Abstract
Marine invertebrate-associated symbiotic bacteria produce a plethora of novel secondary metabolites which may be structurally unique with interesting pharmacological properties. Selection of strains usually relies on literature searching, genetic screening and bioactivity results, often without considering the chemical novelty and abundance of secondary metabolites being produced by the microorganism until the time-consuming bioassay-guided isolation stages. To fast track the selection process, metabolomic tools were used to aid strain selection by investigating differences in the chemical profiles of 77 bacterial extracts isolated from cold water marine invertebrates from Orkney, Scotland using liquid chromatography-high resolution mass spectrometry (LC-HRMS) and nuclear magnetic resonance (NMR) spectroscopy. Following mass spectrometric analysis and dereplication using an Excel macro developed in-house, principal component analysis (PCA) was employed to differentiate the bacterial strains based on their chemical profiles. NMR 1H and correlation spectroscopy (COSY) were also employed to obtain a chemical fingerprint of each bacterial strain and to confirm the presence of functional groups and spin systems. These results were then combined with taxonomic identification and bioassay screening data to identify three bacterial strains, namely Bacillus sp. 4117, Rhodococcus sp. ZS402 and Vibrio splendidus strain LGP32, to prioritize for scale-up based on their chemically interesting secondary metabolomes, established through dereplication and interesting bioactivities, determined from bioassay screening.
Keywords: metabolomics, dereplication, symbiotic bacteria, mass spectrometry, NMR, multivariate analysis, metabolic profiling
1. Introduction
Marine invertebrates such as sponges are a rich source of novel metabolites that are of medicinal interest due to their anti-cancer, anti-tumor, anti-viral and antibacterial properties [1,2,3,4]. However, there is a bottleneck when developing drugs from marine invertebrates. They are largely uncultivable and it is unsustainable to collect large quantities from marine habitats to facilitate the extraction of enough novel marine natural products for the supply chain, making pharmacological development difficult. Sponge-associated endosymbiotic bacteria are highly concentrated within the sponge matrix making up to 50%–60% of the dry weight of the sponge [5]. They are hypothesized to stabilize the sponge skeleton, process metabolic waste and provide chemical defense against environmental stresses such as predators and overgrowth of fouling organisms, by producing a plethora of novel secondary metabolites that may be structurally unique with interesting pharmacological properties [5,6,7], e.g., as antimicrobials [8] or anti-cancer drugs [9].
There is evidence to suggest that these microbes, which live symbiotically with the host organism, are the true source of many bioactive compounds discovered from associated marine invertebrates [5,10,11,12,13,14,15]. Some of these compounds can be produced in large quantities on a biotechnological scale using bacterial fermentation processes without the need to harvest the host organism and are therefore an economically viable and sustainable source of commercial quantities of metabolites of interest [16]. For example, the anti-tumor drug bryostatin 1, isolated from the marine bryozoan Bugula neritina and synthesized by the symbiotic bacterium Candidatus Endobugula sertula [9], is now produced using a large-scale fermentation process to ensure supply [17].
Key to the exploitation of marine bacteria as sources of novel marine natural products has been the implementation of 16S rRNA-based phylogenetic analysis which has been used extensively to provide an insight into sponge-specific microbial communities [18,19]. The development of new analytical technologies and instrumentation has made it possible to rapidly obtain a chemical fingerprint of bacterial extracts to potentially discover new natural products from only a few milligrams of material. Historically, selection of bacterial strains has relied on literature searching, genetic screening and bioactivity results [20]. However, cultivated bacterial strains from the same genus may appear morphologically identical, but may produce different, structurally diverse secondary metabolites [21,22]. In contrast, strains that appear different by morphology and 16S rRNA sequencing often produce the same secondary metabolites, making it difficult to pinpoint interesting bacterial strains before the time-consuming bioassay-guided fractionation and purification stages.
Dereplication is the rapid identification of known metabolites in a sample mixture [23,24,25]. Dereplication uses chromatographic and spectroscopic methods and database searching, for example using the MarinLit [26] and AntiBase [27] databases, to screen samples for known natural products, which saves time and reduces the possibility of redundancy during natural product discovery programs. Common dereplication methods involve using liquid chromatography coupled to a photo diode array (LC-PDA) system or LC-PDA with mass spectrometry (MS) using electrospray ionization (ESI) [28,29] or atmospheric pressure chemical ionization (APCI) as soft-ionization sources. Liquid chromatography mass spectrometry (LC-MS) high resolution instruments such as Quadrupole Time-of-Flight (QTOF) or Orbitrap provide accurate mass data (0.5%–5 ppm) with elemental composition output for a given ion [30]. This enables natural products databases to be queried in a high throughput manner, with fewer candidate metabolite IDs being observed for each feature. With a Quadrupole or an ion trap, data-dependent MS/MS and MSn can also be carried out to provide additional structural information (e.g., using a Q-TOF or LTQ-Orbitrap). TOF-based mass spectrometers enable a higher degree of certainty for identification of elemental compositions on the basis of both mass accuracy and isotope fit [28,31,32,33]. These instruments offer high sensitivity and accuracy in the ng or pg range and, on several newer-generation instruments, spectra can be obtained in positive and negative ionization modes during a single experiment.
Metabolomics is defined as the comprehensive analysis of the small molecules (MW < 1000) in a biological system under a given set of conditions [34]. At the biochemical level, the metabolome is most closely related to the phenotype, providing insight into biological function [35]. Mass spectrometry and nuclear magnetic resonance (NMR)-based metabolomics are readily applicable to natural products research, offering the ability to deal with complex mixtures in a highly efficient manner [36,37,38,39]. Metabolomics methods are combined with chemoinformatics approaches, e.g., unsupervised multivariate analysis methods, to uncover interesting variation amongst groups of samples (e.g., in terms of their m/z values for mass spectrometry data or chemical shifts for NMR data) [40]. Microbial metabolomics is readily applicable to investigate the physical state of cells [41], to investigate intracellular metabolites [40,41] and for the optimization of experimental conditions for the production of pharmacologically active compounds [23,25].
The aims of the study were to utilize metabolomics tools to investigate differences in secondary metabolite production in marine symbiotic bacteria to fast track the strain selection and dereplication processes for natural product drug discovery. LC-HRMS and principal component analysis (PCA) were used to pinpoint strains that were chemically diverse in a high throughput and untargeted manner. LC-HRMS results were then correlated with bioassay screening results to prioritize strains for drug discovery efforts. The study was designed to monitor secondary metabolite production, using extraction methodology optimized for the recovery of secondary metabolites. In comparison with other studies that compared strains from the same species [22,42], we were able to compare chemically diverse, non-related strains from four different phyla, cultured on a variety of growth media. Additionally, an Excel macro, developed in-house, was used to sort and remove features (pairs of m/z ratios and retention times) associated with the different culture media used. This reduced the difficulties in spectral interpretation that are often encountered when comparing bacterial strains grown on different culture media.
It was predicted that bacterial extracts containing the same secondary metabolites would cluster together whilst those extracts with chemically distinct metabolites would be observed as outliers using unsupervised multivariate analysis [23,39], providing a means to focus on chemically diverse extracts during dereplication. Therefore we used a combinatorial approach for strain selection, utilizing a data analysis workflow that encompassed features of dereplication and metabolomics to establish the chemical profiles of bacterial extracts in a high throughput manner. By incorporating metabolomics approaches, dereplication could be focused on chemically diverse bacterial extracts.
2. Results and Discussion
2.1. Diversity of Invertebrate-Associated Bacteria
Several species of cold water marine invertebrates found in Scottish coastal waters (Orkney Islands, Scotland, UK) were swabbed for microbial symbionts. Specimens were then inoculated onto various types of agar media, which yielded a total of 77 isolates (Figure 1 and Table S1 in Supplementary Information). Suberites ficus (sponge) yielded the highest number of isolates (22) followed by sponges Mycale (Carmia) similaris (14), Grantia compressa (12) and an unidentified hydroid (12), followed by sponges Leucosolenia sp. (8) and Sycon ciliatum (4), the soft coral Alcyonium digitatum (4) and sea urchin Diadema (1) (Figure 1a). A variety of isolation media were utilized in this study to maximize the diversity of the isolates obtained. M1 obtained the highest recovery (36 isolates) whilst marine agar recovered only one isolate (Figure 1b). In terms of the diversity of isolates, M1 produced isolates belonging to 15 different genera followed by ISP2 and Luria (seven genera, respectively). Oligo (oligotrophic) media produced isolates from four genera, R2A yielded two genera and marine agar only one genus (Table S1 in Supplementary Information). This variation is consistent with the results of previous studies [43,44]. By 16S rRNA sequencing, the phylogenetic affiliations of 75 of the isolates were determined whilst a further two isolates remained unidentified (Figure 1c). The isolates were grouped to four different phyla representing 23 different identified genera (Figure 1c,d). The most abundant phylum was the Proteobacteria of which 42 were Gammaproteobacteria whilst four were Alpha proteobacteria, followed by the Actinobacteria (23), Bacteriodetes (4) and Firmicutes (2). This is consistent with the observation that it is more successful to culture Gammaproteobacteria than Alphaproteobacteria [45]. The highest numbers of isolates were affiliated to the genus Vibrio (21) followed by uncultured Gammaproteobacteria (12), Psychrobacter (6), Micrococcus (6) and Microbacterium (4) (Figure 1d). High numbers of Vibrio sp. are consistent with previous studies, as they are ubiquitous in the marine environment and are associated with various algae and animals such as sponges and corals [46].
Figure 1.
Classification of the isolates by (a) source invertebrate species; (b) cultivation media; (c) phyla and (d) by genera (if known).
