Abstract
Among the many macromolecular machines involved in eukaryotic gene expression, the spliceosome remains one of the most challenging for structural biologists. Defining features of this highly complex apparatus are its excessive number of individual parts, many of which have been evolutionarily selected for regions of structural disorder, and the remarkable compositional and conformation dynamics it must undertake to complete each round of splicing. Here we review recent advances in our understanding of spliceosome structural dynamics stemming from bioinformatics, deep sequencing, high throughput methods for determining protein-protein, protein-RNA and RNA-RNA interaction dynamics, single molecule microscopy and more traditional structural analyses. Together, these tools are rapidly changing our structural appreciation of this remarkably dynamic machine.
Introduction
In all organisms, gene expression requires coordinate action of multiple macromolecular machines, many with multi-megadalton (MDa) molecular weights. Whereas high-resolution crystal structures have revealed the overall architecture and detailed inner workings of many such machines (e.g., ribosomes and RNA polymerases), one elusive “structure of desire” [1] is the spliceosome. Weighing in at over 3 MDa, the spliceosome is the ribonucleoprotein (RNP) complex responsible for excision of intragenic regions (introns; Box 1) from eukaryotic RNA polymerase II transcripts (precursors to messenger RNAs; pre-mRNAs). The spliceosome must be at once highly accurate – a single nucleotide shift in the site of splicing (splice site; SS) within an open reading frame will result in a non-functional mRNA – and highly malleable to permit alternative splicing, the process by which expressed regions (exons) are spliced together in different arrangements enabling synthesis of many different protein isoforms from a single gene. The proliferation of alternative splicing is the primary reason why organismal complexity is not tightly linked to gene number in the eukaryotic lineage[2,3].
To achieve the right balance between precision and malleability, the spliceosome contains scores of individual parts, many of which are structurally disordered. Working in a highly orchestrated manner, these parts perform incredible feats of molecular gymnastics with each round of splicing. These extremes of complexity and dynamics are no doubt to blame for the spliceosome's recalcitrance to crystallize despite intense efforts by multiple labs over many years. Nonetheless, significant progress is now being made by combining crystal structures of smaller pieces with EM reconstructions of larger assemblages. As detailed in the upcoming review [4], solution of several high-resolution structures containing pieces of Prp8, the massive and highly-conserved protein at heart of the spliceosome, is rapidly transforming our understanding of the catalytic core. In this review, we will focus instead on recent progress in understanding spliceosome evolutionary and structural dynamics.
Evolutionary Dynamics
One of the defining features of pre-mRNA splicing is the sheer number of components that must come and go to accurately identify and excise each new intron (Fig 1A). In budding yeast, this includes five small nuclear RNAs (snRNAs) and ~100 different proteins, whereas mammals utilize nine unique snRNAs and over 300 different proteins [5,6] . Metazoans have more spliceosomal snRNAs because they contain not one, but two spliceosomes: the more abundant “major spliceosome” responsible for removing 99.5% of introns and the “minor spliceosome” excising the other 0.5% [7] (Fig 1B). A long-standing question regarding the function of these minor introns was recently addressed by Younis et al. [8], who showed that under normal growth conditions, their splicing is limited by rapid decay of the key minor snRNA U6atac. In the presence of stress, however, U6atac is stabilized, allowing splicing of preexisting minor intron-containing transcripts, which can then be rapidly translated to help alleviate the stress.
Fig 1. An updated spliceosome assembly cycle.
A. Structural overview of the yeast spliceosome assembly, activation, and disassembly cycle (adapted from [36]). The crystal structures of human U1 and U4 snRNPs [51,52] and EM structures of larger complexes are shown [6] [53] [54] [55],[56] [57], [58].
B. Shared and unique components of the major and minor spliceosomes. Structures of NTC, U1, U4, and U5 snRNPs are as in A; U11/U12 di-snRNP structure is from [59]. NTC complex association with the minor spliceosome has only been inferred to date, not structurally validated. Also shown are the conserved sequences of major and minor introns with exons (boxes), introns (lettering or solid line), the branch adenosine (bold red) indicated (adapted from[7]).
The existence of two spliceosomes is thought to reflect a long ago merging of two eukaryotic genomes that had diverged and separately evolved for prior untold generations. By the time of the merge, so many mutations had accumulated in the separate lineages that the two machineries were no longer fully compatible. Nonetheless, the major and minor spliceosomes do share some key components, and much can be learned about core spliceosome structure from comparing their commonalities and differences (Fig 1B). The largest common component is U5 snRNP, which contains U5 snRNA and 14-15 stably-bound proteins[6]. Of all the snRNAs, U5 has the highest percentage of internal secondary structure and is the least accessible to RNase digestion or nucleotide modification reagents[9], consistent with it being almost entirely coated with proteins. The only region of U5 snRNA making intermolecular RNA-RNA interactions is a U-rich loop that contacts exonic nucleotides to either side of the intron and is thought to help align the exons to facilitate both steps of splicing. Because exon sequences are subject to selective pressures dictated by the encoded protein, the nucleotides at exon ends are not highly conserved; therefore, the contacts made with U5 snRNA are perforce relatively non-specific.
