Abstract
A retrospective analysis of real-time PCR (RT-PCR) results for 151 biopsy samples obtained from 132 patients with proven invasive fungal diseases was performed. PCR-based techniques proved to be fast and sensitive and enabled definitive diagnosis in all cases studied, with detection of a total of 28 fungal species.
TEXT
Most of the methods used in microbiology laboratories for the diagnosis of invasive fungal diseases (IFDs) present limitations (1, 2). Cultures are too slow and ineffective for early diagnosis, although the gold standard methods to prove IFD continue to be isolation in culture from a sterile sample or demonstration of invasion by fungal structures in tissues (3, 4).
Classic histopathological studies have significant drawbacks. Microscopic examinations are quite variable, depending on experience, but the most important limitation of tissue observation is the impossibility of differentiating species, which is essential for determining therapy, given the differences in sensitivity to antifungal agents among fungal species (5).
Molecular methods, such as those based on PCRs, have been used recently for fungal DNA detection in both fresh and paraffin-embedded tissue samples, using different targets (4). The great advantages of these molecular techniques are the determination of the specific agent and greater sensitivity (6). A limitation is the lack of standardization of techniques, which are mostly homemade.
In recent years, the Spanish Mycology Reference Laboratory has developed a number of real-time PCR (RT-PCR) assays in order to improve the diagnosis of IFD (7–11). These techniques serve to confirm the diagnosis of IFD when culture results are negative and to determine the species involved in infections. In this work, a retrospective analysis of RT-PCR results for biopsy samples was performed. The survey included samples analyzed between 2006 and 2013. A total of 151 biopsy specimens from 132 patients with diagnoses of IFD, proven by histopathological examinations, and negative culture results were analyzed. The samples were sent to the Spanish Mycology Reference Laboratory by hospitals throughout Spain and had very heterogeneous origins, depending on the patients' symptoms, with lung, skin, liver, and brain samples being most common.
Fourteen patients had more than one sample for diagnosis; the samples had the same origins (duplicates) for seven patients, and the origins of the samples were different for the rest of them. The biopsy specimens were fresh (n = 92) or embedded in paraffin (n = 59). For samples embedded in paraffin, the blocks were cut into 10-μm sections. Three to 10 cuts were used to extract the DNA, in order to obtain approximately 25 mg of tissue. Biopsy specimens were deparaffinized by lavage with 1.5 ml of xylene (100%) followed by two washes with 1.2 ml of ethanol (96 to 100%). The tissue was incubated at 37°C for about 10 min to evaporate the remaining ethanol.
DNA from fresh and paraffin-embedded tissues was extracted using a QIAmp Tissue DNA minikit (Qiagen, Izasa, Madrid, Spain), following the manufacturer's instructions. Fifty microliters of buffer was used for elution. Two microliters of DNA extracted from each sample was used for each PCR.
The PCR-based assays used in this study were as follows. When there was clear clinical, epidemiological, and histopathological suspicion of a specific fungal disease, such as histoplasmosis, paracoccidioidomycosis, aspergillosis, mucormycosis, scedosporiosis, or fusariosis, a specific PCR test was performed. When there were no conclusive data about the fungal pathogen implicated in the disease, a panfungal assay was performed. Finally, when the initial specific PCR assay results were negative, a panfungal assay was performed (Fig. 1). For the most part, the PCR assays used have been described previously (5, 7–11). In addition, a new multiplex PCR assay able to detect the three main species of Aspergillus was developed. This assay was designed to detect the three most common species of Aspergillus involved in IFD, i.e., Aspergillus fumigatus, Aspergillus flavus, and Aspergillus terreus. Specific molecular beacon probes labeled with different fluorescent dyes (FAM, HEX, and Cyan 500, respectively) were used. The probes were directed to the ITS1 region of ribosomal DNA. All assays were done with a LightCycler 480 RT-PCR unit (Roche, Madrid, Spain) or a CFX96 system (Bio-Rad, Madrid, Spain).
FIG 1.