2.2. Data Processing and Data Clean-Up
Following culturing and chemical extraction, the crude extracts from the 77 bacterial isolates were subjected to metabolomic analysis according to our pre-defined metabolomics workflow pathway (Figure 2). To maximize secondary metabolite detection in this diverse bacterial population (with a range of phylogenetic affiliations and culture media), an Exactive benchtop Orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany) that permitted fast polarity switching was used for untargeted dereplication. The Exactive allows positive and negative mode switching with a maximum scan time of 0.25 s and the instrument always gives good mass accuracy of <3 ppm. The average chromatographic base peak width is about 30 s; therefore, there is adequate time to acquire sufficient scans through the peak in switching mode.
Figure 2.
Metabolomics and dereplication workflow to aid strain selection.
Key features of the metabolomics workflow include data processing in MZmine 2.10 for peak detection, deconvolution, deisotoping, filtering (to narrow the retention time search window to 5–40 min), alignment and gap filling to make multiple data files comparable (Experimental Section 3.5 and Supplementary Information). In order to differentiate between structural isomers that eluted at the same retention time, chromatographic deconvolution using the local minimum search algorithm was applied in MZmine. The isomers were separated into individual LC-HRMS features if their chromatographic 3D resolution was sufficient. However, it is a real challenge to get a perfect setting for this function that would work correctly every time. The raw data were manually validated to confirm the output for outlier and bioactive strains only. The adduct and complex search tools were used for the identification of non-proton adducts and complexes, respectively. This minimized mis-assignment of features such as solvent or salt adducts and complexes such as dimers. The formula prediction tool enabled the possible molecular formulae for each feature to be predicted. The elemental composition output was supported by the heuristic isotopic pattern filter in MZmine [47]. The isotope fit scores were calculated for each isotope ion then combined with the individual fit scores which were weighed by their expected intensities. For each ion peak, the m/z and intensity differences between the expected and the measured patterns were obtained. Those differences were then normalized (normalized deviation values) to the maximum allowed mass and intensity deviation of 0.01%. The relative intensities for the expected and measured values were derived from the isotopic pattern spectra. Each value is a percentage of the isotope’s intensity relative to that of A0. The normalized differences were summed by vector addition of intensity (I) and mass (M) deviations for m/z ions A0 [X], to A1 [X + 1], A2 [X + 2], and A3 [X + 3] [48].
Positive and negative data were then exported as a CSV file for further clean-up. One limitation of MZmine 2.10 is that data obtained in positive and negative ionization modes cannot be combined; therefore, it is not possible to assign the ionization mode for each feature. Thus, a macro was written in Excel that enabled positive and negative ionization mode data files to be processed together. This enabled the features that were observed in either or both positive and negative modes to be merged for further statistical analysis. Hence, this minimized the risk of missing poorly ionizing compounds only detectable in one mode. For example, phenolic and anthraquinone compounds poorly ionize in positive mode but ionize very well in negative mode [25], therefore such compounds were not deleted from the surveyed peaks.
Another complication when analyzing bacterial extracts is that they are cultured on complex growth media which generates multiple peaks in mass spectrometry and NMR datasets. The culture medium is a complex mixture of constituents and unutilized components that could cause interference in the detection of true bacterial secondary metabolites during dereplication. Therefore, a medium blank was analyzed together with the bacterial extracts in LC-HRMS and NMR experiments during data processing. The obtained features from the blank were regarded as interference and subtracted. A threshold intensity ratio of 1/20 was used if ion peaks (MS) were found in both the medium blank and the sample. The Excel macro was then utilized to extract and remove peaks originating from the culture medium by applying an algorithm to calculate the intensity of each m/z in both bacterial and medium extracts. This removed features thought to originate from the medium by only keeping those features with peak intensities 20 times greater in the bacterial samples than in the medium. Bacterial extracts were grouped according to their culture medium and this data clean-up step was carried out for each of the six types of culture medium used.
Using the Excel macro, the data were then recombined into CSV files that were utilized for statistical analysis in SIMCA-P V13.0 (Umetrics, Umeå, Sweden) as well as for dendrogram and heat map generation in the R program (version ×64 2.15.2) (R Foundation for Statistical Computing, Vienna, Austria). The Excel macro was also utilized to dereplicate the samples, matching each m/z found in each bacterial extract with compounds in the AntiMarin database (using a m/z threshold of + or −3 ppm) to provide details on the putative identities of all metabolites and to calculate the number of remaining unidentified features for each extract. This macro contains a function to identify the top 20 features (ranked by peak intensity) and corresponding putative identities in each sample by creating individual CSV files for each extract. Hits from the database were accessed using ChemBioFinder version 13 (PerkinElmer Informatics, Cambridge, UK) and structures were confirmed by tandem mass spectrometry (MS/MS) and two-dimensional 1H-1H correlation NMR spectroscopy (1H-1H COSY). For COSY analysis, spectra from the bacterial extracts were overlaid with spectra from the culture medium to determine signals and cross peaks originating from the culture medium. Three outlier strains were chosen for detailed discussion in this paper to demonstrate the application of different methodologies in dealing with a chemically diverse set of samples showing variation in terms of secondary metabolite production.
2.3. Multivariate Analysis for Strain Selection
Processed data was analyzed using SIMCA-P V 13.0 (Umetrics, Umeå, Sweden) using the unsupervised statistical analysis method, principal component analysis (PCA). PCA was used to identify differing features found in the outlying bacterial strains to aid prioritization of the strains with interesting secondary metabolomes. Four predominant outliers, Bacillus sp. 4117, Rhodococcus sp. ZS402 Vibrio splendidus strain LGP32, and Psychrobacter sp. were observed, indicating that there was variance in the secondary metabolites produced by these bacterial extracts as they lay furthest from the main group of samples in the score plot (Figure 3a). From the primary general screen, the extracts from these isolates were also found to have interesting bioactivities against Trypanosoma brucei brucei and Enterococcus faecalis, as well as in target-based functional assays which includes TRPV1, TRPA1 and TRPM8 (pain and cancer) as well as PTPI1 and PPARα (inflammation, diabetes, metabolic disorders and atherosclerosis) (data not shown). Rhodococcus sp. ZS402 was also identified as NRPS positive from genetic screening, containing the non-ribosomal peptide genes. The PCA loading plot (Figure 3b) illustrates the features (m/z ratios, displayed as green dots) that are responsible for the separation shown in the score plot (Figure 3a), indicating the production of unique secondary metabolites particularly by two bacterial strains, Rhodococcus sp. ZS402 and Vibrio splendidus strain LGP32. Using SIMCA-P it is possible to select any point in the loadings plot to highlight the putative identity of any metabolite and to investigate the peak intensity of this metabolite across the sample set (Figure 3c). Two metabolites (m/z 265.1476 and 279.1631) were found to be abundant in the Rhodococcus sp. compared to the other extracts (Figure 3c); however, they were also observed in the Psychrobacter sp. (which was also an outlier in the PCA scoring plot lying adjacent to the Rhodococcus sp.) with lower peak intensities, suggesting they both produce some similar metabolites not found in the other extracts. However, further study with Psychrobacter sp. was halted due to observed instability in the production of the secondary metabolites leading to disappearance of bioactivity after freezing and thawing of the bacterial isolate. It is also worth mentioning that the outlier strains were repeatedly subjected to MS and NMR analysis every three months prior to scale-up work to evaluate their stability. Heat map analysis was utilized to look at the secondary metabolomes in the 77 bacterial extracts (Figure 4a,b) which is a visual representation of the metabolite diversity in the extracts. Multiple blue bands indicate a rich secondary metabolome with a high diversity of metabolites whilst fewer blue bands indicate that a more limited set of secondary metabolites are being produced. Heat maps were overlaid with dendrograms to relate the chemical profiles to the results of the multivariate analysis (Figure 4a). The heat maps were also arranged by species (Figure 4b) to investigate the chemical diversity amongst strains from the same species, exemplified by the 21 strains of Vibrio sp. that have very different heat map profiles (Figure 4b). It can be seen that several species from different genera have rich chemical profiles whereas other strains do not (Figure 4a,b). Heat map analysis can also be used to gain an overview of the molecular weight range of metabolites as the features were sorted by m/z ratios (Figure 4a,b).
Figure 3.
(a) Principal component score plot analysis of 77 strains clustered according to features (m/z ratios) from mass spectral data (R2 = 0.4). Bioactivities of outliers are represented using symbols; (*) Anti-trypanosomal activity against Trypanosoma brucei brucei, (•) PTP1B, (Δ) TRPV1, (◊) TRPA1, (□) TRPM8, (○) PPARα, and (▪) Enterococcus faecalis. Rhodococcus sp. ZS402 was also found to be NRPS positive (+); (b) Accompanying PCA loading plot of the 77 strains investigated in this study; (c) Variable intensity plot illustrating two metabolites observed as outliers in (b) (m/z 265.1476 and m/z in 279.1631) in Rhodococcus sp. ZS402.
Figure 4.
Heat map based on mass spectrometry data displaying distinct metabolic profiles amongst the 77 bacterial species: (a) dendrogram from multivariate analysis overlaid with heat map; and (b) heat map organized according to species showing differences in the chemical profiles of strains and species. Species observed as outliers from PCA are highlighted and labelled using an asterisk*. (Abbreviations; UBC= uncultured bacterial clone, UGP clone = uncultured gamma proteobacterium, UMB = uncultured marine bacterium).
2.4. Chemical Diversity of Natural Products in Outlying Bacterial Extracts
The limitation of a dereplication study for secondary metabolites, particularly from marine sources, is the difficulty to attain a reference standard for every “hit” from the database. To ensure the correctness of the identification of the basic structure of the identified peaks, UV, MS/MS data and NMR spectral data were used to support the results. However, dereplication through the UV data set was limited only to analyzing chromophore-containing metabolites.