Among all spliceosomal proteins, those comprising the U5 snRNP are the most conserved across species. In Prp8, the largest spliceosomal protein, 61% of its >2300 amino acids are completely conserved from yeast to humans. Such a high level of conservation is indicative of strong structural constraints both internally (to maintain overall folding and activity) and externally (to maintain intermolecular interactions)[10]. Consistent with this, Prp8 is known to directly contact eight other spliceosomal proteins, all three splice site consensus sequences, and U1, U2, U5 and U6 snRNAs[11]. As discussed in the accompanying review [4], Prp8 both supplies amino acids necessary for catalysis and serves as the structural scaffold upon which the entire spliceosome is built. Two other highly conserved U5 snRNP proteins that interact with this scaffold are Brr2 and Snu114, two large NTPases necessary for key structural transitions during spliceosome assembly and disassembly[12].
Each of the other major spliceosomal snRNAs (U1, U2, U4 and U6) has a distinct ortholog in the minor spliceosome (U11, U12, U4atac and U6atac, respectively) (Fig 1B). These snRNAs make key intermolecular base pairs with other snRNAs and/or the splice site consensus sequences (Fig 1B). Because RNA-RNA interactions have simple substitution rules (e.g., A-U → G-C), compensatory mutations could easily accumulate over evolutionary time, resulting in incompatibilities when the two eukaryotic predecessor genomes merged. In comparison, many proteins were apparently less mutable and therefore remained sharable. Other shared proteins between the major and minor spliceosomes include the SF3B proteins required for early spliceosome assembly, the 110K and 65K tri-snRNP proteins[13] and possibly the NTC complex[7]. Among non-shared components, the 20 and 35 kDa proteins in 18S U11/U12 are likely orthologs of U1C and U1-70K [7]. U1C protein recognizes the 5'SS consensus of introns removed by the major spliceosome[14]; the different 5'SS consensus in minor spliceosomal introns may therefore explain the need for a different, but functionally related protein. Interactions between the N-terminal half of U1-70K and SR proteins modulate both exon definition and alternative splicing[15], and this homology region is retained in the U11/U12 35K protein. The unique C-terminal half of U1-70K interacts with polyA polymerase and suppresses pre-mRNA polyadenylation [16]. This additional activity of U1 snRNP was recently shown to be crucial for preventing premature polyadenylation at cryptic polyA sites within introns [17] and to control 3'-UTR length [18]. Consistent with the lack of this domain in U11/U12 35K, no such activity has yet been ascribed to the U11/U12 snRNP.
Evolving Toward Disorder?
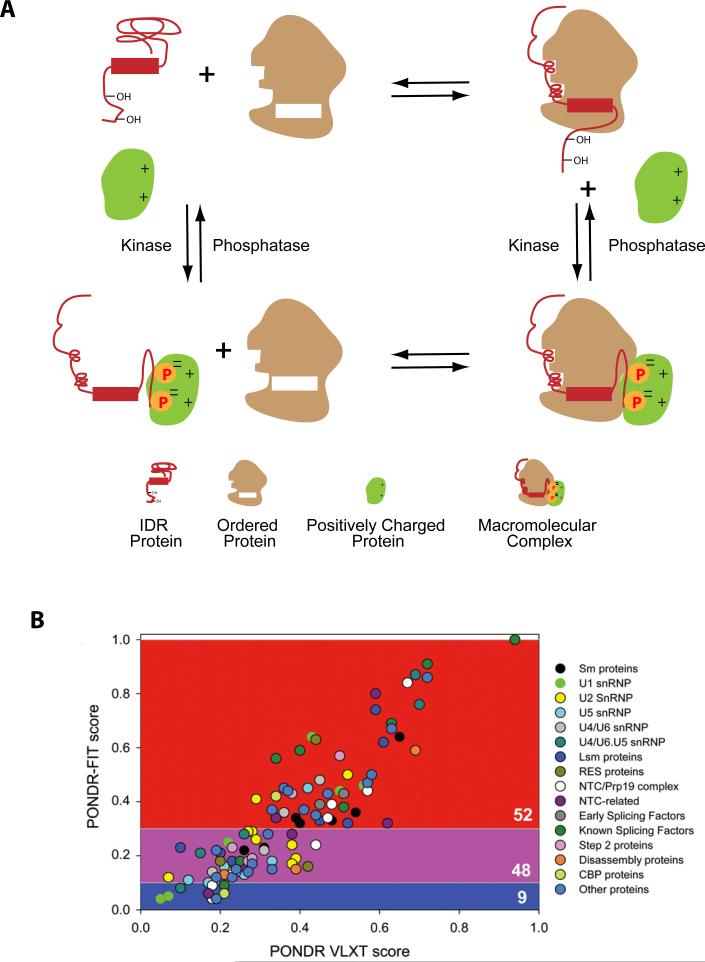
Many spliceosomal proteins contain intrinsically disordered regions (IDRs), polypeptide stretches that in isolation lack stable, well-defined 3D structures. IDRs have a variety of useful functions – they can serve as linkers between structured domains, as sites of post-translational modification, and as sites of protein-protein and protein-RNA recognition [19]. A general feature of IDRs is their ability to transition to a more ordered state upon interaction with a specific binding partner. Many IDR-containing proteins are able to bind multiple targets simultaneously, thereby facilitating larger complex assembly (Fig 2 A). Another important feature, however, is the capacity of some IDRs to adopt different conformations upon interaction with different binding partners. This capacity for multiple mutually exclusive specific interactions (multi-specificity) makes IDRs particularly adept at facilitating structural transitions within larger complexes.
