The ribosome is considered to be the most important molecular machine in the cell. Eukaryotes have two specialized transcription systems—the RNA polymerase I and III systems—for the synthesis of rRNAs and tRNAs. Wang et al. now uncover a specialized RNA polymerase II transcription system for ribosomal protein production in metazoans. Surprisingly, this system uses TRF2 (TBP-related factor 2) rather than the generally used TBP (TATA box-binding protein). This specialized TRF2-based transcription system complements the RNA polymerase I and III systems, as they collectively produce the proteins and RNAs of the ribosome.
Keywords: RNA polymerase II, core promoter, TRF2, TCT motif, ribosomal protein genes
Abstract
The TCT core promoter element is present in most ribosomal protein (RP) genes in Drosophila and humans. Here we show that TBP (TATA box-binding protein)-related factor TRF2, but not TBP, is required for transcription of the TCT-dependent RP genes. In cells, TCT-dependent transcription, but not TATA-dependent transcription, increases or decreases upon overexpression or depletion of TRF2. In vitro, purified TRF2 activates TCT but not TATA promoters. ChIP-seq (chromatin immunoprecipitation [ChIP] combined with deep sequencing) experiments revealed the preferential localization of TRF2 at TCT versus TATA promoters. Hence, a specialized TRF2-based RNA polymerase II system functions in the synthesis of RPs and complements the RNA polymerase I and III systems.
The signals that direct the initiation of transcription ultimately converge at the RNA polymerase II (Pol II) core promoter, which is sometimes referred to as the gateway to transcription (for reviews, see Smale and Kadonaga 2003; Goodrich and Tjian 2010; Juven-Gershon and Kadonaga 2010; Kadonaga 2012). The core promoter comprises the stretch of DNA that is typically from −40 to +40 nucleotides (nt) relative to the +1 start site, which is sufficient for accurate transcription initiation. There are a variety of specific sequence motifs that can contribute to the activity of core promoters. These motifs include the TATA box, initiator (Inr), downstream core promoter element (DPE), motif ten element (MTE), TFIIB recognition elements (BREu and BREd), and polypyrimidine initiator (TCT). There are no universal core promoter elements.
Specific core promoter elements can have important roles in biological networks. For instance, the DPE motif is present in nearly all of the promoters of the Drosophila Hox genes, and Caudal, which is one of the master regulators of the Hox genes, is a DPE-specific transcriptional activator (Juven-Gershon et al. 2008). In addition, the TCT motif is a core promoter element that is found in most of the ribosomal protein (RP) gene core promoters in Drosophila and humans and is important for transcriptional activity (Parry et al. 2010). The TCT motif encompasses the transcription start site from −2 to +6 relative to the +1 start site and is hence located at the same position as the Inr motif. It was found, however, that the TCT motif is functionally distinct from the Inr. For instance, the TCT motif cannot function in lieu of an Inr element and is not recognized by the TBP (TATA box-binding protein)-containing TFIID complex (Parry et al. 2010). These data suggest that there is a distinct transcription system, which does not depend on the canonical TFIID complex, that functions via the TCT motif and is dedicated to the synthesis of RPs. To investigate this question, we examined factors that might mediate transcription from TCT-dependent RP gene promoters and found a requirement for TBP-related factor 2 (TRF2; also known as TLP, TRP, TLF, and TBPL1) (Maldonado 1999; Moore et al. 1999; Ohbayashi et al. 1999 ; Rabenstein et al. 1999; Teichmann et al. 1999; Reina and Hernandez 2007; Goodrich and Tjian 2010; Akhtar and Veenstra 2011) but not TBP. These findings reveal that a specialized TRF2-based transcription system functions in the synthesis of RPs and complements the RNA Pol I and III systems, which produce ribosomal and transfer RNAs.