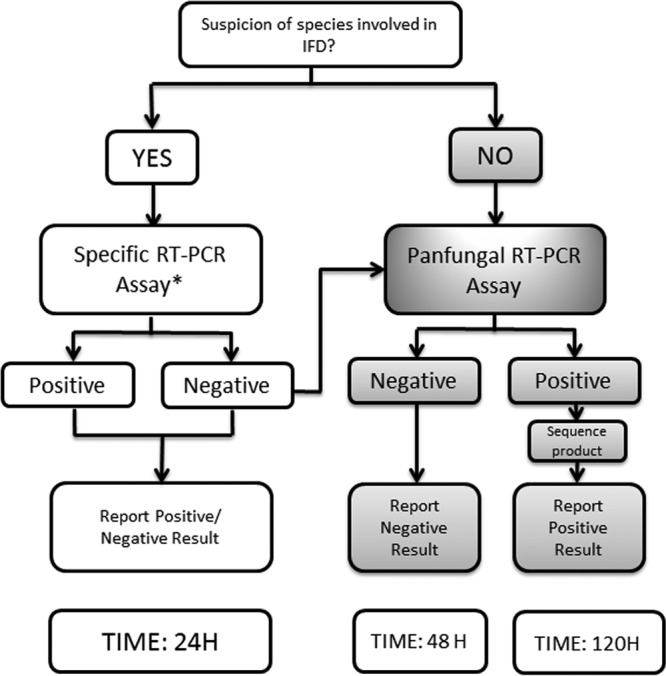
Schematic diagram of procedures performed with biopsy samples, depending on clinical suspicion. When specific assay results were negative, a panfungal assay was performed. *, specific assays detected the following species: A. fumigatus, A. terreus, A. flavus, Histoplasma capsulatum, Paracoccidioides brasiliensis, Rhizopus oryzae, Rhizopus microsporus, Mucor spp., Fusarium spp., Scedosporium prolificans, and Scedosporium apiospermum.
Table 1 shows the main underlying diseases, risk factors, and species detected by using these PCR techniques. In patients with oncohematological conditions (the most common underlying diseases), the main species detected corresponded to the genus Aspergillus (n = 46). Most patients with AIDS (the second most common underlying disease) presented histoplasmosis (n = 13).
TABLE 1.
Underlying diseases and species detected by using PCR techniques
| Clinical condition and subjects | No. (%) of cases | Species detected |
|---|---|---|
| Oncohematological | 71 (54) | |
| 46a | Aspergillus spp. | |
| 5 | Candida spp. | |
| 9b | Mucormycetes | |
| 11 | Other, i.e., other hyaline molds (n = 8) and black molds (n = 3)c | |
| HIV | 19 (14) | |
| 13 | Histoplasma capsulatum | |
| 2 | Aspergillus fumigatus | |
| 1 | Candida albicans | |
| 1 | Lichtheimia corymbifera | |
| 1 | Phoma spp. | |
| 1 | Rhizomucor pusillus | |
| Solid-organ transplant | 6 (4.5) | |
| 1 | Aspergillus fumigatus | |
| 2 | Histoplasma capsulatum | |
| 1 | Scedosporium apiospermum | |
| 1 | Neosartorya pseudofischeri | |
| 1 | Phomopsis longicolla | |
| Respiratory | 3 (2.2) | |
| 3 | Aspergillus fumigatus | |
| Immunocompetent | 25 (19) | |
| Immigrants | 9 | |
| 5 | Paracoccidioides brasiliensis | |
| 1 | Coccidioides immitis | |
| 3 | Histoplasma capsulatum | |
| Travelers | 5 | |
| 1 | Paracoccidioides brasiliensis | |
| 3 | Histoplasma capsulatum | |
| 1 | Candida albicans | |
| Other immunocompetent subjects | 11 | |
| 2 | Candida albicans | |
| 1 | Fusarium oxysporum | |
| 1 | Fonsecaea pedrosoi | |
| 6 | Aspergillus spp. | |
| 1 | Rhizopus oryzae | |
| Other conditions | 8 (6) | |
| Diabetes mellitus | 1 | Rhizopus oryzae |
| Urological disease | 1 | Rhizopus oryzae |
| Neonate | 1 | Candida parapsilosis |
| Vascular surgery | 1 | Candida tropicalis/A. fumigatus |
| Severely burned | 1 | Scedosporium apiospermum |
| Vascular surgery | 1 | Arthrographis kalrae |
| Vascular surgery | 1 | Aspergillus fumigatus |
| Neonate | 1 | Bipolaris spicifera |
Two cases presented mixed infections with two different Aspergillus spp.
One case presented a mixed infection with Aspergillus fumigatus.
Species detected included Fusarium proliferatum, Histoplasma capsulatum, Scedosporium apiospermum, Metarhizium anisopliae, Phialemonium curvatum, Scopulariopsis spp., Fusarium spp., Phoma spp., Alternaria infectoria (n = 2), and Exophiala spp.