Fig 2. Intrinsic disorder among spliceosomal proteins.
A. Illustration of how IDRs can facilitate formation of larger macromolecular assemblies by adopting ordered structures upon binding to a specific partner and/or serving as sites of post-translational modification.
B. Plot showing the correlation between predicted protein disorder of various yeast spliceosomal proteins as calculated by PONDR-FIT (y-axis) and PONDR® VLXT (x-axis) (Reproduced with permission from [21]). These two programs use different algorithms to generate a score for each protein sequence that reflects the fraction of amino acids likely to be in IDRs. This plot shows that both algorithms generate highly correlated outputs with regard to spliceosomal proteins and splicing factors [21]. Using arbitrary PONDRFIT cutoffs, proteins were classified as highly ordered (blue field, 9 proteins), moderately disordered (pink field, 48 proteins) and highly disordered (red field, 52 proteins). Proteins are color coded as indicated to indicate their relationship to different components and complexes.
Remarkably, one recent comprehensive bioinformatics analysis has predicted that IDRs occupy almost half of the combined sequences of abundant human spliceosomal proteins! [20] In S. cerevisiae, ~47% of spliceosomal proteins contain predicted IDRs in comparison to only ~13% of the entire yeast proteome [21]. Within the spliceosome, the distribution of intrinsic order and disorder is highly uneven, with essential catalytic core proteins generally being more ordered than those involved in spliceosome assembly and dynamics. Further, proteins involved in later stages of assembly (e.g., spliceosome activation) tend to be less disordered than components acting at earlier stages (e.g., initial splice site recognition) (Fig 2B). This suggests that over evolutionary time, a preexisting ordered catalytic core gained functionality and flexibility through addition of peripheral and evolutionarily younger IDR-containing proteins [20]. In higher eukaryotes, this increased flexibility likely enabled the spliceosome to integrate more diverse information than just the canonical 5'SS, branch site and 3'SS consensuses, thereby facilitating the proliferation of alternative splicing. Unfortunately for humans, however, the accompanying proliferation of IDR-containing splicing factors also has a cost. Due to their tendency toward disorder, IDR-containing proteins tend to form protein aggregates[22], and aggregates of splicing factors have been implicated in multiple human diseases, especially neurodegeneration[23].
Reversibility is the rule
Another major theme emerging in recent years is that, rather than being the one-way pathway typically drawn in textbooks, almost every step in the spliceosome cycle is readily reversible (Fig 1A). Each required structural and chemical transition is nearly energy neutral, with the overall process being driven in the forward direction by coupling these reversible transitions to an energetically favorable reaction (e.g., ATP hydrolysis by an RNA helicase). One example of this inherent reversibility is the phosphodiester exchange reactions comprising the first and second chemical steps of splicing (lariat formation and exon ligation)[24]. Tseng and Cheng [25] recently showed that not only can the spliceosome catalyze both chemical steps in forward and reverse, it can even convert spliced products (lariat intron and ligated exons) back into unspliced pre-mRNA! Catalysis of the first and second step chemistries requires two different active site configurations between which the spliceosome toggles to favor one step or the other[26,27]. The equilibrium between these two configurations can be altered by mono and divalent metal ion concentrations[28] and by mutations in the pre-mRNA, in U2 and U6 snRNAs, and in Prp8 and Brr2 [29,30], all of which together comprise the catalytic core. The overall process is driven in the forward direction by coupling these active site structural changes to ATP hydrolysis by the RNA helicases Brr2, Prp2, Prp16, and Prp22 [6].
One methodology rapidly rewriting our understanding of splicing dynamics and reversibility is single molecule microscopy. Supporting the idea that the splicing cycle can be thought of as a network of near energy neutral structural transitions separated by relatively low energy barriers, one single molecule fluorescence resonance energy transfer (smFRET) study suggested that pre-mRNA conformation is highly dynamic over the course of spliceosome assembly, with various states interchanging on sec timescales [31]. Using a protein-free system, Guo et al. [32] found evidence for at least three distinct conformations of U2 and U6 snRNA duplexes whose equilibrium and dynamics were functions of Mg2+ concentration and U6 snRNA sequence. These conformational changes bear strong resemblance to those proposed to occur in the transition between the first and second step catalytic states of the intact spliceosome [26] [27] [33].
