Abstract
HCV infection induces autophagy, but how this occurs is unclear. Here, we report the induction of autophagy by the structural HCV core protein and subsequent endoplasmic reticular (ER) stress in Huh7 hepatoma cells. During ER stress, both the EIF2AK3 and ATF6 pathways of the unfolded protein response (UPR) were activated by HCV core protein. Then, these pathways upregulated transcription factors ATF4 and DDIT3. The ERN1-XBP1 pathway was not activated. Through ATF4 in the EIF2AK3 pathway, the autophagy gene ATG12 was upregulated. DDIT3 upregulated the transcription of autophagy gene MAP1LC3B (LC3B) by directly binding to the –253 to –99 base region of the LC3B promoter, contributing to the development of autophagy. Collectively, these data suggest not only a novel role for the HCV core protein in autophagy but also offer new insight into detailed molecular mechanisms with respect to HCV-induced autophagy, specifically how downstream UPR molecules regulate key autophagic gene expression.
Keywords: hepatitis C virus, core, autophagy, ER stress, UPR
Introduction
Hepatitis C virus (HCV), a member of the flaviviridae family of RNA viruses, is a positive-strand that induces hepatotoxicity.1 Four structural (core, E1, E2, and p7) and six nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins are encoded by the 9.6 kb HCV genome.2,3 The major viral particle components are core, E1, and E2. Nonstructural proteins chiefly serve to replicate HCV and regulate pathways of cell signaling that are important in apoptosis, autophagy, and the host immune response.4,5
Autophagy is a cellular process initiated in response to stress (e.g., nutrient deficiency, infections, and ER stress) via formation of a double-membraned autophagosome and subsequent lysosomal fusion leading to degradation.6,7 Some viruses (poliovirus, rhinovirus, and hepatitis virus) can induce autophagy to their benefit by using the autophagosomal membrane as a site for viral replication.8-10
At this time, 31 autophagy-related (ATG) genes have been identified. The products of these genes are reported to regulate the autophagic process.11 Among these is MAP1LC3B/LC3B (microtubule-associated protein 1 light chain 3 β). A soluble LC3B, LC3B-I, is formed by ATG4B-mediated cleavage. LC3B-I is then conjugated to phosphatidylethanolamine to create the lipid-containing LC3B, LC3B-II.12,13 Often, the ratio LC3B-II to LC3B-I can be used as an autophagy marker.
Recently, several groups, including ours, have reported that HCV could induce autophagy in hepatoma cells,14-19 although the mechanism by which this occurs is not fully clear, however some evidence exists that it involves interaction of HCV nonstructural proteins host cell autophagic pathways. For example, HCV NS4B has been reported to induce autophagy by interacting with RAB5 and PIK3C3/Vps34,20 and NS5B is reported to interact with autophagy protein ATG5.9 HCV NS5A is also thought to induce autophagy,21 but whether HCV structural proteins can induce autophagy is unknown.
HCV core protein has numerous functions critical to the viral capsid shell. The core protein modulates cell proliferation, apoptosis, oxidative stress, and host-cell immune regulation.7,11,22,23 The HCV core protein chiefly located on the cytosolic side of the ER membrane where it induces increased expression of HSPA5, HSP90B1, CALR, and ER calcium ATPase. This expression leads to ER stress and multiple pathologies.24 The HCV core protein can localize with mitochondria, droplets of lipids, and the intermediate Golgi compartment.24-27 Our previous study and other reports indicate that HCV can induce ER stress and activate UPR which can regulate autophagy.15,17-19 Therefore, we studied the role of HCV core protein in the induction of autophagy by way of ER stress and the UPR pathways.
Activation of UPR can occur via 3 unique sensors: EIF2AK3 (eukaryotic translation initiation factor 2-α kinase 3), ATF6 (activating transcription factor 6), and ERN1 (endoplasmic reticulum to nucleus signaling 1).14 EIF2AK3, as a serine/threonine protein kinase, phosphorylates EIF2S1 (eukaryotic translation initiation factor 2, subunit 1 α, 35 kDa). EIF2S1 phosphorylation increases ATF4 translation and DDIT3 (DNA-damage-inducible transcript 3) induction. ATF6 is a precursor that is anchored to the ER membrane, where it is retained by HSPA5 chaperone protein.14 After ER stress, ATF6 is released from HSPA5 and transported to the Golgi complex. Here, the N terminus of ATF6 releases from the ER membrane.14 ERN1 is comprised of both serine/threonine kinase and ribonuclease domains. In response to the ER stress, activated ERN1 cleaves 26 nucleotides from the XBP1 (X-box binding protein 1) mRNA to create a spliced XBP1 mRNA. This mRNA encodes the active spliced protein XBP1(s).14,28 Although we (and others) reported that HCV could induce autophagy through all 3 UPR pathways, detailed mechanisms by which ER stress regulates autophagy has not been fully characterized. This modulation may occur by way of key factors in the ER stress pathway that regulate ATG. Rzymski and coworkers report that ATF4 regulates autophagy in response to severe hypoxia by inducing LC3B gene transcription.29 Moreover, DDIT3 is reported to directly bind to the ATG5 promoter and regulate autophagy.30 Others have identified that XBP1 mRNA splicing triggers autophagy in endothelial cells through transcriptional activation of another autophagy protein, BECN1.31
Here, we report that HCV core protein induced autophagy through ER stress, specifically through activation of EIF2AK3 and ATF6 (but not ERN1 or XBP1) pathways. Moreover, we identified a mechanism by which HCV core protein may promote induction of autophagy by upregulating ATG12 through the critical ER stress factor ATF4 and enhancing LC3B expression by DDIT3 directly binding to the LC3B promoter region.
Results
HCV core protein induces autophagy
To investigate whether individual HCV proteins induce autophagy, we transfected flag-tagged HCV core, NS2, NS3, NS3/4A, NS4B, and NS5B expression plasmids into Huh7 cells and used western blot to measure the conversion of LC3B-I to LC3B-II and SQSTM1 degradation, which is a method for evaluating selective autophagy of ubiquitinated aggregate.32 The percentages of transfected cells for HCV core, NS2, NS3, NS3/4A, NS4B, and NS5B were 33%, 38%, 39%, 33%, 33%, and 22% respectively (Fig. S1). Figure 1A shows that the HCV core, NS3/4A, and NS4B proteins induced autophagy. In contrast, NS2, NS3, and NS5B proteins did not induce autophagy. To confirm our observations, we investigated HCV protein-induced autophagy in single cells using an immunofluorescence assay. During autophagy, lipid-conjugated LC3B-II accumulates in autophagosome membranes whereas cytosolic LC3B-I does not.33 Thus, we studied endogenous LC3B puncta formation with confocal microscopy at 48 h after HCV protein transfection. As shown in Figure 1B, LC3B was distributed throughout the cytoplasm in untreated cells and mock- transfected cells, whereas LC3B was distributed in specific puncta in HCV core-, NS3/4A-, and NS4B-transfected cells (Fig. 1B). Quantitative analysis revealed that the number of punctate LC3B structures was significantly higher in cells transfected with HCV core, NS3/4A and NS4B (Fig. 1C), which is consistent with the western blot results.
Figure 1. Multiple HCV proteins induce autophagy in Huh7 cells. (A) Huh7 cells were transfected with empty vectors or various plasmids expressing Flag-tagged HCV core, NS2, NS3, NS3/4A, NS4B, and NS5B proteins. At 48 h post-transfection, cells were harvested and western blotting was performed. Blots are representative of the 3 independent experiments. ACTB was used as sample-loading control. Densitometric LC3B-II/ACTB and SQSTM1/ACTB ratios from at least 3 independent experiments are shown. The value of Huh7 without any treatment was set at 1 for each experiment (**P < 0.01). (B) At 48 h post-transfection, the cells were fixed and analyzed by indirect immunofluorescence using anti-LC3B and anti-Flag antibodies. Patterns of LC3B expression in mock- and HCV protein-transfected cells were visualized with laser confocal microscopy. LC3B (green), HCV proteins (red) staining is shown. Scale bars: 10 μm. (C) Quantitative presentation of punctate LC3B per cell in untreated, mock- and HCV protein-transfected cells.
To confirm autophagy induction by HCV core protein, we measured induction over time after transfection of HCV core protein and over a dose range. As shown in Figure 2A conversion of LC3B-I to LC3B-II and SQSTM1 degradation were observed in HCV core-transfected cells but not in untreated control cells at the corresponding time points. SQSTM1 degradation was more modest at later time points. As HCV core protein expression increased, the conversion of LC3B-I to LC3B-II increased and the degradation of SQSTM1 increased, too (Fig. 2B). 293T cells also exhibited similar behavior (data not shown), indicating that autophagy induction by the HCV core protein did not depend on the cell line tested. To investigate whether HCV core-induced autophagy did not arise from transient HCV core protein overexpression, we measured HCV core protein-induced autophagy in an inducible expression system and in a cell line (Huh7) that stably expressed the HCV core protein (data not shown).
Figure 2. HCV core protein-induced autophagy is further confirmed in Huh7 cells. (A) Huh7 cells were transfected with HCV core protein expression plasmids (Core) or empty vectors (Mock). At different time points (12, 24, 26, and 48 h) post-transfection, cells were harvested and western blotting was performed. (B) Huh7 cells were transfected with various amounts (0.5, 1, 2, and 4 μg) of HCV core protein expression plasmids or empty vectors. At 48 h post-transfection, cells were harvested and western blotting was performed. (A and B) Blots are representative of the 3 independent experiments. ACTB was used as sample-loading control. Densitometric LC3B-II/ACTB and SQSTM1/ACTB ratios from at least 3 independent experiments are shown. The value of Huh7 without any treatment was set at 1 for each experiment (*P < 0.05, **P < 0.01). (C) Huh7 cells were transfected with HCV core protein expression plasmids or empty vectors for 48 h and examined under immunotransmission electron microscopy. Black arrow: autophagosome or autolysosome; White arrow: immunolabeled core protein.