Results and Discussion
TCT-dependent transcription appears to require TRF2 but not TBP
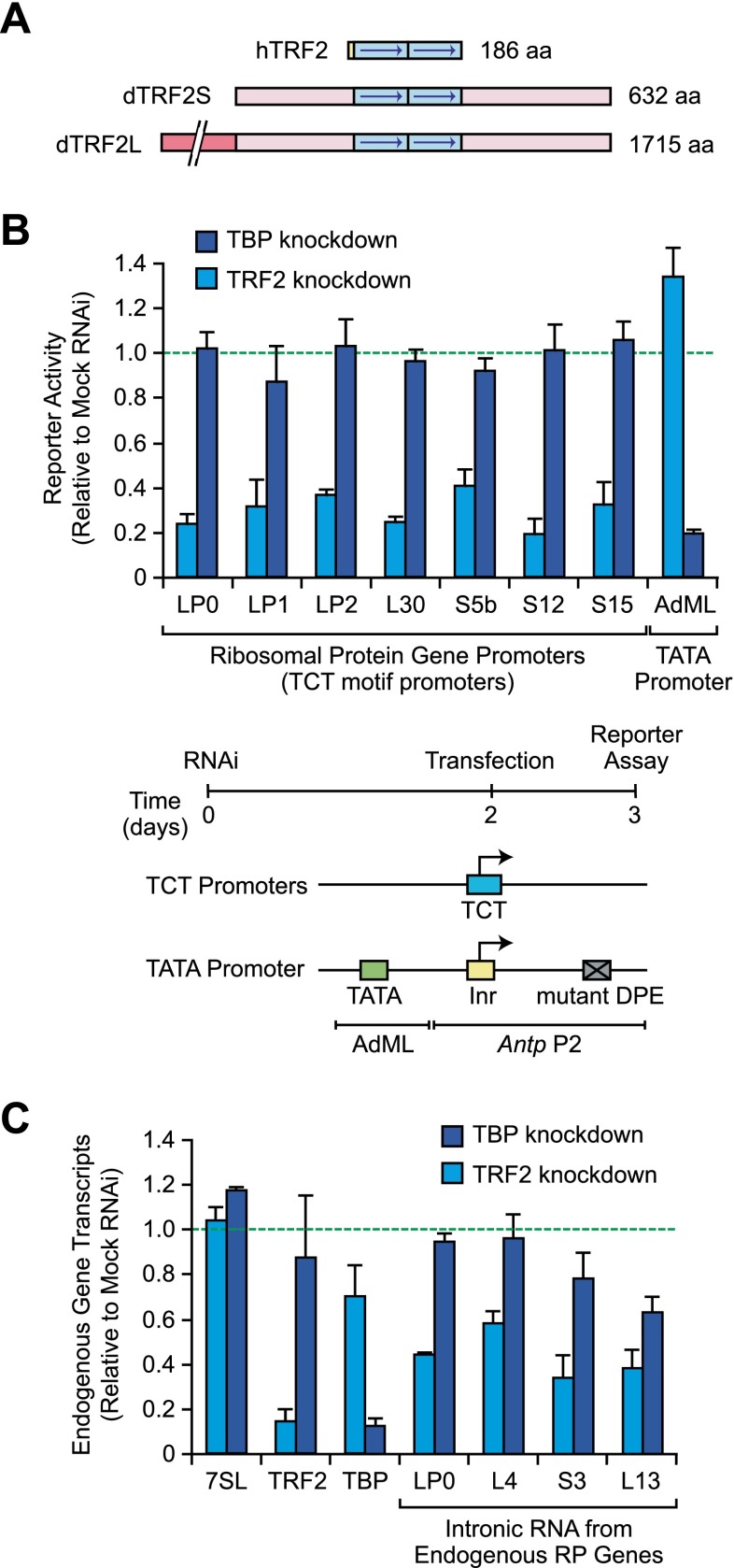
To investigate the factors that are specifically required for transcription from TCT-dependent RP gene promoters, we used an RNAi depletion assay in Drosophila S2 cells to screen candidate proteins for transcriptional activity with TCT-dependent promoters but not a TATA-dependent promoter. Because it appeared that canonical TFIID does not function with the TCT motif (Parry et al. 2010), we were particularly interested in testing the roles of TBP and TBP-related factors in TCT-dependent transcription. Based on its properties, TRF2 was an excellent candidate. TRF2 is widely expressed and has been found to be present in many metazoans (for reviews, see Reina and Hernandez 2007; Goodrich and Tjian 2010; Akhtar and Veenstra 2011). Although TRF2 is related to TBP, it does not bind to TATA sequences, and the DNA sequences, if any, that are directly bound by TRF2 are not known. In Drosophila, there are two forms of TRF2, which we term dTRF2S (for short; also known as p75) and dTRF2L (for long; also known as p175) (Kopytova et al. 2006). dTRF2S is identical to the C-terminal 632-amino-acid residues of dTRF2L and is generated by translation initiation from an internal ribosome entry site (Kopytova et al. 2006). dTRF2S appears to be more closely related to human TRF2 (hTRF2), which lacks the long N-terminal extension that is present in dTRF2L (Fig. 1A). By ChIP–chip analysis (chromatin immunoprecipitation [ChIP] coupled with microarray analysis) with Drosophila S2 cells, dTRF2 (S and/or L) was found to be associated with many RP gene promoters (Isogai et al. 2007). Moreover, RNAi depletion of dTRF2 in Drosophila salivary glands was observed to result in a significant reduction in the sizes of the cells and the glands as well as a decrease in the levels of RP gene transcripts (Isogai et al. 2007), but it was not known whether the decrease in RP transcript levels was due to a transcriptional effect or a general growth defect.
Figure 1.
TCT-dependent transcription appears to require TRF2 but not TBP. (A) Schematic diagrams of hTRF2 and the two forms of Drosophila TRF2 (dTRF2S and dTRF2L). dTRF2S is identical to the C-terminal 632-amino-acid residues of dTRF2L. (B) Depletion of TRF2, but not TBP, reduces RP gene expression. Drosophila S2 cells were depleted of either TRF2 or TBP by RNAi and then transfected with TCT-dependent or TATA-dependent luciferase reporter genes. The experimental scheme and reporter constructs are depicted at the bottom of the figure. The activities of the RNAi-depleted extracts are reported as relative to the activities of mock RNAi-treated control extracts. Error bars represent the standard deviation. (C) Analysis of endogenous transcript levels by qRT–PCR. Drosophila S2 cells were depleted of TRF2 or TBP, as in B. The total RNA was then isolated and analyzed by qRT–PCR. For the RP genes, intronic sequences were used to detect newly synthesized transcripts. We did not analyze RP genes with intronic snoRNA genes, as they could affect the levels of the intronic RNAs. The error bars represent the standard deviation.