Observation of single molecules in biochemically active cellular extracts has recently enabled researchers to directly measure the dynamic comings and goings of individual spliceosomal components, to distinguish on- from off-pathway assembly events and to tie specific conformational changes to individual binding events [34]. Using Colocalization Single Molecule Spectroscopy (CoSMoS), Hoskins et al. [35] demonstrated the reversibility of every major subcomplex addition step along the yeast spliceosome assembly pathway. More recently, Shcherbakova et al. [36] showed that functional A complex formation can occur by either a U1-first or a U2-first pathway (Fig 1A). Intriguingly, if the pre-mRNA contains multiple 5'SS's, there appears to be an ATP-dependent mechanism to ensure that only one U1 snRNP is stably incorporated into complex A [37]. Together with data indicating the likelihood of even more diverse spliceosome assembly pathways in mammalian cells[38,39], the above studies have profound consequences for our understanding of alternative splicing regulation. That is, alternative splicing decisions in vivo likely result from kinetic modulation of competing spliceosome assembly pathways, and the inherent reversibility of these pathways means that such modulation could occur at late as well as early assembly steps. By combining CoSMoS with smFRET, Crawford et al.[40] recently demonstrated that the 5'SS and BS regions remain physically separate until after spliceosome activation. This opens the possibility that the final decision of where to splice might occur much later in the spliceosome cycle than previously thought, further increasing the options for alternative splicing regulation.
Dynamic PPIs and Post-Translational Modifications
In addition to the dynamic RNA-RNA and RNA-protein interactions discussed above, the spliceosome cycle also involves innumerable dynamic protein-protein interactions (PPIs). Recently, Hegele et al. elucidated the complete PPI “wiring diagram” of the human spliceosome [41]. Employing a combination of yeast two-hybrid (Y2H) and coimmunoprecipitation analyses, the authors systematically investigated 632 possible PPIs among core and noncore components. They then used link clustering to integrate 242 positively confirmed core factor interactions with spliceosome subcomplex purification data. The result was a highly expanded understanding of core PPI dynamics during splicing, particularly with regard to mutually exclusive binding partner interactions that help drive the splicing cycle forward. This systematic Y2H and comparative proteomics approach can also be used to identify new splicing factors, as exemplified by the recent discovery of six novel splicing factors in S. pombe [42].
Also driving spliceosome assembly forward are dynamic post-translational modifications. Some spliceosomal proteins are acetylated, and dynamic acetylation/deacetylation is important for spliceosome assembly and rearrangement [6]. Numerous others are subject to reversible methylation and/or phosphorylation, with dynamic SR protein phosphorylation being particularly important for mammalian spliceosome assembly [6][43]. A recent bioinformatics study revealed that sites of dynamic phosphorylation tend to occur at intermolecular binding interfaces, where they can “orthosterically” modulate the strength of protein-protein interactions [44]. Further, phosphorylation, disorder-to-order transitions and formation of new binding partner interactions are all highly coupled (Fig. 2A), with about one quarter of all Ser/Thr/Tyr residues at interfaces being phosphorylated. Among these, phospho-Ser residues are the most likely to occur within intrinsically disordered regions (IDRs). Conversely, phospho-Tyr is more often observed at ordered interfaces (i.e., structures predicted to be ordered even in the unbound state) [45]. The prevalence of Ser phosphorylation on spliceosomal proteins is therefore consistent with the above discussion regarding the remarkable abundance of IDRs within the splicing machinery [20, 21].
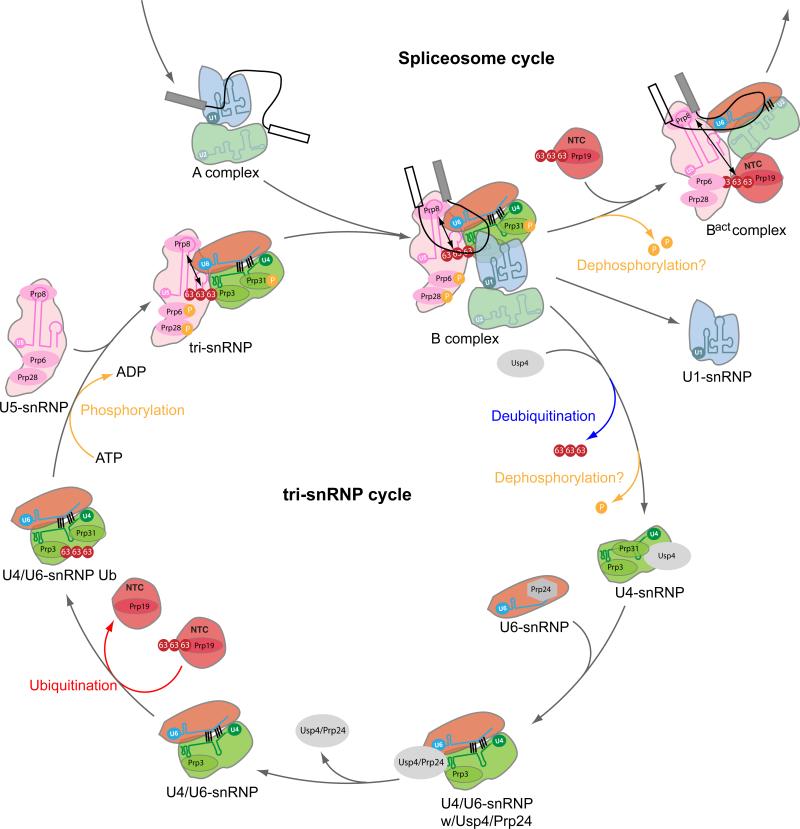
