs were either hypermethylated in both normal samples and tumors (Figs. S2–3), or exhibited low frequency in tumor specimens (Figs. S4–5), these miRNAs were not further evaluated.
The methylation level of miR-203 is examined in endometrioid endometrial specimens including 131 clinical EEC tumor samples and 10 uninvolved normal endometria. Quantitative methylation level of each CpG site is shown in Fig. 3A. The mean methylation level in all measured CpG sites in each specimen was used to compare differences between the tumor and the normal groups. Extensive methylation of miR-203 was found in more than 52% of the primary EEC tumors, but was not seen in the normal tissues (cancer free endometrium, P<0.0001; Fig. 3B), which has not been reported.
Fig. 3.
Methylation of the miR-203 CpG island and clinicopathologic covariates in primary endometrioid endometrial carcinomas (EECs). (A) Methylation profiles of miR-203 in 10 normal endometrial tissues and 131 primary tumors following pyrosequencing analysis. (B) Dot plots demonstrating miR-203 hypermethylation in EEC tumors. (C-D) Dot plots indicating that the methylation level of miR-203 is correlated with MSI status and MLH1 methylation. m: MLH1 methylated; and u: MLH1 unmethylated.
Hypermethylation of miR-203 was significantly associated with microsatellite instability (MSI) status (P=0.04) and MLH1 gene methylation (P=0.01) (Figs. 3C–D). Using linear model analysis, miR-203 methylation was not associated with age, recurrence, body mass index, tumor grade, or tumor stage (Fig. S6).
Hypermethylation of miR-203 is found in subtypes of endometrial primary tumors, but not in ovarian and cervical tumors
Since the methylation of miR-203 has not been extensively examined in primary gynecological tumors, we first determined DNA methylation in 23 paired EEC tissues. Pyrosequencing analysis demonstrated increased methylation levels of miR-203 in tumors relative to those in their adjacent normal counterparts (P<0.05; Fig. 4A). We further found hypermethylation of miR-203 in EEC and clear cell subtypes of endometrial tumors, but not in other gynecological tumors (Fig. 4B). Interestingly, methylation level of miR-203 was very low in both cervical squamous cell and ovarian carcinomas (Fig. 4C). The detailed methylation level of each CpG site is shown in Supplementary Fig. S7. These results suggest that among various gynecological tumors, miR-203 hypermethylation is limited to a few specific subtypes of endometrial carcinomas.
Fig. 4.

Hypermethylation of miR-203 in subtypes of primary gynecological tumors. (A) Methylation analysis of 23 paired endometrioid endometrial (EEC) tissues, measured by pyrosequencing analysis. (B) Dot plots demonstrating miR-203 hypermethylation in another set of primary EEC and clear endometrial tumors, but not in serous, mixed Müllerian (MM), or other mixed (mix) tumors. (C) Dot plots indicating methylation levels of miR-203 in cervical squamous cell carcinomas (CvSCC) and ovarian carcinomas (OvCa).
SOX4 is targeted by miR-203
To further demonstrate that miR-203 regulates SOX4 expression, Ishikawa cancer cells were transiently transfected with miR-203, miR-335 or miR-618 (0–250 pmol) for 24 h. miR-335 was chosen as a positive control because it was reported to target SOX4 in human breast cancer [8]. miR-618 was selected to prove in silico (Figs. 1, S1A) prediction. All of miR-203, miR-335 and miR-618 were found to reduce SOX4 mRNA expression in transfectants (Figs. 5A–C). In addition, Ishikawa cells transfected with miR-203 demonstrated suppressed TP63 mRNA expression (Fig. S8). The TP63 gene has been reported to be regulated by miR-203 in various cancers [14–16] as positive transfection control.
Fig. 5.

Down-regulation of SOX4 mRNA expression by miR-203 (A), miR-335 (B) and miR-618 (C). Cancer cells (Ishikawa) underwent transient transfection with miR-203, miR-335, or miR-618 at the indicated concentrations for 24 h. Gene expression was determined by RT-qPCR analysis and compared to untreated controls. GAPDH was used as an internal control gene. Error bar, SD; and *, P<0.05 compared with untreated cells. (D) A proposed model of miR-203 and SOX4 interactions in normal and cancer cells.