The PCR techniques used enabled the detection of 28 different fungal species (Table 2); 47% of the species detected belonged to the Aspergillus genus, with A. fumigatus being the most frequently observed species. Species causing endemic mycoses represented 21%, followed by mucormycetes (10%) and Candida spp. (8%). Finally, a variety of emerging or rare species represented the remaining 14% of the total, of which hyaline molds represented 7% and black fungi 6% (Table 2). The most frequently observed species in this group belonged to the genera Scedosporium and Fusarium. Also present were species that were rarely described previously as causing infectious diseases, such as Phomopsis longicolla, Metarhizium anisopliae (12), Arthrographis kalrae (13), and Bipolaris spicifera. Four cases of mixed infections were detected; two of those cases involved pediatric oncological patients from the same hospital (14), and the other cases involved an oncohematological patient and a subject addicted to alcohol. The mixed infections always involved Aspergillus spp. mixed with Candida spp. or species of Mucorales.
TABLE 2.
Species distribution in 132 cases of IFD and PCR techniques used to perform diagnoses
| Species detected | Total no. (%) of cases | No. (%) diagnosed by: |
|
|---|---|---|---|
| Specific PCR | Panfungal PCR | ||
| Totala | 136 | 95 (70) | 41 (30) |
| Aspergillus spp. | 64 (47) | 58 (91) | 6 (9) |
| Aspergillus fumigatus | 55 (86) | 54 (98) | 1 (2) |
| Aspergillus flavus | 6 (9.2) | 3 (50) | 3 (50) |
| Aspergillus terreus | 1 (1.5) | 1 (100) | 0 |
| Aspergillus penicillioides | 1 (1.5) | 0 | 1 (100) |
| Neosartorya pseudofischeri | 1 (1.5) | 0 | 1 (100) |
| Endemic mycoses | 29 (21) | 28 (96.5) | 1 (3.5) |
| Histoplasma capsulatum | 22 (76) | 22 (100) | 0 |
| Paracoccidioides brasiliensis | 6 (21) | 6 (100) | 0 |
| Coccidioides immitis | 1 (3.4) | 0 | 1 (100) |
| Mucormycetes | 14 (10) | 2 (14) | 12 (86) |
| Rhizopus oryzae | 4 (28.5) | 2 (50) | 2 (50) |
| Lichtheimia corymbifera | 3 (21) | 0 | 3 (100) |
| Rhizomucor pusillus | 3 (21) | 0 | 3 (100) |
| Mucor spp. | 2 (14) | 0 | 2 (100) |
| Cunninghamella bertholletiae | 2 (14) | 0 | 2 (100) |
| Candida spp. | 11 (8) | 6 (55) | 5 (45) |
| Candida albicans | 8 (73) | 5 (62.5) | 3 (37.5) |
| Candida parapsilosis | 1 (9) | 0 | 1 (100) |
| Candida tropicalis | 2 (18) | 1 (50) | 1 (50) |
| Other | 18 (14) | 1 (5) | 17 (95) |
| Hyaline molds | 11 (7) | 1 (9) | 10 (91) |
| Scedosporium apiospermum | 3 | 1 (33) | 2 (67) |
| Fusarium spp. | 3 | 0 | 3 (100) |
| Phialemonium curvatum | 1 | 0 | 1 (100) |
| Metarhizium anisopliae (12) | 1 | 0 | 1 (100) |
| Scopulariopsis spp. | 1 | 0 | 1 (100) |
| Phomopsis longicolla (15) | 1 | 0 | 1 (100) |
| Arthrographis kalrae (13) | 1 | 1 (100) | |
| Black molds | 7 (6) | 0 | 7 (100) |
| Phoma spp. | 2 (10.5) | 0 | 2 (100) |
| Fonsecaea pedrosoi | 1 | 0 | 1 (100) |
| Bipolaris spicifera | 1 | 0 | 1 (100) |
| Exophiala spp. | 1 | 0 | 1 (100) |
| Alternaria infectoria | 2 (10.5) | 0 | 2 (100) |
The number of species detected is greater than the number of cases because of mixed infections.
Although the species distribution found is biased because it represents cases submitted to a reference center for study and confirmation, these findings prove the existence of emerging and rare species that could be relevant in some clinical settings. Correct identification of the fungal species seems to be essential to adequately treat the diseases, and molecular methods of detection have a potentially high performance level that should be implemented in clinical laboratories.