Another post-translational modification driving spliceosome dynamics is reversible ubiquitination. The best understood example is the polyubiquitination cycle involving the U4 snRNP protein Prp3, the NTC component PRP19, the deubiquitinating enzyme Usp4 and its binding partner in U6 snRNP Prp24/Sart3. Two early studies reported that Prp19 contains a U-box, allowing it to ubiquitinate itself via nonproteolytic K63-linked chains[46], and that dynamic ubiquitination/deubiquitination controls tri-snRNP levels, likely by regulating U4/U6 snRNA winding and unwinding [47]. These observations were recently integrated by Song et al. [48], who found that Prp3 is the downstream target of both Prp19 ubiquitination and Usp4 deubiquitination (Fig 3). Through interaction of the NTC with the U4/U6 di-snRNP, Prp19 transfers its ubiquitin chains to Prp3; this facilitates U4/U6.U5 tri-snRNP reassembly by increasing Prp3's affinity for Prp8, likely via Prp8's ubiquitin-binding JAMM domain. Once the U4/U6.U5 tri-snRNP has joined the spliceosome, Usp4 deubiquitinates Prp3, decreasing its affinity for Prp8. This both facilitates U4 departure, enabling interaction of U6 snRNA with U2 snRNAs to form the catalytic core, and frees up Prp8 to interact with the ubiquitin chains on Prp19 to stabilize NTC addition. Once U4 snRNP has been ejected from the spliceosome, it can bind a molecule of free U6 snRNA facilitated by Prp24/Sart3. Through its own series of structural transitions [49] [50], Prp24/Sart3 promotes U4/U6 snRNA reannealing to reform the U4/U6 snRNP within which Prp3 can once again be ubiquinated by Prp19 to favor U4/U6.U5 trisnRNP formation [48]. Intersecting with this ubiquitination-deubiquitination cycle is a phosphorylation-dephosphorylation cycle on tri-snRNP proteins Prp6, Prp28 and Prp31[6]. This phosphorylation cycle is crucial for both tri-sRNP formation and B complex assembly.
Fig 3.
Ubiquitination and phosphorylation cycles involved in U4/U6.U5 tri-snRNP dynamics (Adapted from [48]). See text for details. Question marks indicate steps that are yet to be elucidated.
Perspective
The ever-increasingly complicated and interconnected cycles of dynamic structural and post-translational changes shown in Figs 1 and 3 illustrate the incredible complexities facing structural biologists bold enough to even contemplate complete structural understanding of the splicing machinery within their lifetimes. Given its remarkable dynamics, a high-resolution crystal structure of any fully assembled spliceosome may yet be years away. Nonetheless, with new tools such as single molecule microscopy, bioinformatics, and high throughput methods for determining protein-protein, protein-RNA and RNA-RNA interaction dynamics increasingly being developed and applied, structural biologists do have much to celebrate. No doubt the splicing machinery has many structural surprises yet to be revealed.
Box 1: Splicing nomenclature.
Exons: Regions of pre-mRNA that are ligated together by the spliceosome.
Introns: Regions of pre-mRNA excised by the spliceosome.
5'SS: 5' splice site; a.k.a. splice donor. The 3'-5' phosphodiester bond at the 5' boundary of an intron. This bond is exchanged for a 2'-5' phosphodiester linking the 5'SS with the BP during the first chemical step of splicing.
BP: Branch point. An intronic adenosine near the 3'SS whose 2'-OH serves as the nucleophile for the first chemical step of splicing.
3'SS: 3' splice site; a.k.a. splice acceptor. The 3'-5' phosphodiester bond at the 3' boundary of an intron. This bond is exchanged for a 3'-5' phosphodiester linking the two exons during the second chemical step of splicing.
Consensus sequence: A region of sequence conservation that helps to define one of the three sites of chemistry.
snRNA: Small nuclear ribonucleic acid. Spliceosomal snRNAs vary in length from ~90 nts to ~1200 nts and are uridine-rich, so are named U1, U2, etc.
snRNP: Small nuclear ribonucleoprotein particle. A complex containing one or more snRNAs plus stably-bound proteins.
NTC: Nineteen complex. A protein-only complex containing Prp19 and other proteins. Stable association of the NTC is the final step in spliceosome assembly, and mediates the transition from the pre-catalytic to the catalytic spliceosome.
First chemical step: Attack by the 2'-OH of the BP adenosine on the 5'SS to form a 2'-5' branched lariat intron and liberate the 5' exon.
Second chemical step: Attack by the 3'-OH of the 5' exon on the 3'SS to join the exons and liberate the lariat intron.
Splicing factor: A protein involved in splicing that is not a stable snRNP component.
Highlights.
