Abstract
Maintenance of organellar quality and quantity is critical for cellular homeostasis and adaptation to variable environments. Emerging evidence demonstrates that this kind of control is achieved by selective elimination of organelles via autophagy, termed organellophagy. Organellophagy consists of three key steps: induction, cargo tagging, and sequestration, which involve signaling pathways, organellar landmark molecules, and core autophagy-related proteins, respectively. In addition, posttranslational modifications such as phosphorylation and ubiquitination play important roles in recruiting and tailoring the autophagy machinery to each organelle. The basic principles underlying organellophagy are conserved from yeast to mammals, highlighting its biological relevance in eukaryotic cells.
Introduction
Organelles are fundamental subunits of eukaryotic cells that possess structurally and functionally distinct characteristics that allow them to perform unique activities crucial for viability. It is thus a matter of the utmost importance for cells to maintain organellar quality and integrity. In addition, cells modulate the quantity of organelles in order to balance organellar activities and cellular demands, which can act as an adaptive mechanism to diverse environmental changes. Both dysfunctional and surplus organelles are cleared from cells through autophagy, a widely conserved self-eating process by which cytoplasmic constituents are sequestered as cargoes by intracellular membranes that fuse with lysosomes for hydrolytic breakdown (Mizushima and Komatsu, 2011; Mizushima et al., 2011; Weidberg et al., 2011).
Although autophagy has primarily been recognized as a nonselective degradation pathway, recent studies reveal that it also plays a vital role in digesting specific cargoes such as proteins and organelles (Mizushima, 2011; Suzuki, 2013). The latter process, termed selective autophagy, includes the following three critical stages: first, signaling from degradation cues induces downstream events specific for a particular target; second, regulation of landmark molecules that tag the target as disposable cargo; third, assembly of core autophagy-related (Atg) proteins to sequester the cargo. In many cases, malfunction in or decreased cellular metabolism related to a protein or organelle leads to expression and activation of a landmark molecule. Core Atg proteins then localize to the cargo via direct or indirect interactions with the landmark molecule and ultimately mediate selective autophagy.
In this short review, we summarize recent findings on organellophagy, autophagy-related pathways selective for organelles such as the peroxisome, mitochondrion, lipid droplet (structure surrounded by a phospholipid monolayer), lysosome, nucleus, ER, and even nonmembraneous structures like the ribosome. Despite the diversity of their degradation cues and landmark molecules, organellophagy seems to be regulated by common basic principles involving protein phosphorylation and ubiquitination. In particular, these two major posttranslational modifications promote targeting of core Atg proteins to the organellar surface. Defects in several organellophagy pathways are associated with various disorders including renal injury, neurodegeneration, obesity, and atherosclerosis (Mizushima and Komatsu, 2011), underscoring their physiological significance in health and disease.
Modes of autophagy in organellophagy
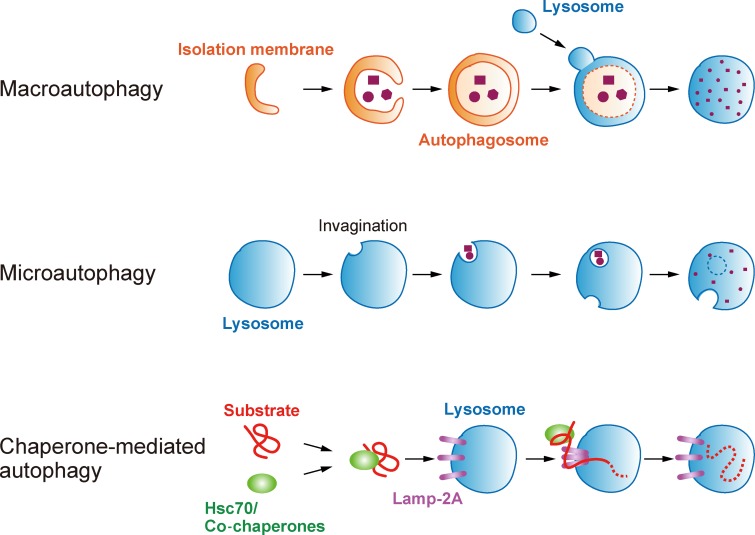
Three morphologically distinct modes of autophagic processes have so far been defined: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA; Fig. 1; Mizushima and Komatsu, 2011; Li et al., 2012; Cuervo and Wong, 2014; Feng et al., 2014). Macro- and microautophagy are conserved from yeast to humans, whereas CMA has been found only in mammals. Upon macroautophagy induction, newly formed double membrane–bound structures enclose proteins and organelles, eventually generating mature vesicles, called autophagosomes. Core Atg proteins play essential roles in autophagosome formation. The engulfed cargoes are then mixed with the lysosomal hydrolases via autophagosome–lysosome fusion and digested into small molecules for recycling. During microautophagy, the lysosomal membrane invaginates to sequester proteins and organelles. In some cases, accompanying membrane structures function in closure of the cargoes, which requires core Atg proteins. The lysosomal lipase then digests the internalized vesicles, leading to breakdown of the cargoes by hydrolases. By contrast, CMA recruits specific protein substrates associated with the molecular chaperone Hsc70 to lysosomes and translocates the substrates one by one into the lysosomal lumen through the receptor protein Lamp-2A in a manner independent on core Atg proteins. Unlike macro- and microautophagy, CMA has been suggested to degrade only proteins but not whole organelles.
Figure 1.
Three distinct modes of autophagy. In macroautophagy, newly generated cup-shaped structures, called isolation membranes, expand to surround cytoplasmic components. The two edges of isolation membranes then fuse to form double membrane–bound autophagosomes. Subsequently, autophagosomes fuse to lysosomes, and the engulfed cargoes are digested by hydrolytic enzymes. In microautophagy, invagination of the lysosomal membrane occurs to sequester proteins and organelles in the cytosol. The resulting vesicular structures are then pinched off and released into the lysosomal lumen for digestion. In chaperone-mediated autophagy (CMA), the Hsc70/co-chaperone complex delivers specific substrate proteins to lysosomes. The substrate polypeptides are then translocated one by one through the lysosomal membrane protein Lamp-2A and digested in the lysosomal lumen. Macro- and microautophagy are conserved from yeast to humans, whereas CMA has been found only in mammals. Unlike macro- and microautophagy, CMA has been suggested to degrade only proteins but not whole organelles.
Morphological classification of organelle-specific autophagy in yeast and mammals is summarized in Table 1. Both macro- and microautophagy mediate selective elimination of the peroxisome (pexophagy; Manjithaya et al., 2010b; Oku and Sakai, 2010), mitochondrion (mitophagy; Ashrafi and Schwarz, 2013; Feng et al., 2013), lipid droplet (lipophagy; Liu and Czaja, 2013), and nucleus (nucleophagy; Mijaljica and Devenish, 2013). These degradation pathways appear to be conserved from yeast to humans. Macroautophagy-related turnover processes specific for lysosome (lysophagy; Hung et al., 2013; Maejima et al., 2013) and ER (reticulophagy/ER-phagy; Bernales et al., 2006) have been found in mammals and yeast, respectively. Although whether ribosome degradation in yeast occurs via macro- or microautophagy remains to be clarified, it seems to be a selective event (ribophagy) because ribosomal subunits are degraded significantly faster than other cytosolic proteins in an autophagy-dependent fashion (Kraft et al., 2008). To date, there has been no evidence suggesting selective degradation of the Golgi apparatus (golgiphagy).
Table 1.