The techniques used depended on the clinical suspicions. In 30% of the cases (41 cases), there was no evidence concerning the species of fungus implicated in the infection; this occurred when histopathological images were not clear and the epidemiological and clinical data for the patients did not allow conclusions regarding the species involved (e.g., for severely burned patients, solid-organ transplant recipients, or neonates). In those cases, a panfungal assay was used and the sequence of the amplicon was analyzed. Specific assays were used in 70% of the cases (96 cases), especially when aspergillosis or endemic mycoses were suspected. In some cases, specific assay results were negative and it was necessary to use the panfungal assay. This happened especially in cases of mucormycosis when other fungi were suspected.
Regarding the results for patients with more than one sample, the results for a sample were negative in just one case; the sample was a cerebral sample from a patient with histoplasmosis. The results for the rest of the duplicate samples were all positive.
By using these methods, diagnostic times were greatly reduced. The samples (fresh or embedded in paraffin) were processed and results were reported within 24 h when specific assays were performed. When panfungal assays were performed, it was necessary to verify the quality of the amplification product and then sequence the product, which increased the response time to 4 or 5 days. Regarding the cost of these techniques and subtracting the investment in the equipment, the average price for these assays was approximately €100 per PCR determination. These techniques are somewhat more expensive than conventional tests but provide significant savings in the time to diagnosis. Moreover, it should be noted that all culture results were negative in this study. These PCR-based procedures proved to be much more sensitive than cultures and allowed for identification at the species level and detection of mixed infections. The only limitation to implementing this type of technique in microbiology laboratories could be a lack of appropriate real-time PCR equipment as well as a sequencing service.
In conclusion, PCR-based techniques were able to identify the species implicated in infections even when two species were involved. Therefore, the transfer of this technology from reference centers to health centers should be a priority, considering that the cost per sample is not prohibitively high and results could be reported in a few hours, allowing for earlier initiation of adequate treatment. Furthermore, the samples could be embedded in paraffin in histopathology laboratories, with minimal loss of sensitivity (10), and then easily sent to the microbiology laboratory.
ACKNOWLEDGMENTS
This work was supported in part by the research project “Utilidad de las determinaciones seriadas de DNA fúngico mediante PCR en Tiempo Real en el diagnóstico de la aspergilosis invasora en niños con enfermedades onco-hematológicas” of the Fundación Mutua Madrileña and also by research project PI11/00412 of the Spanish Fondo de Investigaciones Sanitarias of the Instituto de Salud Carlos III. L.B.-M. has research contracts from Red Española de Investigación en Patología Infecciosa (REIPI) (project MPY 1022/07_1) and from the Instituto de Salud Carlos III, cofinanced by the European Development Regional Fund (A Way to Achieve Europe program), Spanish Network for Research in Infectious Diseases (REIPI project RD06/0008).
In the past 5 years, M.C.-E. has received grant support from Astellas Pharma, bioMérieux, Gilead Sciences, Merck Sharp and Dohme, Pfizer, Schering Plough, Soria Melguizo SA, Ferrer International, the European Union, the ALBAN program, the Spanish Agency for International Cooperation, the Spanish Ministry of Culture and Education, the Spanish Health Research Fund, the Instituto de Salud Carlos III, the Ramon Areces Foundation, and the Mutua Madrileña Foundation. M.C.-E has been an advisor/consultant to the Panamerican Health Organization, Astellas Pharma, Gilead Sciences, Merck Sharp and Dohme, Pfizer, and Schering Plough. He has been paid for talks on behalf of Gilead Sciences, Merck Sharp and Dohme, Pfizer, Astellas Pharma, and Schering Plough.