Thus, to analyze the role of TRF2 in TCT-dependent versus TATA-dependent transcription, we carried out RNAi depletion analyses of dTRF2 (with dsRNAs corresponding to both S and L forms) or dTBP in S2 cells with TCT-dependent or TATA-dependent reporter genes. We achieved efficient depletion of dTRF2 as well as dTBP, each with two nonoverlapping dsRNAs (Supplemental Fig. S1). The depletion of dTRF2 resulted in a decrease in TCT-dependent transcription but not TATA-dependent transcription. Conversely, depletion of TBP caused a decrease in TATA transcription but not TCT transcription (Fig. 1B). To address the possibility of off-target effects, we performed experiments with a separate set of nonoverlapping dsRNAs for dTRF2 and dTBP and obtained essentially the same results (Supplemental Fig. S2). We additionally tested the effect of TRF2 depletion upon endogenous RP gene transcription via quantitative RT–PCR (qRT–PCR) analysis of intronic RNAs as a measure of newly synthesized transcripts. We examined several RP genes (lacking intronic snoRNAs, which could affect intronic RNA levels) and found that depletion of TRF2 resulted in a stronger decrease in RP gene transcription than depletion of TBP (Fig. 1C). Hence, these findings suggest that TCT-dependent core promoters require TRF2 but not TBP.
Purified TRF2 can mediate TCT-dependent but not TATA-dependent transcription in vitro
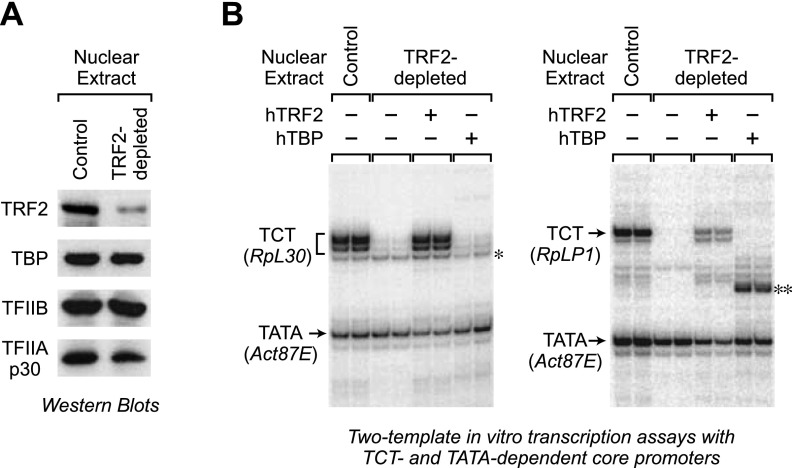
To test the specificity of function of TRF2 protein, we performed in vitro transcription experiments with purified TRF2 at TCT-dependent and TATA-dependent core promoters. For these experiments, we synthesized hTRF2 and hTBP with a wheat germ in vitro translation system (Takai et al. 2010) and purified the proteins to near homogeneity (Supplemental Fig. S3). hTRF2 contains the central conserved region of TRF2 and is smaller than dTRF2S (Fig. 1A). To test the activity of the purified hTRF2, we depleted dTRF2 from Drosophila nuclear extracts with anti-dTRF2 antibodies (Fig. 2A) and then performed two-template (with TCT-dependent and TATA-dependent promoters) in vitro transcription experiments with the TRF2-deficient extracts. As shown in Figure 2B, the depletion of TRF2 results in an essentially complete loss of transcription from two different TCT-dependent promoters (RpL30 and RpLP1) but has little or no effect on transcription from a TATA-dependent promoter (Act87E). We further found that the addition of purified hTRF2 protein to the depleted extracts can partially or nearly fully restore the transcriptional activity that is lost upon depletion of TRF2. In contrast, the addition of purified hTBP did not restore TCT-dependent transcription. We obtained results analogous to those seen in Figure 2B with the RpS12 (TCT), RpS15 (TCT), and hb P2 (TATA) core promoters (Supplemental Fig. S4). It is also important to note that the TRF2-dependent transcription of TCT-dependent promoters is sensitive to low levels of α-amanitin and hence is mediated by RNA Pol II (Supplemental Fig. S5). We additionally found that Drosophila dTRF2S, but not Drosophila TBP, activates RP gene promoters in vitro (Supplemental Figs. S6, S7). These results thus provide biochemical evidence of the specificity of both hTRF2 and dTRF2S proteins for TCT-dependent promoters relative to TATA-dependent promoters. In addition, these experiments further reveal that the central conserved region of TRF2, which is the only segment of TRF2 that is shared by hTRF2 and dTRF2S (Fig. 1A), is sufficient for the specificity of its function for TCT-dependent core promoters.