Immunoelectron microscopy confirmed the identity of autophagosome-like vesicles, which were induced by HCV core protein. As shown in Figure 2C, both autophagosomes and autolysosomes were observed in HCV core-transfected cells. The detection of autolysosomes implied that the autophagy induced by the HCV core is a complete process. Data from electron microscopy analysis confirmed data from western blotting which indicated that HCV core protein induced autophagy in Huh7 cells.
HCV core protein-induced autophagy in Huh7 cells is a complete process
Whether HCV-induced autophagy is a complete process is unclear. Ke and coworkers report that HCV could induce autophagosome formation and fusion to lysosomes, forming autolysosomes.18 This series of events would represent a complete autophagic process. However, Sir and colleagues report that HCV RNA transfection into human Huh7 hepatoma cells induce incomplete autophagy: HCV only induced autophagosome accumulation by activating UPR. It did not enhance autophagic protein degradation.15 Our immunoelectron microscopy results suggest that HCV core protein-induced autophagy may be a complete process (Fig. 2C). To elucidate this, autophagic flux, which is a dynamic process that synthesizes autophagosomes and delivers autophagic substrate to lysosomes, was measured by western blot and confocal immunofluorescent analysis. HCV core-transfected, mock-transfected, and untreated cells were treated with or without bafilomycin A1 (BafA1), which is a specific inhibitor of vacuolar type H+-ATPase (V-ATPase). V-ATPase suppresses autophagosome/lysosome fusion.34 With BafA1 treatment, LC3B-II accumulation increased significantly, and SQSTM1 degradation was inhibited significantly in HCV core-transfected cells when compared with mock-transfected cells. Figure 3A depicts the densitometric analysis of LC3B-II, SQSTM1, and ACTB protein bands in 3 separate experiments. These data show that, in the presence of BafA1, LC3B-II/ACTB ratios increased 22.6% in mock-transfected cells and 66.9% in HCV core protein-transfected cells. SQSTM1/ACTB ratios increased 10.7% in mock-transfected cells and 74.7% in HCV core-transfected cells. These data demonstrate that HCV core protein enhances autophagic flux and induces a complete autophagic process.
Figure 3. HCV core protein-induced autophagy is a complete process. (A) Huh7 cells were transfected with HCV core protein expression plasmids or empty vectors for 36 h and then treated with (+) or without (–) BafA1 (10 nM) for 12 h. Cells were harvested and analyzed by western blotting using the indicated antibodies. Blots are representative of 3 independent experiments. ACTB was used as a sample-loading control. Densitometric LC3B-II/ACTB and SQSTM1/ACTB ratios from at least 3 independent experiments are shown. The value of Huh7 without any treatment was set at 1 for each experiment. aPercentage increased by BafA1 treatment (**P < 0.01). (B) Huh7 cells were cotransfected with either mTagRFP-mWasabi-LC3B plasmids and core protein expression plasmids or empty vectors for 36 h and cells were treated with (+) or without (–) BafA1 (10 nM) for additional 12 h. Cells were fixed and assessed by indirect immunofluorescence analysis using an anti-core antibody. Cells were visualized under laser confocal microscopy. Yellow punctate structures indicate autophagosomes; red punctate structures indicate autolysosomes; and blue punctate structures indicate core protein. Scale bars: 10 μm.
To confirm this observation, we used an improved tandem fluorescent-tagged LC3B construct (mTagRFP-mWasabi-LC3B) to identify autophagic flux.35 This assay is based on fluorescent signals of the tandem fluorescent-tagged LC3B. The acidic lysosomal environment efficiently quenches green fluorescence but not red fluorescence, which is inefficiently quenched. Therefore, when autophagosomes fuse with lysosomes, green fluorescent components from the mTagRFP-mWasabi-LC3B reporter is lost, and red fluorescence can be detected. As shown in Figure 3B, in mock-transfected cells with and without BafA1 treatment, the distribution of mTagRFP-mWasabi-LC3B is mainly diffuse, as evidenced by yellow staining due to merged green and red fluorescence. In HCV core-transfected cells without BafA1 treatment, separate yellow and red puncta appeared, indicating that the formation of autophagosomes and autolysosomes was induced by HCV core protein. Upon treatment with BafA1, green puncta increased, further indicating that HCV core protein induced complete autophagic flux. Viewed together, these data indicate that HCV core protein-induced autophagy is complete.
HCV core protein induces ER stress via activation of UPR signaling of the EIF2AK3 and ATF6 pathways, but not the ERN1-XBP1 pathway
To elucidate how HCV core protein induces autophagy, we studied whether HCV core protein could induce ER stress and the UPR pathway, which has been reported to regulate autophagy.36 HCV core protein synthesis, processing, and folding occur in the ER. Also, in the ER, HCV core protein induces HSPA5, HSP90B1, CALR, and ER calcium ATPase overexpression, and this leads to ER stress.24 Immunofluorescent staining confirmed that HCV core protein was localized in the ER of HCV core-transfected cells and in the ER of HCVcc-infected cells (Fig. 4A). Cells were treated with thapsigargin (Tg, 300 nM) and dithiothreitol (DTT, 10 mM) for 16 h were positive controls; all 3 pathways were activated. HSPA5, an ER chaperone protein, is the chief regulator of the 3 proximal ER stress transducers, which are EIF2AK3, ATF6, and ERN1. These transducers contain a lumenal domain which interacts with HSPA5. After ER stress, transducers release HSPA5. Data show that HCV core protein caused an increased expression of HSPA5. However, Tg and DTT were slightly stronger inducers of HSPA5 (Fig. 4B). These data agree with previous reports.14 Next, we examined whether HCV core protein activated the EIF2AK3 pathway by analyzing the phosphorylation of EIF2AK3 and its downstream effector, EIF2S1, by western blotting. As shown in Figure 4B, higher levels of EIF2AK3 and EIF2S1 phosphorylation were observed in HCV core-transfected cells than in empty vector-transfected cells. This EIF2AK3 and EIF2S1 phosphorylation, in turn, increased ATF4 translation and promoted DDIT3 expression. In response to ER stress, ATF6 (90 kDa), the second sensor, which is translocated from the ER to the Golgi apparatus and cleaved, releases the active N-terminal 50 kDa ATF6. Subsequently, DDIT3 upregulation occurs.37 Data indicate that HCV core protein expression increased degradation of 90-kDa ATF6 precursor, yielding a 50 kDa cleavage product (Fig. 4B). These results indicate that the ATF6 pathway was activated by HCV core protein.
Figure 4. HCV core protein activates EIF2AK3 and ATF6 pathways of UPR. (A) The location of HCV core protein in the ER lumen of HCV core protein-transfected cells and HCVcc-infected cells. Huh7 cells were transfected with core protein expression plasmids or infected with 10 MOI of HCVcc. At 48 h post-transfection or 72 h post-infection, the cells were fixed and assessed by indirect immunofluorescence analysis using anti-core protein and anti-CANX (ER marker) antibodies, and then imaged by laser confocal microscopy. ER marker (green), staining of HCV core protein (red), DAPI staining of nuclei (blue) is shown. Scale bars: 10 μm. (B) HCV core protein activated EIF2AK3 and ATF6 pathways. Huh7 cells were transfected with core protein expression plasmids or empty vectors. Cells treated with 300 nM thapsigargin (Tg, which rapidly elicits ER stress) and 10 mM DL-dithiothreitol (DTT, a known ER stress-inducer) for 16 h before harvesting were used as positive control. At 48 h post-transfection, cells were harvested and expression of HSPA5, EIF2AK3 (p-Thr980), EIF2AK3, EIF2S1 (p-Ser51), EIF2S1, ATF4, DDIT3, and ATF6 were analyzed by western blots using the indicated antibodies. Blots are representative of 3 independent experiments. ACTB was used as a sample-loading control. Densitometric ATF4/ACTB and DDIT3/ACTB ratios from at least 3 independent experiments are shown. The value of Huh7 without any treatment was set at 1 for each experiment (*P < 0.05, **P < 0.01).
Activation of ERN1 by ER stress could cause alternate splicing of XBP1 mRNA with the loss of the Pst I restriction site. As shown in Figure 5A, the 416 bp RT-PCR products from the Tg- and DTT-treated positive control cells were resistant to Pst I digestion, whereas RT-PCR products from HCV core- or mock-transfected cells were digested by Pst I to generate 295 bp and 147 bp fragments. This suggests that HCV core does not activate the ERN1 pathway. This observation was confirmed by western blots and a specific antibody against XBP1, which recognizes both unspliced and spliced forms of XBP1 protein. Data in Figure 5B reveal that the alternatively spliced form of XBP1 protein was only present in the Tg- and DTT-treated cells. To further confirm that HCV core protein did not activate the ERN1-XBP1 pathway, we constructed a Flag-XBP1-FLuc reporter plasmid (Fig. 5C). This plasmid encoded an N-terminal Flag-tagged XBP1 along with a C terminus fused to firefly luciferase (FLuc) at the second ORF of XBP1. The FLuc could be expressed only after splicing of XBP1 and removal of the 26 nt intron. As expected, luciferase activity was only detected in the Tg- and DTT-treated cells but not in mock- or HCV core-transfected cells (Fig. 5D and E). Phosphorylation of MAPK9/MAPK8 is another commonly used indicator of ERN1 activation. Phosphorylation of MAPK9/MAPK8 was measured in cells transfected with HCV core protein or treated with 11′-deoxyverticillin A (C42), a compound that induces MAPK9/MAPK8 phosphorylation,38 by using a specific antibody against MAPK9/MAPK8 (p-Thr183/p-Tyr185). As shown in Figure 5F, phosphorylation of MAPK9/MAPK8 was only observed in C42-treated cells, but not in HCV core-transfected cells. We did observe, using multiple approaches, that HCV core protein did not activate the ERN1-XBP1 pathway (Fig. 5). Collectively, the data show that HCV core protein induces ER stress through activation of UPR signaling of the EIF2AK3 and ATF6 pathways but not the ERN1-XBP1 pathway.