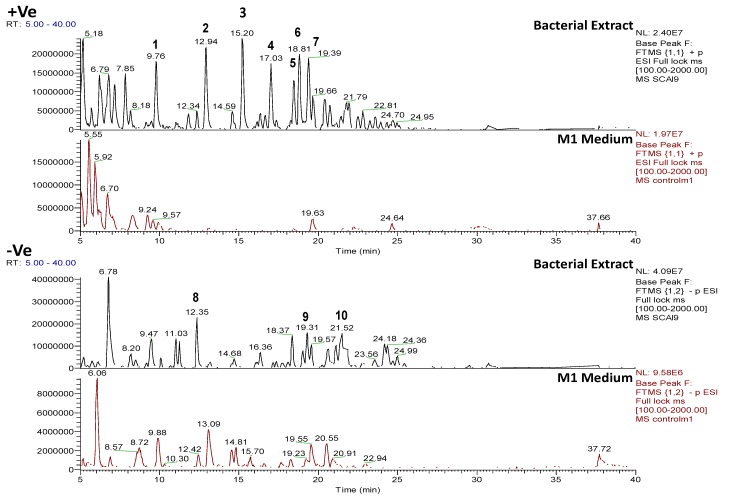
In this study, the mass resolution was 50,000 (at m/z 400), which is high enough to distinguish isobaric compounds with medium molecular weights (<800 Da). The total number of features identified in three of the outlying bacterial extracts by LC-HRMS is documented in Table 1. The highest number of features was detected in the Rhodococcus sp. ZS402, where 1198 features were detected in positive ionization mode and 2361 features were detected in negative ionization mode. By removing features from the ISP2 culture medium, 45% of these features were removed in positive ionization mode, whilst in negative ionization mode 27.4% of these features were removed, leaving 659 in positive mode and 1715 in negative mode. Following dereplication, 28% of features were putatively identified (positive and negative modes combined) whilst 72% were unidentified indicating that this isolate may contain novel compounds (Table 1). The second highest number of features was detected in the Vibrio splendidus strain LGP32, where 2767 were detected in positive ionization mode and 654 features were detected in negative ionization mode. By removing features from the M1 medium, 61.2% of these features were removed in positive ionization mode, whereas in negative ionization mode 5.7% of these features were removed, leaving 1102 in positive mode and 617 in negative mode. Following dereplication, 40.7% of features were putatively identified (positive and negative modes combined), whilst 59.3% were unidentified (Table 1). In the Bacillus sp. 4115, 1220 features were detected in positive ionization mode and 1037 features were detected in negative ionization mode. By removing features from the M1 medium, 71.6% of these features were removed in positive ionization mode, whilst in negative ionization mode 57.8% of these features were removed, leaving 359 in positive mode and 438 in negative mode. Following dereplication, 51.3% of features were putatively identified (positive and negative modes combined), whilst 48.7% were unidentified (Table 1). Base peak chromatograms and tables listing selected interesting secondary metabolites from three of the outlying bacterial species are shown below (Figure 5, Figure 6, Figure 7, Figure 8, Figure 9, Figure 10, Figure 11 and Figure 12 and Table 1, Table 2, Table 3 and Table 4). The putative identities of metabolites, based on hits from the AntiMarin database, are only given if these metabolites have previously been identified from marine bacteria or sponges.
Table 1.
Summary of the number of features detected in the outlying bacterial extracts: (a) total number of features in positive and negative ionization modes (after the removal of features from solvent with intensity >1 × 105); (b) total number of features after removal of features from medium; and (c) total number of features putatively identified by dereplication (from AntiMarin database) and number of unknowns.
| Bacterial Strain | (a) Total number of features (m/z) | (b) Total number of features (m/z) after removal of features (m/z) from medium | (c) Total number of features identified by dereplication with AntiMarin | |||
|---|---|---|---|---|---|---|
| Positive ion mode | Negative ion mode | Positive ion mode | Negative ion mode | Putatively identified (positive and negative modes) | Unidentified (positive and negative modes) | |
| Bacillus sp. 4115 | 1220 | 1037 | 359 (29.4% remaining) |
438 (42.2% remaining) |
270 (51.3%) | 526 (48.7%) |
| Vibrio splendidus strain LGP32 | 2767 | 654 | 1102 (39.8% remaining) |
617 (94.3% remaining) |
699 (40.7%) | 1019 (59.3%) |
| Rhodococcus sp. ZS402 | 1198 | 2361 | 659 (55% remaining) |
1715 (72.6% remaining) |
519 (28%) | 1855 (72%) |
Figure 5.
Positive and negative mode base peak chromatograms from outlying bacterial sample, Bacillus sp. 4115, annotated to indicate metabolites identified in Table 2. NB: several of the metabolites were detected in both positive and negative modes. Positive and negative mode base peak chromatograms from M1 agar medium are shown to indicate that the annotated metabolites are being produced by the bacteria and are not from the medium.
Figure 6.
Mass spectrum for Bacillus sp. 4115 in the positive ionization mode showing the presence of a cluster of features within the RT range of 30–39 min. Those annotated with an asterisk * are sodium ion adducts, [M+Na]+.
Figure 7.
Deconvoluted chromatogram for Bacillus sp. 4115 in the positive ionization mode for extracted ions within the m/z range of 1000–1200 Da.
Figure 8.
2D-NMR COSY spectrum of Bacillus sp. 4115, overlaid with spectrum from M1 medium. Signals in orange are from the sample and signals in grey are from the medium.
Figure 9.
Positive and negative mode base peak chromatograms from outlying bacterial sample, Vibrio splendidus strain LGP32 annotated to indicate metabolites identified in Table 3. Positive and negative mode base peak chromatograms from M1 agar medium are shown to indicate that the annotated metabolites are being produced by the bacteria and are not from the medium.
Figure 10.
2D-NMR COSY spectrum from sample Vibrio splendidus strain LGP32 overlaid with media. Cross signals in orange are from the sample and signals in grey are from the media. Higlighted correlations indicate substructures from oxyplicacetin.
Figure 11.
Positive and negative mode base peak chromatograms from outlying bacterial sample, Rhodococcus sp. ZS402, annotated to indicate metabolites identified in Table 4. Positive and negative mode base peak chromatograms from ISP2 agar medium are shown to indicate that the annotated metabolites are being produced by the bacteria and are not from the ISP2 agar medium.
Figure 12.
2D-NMR COSY spectrum of Rhodococcus sp. ZS402 overlaid with medium. Signals in orange are from the sample and signals in grey are from the medium.
Table 2.
Selected metabolites found in Bacillus sp. 4115. NB: All of these metabolites were also detected in negative ionization mode.
| Peak ID | ESI
Mode |
m/z | Rt (min) | Molecular Formula
(Isotope Fit Score A0 to A3) |
RDB | Hits | Fragmentation Data | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fragment ions MS2 +Ve |
Chemical Formula | RDB | Fragment ions MS3 +Ve |
Molecular Formula | RDB | ||||||||
| 1 | Pos | 445.29092 | 17.5 | C22H40O7N2 (99.49%) |
4 | No hits | 427.27921 399.28485 314.19589 232.15408 214.14343 186.14862 |
C22H39O6N2
C21H39O5N2 C16H28O5N C11H22O4N C11H20O3N C10H20O2N |
5
4 4 2 3 2 |
168.13794
72.08067 |
C10H18ON
C4H10N |
3
1 |
|
| 2 | Pos | 459.30646 | 18.6 | C23H42O7N2 (71.70%) |
4 | No hits | 441.29553 413.30063 328.21176 228.15930 200.16431 |
C23H41O6N2
C22H41O5N2 C17H30O5N C12H22O3N C11H22O2N |
5
4 4 3 2 |
146.11752
86.09630 |
C7H16O2N
C5H12N |
1
1 |
|
| 3 | Pos | 1036.69141 | 30.5 | C53H93O13N7 (99.82%) |
11 | Pumilacidin B// (surfactin-1) or other cyclic peptide |
1018.67596 937.61896 685.44714 667.43732 455.28571 |
C53H92O12N7
C48H85O12N6 C33H61O9N6 C33H59O8N6 C22H39O6N4 |
12
10 7 8 6 |
568.36853
342.20117 |
C28H50O7N5
C16H28O5N3 |
7
5 |
|
| 4 | Pos | 1050.70771 | 31.4 | C54H95O13N7 (99.79%) |
11 | Pumilacidin A// or other cyclic peptide |
1032.69104 937.61823 699.46234 681.45282 455.28555 |
C54H94O12N7
C48H85O12N6 C34H63O9N6 C34H61O8N6 C22H39O6N4 |
12
10 7 8 6 |
568.36816
342.20087 |
C28H50O7N5
C16H28O5N3 |
7
5 |
|
| 5 | Pos | 875.53519 | 33.5 | C43H77O15N3 (98.53%) |
7 | No hits | 710.38348 685.41257 659.46952 654.51534 647.45954 615.44423 610.48905 |
C32H58O15N2
C31H61O14N2 C31H67O12N2 C34H72O10N C34H65O10N C29H63O11N2 C32H68O9N |
5
3 1 1 3 1 1 |
||||
| 6 | Pos | 1078.73917 | 34.5 | C56H99O13N7 (99.80%) |
11 | Pumilacidin C// or other cyclic peptide |
1061.72498 966.65216 699.46283 681.45337 455.28549 |
C55H99O13N6
C49H88O12N7 C34H63O9N6 C34H61O8N6 C22H39O6N4 |
10
10 7 8 6 |
568.36859
342.20135 |
C28H50O7N5C
16H28O5N3 |
7
5 |
|
| 7 | Pos | 889.55163 | 34.5 | C42H76O14N6 (84.86%) |
8 | No hits | 861.55371 817.49072 803.47546 790.47894 776.46429 757.47034 690.39093 676.41254 662.39655 590.33954 563.32874 |
C41H77O13N6
C38H69O13N6 C37H67O13N6 C37H68O13N5 C36H66O13N5 C36H65O11N6 C31H56O12N5 C31H58O11N5 C30H56O11N5 C26H48O10N5 C25H47O10N4 |
7
8 8 7 7 8 7 6 6 6 5 |
449.26096
463.27667 577.34413 477.25619 |
C19H37O8N4
C20H39O8N4 C26H49O10N4 C20H37O9N4 |
4
4 5 5 |
|
| 8 | Pos | 903.56635 | 35.6 | C43H78O14N6 (99.76%) |
8 | No hits | 817.49097 804.49593 790.47988 718.42297 704.40740 690.42803 676.41285 604.35589 590.34010 577.34508 491.27243 463.27740 |
C38H69O13N6
C38H70O13N5 C37H68O13N5 C33H60O13N5 C32H58O12N5 C32H60O11N5 C31H58O11N5 C27H50O10N5 C26H48O10N5 C26H49O10N4 C21H39O9N4 C20H39O8N4 |
8
7 7 7 7 6 6 6 6 5 5 4 |
364.20825 | C15H30O7N3 | 3 | |
Table 3.