Evolutionarily young spliceosomal proteins have high intrinsic disorder
Spliceosome assembly is reversible and can occur by multiple pathways
Reversible post-translational modifications are key to spliceosome dynamics
Acknowledgements
We thank many members of our research group for critical reading of the manuscript, especially J. Braun, I. Shcherbakova and V. Serebrov. This work was supported by funding from HHMI and NIH RO1-GM53007 (MJM). MJM is an HHMI Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published with the period of review, have been highlighted as
* of special interest
** of outstanding interest
- 1.Bhattacharya A. Protein structures: Structures of desire. Nature. 2009;459:24–27. doi: 10.1038/459024a. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends in Genetics. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 4.Nagai K. The upcoming review, Current Opinion in Structural Biology. in press. [Google Scholar]
- 5.Jurica MS, Moore MJ. Pre-mRNA Splicing: Awash in a Sea of Proteins. Molecular Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 6.Will CL, Luhrmann R. Spliceosome Structure and Function. Cold Spring Harbor Perspectives in Biology. 2011;3:a003707–a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turunen JJ, Niemelä EH, Verma B, Frilander MJ. The significant other: splicing by the minor spliceosome. WIREs RNA. 2012;4:61–76. doi: 10.1002/wrna.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Younis I, Dittmar K, Wang W, Foley SW, Berg MG, Hu KY, Wei Z, Wan L, Dreyfuss G, Nilsen T. Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. eLife. 2013;2 doi: 10.7554/eLife.00780. [The authors discovered that the low abundance minor spliceosome's catalytic snRNA, U6atac, is strikingly unstable (t1/2 <2 hr). U6atac levels rapidly increase in response to the cell stress-activated kinase p38MAPK; this increase enhances mRNA expression of hundreds of minor intron-containing genes that are otherwise suppressed by limiting U6atac.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9*.Anokhina M, Bessonov S, Miao Z, Westhof E, Hartmuth K, LUhrmann R. RNA structure analysis of human spliceosomes reveals a compact 3D arrangement of snRNAs at the catalytic core. The EMBO Journal. 2013 doi: 10.1038/emboj.2013.198. doi:10.1038/emboj.2013.198. [Using a variety of chemical modification reagents, the authors examined the RNA structure of affinity-purified human spliceosomes before and after catalytic step 1. They found a stable 3-way junction of the U2/U6 snRNA duplex in active spliceosomes that persists minimally through step 1. Using their experimentally derived structural constraints and the crystal structure of a group II intron, they also generated a model of the RNA network in the step 1 spliceosome.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worth CL, Gong S, Blundell TL. Structural and functional constraints in the evolution of protein families. Nat Rev Mol Cell Biol. 2009;10:709–720. doi: 10.1038/nrm2762. [DOI] [PubMed] [Google Scholar]
- 11.Grainger RJ. Prp8 protein: At the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nancollis V, Ruckshanthi JPD, Frazer LN, O'Keefe RT. The U5 snRNA internal loop 1 is a platform for Brr2, Snu114 and Prp8 protein binding during U5 snRNP assembly. J. Cell. Biochem. 2013;114:2770–2784. doi: 10.1002/jcb.24625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider C, Will CL, Makarova OV, Makarov EM, Luhrmann R. Human U4/U6.U5 and U4atac/U6atac.U5 Tri-snRNPs Exhibit Similar Protein Compositions. Molecular and Cellular Biology. 2002;22:3219–3229. doi: 10.1128/MCB.22.10.3219-3229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du H, Tardiff D, Moore MJ, Rosbash M. Effects of the U1C L13 mutation and temperature regulation of yeast commitment complex formation. PNAS. 2004;101:14841–14846. doi: 10.1073/pnas.0406319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho S, Hoang A, Sinha R, Zhong X-Y, Fu X-D, Krainer AR, Ghosh G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. PNAS. 2011;108:8233–8238. doi: 10.1073/pnas.1017700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP Inhibits Pre-mRNA Polyadenylation through a Direct Interaction between U1 70K and Poly(A) Polymerase. Molecular Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 17.Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. U1 snRNP Determines mRNA Length and Regulates Isoform Expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [Using high-throughput sequencing, the authors found that low U1 levels lead to premature polyadenylation of nascent transcripts, whereas high U1 levels lead to longer 3′-UTRs by a process dubbed “telescripting”. Changes in available U1 snRNP levels can explain much of the 3′ UTR shortening and proximal 3' exon switching characteristic of activated immune and neuronal cells, stem cells, and cancer.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic Disorder and Functional Proteomics. Biophysical Journal. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Korneta I, Bujnicki JM. Intrinsic Disorder in the Human Spliceosomal Proteome. PLoS Comput Biol. 2012;8:e1002641. doi: 10.1371/journal.pcbi.1002641. [Building on previous proteomic, structural and functional data, the authors carried out systematic bioinformatics analysis of intrinsic disorder in the human spliceosome proteome. They predicted intrinsic disorder in almost half of the combined sequences of abundant spliceosomal proteins. Conserved disordered regions in are evolutionarily younger, less widespread proteins than ordered domains in essential spliceosomal core proteins, suggesting that disordered regions were added to a preexistent ordered functional core.