Figure 5. HCV core protein does not activate the ERN1-XBP1 pathway of UPR. (A) Huh7 cells were transfected with core protein expression plasmids or empty vectors. Cells treated with 300 nM Tg or 10 mM DTT for 16 h before harvesting were used as positive controls. At 48 h post-transfection, the cells were harvested and RT-PCR was performed using XBP1-specific primers. Then the PCR products were digested by Pst I, the unspliced XBP1 products were digested into 295 bp and 147 bp fragments, while the 416 bp spliced XBP1 products could not be digested by Pst I. XBP1(u) represents the unspliced XBP1 PCR products, and XBP1(s) represents the spliced XBP1 PCR products. ACTB was used as the loading control. (B) The cells treated as (A) were analyzed by western blotting using the specific anti-XBP1 antibody to detect the unspliced and spliced XBP1 proteins. (C) Frame diagrams of the reporter plasmid designated Flag-XBP1-FLuc encoding an N-terminal Flag-tagged XBP1 along with the C terminus fused to firefly luciferase (FLuc) at the second ORF of XBP1. (D) Huh7 cells were transfected with core protein expression plasmids or empty vectors along with Flag-XBP1-FLuc and Renilla luciferase pRL-SV40 plasmids, and the cells treated with 300 nM Tg or 10 mM DTT for 16 h before harvesting were used as positive controls. At 48 h post-transfection, the cells were harvested and lysed for measuring XBP1 mRNA splicing. The fold-change is expressed relative to mock-transfected control from at least 3 independent experiments (**P < 0.01). (E) Expression of firefly luciferase (FLuc) and core protein in (D) were analyzed by western blotting using the indicated antibodies. (F) Huh7 cells were transfected with core protein expression plasmids or empty vectors. Cells treated for 12 h with 0.25 μM C42 (11'-deoxyverticillin A), and which exhibited phosphorylation of MAPK9/MAPK8, were used as positive controls. At 48 h post-transfection, the cells were harvested and analyzed by western blotting using the indicated antibodies to measure the phosphorylation of MAPK9/MAPK8.
HCV core protein induces autophagy via activation of the EIF2AK3 and ATF6 pathways of UPR
To verify whether HCV core protein-induced autophagy occurs through ER stress and UPR in Huh7 cells, we speculated that knocking down EIF2AK3 or ATF6 would inhibit HCV core protein-induced autophagy. We first knocked down EIF2AK3 and ATF6 in Huh7 cells using lentiviral-based shRNA. Knockdown efficiency varied between 80% and 90% (Fig. 6A and B). Next, we investigated the consequence of EIF2AK3 and ATF6 knockdown on downstream signaling proteins within the EIF2AK3 and ATF6 pathways. As shown in Figure 6A, EIF2AK3 knockdown decreased EIF2AK3 and EIF2S1 phosphorylation and decreased ATF4 and DDIT3 expression in HCV core-transfected cells and Tg-treated cells, compared with control shRNA-transduced cells. Similarly, ATF6 knockdown decreased chaperone protein DDIT3 in cells transfected with HCV core and Tg-treated cells (Fig. 6B). Next, we studied the effect of EIF2AK3 and ATF6 pathway knockdown on HCV core protein-induced autophagy. Western blotting revealed that knockdown of EIF2AK3 or ATF6 reversed Tg- and HCV core protein-induced augmentation of LC3B-II and degradation of SQSTM1, compared with control shRNA-transduced cells. Interestingly, combined knockdown of EIF2AK3 and ATF6 impressively inhibited HCV core-induced autophagy (Fig. 6C). To confirm the results, we analyzed the effect of knocking down EIF2AK3 and ATF6 on punctate LC3B formation in single cells using immunofluorescence and confocal microscopy (Fig. 6D). Quantitative analysis revealed that knockdown of EIF2AK3 or ATF6 decreased the number of LC3B puncta in HCV core-transfected cells and Tg-treated cells (Fig. 6E). Thus, HCV core protein induces autophagy via EIF2AK3 and ATF6 UPR pathway activation.
Figure 6. HCV core protein induces autophagy by activating the EIF2AK3 and ATF6 pathways of UPR. (A and B) Huh7 cells were transfected with HCV core protein expression plasmids or empty vectors. Lentiviral shRNA against EIF2AK3 and ATF6 were added at 6 h post-transfection (MOI = 1) and incubated for 3 h. Cells treated with 300 nM Tg for 16 h were used as positive controls. At 72 h post-transfection, cells were harvested and western blotting was performed using the indicated antibodies. Blots are representative of 3 independent experiments. ACTB was used as sample-loading control. The densitometric ATF4/ACTB and DDIT3/ACTB ratios from at least 3 independent experiments are shown. The value of mock-treated cells with control shRNA was set at 1 each experiment (**P < 0.01). (C) Huh7 cells were transfected with HCV core protein expression plasmids or empty vectors. Lentiviral shRNA against EIF2AK3 and ATF6 were added alone or combined, at 6 h post-transfection (MOI = 1) and incubated for 3 h. Cells treated with 300 nM Tg for 16 h were used as positive controls. At 72 h post-transfection, cells were harvested and expression of LC3B and SQSTM1 were analyzed by western blotting. Blots are representative of 3 independent experiments. ACTB was used as sample-loading control. The densitometric LC3B-II/ACTB and SQSTM1/ACTB ratios from at least 3 independent experiments are shown. The value of mock-treated with control shRNA was set at 1 for each experiment (*P < 0.05, **P < 0.01). (D) The cells in (C) were fixed and analyzed by indirect immunofluorescence using anti-LC3B and anti-core antibodies. Patterns of LC3B expression in mock-transfected and HCV protein-transfected cells were visualized with laser confocal microscopy LC3B (green), HCV core (red) staining is shown. Scale bars: 10 μm. (E) Quantitative presentation of punctate LC3B per cell in (D).
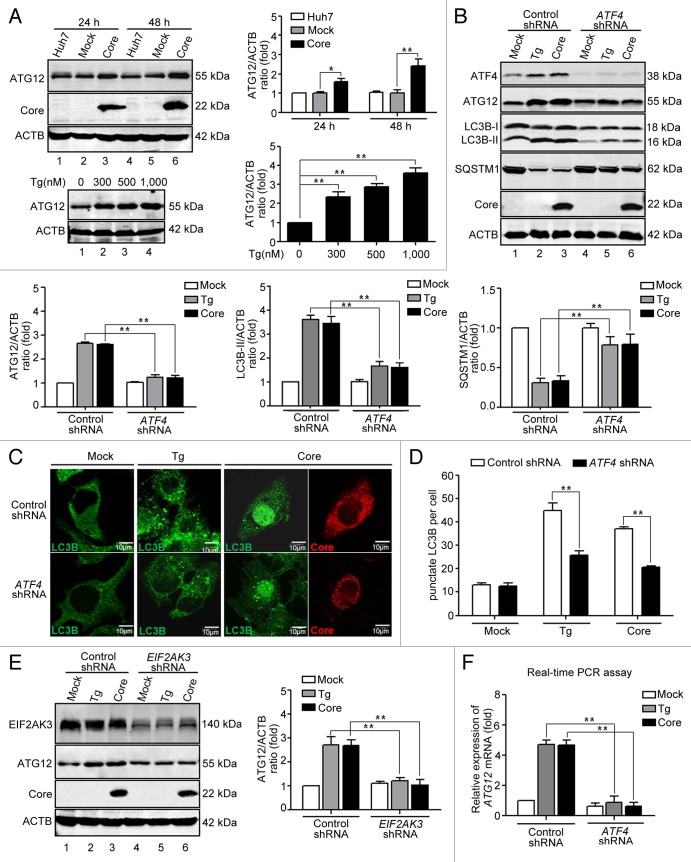
Upregulation of ATG12 by ATF4 is involved in HCV core protein-induced autophagy
After establishing that HCV core protein-induced autophagy occurs via the EIF2AK3 and ATF6 UPR pathways, we investigated mechanisms by which autophagy is promoted by HCV core protein-induced ER stress. The conversion of LC3B-I to LC3B-II is crucial to induction of mammalian cell autophagy, and this process depends on the ATG12-ATG5-ATG16L1 complex. Kouroku and colleagues reported that a polyglutamine72 repeat upregulated ATG12 mRNA through the EIF2AK3-EIF2S1 pathway.30 Considering that HCV core protein could induce phosphorylation of EIF2AK3 and EIF2S1, we studied ATG12 expression in HCV core protein-transfected cells. Figure 7A shows that HCV core transfection upregulated ATG12 expression. Similarly, Tg also increased ATG12 expression in a dose-dependent manner. ATF4 is an important factor downstream of EIF2AK3-EIF2S1 when ER stress is induced. Data indicate that ATF4 knockdown, with a lentiviral-based shRNA in cells transfected with HCV core plasmid or treated with Tg inhibited ATG12 expression and inhibited LC3B-I conversion to LC3B-II and subsequent SQSTM1 degradation (Fig. 7B). Data were confirmed in single cells using immunofluorescence and confocal microscopy (Fig. 7C). Knockdown of ATF4 decreased the number of LC3B puncta in Tg-treated and HCV core-transfected cells (Fig. 7D). In addition, we observed that in HCV core-transfected and Tg-treated cells, EIF2AK3 knockdown reduced ATG12 expression (Fig. 7E), which confirmed that ATG12 is regulated by the EIF2AK3-EIF2S1-ATF4 pathway of UPR in HCV core-transfected cells. Furthermore, real-time PCR revealed that HCV core protein increased ATG12 mRNA expression and knockdown of ATF4 decreased ATG12 mRNA (Fig. 7F). Therefore, we speculated that ATF4 regulates ATG12 transcription, contributing to HCV core protein-induced autophagy. Using the PromoterInspector software (Genomatix software GmbH, Munich, Germany), 4 ATF4 binding sites in ATG12 gene intron regions were predicted. To determine whether ATF4 acts directly at the intron region of ATG12, we performed an ATF4 chromatin immunoprecipitation (ChIP) assay, but we did not observe that ATF4 directly binds to the ATG12 gene (data not shown). Overall, our data indicate that HCV core protein-induced ATG12 expression was regulated by ATF4 by way of a mechanism that is unrelated to direct interaction with the ATG12 gene.