Selected metabolites found in positive and negative ionization modes in Vibrio splendidus strain LGP32. (P = positive mode; N = negative mode).
| Peak ID | ESI Mode |
m/z | Rt (min) | Molecular Formula (Isotope Fit Score A0 to A3) |
RDB | Hits | Fragment Ions MS2 | Molecular Formula | RDB | Fragment ions MS3 | Molecular Formula | RDB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | P | 219.12266 | 9.8 | C10H18O5 (99.97%) |
2 | (4E)-6,7,9-Trihydroxydec-4-enoic acid | 173.08047 133.08556 115.07513 87.04388 73.06467 |
C8H13O4 C6H13O3 C6H11O2 C4H7O2 C4H9O |
3 1 2 2 1 |
|||
| 2 | P | 305.1590 | 12.9 | C14H24O7 (99.99%) |
3 | No hits | 259.11685 219.12209 173.08040 155.06985 133.08549 115.07516 87.04391 |
C12H19O6 C10H19O5 C8H13O4 C8H11O3 C6H13O3 C6H11O2 C4H7O2 |
4 2 3 4 1 3 2 |
|||
| 3 | P | 408.22407 | 15.2 | C18H33O9N (Ammonium adduct of C18H31O9) (60.57%) |
3 | No hits | 392.19974 305.15979 259.11786 219.12292 |
undetermined C14H25O7 C12H19O6 C10H19O5 |
3 4 2 |
|||
| 3 | P | 408.22407 | 15.2 | C19H29O5N5 (99.50%) |
8 | No hits | 392.19969 305.15978 259.11783 219.12292 |
undetermined C15H21O3N4 C13H15O2N4 C10H1905 |
8 9 2 |
173.08086 155.07023 133.08593 115.07541 |
C8H13O4 C8H11O3 C6H1303 C6H11O2 |
3 4 1 2 |
| 4 | P | 494.25967 | 17.0 | C22H39O11N (Ammonium adduct of C22H37O11) (99.97%) |
4 | No hits | 477.23270 | C22H37O11 | 5 | 459.22238 431.19101 373.18582 345.15396 305.15924 259.11740 219.12263 155.07021 |
C22H35O10 C20H31O10 C18H29O8 C16H25O8 C14H25O7 C12H19O6 C10H19O5 C8H11O3 |
6 6 5 5 3 4 2 3 |
| 5 | P | 580.2965 | 18.5 | C26H45O13N (Ammonium adduct of C26H43O13) (99.96%) |
5 | No hits | 563.26880 477.23288 431.19122 345.15424 305.15945 |
C26H43O13 C22H37O11 C20H31O10 C16H25O8 C14H25O7 |
6 5 6 5 3 |
259.11752 219.12265 155.07025 |
C12H19O6
C10H19O5 C8H11O3 |
4
2 4 |
| 6 | P | 448.2180 | 18.8 | C20H33O10N (Ammonium adduct of C20H30O10) (91.50%) |
5 | No hits | 431.18991 345.15372 259.11725 241.10663 155.07002 |
C20H31O10 C16H25O8 C12H19O6 C12H17O5 C8H11O3 |
6 5 4 5 4 |
|||
| 7 | P | 534.2550 | 19.4 | C24H39O12N (Ammonium adduct of C24H37O12) (99.13%) |
6 | No hits | 517.22723 431.19070 345.15402 259.11737 241.10681 155.07013 |
C24H37O12 C20H31O10 C16H25O8 C12H19O6 C12H17O5 C8H11O |
7 6 5 4 5 4 |
|||
| 7 | P | 534.2550 | 19.4 | C25H35O8N5 (71.66%) |
11 | Oxyplicacetin; 3′-Hydroxy-plicacetin |
517.22723 431.19070 345.15402 259.11737 241.10681 |
C25H33O8N4 C21H27O6N4 C17H21O4N4 C10H17O5N3 C10H15O4N3 |
12 11 10 4 5 |
|||
| 8 | N | 269.13940 | 12.3 | C14H22O5 (99.99%) |
5 | No hits | 251.12892 225.14969 |
C14H19O4 C13H21O3 |
6 4 |
|||
| 9 | N | 405.24944 | 19.3 | C20H38O8 (99.92%) |
3 | No hits | 359.24274 267.19690 |
C19H35O6 C16H27O3 |
3 4 |
|||
| 10 | N | 285.20719 | 21.5 | C16H30O4 (88.36%) |
3 | Hexadecanedioic acid/ethyl plakortide Z/ethyl didehydro-seco-plakortide Z | 267.19641 | C16H27O3 | 4 | 125.09721 141.12836 185.11803 223.20638 |
C8H13O C9H17O C10H17O3 C15H27O |
3 2 3 3 |
Table 4.
Selected metabolites found in Rhodococcus sp. ZS402 in positive and negative ionization modes. (P = positive mode; N = negative mode).
| Peak ID | ESI Mode |
m/z | Rt (min) | Molecular Formula (Isotope Fit Score A0 to A3) |
Hits | RDB | Fragment Ions MS2 |
Molecular Formula | RDB | Fragment Ions MS3 |
Molecular Formula | RDB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | P | 230.2481 | 17.3 | C14H31ON (99.77%) |
Xestoaminol C | 1 | 212.23662 | C14H30N | 1 | |||
| 2 | P | 258.2793 | 19.4 | C16H35ON (99.95%) |
No hits | 1 | 240.26793 | C16H34N | 1 | |||
| 3 | P | 597.5208 | 30.1 | C35H68O5N2 (99.01%) |
No hits | 3 | 337.28409 355.29462 351.29974 369.31042 |
C20H37O2N2 C20H39O3N2 C21H39O2N2 C21H41O3N2 |
4 3 4 3 |
319.27350
295.27368 |
C20H35ON2
C18H35ON2 |
5
3 |
| 4 | N | 265.1476 | 17.6 | C12H26O4S (99.25%) |
No hits | 1 | 96.9590 | [HSO4]− | 1 | |||
| 5 | N | 760.54162 | 17.6 | C42H75O5N5S (90.86%) |
No hits | 9 | 531.30280 | C30H45O5NS | 9 | 96.9590 | [HSO4]− | |
| 6 | N | 279.1631 | 19.1 | C13H28O4S (98.04%) |
No hits | 1 | 96.9590 | [HSO4]− | 1 | 96.9590 | ||
| 7 | N | 816.60400 | 19.1 | C46H83O5N5S (87.43%) |
No hits | 9 | 279.16318 | C32H49O5NS C13H27O4S |
9 1 |
96.9590 | [HSO4]− | |
| 8 | N | 309.17358 | 19.7 | C14H30O5S (98.04%) |
No hits | 1 | 96.9590 | [HSO4]− | 1 | |||
| 9 | N | 293.1790 | 21.6 | C14H30O4S (99.86%) |
No hits | 1 | 96.9590 | [HSO4]− | 1 |
2.4.1. Dereplication of Bacillus sp. 4115
The crude ethyl acetate extract of the Bacillus sp. 4115 isolate was active on the initial screen in the target-based functional assay on TRPV1 against pain. Metabolites from the Bacillus sp. 4115 extract were putatively assigned as peptides through dereplication (Table 2). They were eluted within the retention time range of 16–38 mins, when the percentage of organic mobile phase (acetonitrile) was greater than 50% (Figure 5, Figure 6 and Figure 7), and could be detected in both positive and negative ionization modes. Several of these were dereplicated using the AntiMarin 2013 natural products database as pumilacidin peptides (surfactins) which have already been described from the marine bacterium Bacillus pumilus. Pumilacidins have been described to exhibit antiviral activity [49]. Other plausible congeners that could not be found in AntiMarin were structural analogs of pumilacidins with varying numbers/length of alkyl or peptide side chains that could be targeted for isolation work. It has previously been reported that members of the Bacillus genus produce antibiotic peptides as part of their defence mechanism [50]. The fragmentation data reveals the presence of the cyclic and linear moities in the peptides compatible with those of the pumilacidins. The presence of peptides can be further observed by 2D-COSY correlation (Figure 8) of NH signals between 8 and 9 ppm with the alpha protons resonating between 3 and 5 ppm which gave additional cross peaks upfield from 1 to 2 ppm, representing the beta proton in amino acid units.