Classification of organellophagy
| Cargo organelle | Macroautophagy | Microautophagy | ||
| Yeast | Human | Yeast | Human | |
| Peroxisome | ✓ | ✓ | ✓ | ND |
| Mitochondrion | ✓ | ✓ | ✓ | ND |
| Lipid droplet | ND | ✓ | ✓ | ND |
| Nucleus | ND | ✓ | ✓ | ND |
| Lysosome | ND | ✓ | ND | ND |
| Endoplasmic reticulum | ✓ | ND | ND | ND |
| Ribosome | ✓a | ND | ND | ND |
ND, not determined.
Ribophagy seems to depend on macroautophagy rather than microautophagy, although this has not yet been confirmed morphologically.
Common features of organellophagy
Studies on pexophagy and mitophagy have extensively explored molecular mechanisms underlying cargo recognition, implicating two common types, receptor- and ubiquitin-mediated processes (Fig. 2). Both types involve protein phosphorylation that activates or inactivates downstream events.
Figure 2.
Two common mechanisms of organellophagy. Molecular mechanisms underlying cargo recognition in pexophagy and mitophagy have extensively been explored, including two common types, receptor- and ubiquitin-mediated processes. Both types involve protein phosphorylation that activates or inactivates their downstream events. In the receptor-mediated process, membrane-anchored or peripherally associated receptors on the organellar surface interact with Atg8/LC3, ubiquitin-like proteins conjugated to the phospholipid phosphatidylethanolamine and localized to autophagosomes, and Atg11/Atg17, scaffold proteins required for core Atg protein assembly. Protein kinases phosphorylate receptors and regulate receptor interactions with Atg8/LC3 and Atg11/Atg17. In the ubiquitin-mediated process, E3 ubiquitin ligases target to the organelle and ubiquitinate proteins on the organellar surface. The ubiquitin chains then interact with LC3-binding adaptors such as p62/NBR1, or unknown factors (X) that may promote core Atg protein assembly. Protein kinases phosphorylate the ubiquitin ligases and promote targeting and activation of the E3 enzymes.
In the receptor-mediated process, specific proteins membrane-anchored or tightly associated on the organellar surface interact directly, or indirectly via adaptor proteins, with Atg8 (LC3, GABARAP, and GATE-16 in mammalian cells), a highly conserved ubiquitin-like protein essential for all autophagy-related pathways (Shpilka et al., 2011; Rogov et al., 2014; Wild et al., 2014). Notably, receptor proteins contain tetrapeptide consensus sequences called Atg8 family–interacting motif (AIM) and LC3-interacting region (LIR) that consist of W/YxxI/L/V and W/F/YxxL/I/V, respectively (Noda et al., 2010; Birgisdottir et al., 2013). AIM/LIR directly associates with Atg8/LC3 through the side chains of their conserved residues bound deeply into the hydrophobic pocket of Atg8/LC3. Mutations in AIM and LIR impair degradation of cargo organelles, suggesting the significance of these interactions. Because Atg8 is covalently linked to the phospholipid phosphatidylethanolamine and localized predominantly to autophagosomes, the receptor–Atg8/LC3 interactions could assist generation and expansion of cup-shaped structures called isolation membranes surrounding cargo organelles. In yeast, pexophagy and mitophagy receptors also interact with Atg11 or Atg17, scaffold proteins that serve as platforms for core Atg protein assembly (Farré et al., 2008; Kanki et al., 2009; Okamoto et al., 2009; Motley et al., 2012). Importantly, protein kinases and phosphatases modify receptors and appear to play regulatory roles in stabilizing or destabilizing the interactions of receptor proteins with Atg8/LC3 and Atg11/Atg17 (Farré et al., 2008, 2013; Novak et al., 2010; Aoki et al., 2011; Kondo-Okamoto et al., 2012; Liu et al., 2012; Kanki et al., 2013; Zhu et al., 2013).
In the ubiquitin-mediated process, peripheral and/or membrane-anchored proteins on the surface of cargo organelles are ubiquitinated by specific E3 ligases (Shaid et al., 2013). These ubiquitin chains act as “degradation tags” recognized by soluble adaptor proteins such as p62 and NBR1 that also interact with LC3 (Johansen and Lamark, 2011). Targeting of other core Atg proteins to these cargo organelles seems to be independent of p62 and LC3 (Itakura et al., 2012), which may be mediated directly by ubiquitin, or indirectly via unknown ubiquitin-binding proteins. In some cases, mitophagy-specific E3 ligases are regulated by phosphorylation. For example, the protein kinase PINK1 phosphorylates the ubiquitin E3 ligase Parkin to promote mitophagy (Kondapalli et al., 2012; Shiba-Fukushima et al., 2012; Iguchi et al., 2013). This type of mitophagy has so far been found in mammals, but not in yeast. Finally, it should be noted that the receptor- and ubiquitin-mediated processes are not mutually exclusive, as the LC3 receptor Pex14 is also involved in the ubiquitin/NBR1-mediated pexophagy in mammalian cells (Deosaran et al., 2013).
Pexophagy
In response to changes in the intra- and extracellular environments, peroxisome number dynamically increases or decreases in order to maintain the appropriate levels of the metabolic reactions including fatty acid oxidation and H2O2 detoxification (Smith and Aitchison, 2013). For example, the methylotrophic yeasts Pichia pastoris and Hansenula polymorpha can proliferate large peroxisome clusters when they grow in media containing methanol as the sole carbon source (van der Klei et al., 2006). Pexophagy is then drastically triggered upon a shift from methanol to glucose or ethanol media in which the peroxisomal metabolism is not critical for cell growth and viability (Manjithaya et al., 2010b; Oku and Sakai, 2010). Thus, molecular mechanisms underlying the selectivity of pexophagy have mostly been uncovered in these methylotrophic yeasts.
Macro- and microautophagy mediate pexophagy (macro- and micropexophagy, respectively) in P. pastoris that requires the soluble receptor protein Atg30 that interacts with Pex3 and Pex14, two peroxisomal membrane proteins, and recruits Atg8, Atg11, and Atg17 to the surface of peroxisomes (Farré et al., 2008, 2013). Similarly, Atg36 acts as a soluble receptor protein in the budding yeast Saccharomyces cerevisiae, localizes to the peroxisomal surface via Pex3, and binds Atg8 and Atg11 to promote macropexophagy (Motley et al., 2012; Farré et al., 2013). Interestingly, both Atg30 and Atg36 contain AIMs flanked with putative phosphoserine residues (Farré et al., 2013). These amino acids modified by unknown kinase(s) may stabilize the Atg30–Atg8 and Atg36–Atg8 interactions. Additional phosphorylation sites are also required for binding of Atg30 and Atg36 to Atg11 (Farré et al., 2008, 2013). In S. cerevisiae, the MAPK cascade Mid2–Pkc1–Bck1–Mkk1/Mkk2–Slt2 is necessary for peroxisome degradation, but not for pexophagosome formation (Manjithaya et al., 2010a; Mao et al., 2011). Hence, Slt2 is unlikely to regulate the interactions of Atg36 with Atg8 and Atg11. Nonetheless, pexophagy in S. cerevisiae is enhanced in a set of mutants containing dysfunctional peroxisomes through yet-uncharacterized modifications of Atg36 (Nuttall et al., 2014). Despite the common role in recruiting the pexophagy receptors to the peroxisomal surface in P. pastoris and S. cerevisiae, Pex3 in H. polymorpha is degraded via the ubiquitin–proteasome pathway in order to initiate macropexophagy by unknown mechanisms (Bellu et al., 2002; Williams and van der Klei, 2013), suggesting the diversity of peroxisome turnover mechanisms among yeast species. It should also be noted that dynamin-related GTPases, Dnm1 and Vps1, target to peroxisomes, and promote peroxisomal fission, which is a critical step before pexophagy in H. polymorpha and S. cerevisiae (Manivannan et al., 2013; Mao et al., 2014).