Figure 7. HCV core protein upregulation of ATG12 expression depends on ATF4 of the EIF2AK3 pathway. (A) Huh7 cells were transfected with HCV core protein expression plasmids or empty vectors. At 24 and 48 h post-transfection, cells were harvested and western blotting was performed. Huh7 cells treated with 300, 500, 1,000 nM Tg for 16 h, and then western blotting was performed. Expression of ATG12 and HCV core protein was analyzed using the indicated antibodies. Blots are representative of 3 independent experiments. ACTB was used as sample-loading control. The densitometric ATG12/ACTB ratio from at least 3 independent experiments is shown. The value of Huh7 without any treatment was set at 1 for each experiment (*P < 0.05, **P < 0.01). (B) Knockdown of ATF4 inhibited expression of ATG12 and autophagy in HCV core protein-transfected cells. Huh7 cells were transfected with HCV core protein expression plasmids or empty vectors. Lentiviral shRNA against ATF4 was added 6 h post-transfection (MOI = 1) and incubated for 3 h. At 72 h post-transfection, expression of ATF4, ATG12, LC3B, SQSTM1 and HCV core protein were analyzed by western blotting using the indicated antibodies. Blots are representative of 3 independent experiments. ACTB was used as a sample-loading control. The densitometric ATG12/ACTB, LC3B-II/ACTB and SQSTM1/ACTB ratios from at least 3 independent experiments are shown. The value of mock-treated cells with control shRNA is set on 1 each experiment (**P < 0.01). (C) The cells in (B) were fixed and analyzed by indirect immunofluorescence using anti-LC3B and anti-core antibodies. Patterns of LC3B expression in mock-transfected and HCV protein-transfected cells were visualized with laser confocal microscopy. LC3B (green), HCV core (red) staining is shown. Scale bars: 10 μm. (D) Quantitative presentation of punctate LC3B per cell in (C). (E) Knockdown of EIF2AK3 decreased expression of ATG12. Huh7 cells were transfected with HCV core protein expression plasmids or empty vectors. Lentiviral shRNA against EIF2AK3 was added at 6 h post-transfection (MOI = 1) and incubated for 3 h. At 72 h post-transfection, expression of EIF2AK3, ATG12 and HCV core protein were analyzed by western blotting using the indicated antibodies. Blots are representative of 3 independent experiments. ACTB was used as a sample-loading control. The densitometric ATG12/ACTB ratio from at least 3 independent experiments is shown. The value of mock-treated cells with control shRNA was set at 1 for each experiment (**P < 0.01). (F) Knockdown of ATF4 inhibited ATG12 mRNA expression. Huh7 cells were transfected with HCV core protein expression plasmids or empty vectors. Lentiviral shRNA against ATF4 was added 6 h post-transfection (MOI = 1) and incubated for 3 h. At 72 h post-transfection, cells were harvested and real-time PCR was performed. Data are representative of 3 independent experiments (*P < 0.05).
Direct binding of DDIT3 to the LC3B promoter upregulates LC3B expression, contributing to HCV core protein-induced autophagy
DDIT3 is another important factor downstream of both EIF2AK3 and ATF6 during ER stress. Our data show that HCV core transfection upregulated DDIT3 expression (Fig. 4B), and ATF4 knockdown decreased DDIT3 expression in HCV core protein-transfected cells (Fig. S2A). To learn whether DDIT3 could regulate autophagy, DDIT3 gene knockdown was performed with siRNA in Huh7 cells. Data indicate that knockdown of DDIT3 inhibited Tg- and HCV core protein-induced conversion of LC3B-I to LC3B-II and SQSTM1 degradation, in contrast to control siRNA. Of note, DDIT3 knockdown not only inhibited Tg- and HCV core protein-induced conversion of LC3B but also decreased total LC3B expression (Fig. 8A; Fig. S3). The results were confirmed in single cells with immunofluorescence and confocal microscopy. DDIT3 knockdown decreased the number of LC3B puncta in Tg-treated and HCV core-transfected cells (Fig. 8B and C). DDIT3 knockdown also decreased LC3B mRNA levels as detected by real-time PCR analysis in HCV core protein-transfected and Tg-treated cells (Fig. 8D).
Figure 8. DDIT3 upregulates LC3B expression in HCV core-transfected cells. (A) Knockdown of DDIT3 inhibited the conversion of LC3B-I to LC3B-II and degradation of SQSTM1. Huh7 cells were transfected with HCV core protein expression plasmids or empty vectors along with DDIT3 siRNA or control siRNA. Cells treated with 300 nM Tg for 16 h were used as positive controls. At 72 h post-transfection, expression of DDIT3, LC3B, SQSTM1, and HCV core protein were analyzed by western blotting using the indicated antibodies. Blots are representative of 3 independent experiments. ACTB was used as a sample-loading control. The densitometric LC3B-II/ACTB and SQSTM1/ACTB ratios from at least 3 independent experiments are shown (**P < 0.01). (B) The cells in (A) were fixed and analyzed by indirect immunofluorescence using anti-LC3B and anti-core antibodies. Patterns of LC3B expression in mock-transfected and HCV protein-transfected cells were visualized with laser confocal microscopy. LC3B (green), HCV core (red) staining is shown. Scale bars: 10 μm. (C) Quantitative presentation of punctate LC3B per cell in (B). (D) Real-time PCR analysis for LC3B mRNA expression. The cells treated as (A) were harvested for RNA extraction and real-time PCR analysis. The results are representative of 3 independent experiments (*P < 0.05, **P < 0.01).
To study whether DDIT3 regulates LC3B transcription, we investigated whether DDIT3 activates the LC3B promoter using a luciferase assay. Data also show that DDIT3 and HCV core protein activated the LC3B promoter in Huh7 cells (Fig. 9A). Results that were consistent with these findings were also obtained in 293T cells (data not shown). Collectively, these findings suggest that DDIT3 upregulates LC3B expression through binding to the LC3B promoter. To confirm these findings, ChIP assay was performed with a DDIT3-specific antibody. A map of the primer pairs in the LC3B promoter region used for the ChIP assay is illustrated in Figure 9B. As shown in Figure 9C the –253 to –99 (from start codon) primer set amplified specific DNA bands only in HCV core plasmid-transfected and Tg-treated cells, but not in mock-transfected cells. These data show that DDIT3 could bind to the –253 to –99 base region of the LC3B promoter. Real-time quantitative PCR confirmed these ChIP results (Fig. 9D). DNA sequencing of PCR products revealed that the sequence was matched to the LC3B promoter sequence published in NCBI with the exception of a base mutation at –224 in the LC3B promoter (“G” at –224 instead of “A”). This may be caused by the difference in the genome source; our experiments were performed in Huh7 cells, whereas the LC3B sequence from NCBI was from human blood cells. Finally, we investigated the effect of knockdown of DDIT3 on LC3B promoter activity. Data in Figure 9E suggest that DDIT3 knockdown reduced LC3B promoter activity compared with control siRNA in HCV core-transfected and Tg-treated cells. In mock-transfected cells, no significant differences in LC3B promoter activity were observed, irrespective of DDIT3 knockdown. We also analyzed the physiological role of DDIT3 protein in the LC3B expression during infection with HCVcc. The results showed that knockdown of EIF2AK3 and ATF6 decreased the expression of DDIT3 in HCVcc-infected cells, and then knockdown of DDIT3 inhibited LC3B expression both at protein and mRNA level as well as the activating of LC3B promoter (Fig. S4). Thus, DDIT3 is crucial to the expression of LC3B either in HCV core-induced autophagy or during HCVcc infection.