2.4.2. Dereplication of Vibrio splendidus Strain LGP32
The ethyl acetate extract of the Vibrio splendidus strain LGP32 exhibited biological activity against Trypanosoma brucei brucei (marker assay system for trypanosomiasis) and PTP1B. Protein-tyrosine phosphatase 1B (PTP1B) is a novel therapeutic target for type 2 diabetes mellitus, obesity and related states of insulin resistance [51]. Vibrio splendidus strain LGP32 contains many semi-polar metabolites indicated by the retention times of the major peaks which ranged from 8 to 25 min (Figure 9). Its LC-HRMS and MS/MS data (Table 3) depicted a highly oxygenated set of metabolites, with the number of oxygen atoms varying from 4 to 13. The ratio of RDBs (ring-plus-double-bond equivalents) to the number of oxygen atoms ranged from 1:2 to 3:5, thus indicating the aromatic nature of the metabolites. COSY correlations between 3 to 5 ppm and 6 to 8 ppm shown in Figure 10 indicate the presence of a glycosidic-like moiety and an aromatic ring system, respectively. The aromatic signals between 6 and 7 ppm signify a phenolic or aniline system. The specified substructures can be found in oxyplicacetin, first detected in the mass spectral dereplication analysis of the isolate (Table 3). Oxyplicacetin, an anti-coccidal agent, was previously isolated from Streptomyces ramulosus [52,53].
2.4.3. Dereplication of Rhodococcus sp. ZS402
The ethyl acetate extract from Rhodococcus sp. ZS402 isolate was found to be active in several of the target-based functional assays, which included TRPA1 and TRPM8 against pain, PPARα in inflammation, diabetes, or other metabolic disorders, as well as against Enterococcus faecalis. The chromatographic trace from Rhodococcus sp. ZS402, indicated that several interesting features were observed in both positive and negative ionization modes within the retention time range of 16–23 min (Figure 11). Only one of these features was identified using the AntiMarin natural products database during dereplication as xestoaminol C, an unsaturated acyl compound previously described from the sponge Xestospongia sp. Several others could not be dereplicated using AntiMarin but were indicated to be structural derivatives with additional C2H4 on their side chains. Undereplicated features observed in the negative ionization mode specified the presence of sulfated metabolites from the molecular formula identification searches in Xcalibur and MZmine [30], as well as the occurrence of the sulfate fragment ion [HSO4]− at m/z 96.9590 in the MS/MS data (Table 4).
The 2D-COSY spectrum (Figure 12) illustrates that this extract has a rich secondary metabolome. Signals can be seen which correspond to aromatic compounds (6–9 ppm), sugars (4–6 ppm) and sulfated aliphatics (0–4 ppm) and/or olefinics (2–5 ppm). The presence of peptides was also observed by cross peaks exhibited from the NH to the alpha and beta proton, typical for an amino acid. Table 5, summarizes the peptide metabolites that were detected in the positive ionization mode. This supports the presence of NRPS genes in the Rhodococcus sp. ZS402 bacterium. However, MS/MS data was only achieved for one detected metabolite. In Table 5, the presence of peptides can be determined within a range of double-bond equivalences [47] or alternatively calculated where the RDB is equivalent to [(#O − #N)/2 + #N] IF linear; (−1) IF linear: (+1) IF cyclic; (+4) for additional Phe/Tyr; (+6) for additional Trp but account for extra Nitrogen(s) when [(#O − #N)/2 + #N] is less than the found RDB which is also encountered with Arg. Besides following the Nitrogen Rule, approximately every 100 Da represents one amino acid with 1 RDB except for Phe, Tyr, and Trp. However, it was not possible to obtain the fragmentation data for most of the detected ion peaks due to the low intensities of parent ion peaks and/or the conceivable cyclic nature of some of the peptides. Devoid of a chain moiety, in comparison with the pumilacidins found in the Bacillus sp. 4115 isolate, cyclic peptides would need a hydrolysis step to cleave the ring prior to further fragmentation.
Table 5.
Probable peptide metabolites detected in Rhodococcus sp. ZS402 isolate in the positive ionization mode. Calculated RDB = [(#O − #N)/2 + #N] IF linear; (−1) IF linear: (+1) IF cyclic; (+4) for additional Phe/Tyr; (+6) for additional Trp but account for extra N when [(#O − #N)/2 + #N] < found RDB especially with Arg. Approximately 100 Da represents one amino acid with 1 RDB except for Phe, Tyr, and Trp.
| Rt (min) | m/z [M + H]+ | Molecular Formula | Isotope Fit Score A0 to A3 (%) | RDB | Predictions to Calculated RDB |
|---|---|---|---|---|---|
| 19.23 | 462.1727 | C19H23O7N7 | 87.66 | 12 | Cyclic with Phe/Tyr |
| 21.43 | 499.1871 | C18H26O9N8 | 87.64 | 10 | Cyclic |
| 22.15 | 587.2399 | C22H34O11N8 | 82.51 | 10 | Linear |
| 28.14 | 569.4893 | C33H64O5N2 | 99.99 | 3 | Linear |
| 29.46 | 583.5048 | C34H66O5N2 | 99.95 | 3 | Linear |
| 33.77 | 1078.7151 | C59H95O11N7 | 99.86 | 16 | Cyclic with Trp |
| 35.30 | 1118.7461 | C62H99O11N7 | 98.83 | 17 | Cyclic with Trp/Arg |
| 35.64 | 1092.7308 | C60H97O11N7 C55H97O13N9 |
99.91 85.21 |
16 12 |
Cyclic with Trp Cyclic |
| 36.80 | 1106.7460 | C61H99O11N7 C56H99O13N9 |
99.50 99.05 |
16 12 |
Cyclic with Trp Cyclic |
| 37.55 | 849.6953 | C51H88O4N6 C56H88O2N4 |
99.92 58.43 |
11 15 |
Cyclic with Trp/Arg Cyclic with Trp/Arg |
3. Experimental Section
3.1. Sample Collection and Bacterial Isolation
Several species of cold water marine invertebrates found in Scottish coastal waters (Orkney Islands, Scotland, UK) were swabbed for microbial symbionts. Six different media were utilized for the isolation of bacteria: M1 [54], ISP2 medium 2 [55], oligotrophic medium (OLIGO) [56], Luria agar (LA), marine agar (MA) [57], and R2A agar [58]. For medium preparation, starch and glucose monohydrate (Alfa Aesar, Heysham, England), yeast extract and malt extract (Oxoid Limited, Hampshire, England), peptone and tryptone (Fisher Scientific, Hemel Hempstead, UK) and glycerol–phosphate, and R2A isolation agar (Sigma Aldrich, Steinheim, Germany) were purchased. All media contained nutrient agar (Oxoid Limited, Hampshire, England) and were prepared using artificial seawater, prepared using Advanced Pro Formula sea salt mix (23 g/L) (Royal Nature, Nesher, Israel).
Plates were incubated at 12 °C which led to the growth of visually diverse colonies of bacteria after 1–4 weeks. Distinct colony morphotypes were picked and bacterial streaking was utilized until pure bacterial colonies were isolated. Bacteria were then maintained on agar plates for short-term storage or archived for long-term storage. To archive the isolates, 3 mL of sterile artificial seawater was added to each plate and mixed before 400 μL of bacteria and artificial seawater solution was transferred into a 2 mL cryovial that contained 400 μL of 30% glycerol (Fisher Scientific, Hemel Hempstead, UK) using a pipette with sterile filter tips.
3.2. Bacterial Culture and Extraction
Seventy-seven fast-growing bacteria were selected from the archive. When required, bacteria in glycerol from archived cryovials were reinoculated onto agar plates and cultured for seven days in a dark incubator at 12 °C. Bacteria were then reinoculated to fresh agar plates by streaking, using disposable sterile loops and cultured as described above for seven days. This step was carried out to get rid of the glycerol in which the bacteria had been stored. Bacteria and agar from three replica plates were then collected into conical flasks using a sterile scalpel to cut the agar into small pieces. Culture growth was terminated with 200 mL HPLC grade ethyl acetate (Sigma Aldrich, Dorset, UK). After 24 h, samples were individually homogenized with an Ultra-turrax T 18 basic homogenizer (IKA, Staufen, Germany), filtered using a Buchner funnel with 110 mm Fisherbrand filters (Fisher Scientific, Hemel Hempstead, UK), transferred to a 500 mL separating funnel and subjected to liquid–liquid extraction and separation. This procedure involved initially separating the aqueous and ethyl acetate phases and washing the aqueous phase twice more with ethyl acetate. Ethyl acetate fractions were then collected, concentrated, weighed and reconstituted for mass spectrometry (1 mg/mL), NMR (5 mg/600 μL solvent) and bioassay screening (10 mg/mL), respectively. The mentioned fixed concentrations were strictly followed for MS and NMR analysis to normalize the weight of biomass used for each of the individual strains.
3.3. Mass Spectrometry
Methanol (MeOH), dichloromethane (DCM), acetonitrile (MeCN) and formic acid were purchased (Fisher Scientific, Hemel Hempstead, UK). All reagents were of analytical grade. HPLC grade water was obtained in-house from a direct Q-3 water purification system (Millipore, Watford, UK). Samples and medium control samples were prepared at a concentration of 1 mg/mL in 80:20 MeOH: DCM. A solvent blank was also included. Experiments were carried out using an Exactive mass spectrometer with an electrospray ionization source attached to an Accela 600 HPLC pump with Accela autosampler and UV/Vis detector (Thermo Scientific, Bremen, Germany). The mass accuracy was set to less than 3.0 ppm. The Orbitrap mass analyzer is able to limit the mass error within ±3.0 ppm. The instrument was calibrated to maintain a mass accuracy of ±1.0 ppm by applying the lock mass function. The instrument was externally calibrated according to the manufacturer’s instructions before the run and was internally calibrated during the run using lock masses. In positive ion mode, lock masses were m/z 83.06037 (acetonitrile dimer) and m/z 195.08625 (caffeine) and in negative ion mode the lock mass was m/z 91.00368 (formic acid dimer). Mass spectrometry was carried out over a mass range of 100–2000 m/z in positive and negative ionization modes with spray voltage of 4.5 kV and capillary temperature at 270 °C. Ten μL was injected from each vial, at a flow rate of 300 μL/min. The column used was an ACE5 C18 column (5 μm × 75 mm × 3 mm) (Hichrom Limited, Reading, UK). A binary gradient method was utilized. The two solvents were A (water and 0.1% formic acid) and B (MeCN and 0.1% formic acid). The gradient was carried out for 45 minutes and the program followed; at zero minutes A = 90% and B = 10%, at 30 min A = 0% and B = 100% at 36 min A = 90% and B = 10% until end at 45 min. The UV absorption wavelength was set at 254 nm, the sample tray temperature was maintained at 4 °C and the column maintained at 20 °C. The samples were run sequentially, with solvent and media blanks analyzed first. LC-MS data was acquired using Xcalibur version 2.2 (Thermo Scientific, Bremen, Germany).