A study using Chinese hamster ovary cells demonstrates that pexophagy can be induced upon a shift from starvation to nutrient-rich media (Hara-Kuge and Fujiki, 2008). Under this condition, Pex14 interacts with LC3-II, a phosphatidylethanolamine-conjugated form anchored on autophagosomes (Hara-Kuge and Fujiki, 2008). When monoubiquitinated peroxisomal membrane proteins are overexpressed in COS-7 cells, pexophagy occurs in a manner dependent on p62 (Kim et al., 2008). More recently, down-regulation of either p62 or NBR1 has been shown to suppress degradation of peroxisomes in HeLa cells (Deosaran et al., 2013). Overexpression of NBR1, but not p62, can facilitate pexophagy through its LIR, coiled-coil domain (for homo-oligomerization), JUBA domain (for membrane association), and UBA domain (for ubiquitin binding; Deosaran et al., 2013). Notably, an NBR1 mutant defective in p62 interaction is not fully functional for pexophagy (Deosaran et al., 2013). Thus, p62 may not be a major adaptor, but still contributes to pexophagy in cooperation with NBR1. Nonetheless, it seems likely that ubiquitination of peroxisomal proteins promotes recruitment of LC3 to peroxisomes via p62 and NBR1, ultimately leading to pexophagy in mammalian cells. How the core factors of the autophagy machinery are targeted to peroxisomes remains to be clarified.
Mitophagy
Mitochondria are major organelles that are platforms for many important processes including energy conversion, calcium homeostasis, and programmed cell death (Nunnari and Suomalainen, 2012). These organelles concomitantly generate reactive oxygen species (ROS) as hazardous byproducts during respiration. Consequently, accumulation of ROS causes mitochondrial dysfunction. Elimination of damaged mitochondria is therefore critical for cell homeostasis (Okamoto and Kondo-Okamoto, 2012). The other problem related to their energy metabolism is that cells need to maintain the balance between ATP supply and demand. Upon a shift from high to low energy consumption state, surplus mitochondria become vital targets for clearance (Okamoto and Kondo-Okamoto, 2012). Numerous studies demonstrate that mitophagy contributes to mitochondrial quality and quantity control, and that its selectivity is established via common mechanisms (Youle and Narendra, 2011; Jin and Youle, 2012; Narendra et al., 2012; Ashrafi and Schwarz, 2013; Feng et al., 2013).
In the yeast S. cerevisiae, the mitophagy receptor Atg32 is induced in response to oxidative stress and anchored on the surface of mitochondria with its N- and C-terminal regions exposed to the cytosol and mitochondrial intermembrane space (IMS), respectively (Fig. 3 A; Kanki et al., 2009; Okamoto et al., 2009). Atg32 contains an AIM near the N terminus that is embedded into the hydrophobic pocket of Atg8 (Okamoto et al., 2009; Kondo-Okamoto et al., 2012). The C-terminal coiled-coil domain of Atg11 physically associates with Atg32 via a consensus region following the AIM (Aoki et al., 2011). Notably, this Atg11-interacting region contains serine residues that appear to be modified directly by casein kinase-2 (CK2), a housekeeping protein kinase (Kanki et al., 2013). This posttranslational modification stabilizes Atg32–Atg11 interaction (Fig. 3 A; Aoki et al., 2011; Kondo-Okamoto et al., 2012). A recent study suggests that processing of the Atg32 C-terminal region by Yme1, a catalytic subunit of the mitochondrial inner membrane AAA protease facing the IMS, is important for Atg32–Atg11 interaction (Wang et al., 2013), yet the role of Yme1 in mitophagy is currently a matter of debate (Campbell and Thorsness, 1998; Welter et al., 2013). In addition to CK2, the MAPK cascades Wsc1–Pkc1–Bck1–Mkk1/2–Slt2 and Ssk1–Pbs2–Hog1 are important for mitophagy (Aoki et al., 2011; Mao et al., 2011). Phosphorylation of Atg32 depends on Hog1, but not Slt2, while Atg32 is not a substrate for Hog1 (Aoki et al., 2011). Atg1, a protein kinase essential for all autophagy-related processes, is also involved in Atg32 phosphorylation (Kondo-Okamoto et al., 2012), although the molecular function of this modification remains unclear.
Figure 3.
Models for mitophagy in yeast and mammalian cells. (A) Atg32-mediated mitophagy in S. cerevisiae. Under respiratory conditions, the mitophagy receptor Atg32 is induced in response to oxidative stress, targeted, and anchored to the mitochondrial surface. Atg32 recruits Atg8 and Atg11 to mitochondria via distinct domains. CK2 phosphorylates Atg32 to stabilize the interaction between Atg32 and Atg11. This tertiary complex and core Atg proteins cooperatively generate isolation membranes to sequester mitochondria. The protein kinases Slt2 and Hog1 are also critical for mitophagy in yeast, although their targets remain unknown. (B) FUNDC1-mediated mitophagy in mammals. Under normoxic conditions, the mitochondrial outer membrane protein FUNDC1 is phosphorylated by Src and CK2, thereby preventing LC3 binding. Upon hypoxia, the expression of Src is strongly suppressed, and the protein phosphatase PGAM5 dephosphorylates FUNDC1 and promotes LC3 binding. In addition, ULK1, a mammalian Atg1 kinase homologue, interacts with FUNDC1 and phosphorylates the mitophagy receptor. This posttranslational modification also stabilizes the interaction between FUNDC1 and LC3. (C) PINK1/Parkin-mediated mitophagy in mammals. When targeted to healthy mitochondria, PINK1 is partially translocated across the mitochondrial membranes, proteolytically processed, released back to the cytosol, and rapidly degraded. In cells containing damaged mitochondria, PINK1 is stalled in the outer membrane and associated with the TOM complex. Two molecules of PINK1 undergo self-activation via autophosphorylation. Active PINK1 then phosphorylates Parkin and stabilizes the E3 ligase on the surface of mitochondria. Mitochondria-associated Parkin promotes ubiquitination of multiple substrates, ultimately leading to LC3 and p62/NBR1 recruitment and core Atg protein assembly. Ubiquitin chains and these proteins are bridged by an unknown factor (X).
Similar to Atg32-mediated mitophagy in yeast, three mitochondria-anchored receptors, NIX, BNIP3, and FUNDC1, promote autophagic degradation selective for mitochondria in mammalian cells (Schweers et al., 2007; Sandoval et al., 2008; Zhang et al., 2008; Liu et al., 2012). All three proteins contain the LIR consensus sequences that are important for their mitophagy activities (Novak et al., 2010; Liu et al., 2012; Zhu et al., 2013). NIX is highly induced during reticulocyte maturation and interacts with LC3 and GABARAP (Schweers et al., 2007; Novak et al., 2010). BNIP3 is strongly expressed in response to hypoxia and activated by reoxygenation (Zhang et al., 2008; Zhu et al., 2013). Notably, phosphorylation of serine residues near the BNIP3 LIR is crucial for LC3 and GATE-16 binding, and efficient mitophagy (Zhu et al., 2013). Kinases regulating the BNIP3 LIR are currently unknown. Although FUNDC1 is constitutively expressed under normoxic conditions, the tyrosine residue of the LIR is phosphorylated by the Src family kinase, which prevents LC3 binding and mitophagy (Fig. 3 B; Liu et al., 2012). Strikingly, hypoxia strongly suppresses Src expression, leading to dephosphorylation of the FUNDC1 LIR by unknown protein phosphatases, subsequent binding of LC3, and ultimate activation of mitophagy (Fig. 3 B; Liu et al., 2012). Furthermore, a serine residue near the LIR is phosphorylated by CK2 under normal conditions, and conversely dephosphorylated by the mitochondrial phosphatase PGAM5 upon hypoxic stress and mitochondrial membrane potential (ΔΨm) dissipation, leading to efficient LC3 binding and mitophagy activation (Fig. 3 B; Chen et al., 2014). Another recent study reveals that ULK1, a mammalian Atg1 kinase, targets to mitochondria via interaction with FUNDC1 and phosphorylates the mitophagy receptor to stabilize the FUNDC1–LC3 interaction (Fig. 3 B; Wu et al., 2014). It is not certain whether these mitophagy receptors could also recruit core Atg proteins to mitochondria.