Figure 9. HCV core protein upregulation of LC3B expression occurs through DDIT3 directly binding to the LC3B promoter. (A) LC3B promoter activity was analyzed by Luciferase assay. Huh7 cells were transfected with various amounts of ATF4, DDIT3, or HCV core protein expression plasmids along with LC3B promoter plasmids and Renilla luciferase pRL-SV40 plasmids. Cells were treated with various concentrations of DTT and Tg for 16 h. At 48 h post-transfection, cells were harvested and analyzed. Luminescence was measured with a luminometer and the fold-change is expressed relative to the mock-transfected control, and the mock value was set at 1 each for experiment (*P < 0.05, **P < 0.01). Expression of ATF4, DDIT3, and core proteins in the cells were analyzed by western blotting using the indicated antibodies. (B) Location of primer sets in the LC3B promoter region used for PCR after ChIP. The number for primers is relative to the start codon. (C) DDIT3 ChIP assay for the LC3B promoter. Huh7 cells were transfected with HCV core protein expression plasmids or empty vectors. Cells treated with 300 nM Tg or 10 mM DTT for 16 h were used as positive controls. At 48 h post-transfection, the cells were processed for DDIT3 ChIP. The agarose gel electrophoresis was performed to analyze the PCR products. (D) Real-time PCR for the quantitative DDIT3 ChIP assay of the region corresponding to the primer set –253 to –99. The values represent the ratio of PCR signals from the immunoprecipitated samples to the corresponding input (percent of input DNA; **P < 0.01). Data shown are from a single ChIP, in which 3 independent dishes were pooled for each treatment. Three such independent experiments were done. (E) Knockdown of DDIT3 reduced the LC3B promoter activity activated by HCV core protein. Huh7 cells were transfected with core protein expression plasmids or empty vectors and DDIT3 siRNA or control siRNA along with LC3B promoter plasmids and Renilla luciferase pRL-SV40 plasmids. Cells treated with 300 nM Tg for 16 h were used as positive controls. At 72 h post-transfection, the cells were harvested and analyzed. The luminescence was measured using a luminometer. The fold-change is expressed relative to mock-transfected controls and the value of mock-treated cells with control siRNA was set at 1 for each experiment (*P < 0.05). Expression of DDIT3 and core proteins in the cells were analyzed by western blotting using the indicated antibodies.
Overall, our data suggest that HCV core-induced autophagy involves LC3B transcription upregulation via the direct binding of DDIT3 to the LC3B promoter.
Discussion
Others and we have shown that HCV infection induces autophagy via the 3 UPR pathways of ER stress: EIF2AK3, ATF6, and ERN1-XBP1, although the detailed mechanism through which HCV induces autophagy remains unclear.14-19 HCV interacts with host cells via its encoded structural and nonstructural proteins, so investigating the contribution of individual HCV proteins may allow us to elucidate the mechanism of HCV-induced autophagy. Until now, no reports are available to describe the mechanism by which HCV structural proteins participate in autophagy induction, but some studies do suggest that HCV nonstructural proteins have some type of role in this process.9,20,21 Here we report that HCV core, NS3/4A, and NS4B proteins induced autophagy, an observation consistent with Su’s report that HCV core protein and NS4B could promote the transformation of LC3B-I to LC3B-II.20 In contrast to our results, Su’s group does not show that NS3/4A alone induced autophagy; the difference may be that Su’s group has used Huh7.5 cells, which are DDX58/RIG-I deficient.20 Because we used Huh7 cells in our studies, whether DDX58/RIG-I may interfere with HCV protein-induced autophagy is still unclear. Also, recently, Shubham’s laboratory has reported that HCV NS5A induced autophagy in immortalized human hepatocytes (IHH).21 NS5A was not explored in this study because of the lack of this construct. Therefore, future studies are warranted to determine whether NS5A can induce autophagy in Huh7 cells and IHH.
Whether HCV-induced autophagy is a complete process is still elusive. Our earlier report and those of Po-Yuan Ke suggest that HCV infection induces complete autophagic processes via enhancement of autophagic flux.18,19 Sir and coworkers report that autophagic degradation is not increased, so autophagy activated by HCV is incomplete.15 Such conflicting conclusions could be attributed to the differences in the methods of HCV infection. Our group infected Huh7 cells with HCVcc, and another laboratory transfected Huh7.5 cells with transcribed HCV RNA.15,19 Although we could not determine the extent to which core protein induced autophagy is contributed to HCV, in our present work, we clearly demonstrated that HCV core protein-induced autophagy is a complete process through multiple approaches (Fig. 3).
In our work to investigate how HCV core protein induces autophagy, we observed that data from immunofluorescence studies indicated that HCV core protein localizes to the ER in both HCV core-transfected and HCVcc-infected cells. We and others have shown that HCV can induce autophagy through ER stress, but considering the distribution of HCV core in the cells, we hypothesized that HCV core protein might induce autophagy through ER stress-related UPR signaling pathways. We observed that although HCV activates autophagy through all 3 EIF2AK3, ATF6, and ERN1-XBP1 URP pathways, the HCV core protein only activated EIF2AK3 and ATF6 pathways (not the ERN1-XBP1 pathway). Thus, other HCV proteins or HCV RNA may activate the ERN1-XBP1 pathway during HCV infection, and we observed that NS4B could activate the ERN1-XBP1 pathway (Fig. S5B). To measure the specificity of the pathway induced by the HCV core, we compared the effect of HCV NS3/4A and NS4B proteins on the 3 UPR pathways. Data show that NS3/4A could not activate the UPR pathway, and NS4B could activate any of the 3 UPR pathways (Fig. S5).
Also, data reveal that inhibition of UPR by lentiviral-based shRNA against EIF2AK3 and ATF6 decreased the conversion of and decreased the degradation of SQSTM1. Still, induction of autophagy was not fully inhibited, even with the combination of EIF2AK3 and ATF6 pathway knockdown. These results indicate that other signal pathways independent of ER stress may be involved in HCV core protein-induced autophagy.
BECN1 has been reported to have a BCL2 homology 3 (BH3) domain which may bind to BCL2 and BCL2L1 via BH3-BH3 interactions.39 This would have an important functional impact on the induction of autophagy. After inactivation of BECN1 after interaction with any BCL2 family member that contains a BH3 domain, autophagy is blocked.39 HCV core protein also contains a BH3 domain; this protein was reported to regulate apoptosis via specific interactions with MCL1.40 Possibly, HCV core protein may promote autophagy by competitively disrupting interactions between BECN1 and BCL2 or BCL2L1 to create free BECN1. However, additional studies are required to confirm this speculation.
Although how ER stress and UPR induce autophagy is unclear, in our studies, HCV core protein upregulated ATF4 and DDIT3 expression (Fig. 4B). ATF4 and DDIT3 are 2 critical molecules downstream of the UPR pathways. ATF4 is known to facilitate autophagy through directly binding to a cyclic AMP response element in a LC3B promoter region-binding site thereby upregulating LC3B expression.41 We confirmed that knockdown of ATF4 inhibited LC3B expression at the protein and mRNA level in HCV core protein-transfected cells (Fig. 7B; Fig. S2B). In addition, we identified that HCV core protein-induced autophagy depends on increased transcriptional expression of ATG12 regulated by ATF4. An ATF4 ChIP assay to investigate interactions between ATF4 and ATG12 revealed that ATF4 did not directly bind to the ATG12 gene (data not shown). So, ATF4 may upregulate ATG12 expression via other pathways. For example, ATF4 is shown to increase the expression of BH3-only protein like BECN1,39,42 which has been reported to regulate ATG12 expression, contributing to autophagy.43 Also, FOXO3, belonging to the family of mammalian forkhead transcription factors, binds to the ATG12 promoter and activates ATG12 transcription.44 Moreover, DDIT3 is reported to interact with FOXO3 in response to ER stress.45 In our study, we identified that ATF4 upregulated DDIT3 expression in HCV core-transfected cells (Fig. S2A). Thus, ATF4 could upregulate ATG12 expression via FOXO3 upregulation. Moreover, we observed that in HCV core-transfected cells, ATF4 knockdown inhibited ATG5 expression (Fig. S2C), which can be explained by ATF4 inducing the upregulation of DDIT3, which upregulated ATG5 expression in HCV core protein-transfected cells (Fig. S2A and S2D).
We observed that the EIF2AK3-EIF2S1-ATF4 and ATF6 pathways activated DDIT3 expression in HCV core protein-transfected cells. To identify mechanisms behind autophagy induced by DDIT3 upregulation through ER stress, we applied the DDIT3 ChIP assay and observed that DDIT3 upregulated LC3B expression by directly binding to the –253 to –99 base region of the LC3B promoter in HCV core protein-transfected or Tg-treated cells (Fig. 9). A number of transcription factors binding sites in the –253 to –99 base region were identified using Genomatix software. Therefore, DDIT3 may function either alone or synergistically with other factors to induce LC3B. For example, FOXO3 is reported to bind to the LC3B promoter, and Ghosh and colleagues reported that DDIT3 could interact with FOXO3.45,46 Thus, DDIT3 may function with FOXO3 to regulate LC3B transcription. Therefore, although our present results clearly indicated that DDIT3 could directly bind to the LC3B promoter to regulate LC3B expression, we cannot exclude the possibility that other factors may contribute to regulation of LC3B transcription.