Discussion
SOX4 is a member of the SOX family of transcription factors. Its known functions involve the regulation of embryonic development and differentiation to determine cell fate [1]. SOX4 expression was shown to be elevated in a wide variety of tumors, including those of the endometrium [1, 2], suggesting a fundamental role in tumorigenesis. The functions of SOX4 in tumor development and progression could be dependent upon cellular content and tumor origin. In many cancers including those of the endometrium, SOX4 acts as a pro-oncogene and is associated with increased cell proliferation, cell survival, epithelial-to-mesenchymal transition, and metastasis, and with reduced apoptosis [1, 2]. In a subset of cancers such as the bladder and colon, SOX4 was reported to suppress tumor formation [1] by inhibiting tumor initiation and metastasis.
The SOX4 gene is located on chromosome 6p22 and has been reported to be amplified in bladder and lung cancers, resulting in increased SOX4 expression [17, 18]. The expression of this locus can be regulated by signaling pathways, such as the transforming growth factor-β and the WNT pathways, and at the post-translational modifications such as phosphorylation and acetylation on specific residues, or through interaction with specific cofactors such as TCF, syntenin-1 and p53 [1]. In addition, we previously have reported a new cascade regulation of SOX4 expression by miR-129-2, which serves as an upstream regulator. An inverse association between the expression of miR-129-2 and SOX4 was found in endometrial cancer. Silencing of miR-129-2 by an epigenetic event, DNA hypermethylation, resulted in lost expression of this miRNA in endometrial cancer, while also resulting in overexpression of SOX4 [2].
In this study, we have extended our search for potential miRNA effectors by scrutinizing the miRBase database [4] and found that the expression of SOX4 may be regulated by at least 13 putative miRNA loci with upstream canonical CpG islands. Five of these miRNAs (miR-203, miR-335, miR-219-2, miR-596 and miR-618) demonstrated hypermethylation in endometrial cancer cells (Fig. 1). However, only miR-203, miR-335, and miR-618 demonstrated a significant increase in hypermethylation relative to non-cancer endometrium (Figs. 3, S2–5). miR-335 was reported to be lost in primary breast tumors of recurrent patients [8], but its promoter was hypermethylated in both normal endometrium and tumors. Promoter methylation of miR-618 was less extensive in endometrial tumors and was not further evaluated.
miR-203 has been reported to suppress cancer cell proliferation, invasion, and metastasis [5, 19]. It is downregulated in many cancers, including endometrial carcinosarcoma [20]. Modulation of miR-203 expression by genetic and epigenetic silencing was found in hematopoietic tumors [5], resulting in enhancing ABL1 and BCR-ABL1 oncogene expression. In endometrial carcinoma, we identified SOX4 as a potential target of miR-203. Hypermethylation of miR-203 was detected in endometrial cancer cell lines (Fig. 1). When these cancer cells were treated with inhibitors of DNA methyltransferase and histone deacetylase, the expression of miR-203 was reactivated (Fig. 2), suggesting that this miRNA expression is subjected to epigenetic regulation. Further methylation analysis revealed hypermethylation of miR-203 in primary endometrioid endometrial tumors (Fig. 5D). This hypermethylation was significantly associated with microsatellite instability and MLH1 methylation (Fig. 3).
In addition, miR-203 has been reported to target and regulate the expression of TP63 (p63) [14–16]. TP63 is a TP53 (p53) homologue, and its structure and functions are similar to those of p53, particularly in the DNA-binding domain. Certain specific isoforms of p63 are involved in cellular response to stress, development and tumorigenesis. DeltaNp63 isoform may promote tumorigenesis, but TAp63 is a tumor suppressor in the female germline [21]. In a soft tissue sarcoma, Rhabdomyosarcoma, miR-203 functions as a tumor suppressor by targeting p63 and by inhibiting cell migration and promoting terminal myogenic differentiation [15]. In the current study, endometrial cancer cells transfected with miR-203 also showed inhibition of p63 mRNA (delta isoform) expression, which suggests that miR-203 could regulate endometrial cancer differentiation and migration. This observation needs to be further examined with regard to the role of miR-203 and p63 in endometrial tumorigenesis.