Figure 2.
Purified TRF2 is required for in vitro transcription of TCT-dependent genes but not a TATA-dependent gene. (A) Immunodepletion of endogenous dTRF2 from a Drosophila embryo nuclear extract. The levels of TRF2 (dTRF2S), TBP, TFIIB, and TFIIA (p30 subunit) in TRF2-depleted extracts versus control extracts were monitored by Western blot analysis. We were able to detect dTRF2S but not dTRF2L even though the antibodies were raised against a polypeptide that is shared by both proteins. This effect may be due to inefficient transfer of the dTRF2L protein to the blot, the lack of recognition of dTRF2L by the antibodies, or the absence of dTRF2L in the extract used in the Western blot. (B) Purified hTRF2, but not purified hTBP, is able to restore the specific loss of TCT-dependent transcription that occurs upon depletion of TRF2 from a nuclear extract. Two-template in vitro transcription assays were performed with TCT-dependent and TATA-dependent promoters. Reactions were carried out with either TRF2-depleted or control nuclear extracts. Where indicated, purified hTRF2 or hTBP was added to reactions with the TRF2-depleted extracts. The resulting transcripts were detected by primer extension–reverse transcription analysis. The single asterisk denotes a nonspecific transcript. The double asterisk indicates a nonspecific transcript that is observed in the presence of hTBP. This stimulation of nonspecific initiation by hTBP is most likely a consequence of the relatively nonspecific binding of hTBP to DNA. This effect was not observed with dTBP (Supplemental Fig. S7).
Overexpression of TRF2 or TBP has opposite effects on TCT-dependent and TATA-dependent transcription
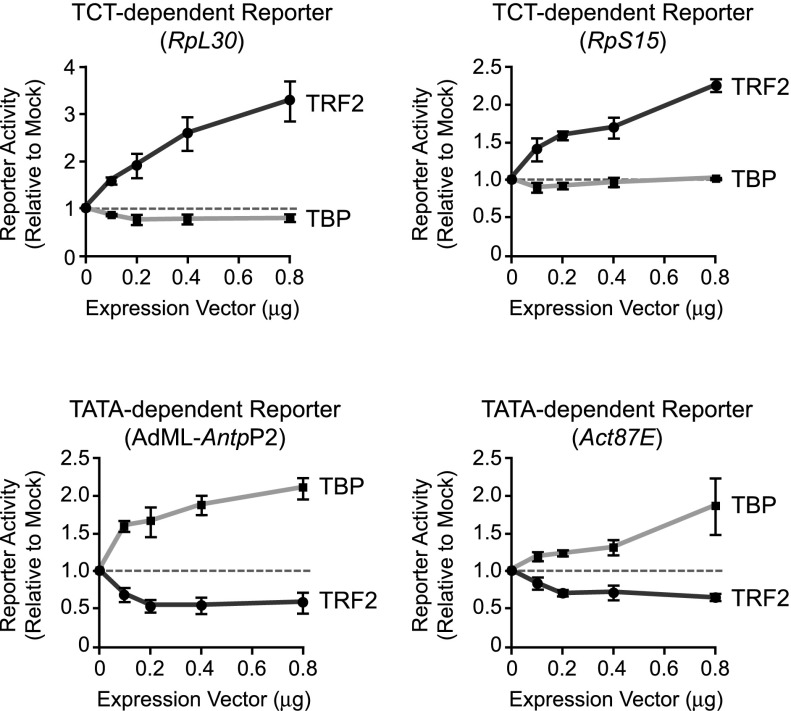
To complement the depletion experiments, we investigated the effects of the overexpression of TRF2 or TBP on the TCT-dependent versus TATA-dependent promoters (Fig. 3). To this end, TRF2 or TBP expression vectors were cotransfected into S2 cells with either TCT-dependent or TATA-dependent reporter constructs (for Western blots, see Supplemental Fig. S8). With TCT-dependent promoters, overexpression of TRF2 increases TCT-dependent transcription in a dose-dependent manner, whereas overexpression of TBP has little or no effect. With TATA-dependent promoters, however, overexpression of TRF2 has a negative effect on TATA-dependent transcription, whereas overexpression of TBP increases TATA-dependent transcription. These findings further support the conclusion that TRF2 has a positive role in TCT-dependent transcription, whereas TBP does not. Moreover, both the cell-based and biochemical experiments show that TRF2 has either no effect or perhaps a slight negative effect on TATA-dependent transcription.
Figure 3.
Overexpression of TRF2 increases TCT-dependent but not TATA-dependent transcription, whereas overexpression of TBP increases TATA-dependent but not TCT-dependent transcription. Drosophila S2 cells were transfected with a TCT-dependent or TATA-dependent reporter construct along with the indicated amounts of expression vector for either dTRF2S or dTBP. The AdML-AntpP2 promoter is identical to the TATA promoter that was used in Figure 1B. Luciferase reporter activities were normalized to those obtained with the empty vector alone. Error bars represent the standard deviation.