In addition to the receptor-driven pathways described above, mammalian cells use the ubiquitin-dependent processes to promote degradation of mitochondria. The best known is the mitophagy involving PINK1, a mitochondrial protein kinase, and Parkin, a cytosolic E3 ubiquitin ligase, two closely related causal factors for autosomal-recessive familial Parkinsonism (Kitada et al., 1998; Valente et al., 2004). When targeted to healthy mitochondria, PINK1 is partially translocated across the mitochondrial outer and inner membranes, cleaved by several enzymes including the matrix-localized mitochondrial-processing peptidase MPP and the inner membrane protease PARL, released back into the cytosol, and rapidly degraded by the proteasome via the N-end rule pathway (Fig. 3 C; Jin et al., 2010; Matsuda et al., 2010; Narendra et al., 2010b; Deas et al., 2011; Meissner et al., 2011; Shi et al., 2011; Greene et al., 2012; Yamano and Youle, 2013). In this situation, Parkin is dispersed throughout the cytosol as an inactive form and is not stably associated with mitochondria (Narendra et al., 2008, 2010b; Matsuda et al., 2010; Chaugule et al., 2011; Chew et al., 2011) (Fig. 3 C). As a result, mitophagy is mostly suppressed in normally respiring cells. Upon mitochondrial dysfunction such as ΔΨm dissipation, PINK1 is stalled in the outer membrane and anchored on the surface of mitochondria (Kawajiri et al., 2010; Matsuda et al., 2010; Narendra et al., 2010b; Rakovic et al., 2010). Subsequently, PINK1 forms a supermolecular complex together with the translocase of the outer membrane (TOM) components (Fig. 3 C; Lazarou et al., 2012; Okatsu et al., 2013). In this supermolecular complex, two molecules of PINK1 undergo intermolecular phosphorylation (Okatsu et al., 2013). PINK1 complex formation is correlated well with its autophosphorylation, which is prerequisite for recruitment of Parkin to damaged mitochondria (Okatsu et al., 2012, 2013). Through these processes, PINK1 becomes more active, efficiently phosphorylating a serine residue of the Parkin ubiquitin-like (Ubl) domain (Fig. 3 C; Kondapalli et al., 2012; Shiba-Fukushima et al., 2012; Iguchi et al., 2013). Phosphorylation of the Ubl domain probably induces a conformational change, at least to some extent, resulting in Parkin self-association and ubiquitin-thioester formation at the RING2 domain, which is essential for the E3 ligase activity (Chaugule et al., 2011; Iguchi et al., 2013; Lazarou et al., 2013; Spratt et al., 2013; Zheng and Hunter, 2013). Importantly, these PINK1-mediated events are consistent with the mechanisms of Parkin inactive–active state transition revealed by recent structural studies (Riley et al., 2013; Trempe et al., 2013; Wauer and Komander, 2013).
Whether specific Parkin targets are required for mitophagy remains controversial (Geisler et al., 2010; Lee et al., 2010; Narendra et al., 2010a; Okatsu et al., 2010). A high-throughput analysis on the Parkin-dependent ubiquitylome demonstrates numerous targets on the surface of depolarized mitochondria including mitofusins (Mfns), large GTPases required for mitochondrial fusion (Sarraf et al., 2013). Parkin is responsible for degradation of mitofusins, preventing refusion of damaged mitochondria and assisting subsequent mitophagy (Gegg et al., 2010; Tanaka et al., 2010; Glauser et al., 2011; Rakovic et al., 2011). The role of Mfns in the PINK1/Parkin pathway is rather intricate, as it has been reported that Mfn2 serves as a Parkin receptor to promote mitochondrial degradation in mouse cardiomyocytes (Chen and Dorn, 2013). Notably, genome-wide siRNA screens uncover additional factors for PINK1/Parkin-mediated mitophagy, including TOMM7, a component of the TOM complex, as essential for stabilizing PINK1 on the outer membrane of depolarized mitochondria (Hasson et al., 2013). Rab GTPase-activating proteins have recently been shown to interact with Fis1, a tail-anchored protein, and LC3/GABARAP family members on the surface of mitochondria where they promote formation of autophagosomes by regulating Rab7 activity during PINK1/Parkin-mediated mitophagy (Yamano et al., 2014). Very recently, three studies demonstrate that PINK1 phosphorylates ubiquitin to activate Parkin in a manner similar to Parkin self-activation via the phosphorylated Ubl domain (Kane et al., 2014; Kazlauskaite et al., 2014; Koyano et al., 2014). Whether ubiquitin/LC3-binding adaptors such as p62 and NBR1 are necessary for the PINK1/Parkin pathway, and how core Atg proteins are recruited to damaged mitochondria remain inconclusive.
In addition to Parkin, two ubiquitin E3 ligases, Gp78 and SMURF1, have been implicated in mammalian mitophagy. The Gp78-mediated process depends on Mfn1, but does not require Parkin (Fu et al., 2013). In contrast, SMURF1 is required for the PINK1/Parkin pathway (Orvedahl et al., 2011). Molecular mechanisms underlying Gp78 and SMURF1 functions have not yet been elucidated.
Lipophagy
Lipid droplets (LDs) consist of a core mainly containing triglycerides and sterol esters surrounded by a phospholipid monolayer and associated with various proteins. They are dynamic organelles that change their size and number in response to diverse conditions, and play key roles in lipid storage and metabolism (Walther and Farese, 2012). In addition to the cytosolic lipases, lysosomal hydrolases catabolize LDs that are transported via lipophagy (Liu and Czaja, 2013). In yeast, LDs are degraded through microautophagy (van Zutphen et al., 2014). By contrast, lipophagy occurs via macroautophagy in mouse hepatocytes and human enterocytes (Singh et al., 2009; Khaldoun et al., 2014). How the selectivity of lipophagy is established needs future studies.
Nucleophagy
Accumulating evidence suggests that portions of the nucleus, nucleus-derived components, or even a whole nucleus, are degraded by selective autophagy in a variety of eukaryotes (Mijaljica and Devenish, 2013). These processes, defined as nucleophagy, can be induced under starvation and other stress conditions such as DNA damage and cell cycle arrest (Mijaljica and Devenish, 2013). In the yeast S. cerevisiae, small teardrop-shaped parts of the nucleus are engulfed by the vacuole, a lytic organelle equivalent to the lysosome, at nucleus–vacuole (NV) junctions (Roberts et al., 2003). This event, termed piecemeal microautophagy of the nucleus (PMN), is induced soon after nutrient deprivation (Roberts et al., 2003). Formation of NV junctions requires Nvj1 in the nuclear envelope and Vac8 on the vacuolar membrane, two physically associated proteins that establish the vacuolar diffusion barrier, invaginate NV junctions, and generate PMN vesicles in a manner dependent on the vacuolar electrochemical gradient and lipid-modifying enzymes (Roberts et al., 2003; Dawaliby and Mayer, 2010). Atg11, Atg17, and other core Atg proteins are indispensable for PMN, as in the case of micropexophagy in the methylotrophic yeasts (Krick et al., 2008). After prolonged starvation, another type of nucleophagy also occurs through unknown mechanisms, which does not require Nvj1, Vac8, and Atg11 (Mijaljica et al., 2012).