Footnotes
Published ahead of print 26 February 2014
REFERENCES
- 1.Cuenca-Estrella M, Bassetti M, Lass-Florl C, Racil Z, Richardson M, Rogers TR. 2011. Detection and investigation of invasive mould disease. J. Antimicrob. Chemother. 66(Suppl 1):i15–i24. 10.1093/jac/dkq438 [DOI] [PubMed] [Google Scholar]
- 2.Cuenca-Estrella M, Bernal-Martinez L, Buitrago MJ, Castelli MV, Gomez-Lopez A, Zaragoza O, Rodriguez-Tudela JL. 2008. Update on the epidemiology and diagnosis of invasive fungal infection. Int. J. Antimicrob. Agents 32(Suppl 2):S143–S147. 10.1016/S0924-8579(08)70016-5 [DOI] [PubMed] [Google Scholar]
- 3.De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Munoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 46:1813–1821. 10.1086/588660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mrazek C, Lass-Florl C. 2011. Biopsy procedures for molecular tissue diagnosis of invasive fungal infections. Curr. Infect. Dis. Rep. 13:504–509. 10.1007/s11908-011-0215-7 [DOI] [PubMed] [Google Scholar]
- 5.Buitrago MJ, Aguado JM, Ballen A, Bernal-Martinez L, Prieto M, Garcia-Reyne A, Garcia-Rodriguez J, Rodriguez-Tudela JL, Cuenca-Estrella M. 2013. Efficacy of DNA amplification in tissue biopsy samples to improve the detection of invasive fungal disease. Clin. Microbiol. Infect. 19:E271–E277. 10.1111/1469-0691.12110 [DOI] [PubMed] [Google Scholar]
- 6.Rickerts V, Mousset S, Lambrecht E, Tintelnot K, Schwerdtfeger R, Presterl E, Jacobi V, Just-Nubling G, Bialek R. 2007. Comparison of histopathological analysis, culture, and polymerase chain reaction assays to detect invasive mold infections from biopsy specimens. Clin. Infect. Dis. 44:1078–1083. 10.1086/512812 [DOI] [PubMed] [Google Scholar]
- 7.Bernal-Martinez L, Buitrago MJ, Castelli MV, Rodriguez-Tudela JL, Cuenca-Estrella M. 2012. Detection of invasive infection caused by Fusarium solani and non-Fusarium solani species using a duplex quantitative PCR-based assay in a murine model of fusariosis. Med. Mycol. 50:270–275. 10.3109/13693786.2011.604047 [DOI] [PubMed] [Google Scholar]
- 8.Bernal-Martinez L, Buitrago MJ, Castelli MV, Rodriguez-Tudela JL, Cuenca-Estrella M. 2013. Development of a single tube multiplex real-time PCR to detect the most clinically relevant Mucormycetes species. Clin. Microbiol. Infect. 19:E1–E7. 10.1111/j.1469-0691.2012.03976.x [DOI] [PubMed] [Google Scholar]
- 9.Buitrago MJ, Berenguer J, Mellado E, Rodriguez-Tudela JL, Cuenca-Estrella M. 2006. Detection of imported histoplasmosis in serum of HIV-infected patients using a real-time PCR-based assay. Eur. J. Clin. Microbiol. Infect. Dis. 25:665–668. 10.1007/s10096-006-0207-y [DOI] [PubMed] [Google Scholar]
- 10.Buitrago MJ, Merino P, Puente S, Gomez-Lopez A, Arribi A, Zancope-Oliveira RM, Gutierrez MC, Rodriguez-Tudela JL, Cuenca-Estrella M. 2009. Utility of real-time PCR for the detection of Paracoccidioides brasiliensis DNA in the diagnosis of imported paracoccidioidomycosis. Med. Mycol. 47:879–882. 10.3109/13693780802713208 [DOI] [PubMed] [Google Scholar]
- 11.Castelli MV, Buitrago MJ, Bernal-Martinez L, Gomez-Lopez A, Rodriguez-Tudela JL, Cuenca-Estrella M. 2008. Development and validation of a quantitative PCR assay for diagnosis of scedosporiosis. J. Clin. Microbiol. 46:3412–3416. 10.1128/JCM.00046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osorio S, de la Camara R, Monteserin MC, Granados R, Ona F, Rodriguez-Tudela JL, Cuenca-Estrella M. 2007. Recurrent disseminated skin lesions due to Metarrhizium anisopliae in an adult patient with acute myelogenous leukemia. J. Clin. Microbiol. 45:651–655. 10.1128/JCM.01502-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Diego Candela J, Forteza A, Garcia D, Prieto G, Bellot R, Villar S, Cortina JM. 2010. Endocarditis caused by Arthrographis kalrae. Ann. Thorac. Surg. 90:e4–e5. 10.1016/j.athoracsur.2010.04.021 [DOI] [PubMed] [Google Scholar]
- 14.Rubio PM, Sevilla J, Gonzalez-Vicent M, Lassaletta A, Cuenca-Estrella M, Diaz MA, Riesco S, Madero L. 2009. Increasing incidence of invasive aspergillosis in pediatric hematology oncology patients over the last decade: a retrospective single centre study. J. Pediatr. Hematol. Oncol. 31:642–646. 10.1097/MPH.0b013e3181acd956 [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Reyne A, López-Medrano F, Morales JM, García Esteban C, Martín I, Eraña I, Meije Y, Lalueza A, Alastruey-Izquierdo A, Rodríguez-Tudela JL, Aguado JM. 2011. Cutaneous infection by Phomopsis longicolla in a renal transplant recipient from Guinea: first report of human infection by this fungus. Transpl. Infect. Dis. 13:204–207. 10.1111/j.1399-3062.2010.00570.x [DOI] [PubMed] [Google Scholar]