DDIT3 is also reported to upregulate expression of ATG5 by binding to the ATG5 promoter.41 We observed similar results wherein knockdown of DDIT3 decreased ATG5 expression in cells transfected with HCV core plasmid (Fig. S2D). These findings emphasize the complex roles of DDIT3 in mediating ER stress-induced autophagy by transcriptional mechanisms. Although our study clearly demonstrated the involvement of DDIT3 and ATF4 in HCV core protein-induced ER stress and autophagy, it is evident that inhibition of these pathways or factor knockdown does not completely abrogate the induction of autophagy. This suggests that additional factors may contribute to autophagy activation during HCV core protein-induced ER stress. This represents the first report, to our knowledge, regarding DDIT3-mediated regulation of LC3B transcription by direct binding to the LC3B promoter. From these data, we propose the schematic illustrated in Figure 10 to summarize our findings. HCV core protein can induce autophagy mainly through activating the EIF2AK3 and ATF6 pathways but not the ERN1-XPB1 pathway of ER stress. HCV core protein promotes autophagy induction by upregulating ATG12 via ATF4 and enhancing LC3B expression by direct binding of DDIT3 to the LC3B promoter region. These data will lay a foundation for future studies to better understand the molecular mechanisms through which HCV induces autophagy, viral persistence, and viral pathogenesis.

Figure 10. Schematic model of the molecular mechanism through which HCV core protein induces autophagy. HCV core protein induces ER stress, which activates the EIF2AK3 and ATF6 pathways of UPR, but not the ERN1-XBP1 pathway. In the EIF2AK3 pathway, HCV core protein activates the phosphorylation of EIF2AK3 and EIF2S1. Phosphorylated EIF2S1 upregulates expression of ATF4, which leads to the upregulation of DDIT3, LC3B, and ATG12. In the ATF6 pathway, ATF6 (90 kDa) is cleaved to release an active form, which leads to the upregulation of DDIT3. DDIT3 increases expression of LC3B and ATG5. LC3B, a specific autophagy marker, plays an important role in the development of autophagy. ATG12 and ATG5 are components of the ATG12-ATG5-ATG16L1 complex, which promotes the conversion of LC3B-I to LC3B-II to induce autophagy.
Materials and Methods
Cells cultures and HCV virus infection
The human hepatoma cell line, Huh7, was obtained from the Chinese Academy of Sciences cell library. HEK293FT cell lines were purchased from Life Technologies. All cell lines were grown in Dulbecco’s Modified Eagle Medium (DMEM, Life Technologies, 11995) supplemented with 100 units/ml penicillin, 100 mg/ml streptomycin, 1% nonessential amino acids (NEAA, Life Technologies, 11140-050) and 10% fetal bovine serum at 37 °C in the presence of 5% CO2. The preparation of JFH1 strain HCV cell culture virus (HCVcc) has been previously described. Briefly, for infections, 1 × 105 Huh7 cells per well in 6-well dishes were infected with HCVcc (MOI = 10) for 12 h and then media was changed.19 The infected cells were analyzed on day 3 post-infection.
Antibodies and reagents
Rabbit anti-LC3B (L7543), rabbit anti-SQSTM1 (P0067), rabbit anti-CANX (C4731), mouse anti-Flag (A8592), mouse anti-ACTB (A5441) antibodies and gold-conjugated anti-mouse IgG (G7652) were purchased from Sigma-Aldrich. Antibodies against HSPA5 (#3183), EIF2AK3 (#3192), Phospho-EIF2AK3 (p-Thr980) (#3179), EIF2S1 (#2103), Phospho-EIF2S1 (p-Ser51) (#3398), DDIT3 (#2895), MAPK9/MAPK8 (#9252), Phospho-MAPK9/MAPK8 (p-Thr183/p-Tyr185) (#4668), ATG5 (#2630), and ATG12 (#4180) were purchased from Cell Signaling Technology. Antibodies against HCV core protein (sc-57800), ATF6 (sc-22799), ATF4 (sc-22800) and XBP1 (sc-81427), as well as DDIT3 siRNA (RC201301) were purchased from Santa Cruz Biotechnology. Goat anti-mouse IgG IRDye 680RD (926-68070) and goat anti-rabbit IgG IRDye 800CW (926-32211) antibodies were obtained from Li-Cor Biosciences. Dithiothreitol (DTT, D9779), Thapsigargin (Tg, T9033), Bafilomycin A1 (BafA1, B1793), polyethylenimine (PEI, 408727), and polybrene (107689) were purchased from Sigma-Aldrich.
Plasmids
The flag-tagged plasmids encoding individual HCV (2a) proteins (core, NS2, NS3, NS3/4A, NS4B, and NS5B) were gifts from Dr Wei Yang (Institute of Pathogen Biology, Chinese Academy Medical Sciences and Peking Union Medical College, Beijing, China).47 To construct the untagged HCV core expression plasmids, core cDNA (coding region is 191 aa) was amplified from HCV JFH1 2a (GenBank accession number: AB047639) genome, and then cloned into the Hind III and EcoR V sites of pcDNA3.1 vector. The improved tandem fluorescent-tagged LC3B plasmids (mTagRFP-mWasabi-LC3B) were generously provided by Dr Jian Lin (Beijing Nuclear Magnetic Resonance Center; College of Chemistry and Molecular Engineering; Peking University; Peking, China).35 A reporter plasmid, Flag-XBP1-FLuc, was constructed with a 74 bp XBP1 fragment at the second open reading frame containing the splicing site into a pcDNA3.1 vector, which has the flag tag at the N-terminal, and a C terminus fused to firefly luciferase (FLuc).
Plasmids for control shRNA (SHC010) and shRNA against EIF2AK3 (TRCN0000001401), ATF6 (TRCN0000017855), and ATF4 (TRCN000013573) were purchased from Sigma-Aldrich. The packaging plasmid used for the preparation of the lentivirus pCMVΔ8.74 and the plasmid encoding the envelope of the vesicular stomatitis virus pCMV-VSVG were obtained from Addgene.
ATF4 expression plasmids (SC119103) and DDIT3 expression plasmids (RC201301) were purchased from Origene. To construct the LC3B promoter luciferase reporter plasmid, the LC3B promoter (−1033 to +25, from start codon) was amplified from the Huh7 cell genome by PCR using the following primers: LC3B sense: 5′-ATCGGAGCTC TGGTTTACGC ACTGCA-3′, LC3B antisense: 5′-ATGCCTCGAG GCTTGAAGGT CTTCTCCGAC G-3′. PCR products were digested by restriction endonucleases Sac I and Xho I, and then cloned into a pGL3-basic vector. Renilla luciferase pRL-SV40 plasmids were stored in our laboratory.
Lentiviral shRNA packaging and transduction
HEK293FT cells were seeded at a cell density of 5 × 106 cells per 10-cm dish in complete DMEM media. After 24 h, the cells in 10-cm dishes were transfected with 6 μg pCMVΔ8.74, 3 μg shRNA, or 1 μg pCMV-VSVG using 30 μl polyethylenimine (PEI, 1 mg/ml). At 10 h post-transfection, the media was replaced with 10 ml of fresh complete DMEM media. After an additional 48 h, the cell culture supernatants were collected by centrifuging at 2,000 g for 10 min at 4 °C. The lentiviral titer was determined using Lenti-X p24 Rapid Titer Kit (Clontech, 632200) according to the manufacturer’s instructions and stored at –80 °C until use. For the experiment, lentiviral shRNA was added onto the cells (MOI = 1) with 4 μg/ml polybrene and incubated for 3 h. The media was replaced with fresh complete media and cells were harvested and analyzed using the indicated assays at 72 h post-transduction.
Reverse transcription-polymerase chain reaction (RT-PCR) and real-time PCR
Total cellular RNA was extracted using Trizol reagent (Life Technologies, 15596-018) according to the manufacturer’s protocol. One microgram of total RNA was reverse transcribed in a 20-μl volume using Improm-IITM reverse transcriptase (Promega, A3500) according to the manufacturer’s instructions. All cDNA samples were stored at –80 °C until further use. To detect the XBP1 splicing status, PCR was performed using 1 μl cDNA and the following primers: XBP1 sense: 5′-CCTTGTAGTT GAGAACCAGG-3′, XBP1 antisense: 5′-GGGGCTTGGT ATATATGTGG-3′; ACTB sense: 5′-ATCTGGCACC ACACCTTCTA CAATGAG-3′, ACTB anti-sense: 5′-CGTCATACTC CTGCTTGCTG ATCC-3′. PCR products were digested by Pst I, and then separated on a 2% agarose gel. ACTB was used as an internal control.
RNA from cell samples was extracted as previously described. Briefly, 1 μl of cDNA was used for real-time PCR with Power SYBR Green PCR master mix (Life Technologies, 4367659). Each experiment was performed in triplicate. The following primers were used for measuring LC3B mRNA: LC3B sense: 5′-AAGGCGCTTA CAGCTCAATG-3′, LC3B antisense: 5′-CTGGGAGGCA TAGACCATGT-3′; ATG12 sense: 5′-TAGAGCGAAC ACGAACCATCC-3′, ATG12 antisense: 5′-CACTGCCAAA ACACTCATAG AGA-3′; ACTB sense: 5′- CACTGTGCCC ATCTACGAGG G-3′, ACTB antisense: 5′- CTCCTTAATG TCACGCACG-3′. The PCR reaction was conducted in a 7900HT Fast Real-Time PCR machine (Life Technologies, Grand Island, NY, USA) and data were analyzed using RQ manager software (Life Technologies).