In mammals, LC3- and several core Atg-positive structures containing nuclear components accumulate in close proximity to the nucleus in cells from nuclear envelopathies (Park et al., 2009). In addition, micronuclei, small structures containing displaced chromosomes or chromosome fragments efficiently generated in cells expose to genotoxic stress, are degraded via autophagy (Rello-Varona et al., 2012). These autophagic micronuclei are p62 positive and exhibit signs of nuclear envelope degradation and DNA damage (Rello-Varona et al., 2012). It has also been suggested that LC3-positive micronuclei represent vesicles containing DNA that has not been repaired (Erenpreisa et al., 2012). Whether macro- and microautophagy could mediate nucleophagy in mammals and how the selectivity is established remain to be clarified.
Lysophagy
Lysosomes are acidic organelles highly enriched with hydrolytic enzymes that digest macromolecules delivered via the endocytic and autophagic pathways. Recent studies demonstrate that lysosomal rupture causes release of hydrolases into the cytosol, ultimately leading to destruction of intracellular structures and functions (Boya and Kroemer, 2008). It is therefore conceivable that cells must use surveillance and quality control systems for lysosomes. Indeed, emerging evidence reveals that damaged lysosomes are selectively sequestered by macroautophagy in mammalian cells (Hung et al., 2013; Maejima et al., 2013). Lysophagy seems to be a ubiquitin-mediated process involving LC3 and p62, which could contribute to recovery of lysosomal activities (Maejima et al., 2013). How ubiquitin and core Atg proteins selectively target to damaged lysosomes awaits further investigations.
Reticulophagy/ER-phagy
ER membranes are most abundant in many cell types, and their lumens serve as major factories for protein folding and modification. Although macroautophagy in yeast under starvation conditions can nonselectively sequester the ER together with other cytoplasmic constituents, ER components are more enriched than cytosolic proteins in autophagic bodies, suggesting a selective feature of this ER turnover (Hamasaki et al., 2005). Strikingly, when yeast cells are challenged with protein folding stress, ER membrane stacks are densely enclosed in autophagosome-like structures (Bernales et al., 2006). Recently, a Ypt/Rab GTPase module containing Atg11 has been reported to regulate reticulophagy/ER-phagy in yeast (Lipatova et al., 2013). These observations raise the possibility that ER turnover occurs via unknown selective mechanisms.
Ribophagy
In yeast, a hallmark of starvation-induced, nonselective macroautophagy is that autophagic bodies in the vacuolar lumen contain myriad ribosomes (Takeshige et al., 1992). However, under the same conditions, ribosomal subunits are degraded faster than other cytosolic proteins (Kraft et al., 2008). Intriguingly, the Rsp5 ubiquitin ligase and the Ubp3/Bre5 ubiquitin protease are involved in this preferential ribosome turnover but not bulk autophagy, supporting the existence of ribophagy (Kraft and Peter, 2008). The Ubp3–Bre5 complex interacts with the AAA ATPase Cdc48 and the ubiquitin-binding Cdc48 adaptor Ufd3 that are also required for ribophagy (Ossareh-Nazari et al., 2010). Recently, the E3 ubiquitin ligase Ltn1 has been suggested to negatively regulate ribophagy through ubiquitinating Rpl25, a 60S ribosomal subunit protein, which is also de-ubiquitinated by Ubp3 in an antagonistic action (Ossareh-Nazari et al., 2014). Whether ribosomes are recognized as disposable cargoes via ubiquitin or unknown receptor(s), and how de-ubiquitination regulates ribophagy remain to be addressed.
During starvation-induced macroautophagy in mammalian cells, the timing of ribosomal degradation is different from those of other proteins and organelles, implying that bulk autophagy can even be intimately regulated in terms of cargo recognition and sequential activation (Kristensen et al., 2008).
Perspectives
Herein, we have highlighted recent progress in our understanding of organelle-specific autophagy pathways. Despite the diversity of their degradation cues and tags, the basic principles underlying organellophagy are similar among different organelles, and are likely to be universal in almost all eukaryotes. However, many of the landmark molecules for recruiting core Atg proteins are still missing, and the molecular details of organellophagy induction and termination are largely unknown.
The origin of autophagosomal membranes is a fundamental, ongoing issue for all autophagy-related processes in unicellular and multicellular eukaryotes (Lamb et al., 2013). Recent imaging studies reveal the ER–mitochondria contacts as autophagosome formation sites for autophagy in mammals (Hamasaki et al., 2013) and mitophagy in yeast (Böckler and Westermann, 2014), whereas others implicate ER exit sites and the ER–Golgi intermediate compartment involving COPII vesicles for autophagy in yeast and mammals (Ge et al., 2013; Graef et al., 2013; Suzuki et al., 2013). In COS-7 cells under starvation conditions, COPII vesicles seem to localize at ER–mitochondria contacts (Tan et al., 2013), raising the possibility that these autophagosome formation sites may not be mutually exclusive. Whether degradation of other organelles utilizes the aforementioned sites for formation of autophagosomes remains to be addressed.
Finally, the challenging attempts will be to decipher whether there is a cross talk between organelle biogenesis and degradation, and how the organellar quality and quantity control pathways regulate higher-order functions such as cell differentiation and development in multicellular organisms. Definitely, more stimulating discoveries are yet to come.
Acknowledgments
We apologize to the many colleagues whose work could not be cited because of space limitations, and hope the readers refer to the cited reviews for more information.
This work was supported in part by the Ministry of Education, Culture, Science, and Technology of Japan Grant-in-Aid for Challenging Exploratory Research grant (25650067), Scientific Research (B) (25291045), Scientific Research on Innovative Areas (26111513), and by grants from the Ono Medical Research Foundation.
The author declares no competing financial interest.