Data-dependent MS2 and MS3 experiments were carried out using a Finnigan LTQ Orbitrap coupled to a Surveyor Plus HPLC pump (Thermo Scientific, Bremen, Germany) and autosampler (Thermo Fisher, Bremen, Germany) in positive and negative ionization modes using a mass range of m/z 100–2000 and 30,000 resolution. The capillary temperature was 270 °C, the ion spray voltage was 4.5 kV, the capillary voltage 35 V, the tube lens voltage 110 V and the sheath and auxiliary gas flow rates were 50 and 15, respectively (units not specified by manufacturer). Multi-fragmentation (MSn) experiments were accomplished on an Orbitrap analyzer, CID (collision-induced dissociation) was utilized with a normalized collision energy of 35%, activation Q of 0.250 ms and activation time of 30,000 ms applied on ions of most intense, 2nd most intense, and 3rd most intense peaks for MS2 and MS3, respectively, at an isolation width of 3 microns with 5 microscans. Resolution was at 15,000 m/Δm50%, while the minimum ion signal threshold was set to 500. Fragment mass tolerance for molecular formula detection was set at ±5 ppm.
3.4. NMR Spectroscopy
Samples were prepared by dissolving 5 mg of bacterial extract (or culture medium extracts as controls) in 600 μL DMSO-d6 (Sigma-Aldrich, Dorset, UK). These were transferred to 5 mm 7″ NMR tubes (Sigma-Aldrich, Dorset, UK). NMR was carried out on a 400 MHz Jeol-LA400 FT-NMR spectrometer system equipped with a 40TH5AT/FG probe (JEOL, Tokyo, Japan). A presaturation sequence was included to suppress the DMSO solvent signal. For presaturation and proton experiments, sixteen scans were recorded while eight scans were recorded for 2D-1H-1H Correlation Spectroscopy (COSY) analysis. Presaturation and COSY spectra were processed using MestReNova (Mnova 8.1.0) software (Mestrelab Research, Santiago de Compostela, Spain). Normalization, baseline correction with Whittaker Smoother, apodization with Gaussian 1 and smoothing with Savitzy-Golay were carried out in MestReNova. For COSY analysis, spectra from the bacterial extracts were overlaid with the corresponding medium spectrum (control) to differentiate correlations from metabolites produced by the bacteria from those of the culture medium.
3.5. Data Analysis Tools for Mass Spectrometry Data
Raw data were initially sliced into two data sets based on the ionization mode (positive and negative modes) using the MassConvert tool from ProteoWizard [59]. The sliced data sets were imported and processed in MZmine 2.10 [30] using predefined settings to extract features from the raw data. The following data processing steps were carried out using MZmine: peak detection, (mass detection and chromatographic builder), deconvolution, deisotoping, filtering, alignment and gap filling. Identification of adducts and complexes and formula prediction steps were carried out to predict possible molecular formulae for each feature and to minimize mis-assignment of features by eliminating adducts and complexes (see Supplementary Information for full details of all settings and procedures utilized to process data in MZmine). Data was then exported as a CSV file for further clean-up.
An algorithm was employed to use the molecular formula data set from Antibase® (February 2013) and Marinlit® (September 2013). These versions are manually curated databases and the given molecular weights do not differentiate between monoisotopic, average, and most abundant masses. The monoisotopic exact masses for each metabolite were then calculated to be used for the customized library. The processed data from MZmine was incorporated into the customized library through the built-in Excel macro for peak identification and dereplication. “Hits” and unidentified peaks were double checked against the MS raw data in Xcalibur 2.2.
Excel macros were written to enable the subtraction of background peaks and to combine positive and negative ionization mode data files generated by MZmine. Peaks originating from the culture medium were extracted. By applying an algorithm to calculate the intensity of each m/z in both bacterial extracts and medium extracts, ion peaks originating from the medium were subtracted while features with peak intensity 20 times greater in the samples than in the medium were retained. Bacterial extracts were grouped according to their culture media and this data clean-up step was carried out for each culture medium used. The positive and negative ionization mode data sets from each of the respective bacterial extracts were combined by the macro enabling ion peaks that were observed in either or both positive and negative modes to be overlaid for further statistical analysis. The Excel macro was used to dereplicate each m/z ion peak with compounds in the customized database (using RT and m/z threshold of ±3 ppm) which provided details on the putative identities of all metabolites in each bacterial extract and sequentially sorted the number of remaining unknowns for each extract. The macro was then utilized to identify the top 20 features (ranked by peak intensity) and corresponding putative identities in each sample by creating a list for each extract. Hits from the database were accessed using ChemBioFinder version 13 (PerkinElmer Informatics, Cambridge, UK). The data was then converted into a CSV file and exported to SIMCA-P V 13.0 Umetrics, Umeå, Sweden), consequentially providing a feature ID number, ionization mode, m/z, retention time, possible molecular formulae and peak intensity for each feature in all 77 samples. The CSV file was also used to generate a heat map. Heat maps were plotted using the programming software R (version ×64 2.15.2) (R Foundation for Statistical Computing, Vienna, Austria) using a script utilizing the g-plot package. The data set was further analyzed using SIMCA-P V 13.0 using the unsupervised statistical analysis method, principal component analysis (PCA). Dendrograms were also created using SIMCA-P V 13.0 package (Umetrics, Umeå,Sweden).
3.6. Molecular Identification
The whole genome DNA of each strain was extracted by scraping bacterial biomass, suspending in 100 μL of sterile water and heating at 95 °C for 10 min before cooling down the lysate on ice and centrifuging at 13,000 rpm for 10 min. The supernatant containing genome DNA was transferred into a new Eppendorf for 16S rRNA gene amplification. For some strains, the genome DNA could not be extracted using the method described above. For these strains, the FastDNA spin kit (MP Biomedicals, Eschwege, Germany) was used to obtain the whole genome DNA according to the manufacturer’s protocol.
Nearly full-length 16S rRNA genes (1542 nucleotide bases) were amplified by polymerase chain reaction (PCR) using primers 27F and 1492R [60]. The reaction mixture consisted of 5 μL of 10× FastDigest green buffer including 20 mM MgCl2 (Fermentas, Vilnius, Lithuania ), 1 μL of 10 mM dNTPs mixture (Fermentas, Vilnius, Lithuania), 1 μL of 25 mM of each primer (Sigma, München, Germany), 0.19 μL of 5 U/μL DreamTaq DNA polymerase (Thermo Scientific, Bremen, Germany), 1 μL of template DNA and 41.81 μL sterile water to make a final volume of 50 μL. The PCR was performed on a thermal cycler (Biometra, Goettingen, Germany) using the following thermal cycling protocol: the initial denaturation temperature was 95 °C for 2 min, followed by 34 cyclers of reaction starting another denaturation at 95 °C for 0.5 min, then primer annealing at 56 °C for 0.5 min and primer extension at 72 °C for 1.5 min, as well as the final primer extension at 72 °C for 10 min. The reaction was stopped by chilling at 16 °C to limit the polymerase activity. Five μL of PCR product was examined on agarose gel electrophoresis at 300 V for 20 min. An equal volume of 0.5 μg/μL Genen Rular 1Kb DNA ladder (Fermentas, Vilnius, Lithuania) was used as the reference object. The successfully amplified 16S rRNA genes presenting a clear single band around 1500 bases compared to the ladder under a Molecular Imager® Gel Doc™ XR System (Bio-Rad laboratories, Berkeley, CA, USA) were purified using NucleoSpin Gel and PCR Clean-up package (MACHEREY-NAGEL, Düren, Germany) following the manufacturer’s protocol. The genes amplified with more than one band were purified by cutting off the right band and extracting from the agarose gel using NucleoSpin Gel and PCR Clean-up package according to the manufacturer’s protocol. The concentration of the purified 16S rRNA genes was determined using a NanoDrop 2000C Spectrophotometer (Thermo Scientific, Bremen, Germany) and adjusted to 30 μg/μL. High quality 16S rRNA genes were sent to LGC Genomics GmbH (Berlin, Germany) for initial sequencing using forward primer 27F. Sequences of good quality (usually between 150 and 900 bases) were chosen and contrasted in the GenBank database [61] using the BLASTn tool to identify the nearest neighbour to the amplified sequence. 98% and 95% were used as the thresholds to discriminate between sequences of the same species and genus, respectively.
3.7. Bioassay Screening
Extracts were prepared as 10 mg/mL solutions in DMSO in 96-well plate format and delivered for bioassays in dry ice. Extracts were stored at −20 °C until use and then appropriately diluted for testing. Assay-specific thresholds were set to determine the putative active hits: any samples which met this criterion were retested and their activity was assessed over a concentration range of the primary hit. When possible, quantitative measurements of activity (e.g., IC50, Ki, MIC) were determined. A full description of assays is presented only for bioactivities observed in extracts from outlier strains.
3.7.1. Anti-Infectives
Bioassays against Trypanosoma brucei brucei (model assay system for trypanosomiasis) were carried out as previously described [62]. Bacterial extracts were dissolved in DMSO to prepare 10 mg/mL stock solutions. DMSO was used as the negative control at a concentration of 1% to 0.002% and suramin was used as the positive control at a concentration range of 1 to 0.008 μM. The results were calculated as percentages of control values.