The hypermethylation status of miR-203 has not been extensively reported on in cervical, endometrial and ovarian malignancies. Our study adds to the current literature on miR-203. In this study, miR-203 hypermethylation not only is found in endometrioid endometrial carcinomas (EECs), but also was demonstrated in clear cell endometrial tumors. In the cervix, previous reports have demonstrated that the expression of miR-203 was downregulated in high-grade cervical intraepithelial neoplasia (CIN) and carcinomas [22–24], and in each case this was associated with hypermethylation of this locus. However, we did not detect any miR-203 methylation in cervical tumors, although it’s possible that our sample size is too small to detect significance. Previous report of expression of miR-203 as upregulated in ovarian cancer [25] is consistent with our finding of very low promoter methylation in this miRNA.
miRNAs have been found to be dysregulated in tissue-specific manners in various cancers [3, 26]. Many studies have explored the potential usefulness of miRNA expression profiles as biomarkers for cancer diagnosis, prognosis, and response to treatment [26]. miRNAs are likely to be useful as noninvasive biomarkers for both solid tumors and hematologic malignancies. Expression of circulating miRNAs in body fluids as serum, plasma, and saliva has confirmed their potential use as diagnostic and prognostic markers. miR-129-2 methylation was detectable in hepatocellular carcinoma (HCC) tumors [27], as well as in plasma from HCC patients [28], but not in healthy individuals or patients with liver cirrhosis. This selectivity implies its potential utility as an early diagnostic marker for HCC. We reported that hypermethylation of miR-129-2 is associated with shorter patient survival time, MSI, and MLH1 methylation in endometrial tumors [2]. This study suggests miR-203 as a potential biomarker to discriminate particular subtypes of endometrial cancer from other gynecological tumors. Such a marker could provide an important diagnostic tool to distinguish specific tumor subtypes if able to be linked with patient outcomes. More work is needed to confirm the use of DNA methylation of circulating miRNAs as a potential biomarker. We are currently examining whether hypermethylation of miR-203 and/or miR-129-2 could serve as a biomarker in plasma from endometrial cancer patients before and after hysterectomy.
Supplementary Material
Acknowledgments
This study was supported by NIH grants, U54 CA113001 (T.H.H.) and P50 CA134254 (P.J.G.), by Froedtert Hospital Foundation (D.S.U.), and by the Women’s Health Research Program, Falk Medical Research Trust, and the Frank L. Weyenberg Charitable Trust at the Medical College of Wisconsin (Y.H.). We thank the Cooperative Human Tissue Network (CHTN) for procuring the specimens, and the Clinical and Translational Scientific Institute (CTSI) of Southeast Wisconsin (NIH grant 8UL 1TR000055) for providing assistance in the preparation of this manuscript.
Footnotes
Conflict of interest statement
The authors have no conflicts of interest to declare.
References
- 1.Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32:3397–409. doi: 10.1038/onc.2012.506. [DOI] [PubMed] [Google Scholar]
- 2.Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–46. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 6.Patz M, Pallasch C, Wendtner C. Critical role of microRNAs in chronic lymphocytic leukemia: overexpression of the oncogene PLAG1 by deregulated miRNAs. Leuk Lymphoma. 2010;51:1379–81. doi: 10.3109/10428194.2010.496016. [DOI] [PubMed] [Google Scholar]
- 7.Shen R, Pan S, Qi S, Lin X, Cheng S. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem Biophys Res Commun. 2010;394:1047–52. doi: 10.1016/j.bbrc.2010.03.121. [DOI] [PubMed] [Google Scholar]
- 8.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C, Cai J, Wang Q, Tang H, Cao J, Wu L, et al. Epigenetic silencing of miR-130b in ovarian cancer promotes the development of multidrug resistance by targeting colony-stimulating factor 1. Gynecol Oncol. 2012;124:325–34. doi: 10.1016/j.ygyno.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Formosa A, Lena AM, Markert EK, Cortelli S, Miano R, Mauriello A, et al. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2013;32:127–34. doi: 10.1038/onc.2012.14. [DOI] [PubMed] [Google Scholar]