Footnotes
Abbreviations used in this paper:
- AIM
- Atg8 family–interacting motif
- Atg
- autophagy related
- CK-2
- casein kinase-2
- LIR
- LC3-interacting region
- Mfn
- mitofusin
- TOM
- translocase of the outer membrane
- Ubl
- ubiquitin-like
References
- Aoki Y., Kanki T., Hirota Y., Kurihara Y., Saigusa T., Uchiumi T., Kang D. 2011. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol. Biol. Cell. 22:3206–3217 10.1091/mbc.E11-02-0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi G., Schwarz T.L. 2013. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20:31–42 10.1038/cdd.2012.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellu A.R., Salomons F.A., Kiel J.A., Veenhuis M., Van Der Klei I.J. 2002. Removal of Pex3p is an important initial stage in selective peroxisome degradation in Hansenula polymorpha. J. Biol. Chem. 277:42875–42880 10.1074/jbc.M205437200 [DOI] [PubMed] [Google Scholar]
- Bernales S., McDonald K.L., Walter P. 2006. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4:e423 10.1371/journal.pbio.0040423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir A.B., Lamark T., Johansen T. 2013. The LIR motif - crucial for selective autophagy. J. Cell Sci. 126:3237–3247 [DOI] [PubMed] [Google Scholar]
- Böckler S., Westermann B. 2014. Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev. Cell. 28:450–458 10.1016/j.devcel.2014.01.012 [DOI] [PubMed] [Google Scholar]
- Boya P., Kroemer G. 2008. Lysosomal membrane permeabilization in cell death. Oncogene. 27:6434–6451 10.1038/onc.2008.310 [DOI] [PubMed] [Google Scholar]
- Campbell C.L., Thorsness P.E. 1998. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J. Cell Sci. 111:2455–2464 [DOI] [PubMed] [Google Scholar]
- Chaugule V.K., Burchell L., Barber K.R., Sidhu A., Leslie S.J., Shaw G.S., Walden H. 2011. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 30:2853–2867 10.1038/emboj.2011.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Han Z., Feng D., Chen Y., Chen L., Wu H., Huang L., Zhou C., Cai X., Fu C., et al. 2014. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell. 54:362–377 10.1016/j.molcel.2014.02.034 [DOI] [PubMed] [Google Scholar]
- Chen Y., Dorn G.W., II 2013. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 340:471–475 10.1126/science.1231031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew K.C., Matsuda N., Saisho K., Lim G.G., Chai C., Tan H.M., Tanaka K., Lim K.L. 2011. Parkin mediates apparent E2-independent monoubiquitination in vitro and contains an intrinsic activity that catalyzes polyubiquitination. PLoS ONE. 6:e19720 10.1371/journal.pone.0019720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A.M., Wong E. 2014. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 24:92–104 10.1038/cr.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawaliby R., Mayer A. 2010. Microautophagy of the nucleus coincides with a vacuolar diffusion barrier at nuclear-vacuolar junctions. Mol. Biol. Cell. 21:4173–4183 10.1091/mbc.E09-09-0782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas E., Plun-Favreau H., Gandhi S., Desmond H., Kjaer S., Loh S.H., Renton A.E., Harvey R.J., Whitworth A.J., Martins L.M., et al. 2011. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet. 20:867–879 10.1093/hmg/ddq526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deosaran E., Larsen K.B., Hua R., Sargent G., Wang Y., Kim S., Lamark T., Jauregui M., Law K., Lippincott-Schwartz J., et al. 2013. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 126:939–952 10.1242/jcs.114819 [DOI] [PubMed] [Google Scholar]
- Erenpreisa J., Huna A., Salmina K., Jackson T.R., Cragg M.S. 2012. Macroautophagy-aided elimination of chromatin: sorting of waste, sorting of fate? Autophagy. 8:1877–1881 10.4161/auto.21610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré J.C., Manjithaya R., Mathewson R.D., Subramani S. 2008. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell. 14:365–376 10.1016/j.devcel.2007.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré J.C., Burkenroad A., Burnett S.F., Subramani S. 2013. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 14:441–449 10.1038/embor.2013.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D., Liu L., Zhu Y., Chen Q. 2013. Molecular signaling toward mitophagy and its physiological significance. Exp. Cell Res. 319:1697–1705 10.1016/j.yexcr.2013.03.034 [DOI] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z., Klionsky D.J. 2014. The machinery of macroautophagy. Cell Res. 24:24–41 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M., St-Pierre P., Shankar J., Wang P.T., Joshi B., Nabi I.R. 2013. Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Mol. Biol. Cell. 24:1153–1162 10.1091/mbc.E12-08-0607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Melville D., Zhang M., Schekman R. 2013. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2:e00947 10.7554/eLife.00947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg M.E., Cooper J.M., Chau K.Y., Rojo M., Schapira A.H., Taanman J.W. 2010. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 19:4861–4870 10.1093/hmg/ddq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. 2010. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12:119–131 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- Glauser L., Sonnay S., Stafa K., Moore D.J. 2011. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J. Neurochem. 118:636–645 10.1111/j.1471-4159.2011.07318.x [DOI] [PubMed] [Google Scholar]
- Graef M., Friedman J.R., Graham C., Babu M., Nunnari J. 2013. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell. 24:2918–2931 10.1091/mbc.E13-07-0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene A.W., Grenier K., Aguileta M.A., Muise S., Farazifard R., Haque M.E., McBride H.M., Park D.S., Fon E.A. 2012. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 13:378–385 10.1038/embor.2012.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Noda T., Baba M., Ohsumi Y. 2005. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic. 6:56–65 10.1111/j.1600-0854.2004.00245.x [DOI] [PubMed] [Google Scholar]
- Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. 2013. Autophagosomes form at ER-mitochondria contact sites. Nature. 495:389–393 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Hara-Kuge S., Fujiki Y. 2008. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Exp. Cell Res. 314:3531–3541 10.1016/j.yexcr.2008.09.015 [DOI] [PubMed] [Google Scholar]
- Hasson S.A., Kane L.A., Yamano K., Huang C.H., Sliter D.A., Buehler E., Wang C., Heman-Ackah S.M., Hessa T., Guha R., et al. 2013. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 504:291–295 10.1038/nature12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung Y.H., Chen L.M., Yang J.Y., Yang W.Y. 2013. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun. 4:2111 10.1038/ncomms3111 [DOI] [PubMed] [Google Scholar]
- Iguchi M., Kujuro Y., Okatsu K., Koyano F., Kosako H., Kimura M., Suzuki N., Uchiyama S., Tanaka K., Matsuda N. 2013. Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. J. Biol. Chem. 288:22019–22032 10.1074/jbc.M113.467530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., Koyama-Honda I., Mizushima N. 2012. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J. Cell Sci. 125:1488–1499 10.1242/jcs.094110 [DOI] [PubMed] [Google Scholar]
- Jin S.M., Youle R.J. 2012. PINK1- and Parkin-mediated mitophagy at a glance. J. Cell Sci. 125:795–799 10.1242/jcs.093849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S.M., Lazarou M., Wang C., Kane L.A., Narendra D.P., Youle R.J. 2010. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191:933–942 10.1083/jcb.201008084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T., Lamark T. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 7:279–296 10.4161/auto.7.3.14487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane L.A., Lazarou M., Fogel A.I., Li Y., Yamano K., Sarraf S.A., Banerjee S., Youle R.J. 2014. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205:143–153 10.1083/jcb.201402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Wang K., Cao Y., Baba M., Klionsky D.J. 2009. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 17:98–109 10.1016/j.devcel.2009.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Kurihara Y., Jin X., Goda T., Ono Y., Aihara M., Hirota Y., Saigusa T., Aoki Y., Uchiumi T., Kang D. 2013. Casein kinase 2 is essential for mitophagy. EMBO Rep. 14:788–794 10.1038/embor.2013.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawajiri S., Saiki S., Sato S., Sato F., Hatano T., Eguchi H., Hattori N. 2010. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 584:1073–1079 10.1016/j.febslet.2010.02.016 [DOI] [PubMed] [Google Scholar]
- Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D.G., Ritorto M.S., Hofmann K., Alessi D.R., Knebel A., Trost M., Muqit M.M. 2014. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460:127–139 10.1042/BJ20140334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldoun S.A., Emond-Boisjoly M.A., Chateau D., Carrière V., Lacasa M., Rousset M., Demignot S., Morel E. 2014. Autophagosomes contribute to intracellular lipid distribution in enterocytes. Mol. Biol. Cell. 25:118–132 10.1091/mbc.E13-06-0324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.K., Hailey D.W., Mullen R.T., Lippincott-Schwartz J. 2008. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. USA. 105:20567–20574 10.1073/pnas.0810611105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 392:605–608 10.1038/33416 [DOI] [PubMed] [Google Scholar]
- Kondapalli C., Kazlauskaite A., Zhang N., Woodroof H.I., Campbell D.G., Gourlay R., Burchell L., Walden H., Macartney T.J., Deak M., et al. 2012. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2:120080 10.1098/rsob.120080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo-Okamoto N., Noda N.N., Suzuki S.W., Nakatogawa H., Takahashi I., Matsunami M., Hashimoto A., Inagaki F., Ohsumi Y., Okamoto K. 2012. Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy. J. Biol. Chem. 287:10631–10638 10.1074/jbc.M111.299917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., et al. 2014. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 10.1038/nature13392 [DOI] [PubMed] [Google Scholar]
- Kraft C., Peter M. 2008. Is the Rsp5 ubiquitin ligase involved in the regulation of ribophagy? Autophagy. 4:838–840 [DOI] [PubMed] [Google Scholar]
- Kraft C., Deplazes A., Sohrmann M., Peter M. 2008. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10:602–610 10.1038/ncb1723 [DOI] [PubMed] [Google Scholar]
- Krick R., Muehe Y., Prick T., Bremer S., Schlotterhose P., Eskelinen E.L., Millen J., Goldfarb D.S., Thumm M. 2008. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell. 19:4492–4505 10.1091/mbc.E08-04-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen A.R., Schandorff S., Høyer-Hansen M., Nielsen M.O., Jäättelä M., Dengjel J., Andersen J.S. 2008. Ordered organelle degradation during starvation-induced autophagy. Mol. Cell. Proteomics. 7:2419–2428 10.1074/mcp.M800184-MCP200 [DOI] [PubMed] [Google Scholar]
- Lamb C.A., Yoshimori T., Tooze S.A. 2013. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14:759–774 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- Lazarou M., Jin S.M., Kane L.A., Youle R.J. 2012. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell. 22:320–333 10.1016/j.devcel.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M., Narendra D.P., Jin S.M., Tekle E., Banerjee S., Youle R.J. 2013. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J. Cell Biol. 200:163–172 10.1083/jcb.201210111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Nagano Y., Taylor J.P., Lim K.L., Yao T.P. 2010. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J. Cell Biol. 189:671–679 10.1083/jcb.201001039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.W., Li J., Bao J.K. 2012. Microautophagy: lesser-known self-eating. Cell. Mol. Life Sci. 69:1125–1136 10.1007/s00018-011-0865-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z., Shah A.H., Kim J.J., Mulholland J.W., Segev N. 2013. Regulation of ER-phagy by a Ypt/Rab GTPase module. Mol. Biol. Cell. 24:3133–3144 10.1091/mbc.E13-05-0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Czaja M.J. 2013. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 20:3–11 10.1038/cdd.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P., Ma Q., Zhu C., Wang R., Qi W., et al. 2012. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14:177–185 10.1038/ncb2422 [DOI] [PubMed] [Google Scholar]
- Maejima I., Takahashi A., Omori H., Kimura T., Takabatake Y., Saitoh T., Yamamoto A., Hamasaki M., Noda T., Isaka Y., Yoshimori T. 2013. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32:2336–2347 10.1038/emboj.2013.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manivannan S., de Boer R., Veenhuis M., van der Klei I.J. 2013. Lumenal peroxisomal protein aggregates are removed by concerted fission and autophagy events. Autophagy. 9:1044–1056 10.4161/auto.24543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R., Jain S., Farré J.C., Subramani S. 2010a. A yeast MAPK cascade regulates pexophagy but not other autophagy pathways. J. Cell Biol. 189:303–310 10.1083/jcb.200909154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R., Nazarko T.Y., Farré J.C., Subramani S. 2010b. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 584:1367–1373 10.1016/j.febslet.2010.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Wang K., Zhao M., Xu T., Klionsky D.J. 2011. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J. Cell Biol. 193:755–767 10.1083/jcb.201102092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Liu X., Feng Y., Klionsky D.J. 2014. The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy. 10:652–661 10.4161/auto.27852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189:211–221 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner C., Lorenz H., Weihofen A., Selkoe D.J., Lemberg M.K. 2011. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J. Neurochem. 117:856–867 10.1111/j.1471-4159.2011.07253.x [DOI] [PubMed] [Google Scholar]
- Mijaljica D., Devenish R.J. 2013. Nucleophagy at a glance. J. Cell Sci. 126:4325–4330 10.1242/jcs.133090 [DOI] [PubMed] [Google Scholar]
- Mijaljica D., Prescott M., Devenish R.J. 2012. A late form of nucleophagy in Saccharomyces cerevisiae. PLoS ONE. 7:e40013 10.1371/journal.pone.0040013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. 2011. Autophagy in protein and organelle turnover. Cold Spring Harb. Symp. Quant. Biol. 76:397–402 10.1101/sqb.2011.76.011023 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Komatsu M. 2011. Autophagy: renovation of cells and tissues. Cell. 147:728–741 10.1016/j.cell.2011.10.026 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Ohsumi Y. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27:107–132 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Motley A.M., Nuttall J.M., Hettema E.H. 2012. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 31:2852–2868 10.1038/emboj.2012.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., Youle R.J. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Kane L.A., Hauser D.N., Fearnley I.M., Youle R.J. 2010a. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 6:1090–1106 10.4161/auto.6.8.13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. 2010b. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8:e1000298 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Walker J.E., Youle R. 2012. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb. Perspect. Biol. 4:a011338 10.1101/cshperspect.a011338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N.N., Ohsumi Y., Inagaki F. 2010. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584:1379–1385 10.1016/j.febslet.2010.01.018 [DOI] [PubMed] [Google Scholar]
- Novak I., Kirkin V., McEwan D.G., Zhang J., Wild P., Rozenknop A., Rogov V., Löhr F., Popovic D., Occhipinti A., et al. 2010. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11:45–51 10.1038/embor.2009.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., Suomalainen A. 2012. Mitochondria: in sickness and in health. Cell. 148:1145–1159 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttall J.M., Motley A.M., Hettema E.H. 2014. Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy. 10:835–845 10.4161/auto.28259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Kondo-Okamoto N. 2012. Mitochondria and autophagy: critical interplay between the two homeostats. Biochim. Biophys. Acta. 1820:595–600 10.1016/j.bbagen.2011.08.001 [DOI] [PubMed] [Google Scholar]
- Okamoto K., Kondo-Okamoto N., Ohsumi Y. 2009. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell. 17:87–97 10.1016/j.devcel.2009.06.013 [DOI] [PubMed] [Google Scholar]
- Okatsu K., Saisho K., Shimanuki M., Nakada K., Shitara H., Sou Y.S., Kimura M., Sato S., Hattori N., Komatsu M., et al. 2010. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 15:887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K., Oka T., Iguchi M., Imamura K., Kosako H., Tani N., Kimura M., Go E., Koyano F., Funayama M., et al. 2012. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat Commun. 3:1016 10.1038/ncomms2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K., Uno M., Koyano F., Go E., Kimura M., Oka T., Tanaka K., Matsuda N. 2013. A dimeric PINK1-containing complex on depolarized mitochondria stimulates Parkin recruitment. J. Biol. Chem. 288:36372–36384 10.1074/jbc.M113.509653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M., Sakai Y. 2010. Peroxisomes as dynamic organelles: autophagic degradation. FEBS J. 277:3289–3294 10.1111/j.1742-4658.2010.07741.x [DOI] [PubMed] [Google Scholar]