The in vitro antimicrobial testing against Enterococcus faecalis strain JH212 was carried out using the standard disk diffusion assay [63]. Sterile filter disks were impregnated with the bacterial extracts and placed on agar plates that had been inoculated with the pathogen. After incubation for 24 h, the antimicrobial potential was quantitatively assessed from the diameter of the inhibition zone.
3.7.2. Metabolic Disease and Inflammation
In search for potential drugs against metabolic disorders involving regulation of glucose metabolism, particularly diabetes mellitus and obesity, samples were tested in a protein–tyrosine phosphatase 1B (PTP1B) assay. Samples were tested at 30 μg/mL in duplicate.
3.7.3. Cell-based Functional Assays
Samples were tested at 30 μg/mL in quadruplicate in 384 well plate format. A Z factor computation value >0.4 was used to establish primary hits on the initial screen. Cell-based functional assays were carried out on the ion channels involving TRPA1 and TRPV1 (pain), and TRPM8 (pain, cancer) genes, whereas PPARα gene (inflammation, diabetes, metabolic disorders and atherosclerosis) targets a nuclear hormone receptor. The fluorescence readouts for TRPA1, TRPV1 and TRPM8 were measured on a Ca2+ sensitive dye as based on Molecular Devices™ [64], while the activity on PPARα was measured against the luminescence on GAL4-UAS luciferase.
4. Conclusions
LC-HRMS and multivariate analysis by principal component analysis (PCA) were used to successfully compare the secondary metabolite profiles of crude extracts from 77 respective marine invertebrate-associated bacterial symbionts. PCA was shown to be an effective tool to differentiate bacterial strains based on their chemical diversity and novelty of metabolites, providing a means to select bacterial isolates with diverse chemistry without having to carry out full isolation work on each extract. PCA was used to reveal bacterial species producing similar chemical groups of metabolites grouped together whilst those producing distinct secondary metabolomes were observed as outliers. By using an Exactive mass spectrometer, which enabled fast-polarity switching, it was possible to obtain efficient and greater metabolite coverage in a single experiment, greatly speeding up analysis times. The development of a comprehensive metabolomics workflow pathway including an in-house developed Excel macro embedded with the AntiMarin database made it possible to rapidly dereplicate the 77 strains, providing putative identities of known metabolites in each extract. It was also possible to calculate the number of unknowns in each extract and to produce data files ranking the “top 20 metabolite hits” (ranked by peak intensity) from each strain. This Excel macro also removed peaks associated with the culture medium, making it possible to compare bacterial strains cultured on different types of growth medium and provided data output for statistical analysis. NMR 1H and 2D-COSY data was also utilized to confirm the dereplication results obtained from the LC-HRMS data. Additionally, we have shown through PCA and heat map analysis that strains with nearly identical 16S rRNA sequences do not necessarily produce the same secondary metabolites. It is also shown that the dereplication results can also be correlated with bioassay screening results to support drug discovery efforts with the objective of both finding a bacterial isolate that has a unique diverse chemistry and is biologically active. Our approach is to use high resolution MS and NMR in parallel to efficiently detect and confirm the dereplication results. Overall, this shows that metabolomics approaches are worthwhile for the selection of strains for the isolation of novel natural products and that this methodology has the potential to reduce redundancy in drug discovery programs.
Acknowledgments
This work was supported by the SeaBioTech project that is funded by the European Commission within its FP7 Programme, under the thematic area KBBE.2012.3.2-01 with Grant Number 311932. I.J.M was supported by an Erasmus Mundus scholarship. Bioassay screening was carried out by C. Clements, L. Young and G. Abbott at the Strathclyde Institute for Drug Research (S.I.D.R) at the University of Strathclyde as well as by L. Stucchi and D. Carettoni at AXXAM. We also acknowledge J. Porter and M. Winson from Heriot-Watt University for organizing the Orkney expedition and providing C.C. and C.V. the initial training in bacterial isolation work on field.
Supplementary Files
Supplementary Information (PDF, 271 KB)
Author Contributions
L.M. optimized the dereplication study protocol, carried out mass spectrometry and all associated data analysis and wrote the manuscript. T.Z. prepared, optimized and validated all algorithms for statistical analytical work. C.V. did the collection from Orkney and isolation of bacteria as well as media optimization to validate stability of metabolite production of outlier strains. I.J.M. carried out the re-cultivation and extraction of isolated strains for dereplication work as well as the NMR data algorithm for dereplication analysis. C.C. carried out the collection from Orkney and isolation of bacteria as well as 16S rRNA gene sequencing of 22 samples. C.D. carried out the re-cultivation, extraction, preparation and database input for dereplication analysis. U.R.A. carried out the 16S rRNA gene sequencing of 20 samples. C.G carried out the 16S rRNA gene sequencing of 35 samples. U.H. supervised the 16S rRNA gene sequencing activity. R.E.-E. coordinated, designed, and supervised the entire project activity and edited the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Miller J.H., Singh A.J., Northcote P.T. Microtubule-stabilizing drugs from marine sponges: Focus on peloruside A and zampanolide. Mar. Drugs. 2010;8:1059–1079. doi: 10.3390/md8041059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molinski T.F., Dalisay D.S., Lievens S.L., Saludes J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009;8:69–85. doi: 10.1038/nrd2487. [DOI] [PubMed] [Google Scholar]
- 3.Newman D.J., Cragg G.M. Marine-sourced anti-cancer and cancer pain control agents in clinical and late preclinical development. Mar. Drugs. 2014;12:255–278. doi: 10.3390/md12010255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinothkumar S., Parameswaran P.S. Recent advances in marine drug research. Biotechnol. Adv. 2013;31:1826–1845. doi: 10.1016/j.biotechadv.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Wang G. Diversity and biotechnological potential of the sponge-associated microbial consortia. J. Ind. Microbiol. Biotechnol. 2006;33:545–551. doi: 10.1007/s10295-006-0123-2. [DOI] [PubMed] [Google Scholar]
- 6.Hentschel U., Hopke J., Horn M., Friedrich A.B., Wagner M., Hacker J., Moore B.S. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 2002;68:4431–4440. doi: 10.1128/AEM.68.9.4431-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berdy J. Bioactive microbial metabolites. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy J., Baker P., Piper C., Cotter P.D., Walsh M., Mooij M.J., Bourke M.B., Rea M.C., O’Connor P.M., Ross R.P., et al. Isolation and analysis of bacteria with antimicrobial activities from the marine sponge Haliclona simulans collected from Irish waters. Mar. Biotechnol. 2009;11:384–396. doi: 10.1007/s10126-008-9154-1. [DOI] [PubMed] [Google Scholar]
- 9.Hildebrand M., Waggoner L.E., Liu H., Sudek S., Allen S., Anderson C., Sherman D.H., Haygood M. bryA: An unusual modular polyketide synthase gene from the uncultivated bacterial symbiont of the marine bryozoan Bugula neritina. Chem. Biol. 2004;11:1543–1552. doi: 10.1016/j.chembiol.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., An R., Wang J., Sun N., Zhang S., Hu J., Kuai J. Exploring novel bioactive compounds from marine microbes. Curr. Opin. Microbiol. 2005;8:276–281. doi: 10.1016/j.mib.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Thomas T.R., Kavlekar D.P., LokaBharathi P.A. Marine drugs from sponge-microbe association—A review. Mar. Drugs. 2010;8:1417–1468. doi: 10.3390/md8041417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proksch P., Edrada R.A., Ebel R. Drugs from the seas—Current status and microbiological implications. Appl. Microbiol. Biotechnol. 2002;59:125–134. doi: 10.1007/s00253-002-1006-8. [DOI] [PubMed] [Google Scholar]
- 13.Perry N.B., Blunt J.W., Munro M.H.G. Mycalamide A, an antiviral compound from a New Zealand sponge of the genus Mycale. J. Am. Chem. Soc. 1988;110:4850–4851. doi: 10.1021/ja00222a067. [DOI] [Google Scholar]
- 14.Newman D.J., Hill R.T. New drugs from marine microbes: The tide is turning. J. Ind. Microbiol. Biotechnol. 2006;33:539–544. doi: 10.1007/s10295-006-0115-2. [DOI] [PubMed] [Google Scholar]
- 15.Martens T., Gram L., Grossart H.P., Kessler D., Muller R., Simon M., Wenzel S.C., Brinkhoff T. Bacteria of the Roseobacter clade show potential for secondary metabolite production. Microb. Ecol. 2007;54:31–42. doi: 10.1007/s00248-006-9165-2. [DOI] [PubMed] [Google Scholar]
- 16.Piel J. Bacterial symbionts: Prospects for the sustainable production of invertebrate-derived pharmaceuticals. Curr. Med. Chem. 2006;13:39–50. doi: 10.2174/092986706775197944. [DOI] [PubMed] [Google Scholar]
- 17.Sudek S., Lopanik N.B., Waggoner L.E., Hildebrand M., Anderson C., Liu H., Patel A., Sherman D.H., Haygood M.G. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J. Nat. Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- 18.Hentschel U., Schmid M., Wagner M., Fieseler L., Gernert C., Hacker J. Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 2001;35:305–312. doi: 10.1111/j.1574-6941.2001.tb00816.x. [DOI] [PubMed] [Google Scholar]
- 19.Hentschel U., Usher K.M., Taylor M.W. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 2006;55:167–177. doi: 10.1111/j.1574-6941.2005.00046.x. [DOI] [PubMed] [Google Scholar]