- 11.Incoronato M, Urso L, Portela A, Laukkanen MO, Soini Y, Quintavalle C, et al. Epigenetic regulation of miR-212 expression in lung cancer. PLoS ONE. 2011;6:e27722. doi: 10.1371/journal.pone.0027722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Q, Zhang J, Cao W, Wang X, Xu Q, Yan M, et al. Dysregulated miR-363 affects head and neck cancer invasion and metastasis by targeting podoplanin. Int J Biochem Cell Biol. 2013;45:513–20. doi: 10.1016/j.biocel.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Endo H, Muramatsu T, Furuta M, Uzawa N, Pimkhaokham A, Amagasa T, et al. Potential of tumor-suppressive miR-596 targeting LGALS3BP as a therapeutic agent in oral cancer. Carcinogenesis. 2013;34:560–9. doi: 10.1093/carcin/bgs376. [DOI] [PubMed] [Google Scholar]
- 14.Lena AM, Shalom-Feuerstein R, di Val Cervo PR, Aberdam D, Knight RA, Melino G, et al. miR-203 represses ‘stemness’ by repressing deltaNp63. Cell Death Differ. 2008;15:1187–95. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 15.Diao Y, Guo X, Jiang L, Wang G, Zhang C, Wan J, et al. miR-203, a tumor suppressor frequently down-regulated by promoter hypermethylation in Rhabdomyosarcoma. J Biol Chem. 2014;289:529–39. doi: 10.1074/jbc.M113.494716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei T, Orfanidis K, Xu N, Janson P, Stahle M, Pivarcsi A, et al. The expression of microRNA-203 during human skin morphogenesis. Exp Dermatol. 2010;19:854–6. doi: 10.1111/j.1600-0625.2010.01118.x. [DOI] [PubMed] [Google Scholar]
- 17.Medina PP, Castillo SD, Blanco S, Sanz-Garcia M, Largo C, Alvarez S, et al. The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum Mol Genet. 2009;18:1343–52. doi: 10.1093/hmg/ddp034. [DOI] [PubMed] [Google Scholar]
- 18.Bruch J, Schulz WA, Haussler J, Melzner I, Bruderlein S, Moller P, et al. Delineation of the 6p22 amplification unit in urinary bladder carcinoma cell lines. Cancer Res. 2000;60:4526–30. [PubMed] [Google Scholar]
- 19.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–76. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 20.Castilla MA, Moreno-Bueno G, Romero-Perez L, De Vijver KV, Biscuola M, Lopez-Garcia MA, et al. Micro-RNA signature of the epithelial–mesenchymal transition in endometrial carcinosarcoma. J Pathol. 2011;223:72–80. doi: 10.1002/path.2802. [DOI] [PubMed] [Google Scholar]
- 21.Bergholz J, Xiao ZX. Role of p63 in development, tumorigenesis and cancer progression. Cancer Microenviron. 2012;5:311–22. doi: 10.1007/s12307-012-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung TH, Man KM, Yu MY, Yim SF, Siu NS, Lo KW, et al. Dysregulated microRNAs in the pathogenesis and progression of cervical neoplasm. Cell Cycle. 2012;11:2876–84. doi: 10.4161/cc.21278. [DOI] [PubMed] [Google Scholar]
- 23.Wilting SM, Verlaat W, Jaspers A, Makazaji NA, Agami R, Meijer CJLM, et al. Methylation-mediated transcriptional repression of microRNAs during cervical carcinogenesis. Epigenetics. 2013;8:220–8. doi: 10.4161/epi.23605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu X, Er K, Mao C, Yan Q, Xu H, Zhang Y, et al. miR-203 suppresses tumor growth and angiogenesis by targeting VEGFA in cervical cancer. Cell Physiol Biochem. 2013;32:64–73. doi: 10.1159/000350125. [DOI] [PubMed] [Google Scholar]
- 25.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 26.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anwar SL, Albat C, Krech T, Hasemeier B, Schipper E, Schweitzer N, et al. Concordant hypermethylation of intergenic microRNA genes in human hepatocellular carcinoma as new diagnostic and prognostic marker. Int J Cancer. 2013;133:660–70. doi: 10.1002/ijc.28068. [DOI] [PubMed] [Google Scholar]
- 28.Lu CY, Lin KY, Tien MT, Wu CT, Uen YH, Tseng TL. Frequent DNA methylation of miR-129-2 and its potential clinical implication in hepatocellular carcinoma. Genes Chromosomes Cancer. 2013;52:636–43. doi: 10.1002/gcc.22059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.