TRF2 is localized preferentially to TCT promoters relative to TATA promoters
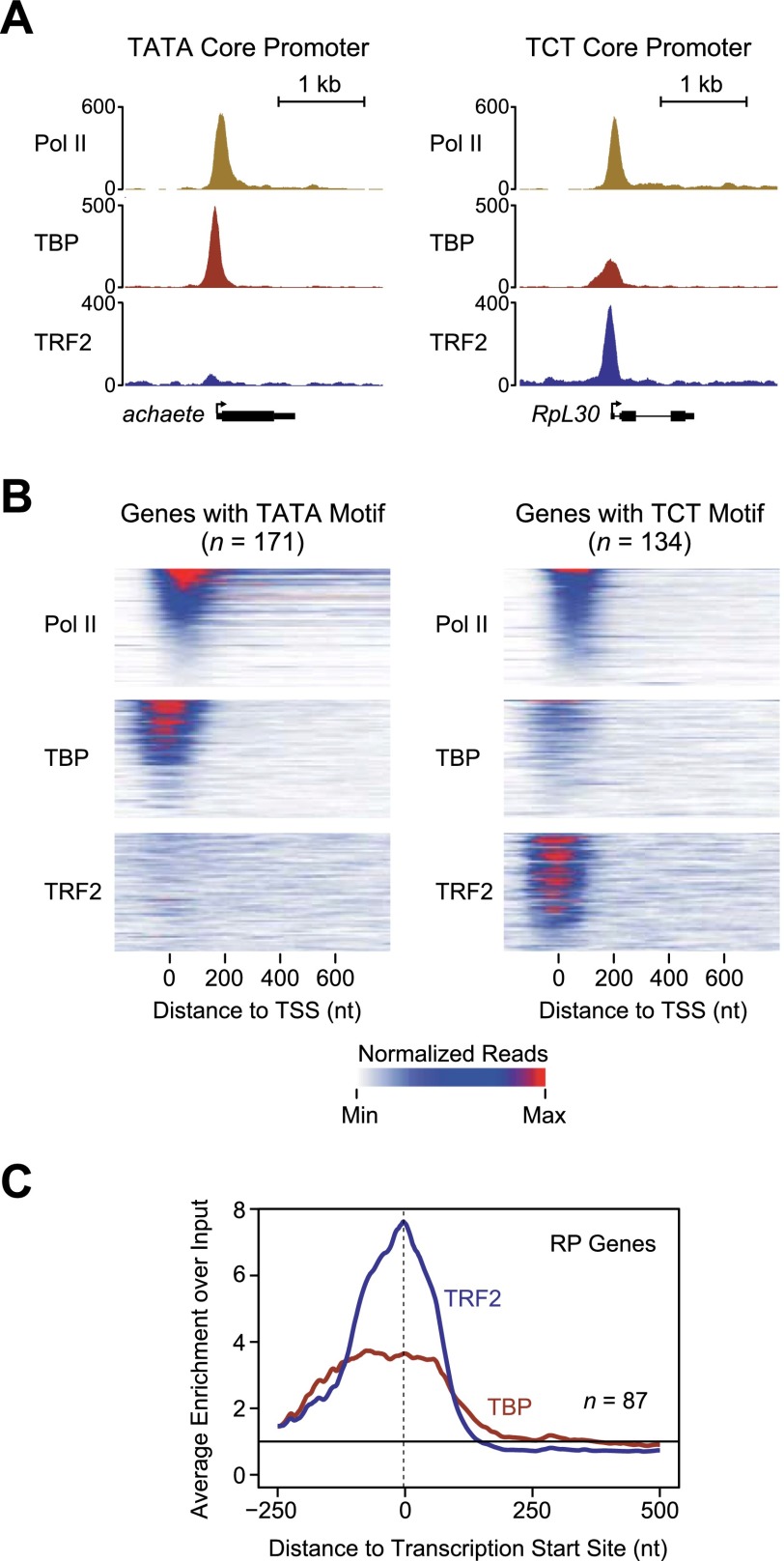
We next sought to determine whether the genome-wide localization of TRF2 in the organism is consistent with its function in TCT-dependent transcription. We therefore performed parallel ChIP-seq (ChIP combined with deep sequencing) analyses of TRF2 and TBP in early Drosophila embryos. At a representative example of a TATA promoter and a TCT promoter with comparable levels of RNA Pol II occupancy, there is a distinct preference for the localization of TRF2 at the TCT core promoter (Fig. 4A). TBP, on the other hand, exhibits a strong peak at the TATA promoter and a weaker, less focused peak at the TCT promoter. This pattern is observed genome-wide, as shown in the heat maps of 171 TATA-containing promoters and 134 TCT-containing promoters (Fig. 4B). There is a sharp preference for TRF2 at TCT-containing promoters and a strong but less absolute preference for TBP at TATA-containing promoters. The analysis of the TRF2 and TBP occupancy at the 87 RP genes additionally revealed a peak of TRF2 near the +1 transcription start site as well as a weaker and broader peak of TBP over the region encompassing the core promoter (Fig. 4C). In all, the ChIP-seq data indicate a strong preference for the localization of TRF2 at TCT core promoters and TBP at TATA core promoters. These findings further reinforce the conclusion that TRF2, but not TBP, functions at TCT-containing promoters.