- Orvedahl A., Sumpter R., Jr, Xiao G., Ng A., Zou Z., Tang Y., Narimatsu M., Gilpin C., Sun Q., Roth M., et al. 2011. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 480:113–117 10.1038/nature10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Bonizec M., Cohen M., Dokudovskaya S., Delalande F., Schaeffer C., Van Dorsselaer A., Dargemont C. 2010. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 11:548–554 10.1038/embor.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Niño C.A., Bengtson M.H., Lee J.W., Joazeiro C.A., Dargemont C. 2014. Ubiquitylation by the Ltn1 E3 ligase protects 60S ribosomes from starvation-induced selective autophagy. J. Cell Biol. 204:909–917 10.1083/jcb.201308139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.E., Hayashi Y.K., Bonne G., Arimura T., Noguchi S., Nonaka I., Nishino I. 2009. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 5:795–804 [DOI] [PubMed] [Google Scholar]
- Rakovic A., Grünewald A., Seibler P., Ramirez A., Kock N., Orolicki S., Lohmann K., Klein C. 2010. Effect of endogenous mutant and wild-type PINK1 on Parkin in fibroblasts from Parkinson disease patients. Hum. Mol. Genet. 19:3124–3137 10.1093/hmg/ddq215 [DOI] [PubMed] [Google Scholar]
- Rakovic A., Grünewald A., Kottwitz J., Brüggemann N., Pramstaller P.P., Lohmann K., Klein C. 2011. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS ONE. 6:e16746 10.1371/journal.pone.0016746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rello-Varona S., Lissa D., Shen S., Niso-Santano M., Senovilla L., Mariño G., Vitale I., Jemaá M., Harper F., Pierron G., et al. 2012. Autophagic removal of micronuclei. Cell Cycle. 11:170–176 10.4161/cc.11.1.18564 [DOI] [PubMed] [Google Scholar]
- Riley B.E., Lougheed J.C., Callaway K., Velasquez M., Brecht E., Nguyen L., Shaler T., Walker D., Yang Y., Regnstrom K., et al. 2013. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 4:1982 10.1038/ncomms2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O’Toole E., Winey M., Goldfarb D.S. 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:129–141 10.1091/mbc.E02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V., Dötsch V., Johansen T., Kirkin V. 2014. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 53:167–178 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- Sandoval H., Thiagarajan P., Dasgupta S.K., Schumacher A., Prchal J.T., Chen M., Wang J. 2008. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 454:232–235 10.1038/nature07006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf S.A., Raman M., Guarani-Pereira V., Sowa M.E., Huttlin E.L., Gygi S.P., Harper J.W. 2013. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 496:372–376 10.1038/nature12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers R.L., Zhang J., Randall M.S., Loyd M.R., Li W., Dorsey F.C., Kundu M., Opferman J.T., Cleveland J.L., Miller J.L., Ney P.A. 2007. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. USA. 104:19500–19505 10.1073/pnas.0708818104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaid S., Brandts C.H., Serve H., Dikic I. 2013. Ubiquitination and selective autophagy. Cell Death Differ. 20:21–30 10.1038/cdd.2012.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G., Lee J.R., Grimes D.A., Racacho L., Ye D., Yang H., Ross O.A., Farrer M., McQuibban G.A., Bulman D.E. 2011. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson’s disease. Hum. Mol. Genet. 20:1966–1974 10.1093/hmg/ddr077 [DOI] [PubMed] [Google Scholar]
- Shiba-Fukushima K., Imai Y., Yoshida S., Ishihama Y., Kanao T., Sato S., Hattori N. 2012. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2:1002 10.1038/srep01002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T., Weidberg H., Pietrokovski S., Elazar Z. 2011. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 12:226 10.1186/gb-2011-12-7-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., Czaja M.J. 2009. Autophagy regulates lipid metabolism. Nature. 458:1131–1135 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.J., Aitchison J.D. 2013. Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 14:803–817 10.1038/nrm3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt D.E., Martinez-Torres R.J., Noh Y.J., Mercier P., Manczyk N., Barber K.R., Aguirre J.D., Burchell L., Purkiss A., Walden H., Shaw G.S. 2013. A molecular explanation for the recessive nature of parkin-linked Parkinson’s disease. Nat Commun. 4:1983 10.1038/ncomms2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. 2013. Selective autophagy in budding yeast. Cell Death Differ. 20:43–48 10.1038/cdd.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Akioka M., Kondo-Kakuta C., Yamamoto H., Ohsumi Y. 2013. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 126:2534–2544 10.1242/jcs.122960 [DOI] [PubMed] [Google Scholar]
- Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119:301–311 10.1083/jcb.119.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D., Cai Y., Wang J., Zhang J., Menon S., Chou H.T., Ferro-Novick S., Reinisch K.M., Walz T. 2013. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl. Acad. Sci. USA. 110:19432–19437 10.1073/pnas.1316356110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M., Youle R.J. 2010. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191:1367–1380 10.1083/jcb.201007013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe J.F., Sauvé V., Grenier K., Seirafi M., Tang M.Y., Ménade M., Al-Abdul-Wahid S., Krett J., Wong K., Kozlov G., et al. 2013. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 340:1451–1455 10.1126/science.1237908 [DOI] [PubMed] [Google Scholar]
- Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. 2004. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 304:1158–1160 10.1126/science.1096284 [DOI] [PubMed] [Google Scholar]
- van der Klei I.J., Yurimoto H., Sakai Y., Veenhuis M. 2006. The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim. Biophys. Acta. 1763:1453–1462 10.1016/j.bbamcr.2006.07.016 [DOI] [PubMed] [Google Scholar]
- van Zutphen T., Todde V., de Boer R., Kreim M., Hofbauer H.F., Wolinski H., Veenhuis M., van der Klei I.J., Kohlwein S.D. 2014. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 25:290–301 10.1091/mbc.E13-08-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Farese R.V., Jr 2012. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81:687–714 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Jin M., Liu X., Klionsky D.J. 2013. Proteolytic processing of Atg32 by the mitochondrial i-AAA protease Yme1 regulates mitophagy. Autophagy. 9:1828–1836 10.4161/auto.26281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T., Komander D. 2013. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 32:2099–2112 10.1038/emboj.2013.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Shvets E., Elazar Z. 2011. Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80:125–156 10.1146/annurev-biochem-052709-094552 [DOI] [PubMed] [Google Scholar]
- Welter E., Montino M., Reinhold R., Schlotterhose P., Krick R., Dudek J., Rehling P., Thumm M. 2013. Uth1 is a mitochondrial inner membrane protein dispensable for post-log-phase and rapamycin-induced mitophagy. FEBS J. 280:4970–4982 10.1111/febs.12468 [DOI] [PubMed] [Google Scholar]
- Wild P., McEwan D.G., Dikic I. 2014. The LC3 interactome at a glance. J. Cell Sci. 127:3–9 10.1242/jcs.140426 [DOI] [PubMed] [Google Scholar]
- Williams C., van der Klei I.J. 2013. Pexophagy-linked degradation of the peroxisomal membrane protein Pex3p involves the ubiquitin-proteasome system. Biochem. Biophys. Res. Commun. 438:395–401 10.1016/j.bbrc.2013.07.086 [DOI] [PubMed] [Google Scholar]
- Wu W., Tian W., Hu Z., Chen G., Huang L., Li W., Zhang X., Xue P., Zhou C., Liu L., et al. 2014. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 15:566–575 10.1002/embr.201438501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Youle R.J. 2013. PINK1 is degraded through the N-end rule pathway. Autophagy. 9:1758–1769 10.4161/auto.24633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Fogel A.I., Wang C., van der Bliek A.M., Youle R.J. 2014. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. Elife. 3:e01612 10.7554/eLife.01612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R.J., Narendra D.P. 2011. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12:9–14 10.1038/nrm3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Bosch-Marce M., Shimoda L.A., Tan Y.S., Baek J.H., Wesley J.B., Gonzalez F.J., Semenza G.L. 2008. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 283:10892–10903 10.1074/jbc.M800102200 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zheng X., Hunter T. 2013. Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res. 23:886–897 10.1038/cr.2013.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Massen S., Terenzio M., Lang V., Chen-Lindner S., Eils R., Novak I., Dikic I., Hamacher-Brady A., Brady N.R. 2013. Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J. Biol. Chem. 288:1099–1113 10.1074/jbc.M112.399345 [DOI] [PMC free article] [PubMed] [Google Scholar]