Reporter assay
Huh7 cells were seeded in 24-well plates at a cell density of 1 × 105 cells per well for 24 h. For the XBP1 splicing assay, Huh7 cells were transfected with empty plasmids or plasmids expressing HCV core protein along with Flag-XBP1-FLuc reporter plasmids and Renilla luciferase pRL-SV40 plasmids using Lipofectamine 2000 (Life Technologies, 11668-019) according to the manufacturer’s instructions. Huh7 cells treated with 300 nM Tg and 10 mM DTT for 16 h served as positive controls. For the LC3B promoter activity assay, Huh7 cells were transfected with empty vectors, ATF4, DDIT3, or core protein expression plasmids along with LC3B promoter plasmids and Renilla luciferase pRL-SV40 plasmids. Cells were treated with various concentrations of DTT and Tg for 16 h before harvesting. Total DNA was kept constant by adding empty control vectors. At 48 h post-transfection, cells were harvested and lysed. Luciferase activity was measured using a dual-Luciferase® reporter assay system (Promega, E1960) according to the manufacturer’s instructions.
SDS-PAGE and western blot
Cells were lysed in lysis buffer containing 150 mM NaCl, 25 mM Tris-Cl (pH 7.4), 1% NP-40, 0.25% sodium deoxycholate, and 1 mM EDTA with a protease inhibitor cocktail (Roche, 04693132001) and a phosphatase inhibitor cocktail (Roche, 04906845001). The total protein concentration was determined using a bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, 23225). Equal amounts of total proteins were separated with SDS-PAGE (8%) for Phospho-EIF2AK3, and EIF2AK3 detection, 10% gels for SQSTM1, HSPA5, Phospho-EIF2S1, EIF2S1, ATF4, DDIT3, ATF6, Phospho-MAPK9/MAPK8, MAPK9/MAPK8, and ATG12 detection, and 15% gels for LC3B and core protein detection, and then transferred to nitrocellulose membranes (Pall, 66485). Membranes were first incubated with different primary antibodies overnight at 4 °C and incubated with IRDye 680 or 800-labeled secondary antibodies at room temperature for 2 h. Protein bands on the membrane were scanned using a Li-Cor Odyssey system (LI-COR Biosciences, Lincoln, NE, USA). Photodensitometric data from protein bands were analyzed and quantified with Quantity One software (Bio-Rad).
Immunofluorescence and confocal microscopy
After transfection, Huh7 cells were fixed with 4% paraformaldehyde (Sigma-Aldrich, 16005) in PBS for 15 min and permeabilized using 0.3% Triton X-100 (Sigma-Aldrich, T8787) and 3% BSA in PBS for 30 min. To detect LC3B puncta formation, the fixed cells were incubated with mouse anti-flag antibody (1: 500 dilution in 3% BSA) or anti-core antibody (1: 50 dilution in 3% BSA) for 2 h at room temperature, and rabbit anti-LC3B antibody (1: 200 dilution in 3% BSA) for additional 2 h at room temperature. After 3 washes with PBST, cells were incubated with secondary TRITC-conjugated anti-mouse (Jackson ImmunoResearch, 115-025-003) and FITC-conjugated anti-rabbit (Jackson ImmunoResearch, 111-095-003) antibodies for 1 h. To identify localization of HCV core protein to the ER, cells were fixed and stained with primary mouse anti-core antibody and rabbit anti-CANX antibody, and then secondary TRITC-conjugated anti-mouse and FITC-conjugated anti-rabbit antibodies followed by incubation with DAPI (4’, 6-diamidino-2-phenylindole, Life Technologies, D1306) for nuclear staining. For analysis of autophagy flux, Huh7 cells were transfected with empty vector, HCV core, and mTagRFP-mWasabi-LC3B plasmids. Cells were then fixed for immunostaining. After blocking, cells were incubated with mouse anti-core antibody and then Alexa Fluor 405-conjugated anti-mouse antibody (Jackson ImmunoResearch, 715-475-150). Images were analyzed using a TCS SP5 Laser Scanning Confocal Microscope and its software (Leica Microsystems CMS GmbH, Mannheim, Germany)
Immunoelectron microscopy
HCV core-transfected and mock-transfected cells were harvested and washed 3 times with PBS. Cells were fixed in 4% paraformaldehyde supplemented with 0.01% glutaraldehyde (in 0.1 M PBS) for 2 h at 4 °C and then processed for ultracryomicrotomy. In brief, cells were scraped and spun down in 12% gelatin. After immersion in 2.3 M sucrose (in 0.1 M PBS) overnight at 4 °C, samples were rapidly frozen in liquid nitrogen. Ultrathin (70-nm thick) cryosections were prepared with an ultracryomicrotome (Leica EM FC6, Vienna, Austria) and mounted on nickel grids. After washing with buffer (2% BSA and 0.15% glycine in PBS), samples were blocked with 2% BSA and 5% normal goat serum in PBS for 1 h at room temperature. Then cryosections were incubated with anti-core protein antibody (1:10 dilution in 2% BSA) at room temperature for 2 h, rinsed, and incubated for 10 nm with gold-conjugated anti-mouse antibody (1:10 dilution in 2% BSA) at room temperature for 1 h. Finally, samples were fixed for 5 min with 2.5% glutaraldehyde and were stained with 2% methylcellulose for 10 min on ice. After air-drying, sections were examined under a transmission electron microscope (FEI Tecnai G2 Spirit 120KV, Oregon, USA).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using the EZ ChIP Chromatin Immunoprecipitation Kit (Millipore, 17-371) according to the manufacturer’s instructions. DNA was crosslinked with formaldehyde (1% final concentration for 10 min) and cells were lysed in the presence of protease inhibitors. Lysates were sonicated with a Bioruptor from Diagenode (UCD-200, Liège, Belgium) to shear DNA (average size 200 to 600 bp). Immunoprecipitations were performed with 5 μg of mouse anti-DDIT3 and 5 μg of normal mouse IgG (included in EZ ChIP). Normal IgG was used as a negative control. Immunoprecipitated and input DNA were subjected to PCR and real-time PCR using primers as described in Table S1.
Statistical analysis
All the data are presented as mean ± standard deviations (SD). All assays were performed in triplicate for all experiments, and all experiments were repeated at least 3 times. One-way analysis of variance (ANOVA) and Dunnett and Tukey post-hoc tests and 2-way ANOVA were used to analyze data in the corresponding experiments. Statistical significance was set at P < 0.05.
Supplementary Material
Glossary
Abbreviations:
- ACTB
actin, beta
- ATF6
activating transcription factor 6
- ATG
autophagy-related
- BafA1
bafilomycin A1
- C42
11′-deoxyverticillin A
- CALR
calreticulin
- ChIP
chromatin immunoprecipitation
- DDIT3
DNA-damage-inducible transcript 3
- DTT
dithiothreitol
- EIF2AK3
eukaryotic translation initiation factor 2-alpha kinase 3
- EIF2S1
eukaryotic translation initiation factor 2, subunit 1 alpha, 35 kDa
- ER
endoplasmic reticulum
- ERN1
endoplasmic reticulum to nucleus signaling 1
- HCV
hepatitis C virus
- HSPA5
heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa)
- HSP90B1
heat shock protein 90 kDa beta (Grp94), member 1
- MAP1LC3B/LC3B
microtubule-associated protein 1 light chain 3 beta
- LC3B-I
unlipidated form of LC3B
- LC3B-II
phosphatidylethanolamine-conjugated form of LC3B
- PI
propidium iodide
- SQSTM1
sequestosome 1
- Tg
thapsigargin
- UPR
unfolded protein response
- XBP1
X-box binding protein 1
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the China Postdoctoral Science Foundation (20100470243), an Intramural Grant from Institute of Pathogen Biology, the Chinese Academy of Medical Sciences (2010IPB107), Grant (2009ZX10004-303) Eleven-Fifth Mega-Scientific project on “Prevention and Treatment of AIDS, Viral Hepatitis and other Infectious Diseases” from PR China, and a Grant (2013ZX10004601-002) from the Twelve-Fifth Mega-Scientific project from the People’s Republic of China.
We thank the Center for Biological Imaging (CBI), Institute of Biophysics, Chinese Academy of Science for our Electron Microcopy work and we are grateful to Lei Sun for her help in making EM samples.
We thank Professor Jian Lin for generously providing the improved tandem fluorescent-tagged LC3B (mTagRFP-mWasabi-LC3B) plasmids. Many thanks to Dr Aimin Zhou for critically reading and revising the manuscript.