Figure 4.
TRF2 is enriched at TCT-dependent promoters in vivo. The occupancy of TRF2 and TBP was analyzed by ChIP-seq experiments with Drosophila embryos collected from 2 to 4 h after egg deposition. (A) Differential TRF2 occupancy at a TATA promoter and a TCT promoter. Read counts across a representative TATA-containing promoter (achaete) and a representative TCT-containing promoter (RpL30) with comparable levels of RNA Pol II show that TRF2 is bound at higher levels to the TCT promoter relative to the TATA promoter. (B) Heat maps of the ChIP occupancy of Pol II, TRF2, and TBP at 171 genes with a predicted TATA box and at 134 genes with a predicted TCT motif. These two sets of genes were sorted in descending order of Pol II occupancy and are shown in a window from −200 to + 800 nt relative to the +1 transcription start site. The same genes and their order are shown for Pol II, TBP, and TRF2 occupancy. (C) TRF2 occupancy is highest near the transcription start site of RP genes. The graph depicts the average enrichments of the TRF2 and TBP ChIP-seq signals over input from −250 to +500 nt relative to the transcription start site for the 87 known RP genes. The dashed line indicates the peak TRF2 signal at −3 nt relative to the transcription start site.
A specialized TRF2-based transcription system for TCT-dependent transcription
This study reveals that the transcription of TCT-dependent genes uses a TRF2-based transcription system that is distinct from the well-known TBP-based transcription systems. The existence of a specialized transcription system for the TCT-containing RP gene promoters suggests that this system, which functions in the synthesis of RPs, is the complement to the RNA Pol I and RNA Pol III systems, which synthesize ribosomal and transfer RNAs.
The TCT motif is known to be present in Drosophila (Parry et al. 2010), zebrafish (Nepal et al. 2013), mice (Perry 2005), and humans (Perry 2005; Parry et al. 2010), and TRF2 is generally present in metazoans (for example, see Reina and Hernandez 2007; Goodrich and Tjian 2010; Akhtar and Veenstra 2011). In contrast, neither TRF2 nor the TCT motif appears to be present in the yeast Saccharomyces cerevisiae (for example, see Reina and Hernandez 2007; Goodrich and Tjian 2010; Akhtar and Veenstra 2011; Bosio et al. 2011). It therefore seems likely that the TRF2–TCT system is widely used among metazoans. As might be expected for a protein that is important for RP gene expression, the loss of TRF2 is embryonic-lethal in Caenorhabditis elegans (Dantonel et al. 2000; Kaltenbach et al. 2000), Drosophila (Kopytova et al. 2006), Xenopus (Veenstra et al. 2000), and zebrafish (Müller et al. 2001). However, in mice, TRF2 is not essential, although it is required for spermiogenesis (Martianov et al. 2001; Zhang et al. 2011; Zhou et al. 2013). It is possible, for instance, that another related protein can compensate for the loss of TRF2 in mice.
Does TRF2 bind to DNA? The ChIP-seq data indicate a peak of TRF2 occupancy in the vicinity of the transcription start site (Fig. 4C) and are hence suggestive of a sequence-specific DNA-binding activity. Nevertheless, to date, sequence-specific DNA binding by TRF2 has not yet been seen. Moreover, with the purified TRF2 that is active for transcription in vitro (Fig. 2), we did not observe sequence-specific DNA binding under an extensive range of conditions with many different template DNAs and methodologies in the absence or presence of different combinations of purified TFIIA and TFIIB. Thus, TRF2 may not bind directly to DNA or, alternatively, may bind to DNA under specific conditions that have not yet been tested.
Last, it is important to note that TRF2 may function in different transcription systems, as such a precedent has been observed with the participation of TBP in RNA Pol I, II, and III transcription systems (for example, see Cormack and Struhl 1992; Hernandez 1993). Thus, the function of TRF2 is probably not restricted to TCT-containing promoters. In support of this hypothesis, it is known that TRF2 is important for the transcription of the histone H1 gene (Isogai et al. 2007), but the histone H1 promoter does not appear to contain a TCT motif. Moreover, TRF2 preferentially occupies the histone H1 gene promoter relative to the core histone gene promoters (Supplemental Fig. 9; Isogai et al. 2007). Ultimately, it is likely that we will find that there are many different transcriptional systems that involve TBP, TRF2, and other factors and that each of these networks serves a specific and important biological function.