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Ribeiro M, Espinosa J, Islam S, Martinez O, Thanki JJ, Mazariegos S, Nguyen T, Larina M, Xue B, Uversky VN. Malleable ribonucleoprotein machine: protein intrinsic disorder in the Saccharomyces cerevisiae spliceosome. PeerJ. 2013;1:1–58. doi: 10.7717/peerj.2. DOI 10.7717/peerj.2. [The authors studied the prevalence of intrinsically disordered proteins in the yeast spliceosome using a wide array of bioinformatics methods. Their study revealed that similar to the proteins associated with human spliceosomes [20], proteins found in the yeast spliceosome are highly enriched in intrinsic disorder.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gsponer J, Babu MM. Cellular Strategies for Regulating Functional and Nonfunctional Protein Aggregation. Cell Reports. 2012;2:1425–1437. doi: 10.1016/j.celrep.2012.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polymenidou M, Lagier-Tourenne C, Hutt KR, Bennett CF, Cleveland DW, Yeo GW. Misregulated RNA processing in amyotrophic lateral sclerosis. Brain Res. 2012;1462:3–15. doi: 10.1016/j.brainres.2012.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore MJ, Sharp PA. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nat Meth. 1993;365:364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- 25.Tseng CK, Cheng SC. Both Catalytic Steps of Nuclear Pre-mRNA Splicing Are Reversible. Science. 2008;320:1782–1784. doi: 10.1126/science.1158993. [DOI] [PubMed] [Google Scholar]
- 26.Hahn D, Kudla G, Tollervey D, Beggs JD. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes & Development. 2012;26:2408–2421. doi: 10.1101/gad.199307.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konarska MM, Vilardell J, Query CC. Repositioning of the Reaction Intermediate within the Catalytic Center of the Spliceosome. Molecular Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 28*.Tseng CK, Cheng SC. The spliceosome catalyzes debranching in competition with reverse of the first chemical reaction. RNA. 2013;19:971–981. doi: 10.1261/rna.038638.113. [By arresting splicing after the first catalytic step, the authors showed that purified spliceosomes can catalyze debranching of lariat-intron-exon 2. This debranching reaction is in competition with the reverse step 1 reaction and is influenced by the ionic environment and the structure of components binding near the catalytic center. This suggests that the catalytic center can switch between different conformations to direct different chemical reactions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn D, Beggs JD. Brr2p RNA helicase with a split personality: insights into structure and function. Biochem. Soc. Trans. 2010;38:1105–1109. doi: 10.1042/BST0381105. [DOI] [PubMed] [Google Scholar]
- 30.Liu L, Query CC, Konarska MM. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nature Structural & Molecular Biology. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 31.Abelson J, Blanco M, Ditzler MA, Fuller F, Aravamudhan P, Wood M, Villa T, Ryan DE, Pleiss JA, Maeder C, et al. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nature Structural & Molecular Biology. 2010;17:504–512. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo Z, Karunatilaka KS, Rueda D. Single-molecule analysis of protein-free U2-U6 snRNAs. Nature Structural & Molecular Biology. 2009;16:1154–1159. doi: 10.1038/nsmb.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mefford MA, Staley JP. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA. 2009;15:1386–1397. doi: 10.1261/rna.1582609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoskins AA, Gelles J, Moore MJ. New insights into the spliceosome by single molecule fluorescence microscopy. Current Opinion in Chemical Biology. 2011;15:864–870. doi: 10.1016/j.cbpa.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Ordered and Dynamic Assembly of Single Spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [The authors combined yeast genetic engineering, chemical biology, and multiwavelength fluorescence microscopy to follow assembly of single spliceosomes in real time in whole-cell extracts. They found that association of every subcomplex is reversible. This experimental strategy should prove widely useful for mechanistic analysis of many other macromolecular machines in environments approaching the complexity of living cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36*.Shcherbakova I, Hoskins AA, Friedman LJ, Serebrov V, Corrêa IR, Jr., Xu M-Q, Gelles J, Moore MJ. Alternative Spliceosome Assembly Pathways Revealed by Single- Molecule Fluorescence Microscopy. Cell Reports. 2013;5:151–165. doi: 10.1016/j.celrep.2013.08.026. [By using colocalization single-molecule spectroscopy to follow initial spliceosome assembly on eight different S. cerevisiae pre-mRNAs, the authors demonstrate that active yeast spliceosomes can form by both U1-first and U2-first pathways.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodson MJ, Hudson AJ, Cherny D, Eperon IC. The transition in spliceosome assembly from complex E to complex A purges surplus U1 snRNPs from alternative splice sites. Nucleic Acids Research. 2012;40:6850–6862. doi: 10.1093/nar/gks322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.David CJ, Boyne AR, Millhouse SR, Manley JL. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes & Development. 2011;25:972–983. doi: 10.1101/gad.2038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider M, Will CL, Anokhina M, Tazi J, Urlaub H, LUhrmann R. Exon Definition Complexes Contain the Tri-snRNP and Can Be Directly Converted into B-like Precatalytic Splicing Complexes. Molecular Cell. 2010;38:223–235. doi: 10.1016/j.molcel.