- 20.Handelsman J., Rondon M.R., Brady S.F., Clardy J., Goodman R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998;5:R245–R249. doi: 10.1016/S1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 21.Ritacco F.V., Haltli B., Janso J.E., Greenstein M., Bernan V.S. Dereplication of Streptomyces soil isolates and detection of specific biosynthetic genes using an automated ribotyping instrument. J. Ind. Microbiol. Biotechnol. 2003;30:472–479. doi: 10.1007/s10295-003-0038-0. [DOI] [PubMed] [Google Scholar]
- 22.Jensen P.R., Williams P.G., Oh D.C., Zeigler L., Fenical W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microbiol. 2007;73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tawfike A.F., Viegelmann C., Edrada-Ebel R. Metabolomics and dereplication strategies in natural products. Methods Mol. Biol. 2013;1055:227–244. doi: 10.1007/978-1-62703-577-4_17. [DOI] [PubMed] [Google Scholar]
- 24.Bobzin S.C., Yang S., Kasten T.P. LC-NMR: A new tool to expedite the dereplication and identification of natural products. J. Ind. Microbiol. Biotechnol. 2000;25:342–345. doi: 10.1038/sj.jim.7000057. [DOI] [PubMed] [Google Scholar]
- 25.Abdelmohsen U.R., Cheng C., Viegelmann C., Zhang T., Grkovic T., Ahmed S., Quinn R.J., Hentschel U., Edrada-Ebel R. Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins A and B from a marine sponge associated-Actinokineospora sp. EG49. Mar. Drugs. 2014;12:1220–1244. doi: 10.3390/md12031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blunt J.W. MarinLit. University of Canterbury; Christchurch, New Zealand: 2012. [Google Scholar]
- 27.Laatsch H. Antibase Version 4.0—The Natural Compound Identifier. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2012. [Google Scholar]
- 28.Nielsen K.F., Mansson M., Rank C., Frisvad J.C., Larsen T.O. Dereplication of microbial natural products by LC-DAD-TOFMS. J. Nat. Prod. 2011;74:2338–2348. doi: 10.1021/np200254t. [DOI] [PubMed] [Google Scholar]
- 29.Blunt J.W., Copp B.R., Keyzers R.A., Munro M.H., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2013;30:237–323. doi: 10.1039/c2np20112g. [DOI] [PubMed] [Google Scholar]
- 30.Pluskal T., Castillo S., Villar-Briones A., Oresic M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klitgaard A., Iversen A., Andersen M.R., Larsen T.O., Frisvad J.C., Nielsen K.F. Aggressive dereplication using UHPLC-DAD-QTOF: Screening extracts for up to 3000 fungal secondary metabolites. Anal. Bioanal. Chem. 2014;406:1933–1943. doi: 10.1007/s00216-013-7582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kind T., Fiehn O. Metabolomic database annotations via query of elemental compositions: Mass accuracy is insufficient even at less than 1 ppm. BMC Bioinform. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erve J.C., Gu M., Wang Y., DeMaio W., Talaat R.E. Spectral accuracy of molecular ions in an LTQ/Orbitrap mass spectrometer and implications for elemental composition determination. J. Am. Soc. Mass Spectrom. 2009;20:2058–2069. doi: 10.1016/j.jasms.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 34.Fiehn O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp. Funct. Genomics. 2001;2:155–168. doi: 10.1002/cfg.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Werf M.J., Jellema R.H., Hankemeier T. Microbial metabolomics: Replacing trial-and-error by the unbiased selection and ranking of targets. J. Ind. Microbiol. Biotechnol. 2005;32:234–252. doi: 10.1007/s10295-005-0231-4. [DOI] [PubMed] [Google Scholar]
- 36.Yang J.Y., Sanchez L.M., Rath C.M., Liu X., Boudreau P.D., Bruns N., Glukhov E., Wodtke A., de Felicio R., Fenner A., et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013;76:1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinette S.L., Bruschweiler R., Schroeder F.C., Edison A.S. NMR in metabolomics and natural products research: Two sides of the same coin. Acc. Chem. Res. 2012;45:288–297. doi: 10.1021/ar2001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kjer J., Debbab A., Aly A.H., Proksch P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc. 2010;5:479–490. doi: 10.1038/nprot.2009.233. [DOI] [PubMed] [Google Scholar]
- 39.Hou Y., Braun D.R., Michel C.R., Klassen J.L., Adnani N., Wyche T.P., Bugni T.S. Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal. Chem. 2012;84:4277–4283. doi: 10.1021/ac202623g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boroujerdi A.F., Vizcaino M.I., Meyers A., Pollock E.C., Huynh S.L., Schock T.B., Morris P.J., Bearden D.W. NMR-based microbial metabolomics and the temperature-dependent coral pathogen Vibrio coralliilyticus. Environ. Sci. Technol. 2009;43:7658–7664. doi: 10.1021/es901675w. [DOI] [PubMed] [Google Scholar]
- 41.Tang J. Microbial metabolomics. Curr. Genomics. 2011;12:391–403. doi: 10.2174/138920211797248619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vynne N.G., Mansson M., Nielsen K.F., Gram L. Bioactivity, chemical profiling, and 16S rRNA-based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Mar. Biotechnol. 2011;13:1062–1073. doi: 10.1007/s10126-011-9369-4. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H., Zhang W., Jin Y., Jin M., Yu X. A comparative study on the phylogenetic diversity of culturable actinobacteria isolated from five marine sponge species. Antonie Van Leeuwenhoek. 2008;93:241–248. doi: 10.1007/s10482-007-9196-9. [DOI] [PubMed] [Google Scholar]
- 44.Abdelmohsen U.R., Pimentel-Elardo S.M., Hanora A., Radwan M., Abou-El-Ela S.H., Ahmed S., Hentschel U. Isolation, phylogenetic analysis and anti-infective activity screening of marine sponge-associated actinomycetes. Mar. Drugs. 2010;8:399–412. doi: 10.3390/md8030399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joint I., Muhling M., Querellou J. Culturing marine bacteria—An essential prerequisite for biodiscovery. Microb. Biotechnol. 2010;3:564–575. doi: 10.1111/j.1751-7915.2010.00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson F.L., Iida T., Swings J. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 2004;68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kind T., Fiehn O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinform. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pluskal T., Uehara T., Yanagida M. Highly accurate chemical formula prediction tool utilizing high-resolution mass spectra, MS/MS fragmentation, heuristic rules, and isotope pattern matching. Anal. Chem. 2012;84:4396–4403. doi: 10.1021/ac3000418. [DOI] [PubMed] [Google Scholar]
- 49.Naruse N., Tenmyo O., Kobaru S., Kamei H., Miyaki T., Konishi M., Oki T. Pumilacidin, a complex of new antiviral antibiotics. Production, isolation, chemical properties, structure and biological activity. J. Antibiot. 1990;43:267–280. doi: 10.7164/antibiotics.43.267. [DOI] [PubMed] [Google Scholar]
- 50.Stein T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005;56:845–857. doi: 10.1111/j.1365-2958.2005.04587.x. [DOI] [PubMed] [Google Scholar]
- 51.Goldstein B.J. Protein-tyrosine phosphatase 1B (PTP1B): A novel therapeutic target for type 2 diabetes mellitus, obesity and related states of insulin resistance. Curr. Drug Targets Immune Endocr. Metab. Disord. 2001;1:265–275. doi: 10.2174/1568008013341163. [DOI] [PubMed] [Google Scholar]
- 52.Shiomi K., Haneda K., Tomoda H., Iwai Y., Omura S. Cytosaminomycins, new anticoccidial agents produced by Streptomyces sp. KO-8119. II. Structure elucidation of cytosaminomycins A, B, C and D. J. Antibiot. 1994;47:782–786. doi: 10.7164/antibiotics.47.782. [DOI] [PubMed] [Google Scholar]
- 53.Haneda K., Shinose M., Seino A., Tabata N., Tomoda H., Iwai Y., Omura S. Cytosaminomycins, new anticoccidial agents produced by Streptomyces sp. KO-8119. I. Taxonomy, production, isolation and physico-chemical and biological properties. J. Antibiot. 1994;47:774–781. doi: 10.7164/antibiotics.47.774. [DOI] [PubMed] [Google Scholar]
- 54.Mincer T.J., Jensen P.R., Kauffman C.A., Fenical W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 2002;68:5005–5011. doi: 10.1128/AEM.68.10.5005-5011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shirling E.B., Gottlieb D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966;16:313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- 56.Olson J.B., Lord C.C., McCarthy P.J. Improved recoverability of microbial colonies from marine sponge samples. Microb. Ecol. 2000;40:139–147. doi: 10.1007/s002480000058. [DOI] [PubMed] [Google Scholar]
- 57.Weiner R.M., Segall A.M., Colwell R.R. Characterization of a Marine Bacterium Associated with Crassostrea virginica (the Eastern Oyster) Appl. Environ. Microbiol. 1985;49:83–90. doi: 10.1128/aem.49.1.83-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reasoner D.J., Geldreich E.E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.ProteoWizard Software Foundation ProteoWizard Homepage. [(accessed on 15 January 2014)]. Available online: http://proteowizard.sourceforge.net/
- 60.Lane D.J. 16S/23S rRNA Sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley; Chichester, UK: 1991. pp. 115–175. [Google Scholar]
- 61.Benson D.A., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. Genbank. Nucleic Acids Res. 2014;42:D32–D37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viegelmann C., Parker J., Ooi T., Clements C., Abbott G., Young L., Kennedy J., Dobson A.D.W., Edrada-Ebel R. Isolation and Identification of Antitrypanosomal and Antimycobacterial Active Steroids from the Sponge Haliclona simulans. Mar. Drugs. 2014;12:2937–2952. doi: 10.3390/md12052937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inderlied C.B., Salfinger M. Antimicrobial agents and susceptibility tests: Mycobacteria. In: Murray P.R., Baron E.J., Pfaller M.A., Tenover F.C., Yolken R.H., editors. Manual of Clinical Microbiology. 6th ed. American Society for Microbiology; Washington, DC, USA: 1995. pp. 1385–1404. [Google Scholar]
- 64.Molecular Devices Home Page. [(accessed on 1 March 2014)]. Available online: http://info.moleculardevices.com/acton/attachment/2560/f-0bc0/0/-/-/-/-/file.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information (PDF, 271 KB)