Materials and methods
Depletion and overexpression assays in Drosophila S2 cells
For reporter assays involving RNAi depletion of TRF2 or TBP, Drosophila S2 cells were seeded at 0.2 × 106 cells per well in a 24-well plate, and then 5 μg of dsRNA was added to each well. After 2 d, cells were transfected with 400 ng of reporter plasmids containing the indicated core promoters and the firefly luciferase gene by using Effectene (Qiagen). The dsRNA sequences used to deplete dTBP were described previously (Hsu et al. 2008). The dsRNA sequences used to deplete dTRF2 correspond to positions 3272–3871 (TRF2-A) and 4361–4986 (TRF2-B) relative to the upstream initiating ATG of TRF2 (Kopytova et al. 2006). For overexpression assays, cells were seeded at 0.6 × 106 cells per well in a 24-well plate. After 24 h, cells were transfected with the indicated amounts of expression vector together with 100 ng of the reporter plasmids containing the indicated core promoters and the firefly luciferase gene by using Effectene (Qiagen). When necessary, the total mass of transfected vector was maintained at a constant level by the addition of the compensatory amount of empty vector (pAc5.1) to give a total of 0.8 μg of expression vector per transfection. For depletion and overexpression assays involving reporter genes, cells were harvested 24 h after transfection, and the lysates were assayed for luciferase activity by using Luciferase Assay Reagent II (Promega). The protein concentration of cell lysates was measured by using the Bradford assay (Bio-Rad). To ensure reproducibility of the data, each experimental condition was performed (in triplicate) a minimum of three times.
For the qRT–PCR analysis of RNAs, TRF2 and TBP were depleted in Drosophila S2 cells, as above. Total RNA was isolated by using TRIzol reagent (Life Technologies) and then subjected to reverse transcription with the iScript cDNA synthesis kit (Bio-Rad), as recommended by the manufacturers. The resulting cDNAs were analyzed by qPCR by using the Opticon 2 instrument (Bio-Rad). TRF1-dependent, RNA Pol III-synthesized 5S rRNA transcripts were used as a reference for normalization. Each experimental condition was performed independently at least two times in triplicate.
ChIP-seq
ChIPs were performed essentially as previously described (Chen et al. 2013). A detailed description is included in the Supplemental Material. TRF2 and TBP ChIP-seq samples were single-end-sequenced on an Illumina HiSeq 2500 at 51 base pairs (bp). All reads passing the standard Illumina quality filter were aligned to the University of California at Santa Cruz Drosophila dm3 reference genome using Bowtie version 1.0.0. Only reads with unique alignments and a maximum of two mismatches were kept. Reads were extended to 110 bp (the estimated insert size of both libraries as determined by a Bioanalyzer), and genome-wide per-base coverage was calculated using R/Bioconductor. The Pol II ChIP-seq data were previously published (Chen et al. 2013). These data were aligned in the same way, and reads were extended to 78 bp.
TATA and TCT gene heat map
Figure 4B used genes with a Pol II enrichment of at least threefold above input in a region from the +1 transcription start site to +100 nt. Predicted TATA-containing genes (171 genes) were selected by the presence of a match to the TATA consensus STATAWAWR (between −60 and the +1 start site). Predicted TCT-containing genes (134 genes) were identified by the existence of a match to the TCT consensus of YYCTTTYY (between −10 and +20 relative to the +1 start site). Pol II, TBP, and TRF2 ChIP-seq signals were plotted (one row per gene) by aligning the genes at the transcription start site in a 5′ (left) to 3′ (right) orientation. The genes were sorted by decreasing total Pol II occupancy in the first 100 nt. The scales for the three factors were independently normalized such that 0 represents no signal and 1 is the signal value at the 99th percentile for the 305 genes plotted.
Average gene analysis
For Figure 4C, the 87 known Drosophila RP genes from the Ribosomal Protein Gene Database (Nakao et al. 2004) were matched to their corresponding FlyBase release 5.51 genes and aligned at their annotated transcription start sites. The average enrichment for TRF2 and TBP over a previously published Drosophila 2- to 4-h after egg deposition whole-cell extract sample (He et al. 2011) was calculated for each base after normalizing for differences in read count and fragment size. The results were smoothened by using a 9-bp sliding window.
Accession number
TBP and TRF2 ChIP-seq data are available from Gene Expression Omnibus (GEO) under the accession number GSE52029. In addition, a list of the ChIP-seq signals of TBP and TRF2 at each annotated transcript is provided in Supplemental Table 1.
Additional Materials and Methods are included in the Supplemental Material.
Acknowledgments
We thank Dr. Tamar Juven-Gershon, Dr. Barbara Rattner, Dr. Jia Fei, Mai Khuong, and James Gucwa for critical reading of the manuscript, as well as Dr. Nicholas Baker for advice on the analysis of RP gene transcription. We are indebted to Dr. Yaeta Endo (Ehime University; Japan) for his invaluable advice and guidance in the cell-free synthesis of TRF2 and TBP. We thank Dr. Tamar Juven-Gershon for the gift of the dTRF2S expression vector. We are also very grateful to Dr. Ernest Martinez (University of California at Riverside) for the use of his luminometer, Dr. Ana Lilia Torres-Machorro for technical advice, and Dr. Lorraine Pillus for the use of her qPCR system. J.T.K. is the Amylin Chair in the Life Sciences. S.H.C.D. is the recipient of a University of California at San Diego Molecular Biology/Cancer Center Fellowship. This work was supported by National Institutes of Health grants 1DP2OD004561-01 (J.Z.) and R01 GM041249 (J.T.K.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.245662.114.