References
- 1.Ray Kim W. . Global epidemiology and burden of hepatitis C. Microbes Infect 2002; 4:1219 - 25; http://dx.doi.org/ 10.1016/S1286-4579(02)01649-0; PMID: 12467763 [DOI] [PubMed] [Google Scholar]
- 2.Moradpour D, Penin F, Rice CM. . Replication of hepatitis C virus. Nat Rev Microbiol 2007; 5:453 - 63; http://dx.doi.org/ 10.1038/nrmicro1645; PMID: 17487147 [DOI] [PubMed] [Google Scholar]
- 3.Gale M Jr., Foy EM. . Evasion of intracellular host defence by hepatitis C virus. Nature 2005; 436:939 - 45; http://dx.doi.org/ 10.1038/nature04078; PMID: 16107833 [DOI] [PubMed] [Google Scholar]
- 4.Egger D, Wölk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. . Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol 2002; 76:5974 - 84; http://dx.doi.org/ 10.1128/JVI.76.12.5974-5984.2002; PMID: 12021330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. . Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol 2003; 77:5487 - 92; http://dx.doi.org/ 10.1128/JVI.77.9.5487-5492.2003; PMID: 12692249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreux M, Gastaminza P, Wieland SF, Chisari FV. . The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A 2009; 106:14046 - 51; http://dx.doi.org/ 10.1073/pnas.0907344106; PMID: 19666601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima N. . Autophagy: process and function. Genes Dev 2007; 21:2861 - 73; http://dx.doi.org/ 10.1101/gad.1599207; PMID: 18006683 [DOI] [PubMed] [Google Scholar]
- 8.Klein KA, Jackson WT. . Human rhinovirus 2 induces the autophagic pathway and replicates more efficiently in autophagic cells. J Virol 2011; 85:9651 - 4; http://dx.doi.org/ 10.1128/JVI.00316-11; PMID: 21752910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guévin C, Manna D, Bélanger C, Konan KV, Mak P, Labonté P. . Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology 2010; 405:1 - 7; http://dx.doi.org/ 10.1016/j.virol.2010.05.032; PMID: 20580051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor MP, Jackson WT. . Viruses and arrested autophagosome development. Autophagy 2009; 5:870 - 1; PMID: 19502808 [DOI] [PubMed] [Google Scholar]
- 11.Reggiori F. . 1. Membrane origin for autophagy. Curr Top Dev Biol 2006; 74:1 - 30; http://dx.doi.org/ 10.1016/S0070-2153(06)74001-7; PMID: 16860663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008; 4:151 - 75; PMID: 18188003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima N, Yoshimori T. . How to interpret LC3 immunoblotting. Autophagy 2007; 3:542 - 5; PMID: 17611390 [DOI] [PubMed] [Google Scholar]
- 14.Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. . Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol 2008; 82:2241 - 9; http://dx.doi.org/ 10.1128/JVI.02093-07; PMID: 18077704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. . Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 2008; 48:1054 - 61; http://dx.doi.org/ 10.1002/hep.22464; PMID: 18688877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sir D, Liang C, Chen WL, Jung JU, Ou JH. . Perturbation of autophagic pathway by hepatitis C virus. Autophagy 2008; 4:830 - 1; PMID: 18635950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estrabaud E, De Muynck S, Asselah T. . Activation of unfolded protein response and autophagy during HCV infection modulates innate immune response. J Hepatol 2011; 55:1150 - 3; http://dx.doi.org/ 10.1016/j.jhep.2011.04.025; PMID: 21723841 [DOI] [PubMed] [Google Scholar]
- 18.Ke PY, Chen SS. . Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest 2011; 121:37 - 56; http://dx.doi.org/ 10.1172/JCI41474; PMID: 21135505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang H, Kang R, Wang J, Luo G, Yang W, Zhao Z. . Hepatitis C virus inhibits AKT-tuberous sclerosis complex (TSC), the mechanistic target of rapamycin (MTOR) pathway, through endoplasmic reticulum stress to induce autophagy. Autophagy 2013; 9:175 - 95; http://dx.doi.org/ 10.4161/auto.22791; PMID: 23169238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su WC, Chao TC, Huang YL, Weng SC, Jeng KS, Lai MM. . Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J Virol 2011; 85:10561 - 71; http://dx.doi.org/ 10.1128/JVI.00173-11; PMID: 21835792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shrivastava S, Bhanja Chowdhury J, Steele R, Ray R, Ray RB. . Hepatitis C virus upregulates Beclin1 for induction of autophagy and activates mTOR signaling. J Virol 2012; 86:8705 - 12; http://dx.doi.org/ 10.1128/JVI.00616-12; PMID: 22674982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siavoshian S, Abraham JD, Thumann C, Kieny MP, Schuster C. . Hepatitis C virus core, NS3, NS5A, NS5B proteins induce apoptosis in mature dendritic cells. J Med Virol 2005; 75:402 - 11; http://dx.doi.org/ 10.1002/jmv.20283; PMID: 15648076 [DOI] [PubMed] [Google Scholar]
- 23.Ray RB, Meyer K, Ray R. . Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology 2000; 271:197 - 204; http://dx.doi.org/ 10.1006/viro.2000.0295; PMID: 10814584 [DOI] [PubMed] [Google Scholar]
- 24.Benali-Furet NL, Chami M, Houel L, De Giorgi F, Vernejoul F, Lagorce D, Buscail L, Bartenschlager R, Ichas F, Rizzuto R, et al. . Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene 2005; 24:4921 - 33; http://dx.doi.org/ 10.1038/sj.onc.1208673; PMID: 15897896 [DOI] [PubMed] [Google Scholar]
- 25.Coller KE, Heaton NS, Berger KL, Cooper JD, Saunders JL, Randall G. . Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog 2012; 8:e1002466; http://dx.doi.org/ 10.1371/journal.ppat.1002466; PMID: 22241992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. . Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology 2002; 122:366 - 75; http://dx.doi.org/ 10.1053/gast.2002.30983; PMID: 11832451 [DOI] [PubMed] [Google Scholar]
- 27.McLauchlan J, Lemberg MK, Hope G, Martoglio B. . Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J 2002; 21:3980 - 8; http://dx.doi.org/ 10.1093/emboj/cdf414; PMID: 12145199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. . XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001; 107:881 - 91; http://dx.doi.org/ 10.1016/S0092-8674(01)00611-0; PMID: 11779464 [DOI] [PubMed] [Google Scholar]
- 29.Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, Pires I, Hammond E, Ragoussis I, Harris AL. . Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 2010; 29:4424 - 35; http://dx.doi.org/ 10.1038/onc.2010.191; PMID: 20514020 [DOI] [PubMed] [Google Scholar]
- 30.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. . ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ 2007; 14:230 - 9; http://dx.doi.org/ 10.1038/sj.cdd.4401984; PMID: 16794605 [DOI] [PubMed] [Google Scholar]
- 31.Margariti A, Li H, Chen T, Martin D, Vizcay-Barrena G, Alam S, Karamariti E, Xiao Q, Zampetaki A, Zhang Z, et al. . XBP1 mRNA splicing triggers an autophagic response in endothelial cells through BECLIN-1 transcriptional activation. J Biol Chem 2013; 288:859 - 72; http://dx.doi.org/ 10.1074/jbc.M112.412783; PMID: 23184933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. . p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171:603 - 14; http://dx.doi.org/ 10.1083/jcb.200507002; PMID: 16286508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. . LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720 - 8; http://dx.doi.org/ 10.1093/emboj/19.21.5720; PMID: 11060023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. . Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 1998; 23:33 - 42; http://dx.doi.org/ 10.1247/csf.23.33; PMID: 9639028 [DOI] [PubMed] [Google Scholar]
- 35.Zhou C, Zhong W, Zhou J, Sheng F, Fang Z, Wei Y, Chen Y, Deng X, Xia B, Lin J. . Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy 2012; 8:1215 - 26; http://dx.doi.org/ 10.4161/auto.20284; PMID: 22647982 [DOI] [PubMed] [Google Scholar]
- 36.Verfaillie T, Salazar M, Velasco G, Agostinis P. . Linking ER Stress to Autophagy: Potential Implications for Cancer Therapy. Int J Cell Biol 2010; 2010:930509; http://dx.doi.org/ 10.1155/2010/930509; PMID: 20145727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y, Brewer JW, Diehl JA, Hendershot LM. . Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol 2002; 318:1351 - 65; http://dx.doi.org/ 10.1016/S0022-2836(02)00234-6; PMID: 12083523 [DOI] [PubMed] [Google Scholar]
- 38.Zhang N, Chen Y, Jiang R, Li E, Chen X, Xi Z, Guo Y, Liu X, Zhou Y, Che Y, et al. . PARP and RIP 1 are required for autophagy induced by 11′-deoxyverticillin A, which precedes caspase-dependent apoptosis. Autophagy 2011; 7:598 - 612; http://dx.doi.org/ 10.4161/auto.7.6.15103; PMID: 21460625 [DOI] [PubMed] [Google Scholar]
- 39.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al. . Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J 2007; 26:2527 - 39; http://dx.doi.org/ 10.1038/sj.emboj.7601689; PMID: 17446862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohd-Ismail NK, Deng L, Sukumaran SK, Yu VC, Hotta H, Tan YJ. . The hepatitis C virus core protein contains a BH3 domain that regulates apoptosis through specific interaction with human Mcl-1. J Virol 2009; 83:9993 - 10006; http://dx.doi.org/ 10.1128/JVI.00509-09; PMID: 19605477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, et al. . The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 2010; 120:127 - 41; http://dx.doi.org/ 10.1172/JCI40027; PMID: 20038797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pike LR, Phadwal K, Simon AK, Harris AL. . ATF4 orchestrates a program of BH3-only protein expression in severe hypoxia. Mol Biol Rep 2012; 39:10811 - 22; http://dx.doi.org/ 10.1007/s11033-012-1975-3; PMID: 23090478 [DOI] [PubMed] [Google Scholar]
- 43.Ravikumar B, Imarisio S, Sarkar S, O’Kane CJ, Rubinsztein DC. . Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci 2008; 121:1649 - 60; http://dx.doi.org/ 10.1242/jcs.025726; PMID: 18430781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sengupta A, Molkentin JD, Yutzey KE. . FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem 2009; 284:28319 - 31; http://dx.doi.org/ 10.1074/jbc.M109.024406; PMID: 19696026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh AP, Klocke BJ, Ballestas ME, Roth KA. . CHOP potentially co-operates with FOXO3a in neuronal cells to regulate PUMA and BIM expression in response to ER stress. PLoS One 2012; 7:e39586; http://dx.doi.org/ 10.1371/journal.pone.0039586; PMID: 22761832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, et al. . FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 2007; 6:458 - 71; http://dx.doi.org/ 10.1016/j.cmet.2007.11.001; PMID: 18054315 [DOI] [PubMed] [Google Scholar]
- 47.Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. . Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol 2009; 83:2011 - 4; http://dx.doi.org/ 10.1128/JVI.01888-08; PMID: 19052094 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.