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 40*.Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. Single-molecule colocalization FRET evidence that spliceosome activation precedes stable approach of 5′ splice site and branch site. PNAS. 2013;110:6783–6788. doi: 10.1073/pnas.1219305110. [The authors investigated the process by which intron ends are brought together using FRET-CoSMoS, a combination of methods that can directly reveal how conformational transitions in macromolecular machines are coupled to specific assembly and disassembly events. They found that the 5′ splice site and branch site only approach one another after the spliceosome is activated for catalysis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, LUhrmann R, Stelzl U. Dynamic Protein-Protein Interaction Wiring of the Human Spliceosome. Molecular Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [Using a comprehensive Y2H interaction matrix screen, the authors generated a protein interaction map comprising 632 interactions between 196 human spliceosomal proteins. Dynamic changes in protein interactions were revealed by integrating spliceosomal complex purification information with the new interaction data.] [DOI] [PubMed] [Google Scholar]
- 42.Ren L, McLean JR, Hazbun TR, Fields S, Vander Kooi C, Ohi MD, Gould KL. Systematic Two-Hybrid and Comparative Proteomic Analyses Reveal Novel Yeast Pre-mRNA Splicing Factors Connected to Prp19. PLoS ONE. 2011;6:e16719. doi: 10.1371/journal.pone.0016719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu MC. The Role of Protein Arginine Methylation in mRNP Dynamics. Molecular Biology International. 2011;2011:1–10. doi: 10.4061/2011/163827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishi H, Hashimoto K, Panchenko AR. Phosphorylation in Protein-Protein Binding: Effect on Stability and Function. Structure. 201119:1807–1815. doi: 10.1016/j.str.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishi H, Fong JH, Chang C, Teichmann SA, Panchenko AR. Regulation of protein- protein binding by coupling between phosphorylation and intrinsic disorder: analysis of human protein complexes. Mol Biosyst. 2013;9:1620–1626. doi: 10.1039/c3mb25514j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellare P, Small EC, Huang X, Wohlschlegel JA, Staley JP, Sontheimer EJ. A role for ubiquitin in the spliceosome assembly pathway. Nature Structural & Molecular Biology. 2008;15:444–451. doi: 10.1038/nsmb.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48*.Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW, Rape M. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes & Development. 2010;24:1434–1447. doi: 10.1101/gad.1925010. [The authors report that the NTC modifies U4 snRNP protein Prp3 with nonproteolytic K63-linked ubiquitin chains. The K63-linked chains increase the affinity of Prp3 for the U5 snRNP component Prp8, thereby allowing for the stabilization of the U4/U6.U5 snRNP. Prp3 is deubiquitinated by Usp4 and its substrate targeting factor, the U4/U6 recycling protein Sart3, which likely facilitates ejection of U4 proteins from the spliceosome during activation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin-Tumasz S, Reiter NJ, Brow DA, Butcher SE. Structure and functional implications of a complex containing a segment of U6 RNA bound by a domain of Prp24. RNA. 2010;16:792–804. doi: 10.1261/rna.1913310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin-Tumasz S, Richie AC, Clos LJ, Brow DA, Butcher SE. A novel occluded RNA recognition motif in Prp24 unwinds the U6 RNA internal stem loop. Nucleic Acids Research. 2011;39:7837–7847. doi: 10.1093/nar/gkr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krummel DAP, Oubridge C, Leung AKW, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leung AKW, Nagai K, Li J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature. 2011 doi: 10.1038/nature09956. doi:10.1038/nature09956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. The EMBO Journal. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf E, Kastner B, Deckert J, Merz C, Stark H, hrmann RLU. Exon, intron and splice site locations in the spliceosomal B complex. The EMBO Journal. 2009;28:2283–2292. doi: 10.1038/emboj.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bessonov S, Anokhina M, Krasauskas A, Golas MM, Sander B, Will CL, Urlaub H, Stark H, Luhrmann R. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA. 2010;16:2384–2403. doi: 10.1261/rna.2456210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grote M, Wolf E, Will CL, Lemm I, Agafonov DE, Schomburg A, Fischle W, Urlaub H, Luhrmann R. Molecular Architecture of the Human Prp19/CDC5L Complex. Molecular and Cellular Biology. 2010;30:2105–2119. doi: 10.1128/MCB.01505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jurica MS, Sousa D, Moore MJ, Grigorieff N. Three-dimensional structure of C complex spliceosomes by electron microscopy. Nature Structural & Molecular Biology. 2004;11:265–269. doi: 10.1038/nsmb728. [DOI] [PubMed] [Google Scholar]
- 58.Ohi MD, Ren L, Wall JS, Gould KL, Walz T. Structural characterization of the fission yeast U5.U2/U6 spliceosome complex. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3195–3200. doi: 10.1073/pnas.0611591104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Golas MM, Sander B, Will CL, LUhrmann R, Stark H. Major Conformational Change in the Complex SF3b upon Integration into the Spliceosomal U11/U12 di-snRNP as Revealed by Electron Cryomicroscopy. Molecular Cell. 2005;17:869–883. doi: 10.1016/j.molcel.2005.02.016. [DOI] [PubMed] [Google Scholar]