References
- Akhtar W, Veenstra GJ 2011. TBP-related factors: a paradigm of diversity in transcription initiation. Cell Biosci 1: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio MC, Negri R, Dieci G 2011. Promoter architectures in the yeast ribosomal expression program. Transcription 2: 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Johnston J, Shao W, Meier S, Staber C, Zeitlinger J 2013. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. eLife 2: e00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Struhl K 1992. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell 69: 685–696 [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Quintin S, Lakatos L, Labouesse M, Tora L 2000. TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Mol Cell 6: 715–722 [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R 2010. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet 11: 549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Bardet AF, Patton B, Purvis J, Johnston J, Paulson A, Gogol M, Stark A, Zeitlinger J 2011. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet 43: 414–420 [DOI] [PubMed] [Google Scholar]
- Hernandez N 1993. TBP, a universal eukaryotic transcription factor? Genes Dev 7: 1291–1308 [DOI] [PubMed] [Google Scholar]
- Hsu JY, Juven-Gershon T, Marr MT 2nd, Wright KJ, Tjian R, Kadonaga JT 2008. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev 22: 2353–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y, Keles S, Prestel M, Hochheimer A, Tjian R 2007. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev 21: 2936–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Kadonaga JT 2008. Caudal, a key developmental regulator, is a DPE-specific transcription factor. Genes Dev 22: 2823–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT 2012. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip Rev Dev Biol 1: 40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach L, Horner MA, Rothman JH, Mango SE 2000. The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol Cell 6: 705–713 [DOI] [PubMed] [Google Scholar]
- Kopytova DV, Krasnov AN, Kopantceva MR, Nabirochkina EN, Nikolenko JV, Maksimenko O, Kurshakova MM, Lebedeva LA, Yerokhin MM, Simonova OB, et al. 2006. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol Cell Biol 26: 7492–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado E 1999. Transcriptional functions of a new mammalian TATA-binding protein-related factor. J Biol Chem 274: 12963–12966 [DOI] [PubMed] [Google Scholar]
- Martianov I, Fimia GM, Dierich A, Parvinen M, Sassone-Corsi P, Davidson I 2001. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol Cell 7: 509–515 [DOI] [PubMed] [Google Scholar]
- Moore PA, Ozer J, Salunek M, Jan G, Zerby D, Campbell S, Lieberman PM 1999. A human TATA binding protein-related protein with altered DNA binding specificity inhibits transcription from multiple promoters and activators. Mol Cell Biol 19: 7610–7620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F, Lakatos L, Dantonel J, Strähle U, Tora L 2001. TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr Biol 11: 282–287 [DOI] [PubMed] [Google Scholar]
- Nakao A, Yoshihama M, Kenmochi N 2004. RPG: the ribosomal protein gene database. Nucleic Acids Res 32: D168–D170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepal C, Hadzhiev Y, Previti C, Haberle V, Li N, Takahashi H, Suzuki AM, Sheng Y, Abdelhamid RF, Anand S, et al. 2013. Dynamic regulation of the transcription initiation landscape at single nucleotide resolution during vertebrate embryogenesis. Genome Res 23: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi T, Makino Y, Tamura TA 1999. Identification of a mouse TBP-like protein (TLP) distantly related to the Drosophila TBP-related factor. Nucleic Acids Res 27: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry TJ, Theisen JW, Hsu JY, Wang YL, Corcoran DL, Eustice M, Ohler U, Kadonaga JT 2010. The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev 24: 2013–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP 2005. The architecture of mammalian ribosomal protein promoters. BMC Evol Biol 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenstein MD, Zhou S, Lis JT, Tjian R 1999. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc Natl Acad Sci 96: 4791–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina JH, Hernandez N 2007. On a roll for new TRF2 targets. Genes Dev 21: 2855–2860 [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT 2003. The RNA polymerase II core promoter. Annu Rev Biochem 72: 449–479 [DOI] [PubMed] [Google Scholar]
- Takai K, Sawasaki T, Endo Y 2010. Practical cell-free protein synthesis system using purified wheat embryos. Nat Protoc 5: 227–238 [DOI] [PubMed] [Google Scholar]
- Teichmann M, Wang Z, Martinez E, Tjernberg A, Zhang D, Vollmer F, Chait BT, Roeder RG 1999. Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc Natl Acad Sci 96: 13720–13725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, Weeks DL, Wolffe AP 2000. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science 290: 2312–2315 [DOI] [PubMed] [Google Scholar]
- Zhang D, Penttila TL, Morris PL, Teichmann M, Roeder RG 2001. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292: 1153–1155 [DOI] [PubMed] [Google Scholar]
- Zhou H, Grubisic I, Zheng K, He Y, Wang PJ, Kaplan T, Tjian R 2013. Taf7l cooperates with Trf2 to regulate spermiogenesis. Proc Natl Acad Sci 110: 16886–16891 [DOI] [PMC free article] [PubMed] [Google Scholar]